Abstract
Findings from PCR-based methods have suggested a role of Felis catus papillomavirus 2 (FcaPV-2) in development of feline cutaneous squamous cell carcinoma (SCC). However, as PCR cannot localize DNA or RNA within the lesion, it is difficult to differentiate a coincidental FcaPV-2 infection and a causative association. As a key event in the pathogenesis of human papillomavirus-induced cancer is the expression of viral E6 and E7 oncogenes, localization of FcaPV-2 E6 and E7 transcription within neoplastic cells in feline SCCs would support a causative role for this papillomavirus. Therefore, RNAscope in situ hybridization (ISH) was used to localize FcaPV-2 E6/E7 transcripts in eighteen formalin-fixed paraffin-embedded samples of cutaneous SCC. Positive signals were present within 5 of 9 (55%) samples from ultraviolet (UV)-protected sites and 0 of 9 samples from UV-exposed sites. In the 4 ISH-positive samples that contained adjacent hyperplastic skin, hybridization patterns in these regions were characterized by intense nuclear signals within the superficial epidermis and punctate signals within the basal epithelial layers. However, within the five SCCs, punctate signals were present within all layers of the epidermis, with progressive loss of intense nuclear signals within the superficial epidermis. This hybridization pattern is consistent with unregulated E6/E7 transcription and decreased viral replication and is similar to the pattern observed in human papillomavirus-induced cancers as they progress from hyperplastic lesions containing productive infections to non-productive neoplasms. These findings support a causative role for FcaPV-2 in the pathogenesis of feline SCC.
Keywords: feline, skin, papillomavirus, squamous cell carcinoma, in situ hybridization
Cutaneous squamous cell carcinoma (SCC) is the most common malignant neoplasm of the skin of cats accounting for 15– 48% of feline skin tumors.7 The majority of SCCs are thought to be caused by chronic ultraviolet (UV) light exposure and develop due to progression from actinic keratosis. These cancers are most common in lightly haired areas such as the pinnae, nasal planum, and eyelids.7,19 A smaller proportion of feline cutaneous SCC develop at UV-protected sites. These have been hypothesized to develop as a progression of bowenoid in situ carcinomas (BISCs).7,9 As BISCs in cats are thought to be caused by papillomavirus infection, this suggests that a proportion of feline SCCs could develop as a result of papillomavirus infection. Papillomaviruses are a well-recognized cause of cancer in humans, especially cancers of the cervix and oral cavity. Papillomaviruses are also recognized to cause some human skin SCCs, usually in association with chronic exposure to UV light or immunosuppression, such as subsequent to organ transplantation.4,22
Papillomaviruses are double-stranded DNA viruses that infect keratinocytes at both mucosal and cutaneous sites.5 These are highly host- and tissue-specific viruses that have been identified from a wide variety of species.20 Most often papillomavirus infections are asymptomatic, although a small proportion of PV types can result in the development of hyperplastic papillomas (warts) or cutaneous plaques. Rarely, these lesions can progress to cancer.5 In humans, progression from a pre-neoplastic lesion to cancer is attributed to unregulated expression of the viral oncogenes, E6 and E7.5 These viral genes are responsible for driving terminally differentiated keratinocytes back into the cell cycle and preventing apoptosis.5 Thus, unregulated expression of viral E6 and E7 oncogenes is a key event in the pathogenesis of human papillomavirus-driven cancer.5
There is growing evidence that PV infection may promote the development of feline cutaneous SCCs.10–13,15,16 A papillomaviral etiology appears more common in SCCs from UV-protected skin, where PV DNA was detected in 9 of 11 SCCs developing at UV-protected sites compared to just 6 of 19 from UV-exposed skin.14 Felis catus PV type 2 (FcaPV-2) is thought to cause feline viral plaques and BISCs and is also the PV type most often identified within SCC.9,16,17 This virus was identified in 17 of 20 cutaneous SCCs using specific primers for FcaPV-215 and recently, abundant FcaPV-2 E6/E7 transcripts and high PV DNA copy numbers were identified within 20 of 60 feline cutaneous SCCs.21 These studies, however, all relied on PCR-based methods to detect PVs, which does not allow localization of the viral DNA and/or RNA within the sample.
Therefore, while these data are quite compelling, the question of causality remains. Since cats are often subclinically infected with PVs18 and PVs can be amplified from non-SCC skin15, PCR alone cannot distinguish between a bystander contaminant, a transient infection, or a biologically relevant infection that drives carcinogenesis. In humans, in order for a human papillomavirus to be causally associated with SCC, there needs to be evidence of (a) viral DNA within tumor samples, (b) active viral oncogene transcription in tumor cells, and (c) interaction of viral oncoproteins with tumor suppressor genes.3 So far, we have evidence for some of these requirements in feline cutaneous SCC, including demonstration of viral DNA within tumor samples, viral oncogene transcription in SCC samples (but not yet localized to tumor cells), and interaction of FcPV2 E6 and E7 with tumor suppressor genes2.
This study was initiated to investigate whether or not active viral oncogene transcription was present in the tumor cells. Therefore, the aim of this study was to detect and localize FcaPV-2 E6 and E7 mRNA within feline SCCs. This was done using colorimetric RNAscope in situ hybridization (ISH). Demonstration of active E6/E7 gene transcription within tumor cells would add to the growing support for a causal role of FcaPV-2 in the development of feline cutaneous SCCs. Furthermore, the hybridization patterns of FcaPV-2 E6 and E7 within the tumors were compared to the patterns observed in human PV-induced cervical lesions.
Methods:
Case selection and histopathology:
Formalin-fixed paraffin-embedded (FFPE) tissue samples of feline cutaneous SCCs and non-SCC skin were selected from archived biopsies from North Carolina State College of Veterinary Medicine Anatomic Pathology Service. Cases were selected based upon location on the body, quality of specimen (lack of autolysis and adequate sample remaining) and presence of intact bordering skin. Histological confirmation of SCCs was determined on hematoxylin and eosin (HE) stained sections from each case by a board-certified veterinary pathologist (JL). SCCs were characterized by disorderly keratinocyte differentiation containing markedly dysplastic keratinocytes with invasion through the basement membrane. In addition to the SCC, the tissue sections were also evaluated for the presence of adjacent hyperplastic skin, characterized by epidermal hyperplasia with orderly differentiation and no to minimal epithelial dysplasia.
PCR for FcaPV-2:
As FcaPV-2 is the PV type that is detected most frequently within feline cutaneous SCC,15 primers specific for FcaPV-2 were used to maximize the sensitivity of the PCR. Conventional PCR was performed using extracted genomic DNA (gDNA) from FFPE tissues. Two 25 μm scrolls were cut from the FFPE tissues, ensuring that the microtome blade was cleaned or replaced between samples to avoid cross contamination. The gDNA was extracted using a commercially available kit following their recommended protocol (DNeasy Blood and Tissue Kit, Qiagen, Valencia, CA) and as previously described.8
The primers JMPF (5’-GTG TCC TGT AGT TCC TAT AC-3’) and JMPR (5’-GTG CCG AAG GTC TCC TCT TC-3’) were used to amplify FcaPV-2 DNA, as previously described.14,15 The positive control was gDNA extracted from a feline viral plaque. A sample without template DNA served as the negative control. PCR reactions were performed as previously described.14,15 PCR for GAPDH was performed to verify adequate quality of extracted gDNA. The primers were felGAPDH-For (5’-GAT TGT CAG CAA TGC CTC CT-3’) and felGAPDH-Rev (5’-AAG CAG GGA TGA TGT TCT GG-3’). Samples of genomic DNA were diluted with distilled water to yield a concentration of 10 ng/µl. The DNA sample (100 ng) was amplified by PCR using a commercially available kit (HotStar Taq DNA polymerase, Qiagen). The reaction mixture contained 1 X buffer, 1.5 mM MgCl2, 200 μM deoxynucleoside triphasphate (dNTP), 1 unit (0.2 µl) taq polymerase, and 1 μM of each primer for a total reaction volume of 50 µl. All PCR reactions were performed on an Applied Biosystems GeneAmp PCR system 2700 thermocycler (Foster City, CA, USA). An initial activation step of 95° C for 10 minutes was followed by 40 cycles of 1) 30s denaturation at 95° C 2) 1 minute annealing at 58° C, and 3) 1 minute elongation at 72° C. There was a hold for 7 minutes at 72° C and a final hold at 4° C. Omission of the template DNA served as the negative control. All PCR products were separated by electrophoresis through 1% agarose at 80 volts for one hour and then stained with the DNA stain GelRed (Fisher Scientific, Pittsburg, PA).
Colorimetric in situ hybridization:
Colorimetric in situ hybridization (ISH) was performed using proprietary RNAscope ISH method (Advanced Cellular Diagnostics [ACD]). A 13ZZ probe named V-FPV-E6-E7 was designed and synthesized. This probe targets the sequence between nucleotides 461–1154 (E6 and E7 genes) of FcaPV-2 (Genbank EU796884.1). A probe to the bacterial gene dihydrodipicolinate reductase (DapB) served as the negative control. In situ hybridization was performed on 4-μm-thick FFPE tissues using the RNAscope 2.5 detection kit according to manufacturer’s protocol. Successful hydridization requires binding of adjacent probe pairs on the targeted nucleic acid, initiating a cascade that leads to deposition of diaminobenzidine (DAB). The final deposit is visualized as brown, punctate precipitate. In one sample, the amount of pigment dispersal precluded definitive localization of hybridization signals. In this case, ISH was performed using the RNAscope 2.5 Red detection kit, which results in deposition of red precipitate.
Characterization of the ISH hybridization patterns was based on an evaluation of HPV E6/E7 hybridization patterns in human cervical papillomavirus-induced pre-neoplastic lesions.6 Hybridization patterns were categorized based on the location of positive ISH signals within the neoplasm and within adjacent hyperplastic epidermis. The hybridization patterns were assessed as one of the following categories: dot-like hybridization signals from the basal layer to mid-epithelium, dot-like hybridization signals throughout the full thickness of the epithelium, and intense diffuse nuclear hybridization signals in the most superficial layer of the epidermis.
Colorimetric in situ hybridization control assay:
Two samples with positive FcaPV-2 hybridization signals were used to investigate the contribution of viral DNA and viral mRNA to the hybridization signal. ISH was performed as above, with the addition of a pretreatment with either RNase A [(QIAGEN Inc., Valencia, CA) 10 mg/mL in PBS, 30 minutes 40°C] or DNase I [(Sigma-Aldrich Corp., St. Louis, MO) diluted 1:50 in 10 mM TrisCl, 2.5 mM MgCl2, 0.5 mM CaCl2, pH7.5, 30 minutes 40°C)]. The samples were then washed in water five times before addition of the targeting probes. Targeting probes included FcaPV-2 E6/E7 probes and a predesigned control probe to the reference gene human peptidylprolyl isomerase B (PPIB) (ACD). ISH was performed using the RNAscope 2.5 Red detection kit, as it was easier to compare signals between RNase and DNase treated samples using this method.
Results:
Samples
This study comprised 18 SCCs including 9 from areas of the skin classified as UV-protected and 9 from areas of the skin classified as UV-exposed (Table 1). The mean age for cats with lesions from UV-protected regions was 14 years (range, 11 to 18 years). The age for two cats was not unknown. The mean age for cats with lesions from UV-exposed regions was 12 years (range, 3 to 18 years). Ten non-SCC cases, including inflamed or hyperplastic skin from various locations, were included (cases 19–28, Suppl. Table S1).
Table 1:
Signalment, lesion location, and detection of Felis catus papillomavirus 2 (FcaPV-2) nucleic acid for selected cases of feline cutaneous squamous cell carcinoma.
| Case | UV-Exposed | Breed | Age | Lesion Location | ISH for E6/E7 | PCR for FcaPV-2 |
|---|---|---|---|---|---|---|
| 1 | No | DSH | 12Y | Subungual | Neg | Neg |
| 2 | No | DSH | 15Y | Dorsal neck | Pos | Pos |
| 3 | No | Abyssinian | 18Y | Subungual | Pos | Pos |
| 4 | No | UNK | UNK | Thoracic limb | Pos | Pos |
| 5 | No | DSH | 11Y | Shoulder, left | Neg | Pos |
| 6 | No | DSH | 14Y | Dorsal head; lateral thorax | Pos | Pos |
| 7 | No | DSH | 16Y | Rostral mandible; left neck | Pos | Pos |
| 8 | No | DLH | 12Y | Subungual, hindlimb D3 | Neg | Neg |
| 9 | No | UNK | UNK | Paw; flank | Neg | Pos |
| 10 | Yes | DSH | 13Y | Pinna | Neg | Neg |
| 11 | Yes | DLH | 12Y | Pinna | Neg | Neg |
| 12 | Yes | DLH | 15Y | Pinna | Neg | Neg |
| 13 | Yes | DSH | 18Y | Third Eyelid | Neg | Neg |
| 14 | Yes | DMH | 3Y | Pinna | Neg | Neg |
| 15 | Yes | DSH | 7Y | Pinna | Neg | Pos |
| 16 | Yes | DSH | 16Y | Pinna | Neg | Pos |
| 17 | Yes | DSH | 11Y | Pinna | Neg | Neg |
| 18 | Yes | DSH | 13Y | Face/Lip | Neg | Neg |
ISH= in situ hybridization; SCC= squamous cell carcinoma; Pos= positive; Neg= negative; UNK= unknown; UV= ultraviolet
PCR for FcaPV-2:
GAPDH DNA was amplified from all 18 SCCs indicating that all samples contained amplifiable DNA. FcaPV-2 DNA was amplified from 9 of 18 (50%) SCCs (Table 1). This included 7 of 9 (78%) SCCs from UV-protected sites and 2 of 9 (22%) from UV- exposed sites.
Colorimetric in situ hybridization
Colorimetric in situ hybridization was performed in order to identify the cellular localization of FcaPV-2 E6 and E7 nucleic acid. Positive hybridization signals were present within 5 of 9 (56%) SCCs from UV-protected sites, while none of the SCCs from UV-exposed sites were positive. All of the 5 SCCs with positive ISH hybridization signals also contained FcaPV-2 DNA amplified by PCR. Four SCCs were PCR-positive for FcaPV-2 DNA but were negative using ISH. These included 2 SCCs from UV-protected sites and 2 SCC from UV-exposed sites. There were 9 SCCs that did not have either ISH or PCR evidence of FcaPV-2 infection. No FcaPV-2 hybridization signals were detected in the ten non-SCC skin samples tested (cases 19–28, Suppl. Table S1).
In four of the five ISH-positive SCCs, the invasive cancer was surrounded by hyperplastic epidermis characterized by focally extensive areas of epithelial hyperplasia with variable hyperkeratosis (Figures 1A and 2A). The hybridization signal pattern in all four regions of hyperplasia was characterized by intense nuclear hybridization signals within the superficial epidermis and punctate nuclear and cytoplasmic hybridization signals limited to the basal epithelial layers (Table 2; Figure 1B, 1C and 2B). In contrast to these areas of hyperplasia, the hybridization pattern within the 5 SCCs was consistently cytoplasmic and nuclear punctate signals within all layers of the epidermis, with progressive loss of intense diffuse nuclear signals within the superficial epidermal layers (Table 2; Figures 3 and 4). Adjacent histologically normal skin and the negative control probe were negative in all cases. A SCC that did not show FcaPV-2 hybridization signals is shown in Figure 5.
Figures 1 and 2.
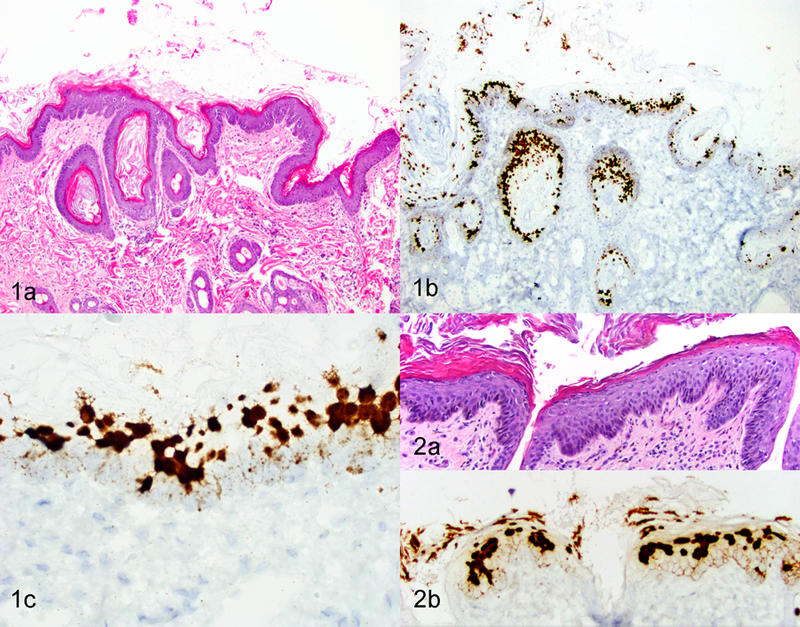
Epidermal hyperplasia adjacent to squamous cell carcinoma (SCC), skin, cat. Figure 1. Case 7. (a) Focally extensive epidermal hyperplasia with hyperkeratosis. Hematoxylin and eosin (HE). (b and c) Intense diffuse hybridization signals are evident within keratinocyte nuclei in upper layers of the epidermis. Faint dot-like signals are evident within basal and suprabasal keratinocytes. In situ hybridization (ISH) for FcaPV-2 E6/E7. Figure 2. Case 2. (a) Focally extensive epithelial hyperplasia with hyperkeratosis. HE. (b) Strong diffuse, hybridization signals are evident within keratinocyte nuclei in upper layers of epidermis. Faint dot-like signals are evident within basal and suprabasal keratinocytes. ISH for FcaPV-2 E6/E7.
Table 2:
Scoring system categorization for ISH positive samples.
| Hyperplastic | SCC | |||||
|---|---|---|---|---|---|---|
| Case No. | ISH Results | Solar Change | SupD | ≤BME/BME+ | SupD | ≤BME/BME+ |
| 2 | POS | NO | + | ≤BME | +/− | BME+ |
| 3 | POS | NO | + | ≤BME | +/− | BME+ |
| 4 | POS | NO | N/A | N/A | + | BME+ |
| 6 | POS | NO | + | ≤BME | +/− | BME+ (faint) |
| 7 | POS | NO | + | ≤BME | +/− | BME+ |
N/A not present; POS= positive; SCC= squamous cell carcinoma; ≤BME= hybridization signals occuring in the basal and suprabasal layers; BME+= hybridization signals occuring in all epithelial layers; SupD= intense nuclear hybridization signals
Figures 3–5.
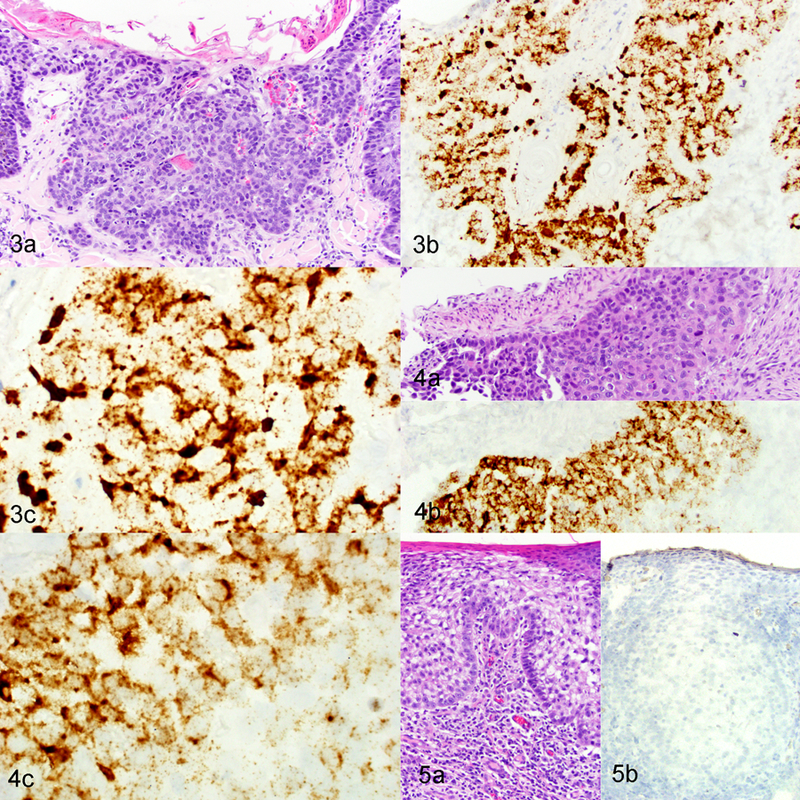
Squamous cell carcinoma (SCC), skin, cat. Figure 3. Case 2. (a) Region of SCC with increasing epithelial disorganization and invasion into superficial dermis. HE. (b and c) Strong, dot-like signals are evident within invasive SCC. In situ hybridization (ISH) for FcaPV-2 E6/E7. Figure 4. Case 3. (a) Neoplastic squamous epithelial cells surrounding a vessel within the deep dermis. HE. (b and c) Strong, dot-like signals are evident within the invasive SCC cells. ISH for FcaPV-2 E6/E7. Figure 5. SCC, pinnae (an area of ultraviolet light exposure), case 14. (a) Region of SCC with epithelial disorganization and invasion into superficial dermis. HE. (b) Hybridization signals are absent from the normal epidermis and SCC. ISH for FcaPV-2 E6/E7.
Colorimetric in situ hybridization control assay:
To determine if the hybridization signals were due to detection of mRNA or DNA, two tissues were pretreated with RNase A or DNase I prior to hybridization with targeting probes. In a region of epithelial hyperplasia merging into dysplasia, there were strong diffuse nuclear signals present within the upper epidermal layers and punctate cytoplasmic and nuclear signals throughout all layers of the epidermis (Figure 6A). Pretreatment with DNase I resulted in loss of a portion of the diffuse nuclear hybridization signals but the punctate signals remained (Figure 6B). Pretreatment with RNase A resulted in the loss of most punctate cytoplasmic and nuclear signals but the diffuse nuclear staining remained (Figure 6C). Islands of neoplastic SCC revealed strong punctate cytoplasmic and nuclear signals, which were largely removed with pretreatment with RNase A but not with DNase I (Figure 7). RNase A pretreatment of samples hybridized for mRNA detection of the reference gene peptidylprolyl isomerase B resulted in loss of most punctate cytoplasmic and nuclear signals (Figure 8).
Figures 6–8.
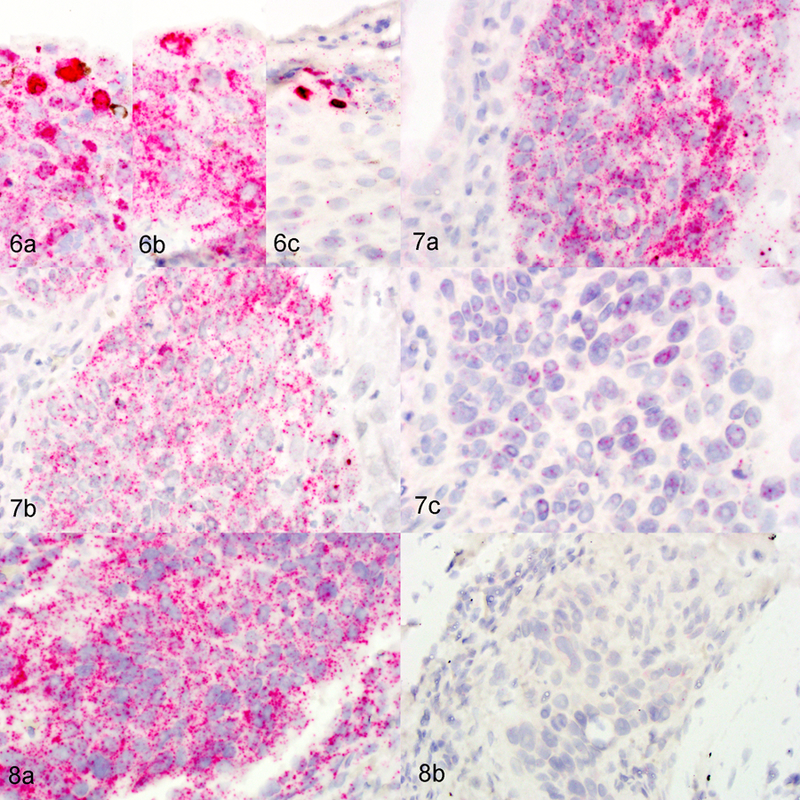
Skin cat, case 3. In situ hybridization for FcaPV-2 E6/E7 with or without nuclease pretreatment. Figure 6. Hyperplastic/ dysplastic skin. (a) Strong diffuse, hybridization signals are evident within keratinocyte nuclei in upper layers of the epidermis. Faint dot-like signals are evident within all layers of the epidermis. No pretreatment. (b) Strong diffuse, hybridization signals are absent but faint dot-like signals remain. Pretreatment with DNase I. (c) Strong diffuse, hybridization signals are present but faint dot-like signals are absent. Pretreatment with RNase A. FcaPV-2 E6/E7 ISH. Figure 7. Squamous cell carcinoma (SCC). (a) Strong, dot-like signals are evident within the invasive SCC cells. No pretreatment. (b) Strong, dot-like signals are evident within the invasive SCC cells. Pretreatment with DNase I. (c) The majority of strong, dot-like signals are absent from the invasive SCC cells, although occasional signals can be detected. Pretreatment with RNase A. Figure 8. SCC. (a) Strong, dot-like signals are evident within the invasive SCC cells. No pretreatment. (b) The majority of strong, dot-like signals are absent. Pretreatment with RNase A. ISH for the reference gene peptidylprolyl isomerase B.
Discussion
FcaPV-2 E6/E7 mRNA was detected within the neoplastic cells of 5 of 18 (28%) feline cutaneous SCCs. These results are in accord with a previous study that identified FcaPV-2 E6/ E7 mRNA in 33% of 60 feline cutaneous SCCs using reverse transcription quantitative PCR (RT-qPCR).21 Another study, however, detected FcaPV-2 E6 and E7 mRNA within 4 of 6 (67%) feline cutaneous SCCs.2 This higher detection rate may reflect differences in sample location (UV-exposed versus UV-protected regions) or the comparatively small sample size.2 In the present study, 5 of 9 (55%) SCCs from UV-protected skin contained FcaPV-2 mRNA, while FcaPV-2 RNA was not detected in any SCCs from UV-exposed sites. Similarly, in an earlier study, 4 of 6 (67%) SCCs from UV-protected sites were found to contain FcaPV-2 E6/E7 gene transcription by RT-qPCR.21 The detection of E6/E7 mRNA within the SCCs suggests that FcaPV-2 was influencing the growth and regulation of these cells. Therefore, these results provide additional evidence that FcaPV-2 may influence the development and growth of feline cutaneous SCCs, especially those that develop in haired, pigmented skin.
All of the ISH-positive cases contained amplifiable FcaPV-2 DNA, consistent with an active infection. However, FcaPV-2 DNA was also amplified by PCR from four cases that did not show any hybridization. These cases may have represented latent infections, where DNA could be detected using PCR, but no viral transcription was present. Alternatively, low levels of viral transcription could have been present, but this low level transcription was not detected by ISH.
Two of the SCCs from UV-protected sites did not contain PV DNA or FcaPV-2 hybridization signals. These SCCs appear unlikely to have been caused by FcaPV-2 infection. However, as the techniques used only detect FcaPV-2, it remains possible that other PV types could have influenced the development of these SCCs. Alternatively, non-viral factors may promote the development of a minority of non-UV-induced SCCs in cats.
Only 2 of 9 (22%) SCCs from UV-exposed skin contained FcaPV-2 DNA. The rates of detection were lower than previously observed in a study of 45 UV-exposed SCCs in which PV DNA was detected in 42% of the samples.14 In the previous study, the highest rates of detection of PV DNA were in SCCs from the nasal planum. It is therefore possible that the inclusion in the present study of mainly pinnal and eyelid SCCs, which were previously identified to have the lowest rate of PV infection, is responsible for the lower rates of detection. None of the SCCs from UV-exposed sites contained detectable FcaPV-2 E6/E7 RNA. In contrast, E6/E7 RNA has been previously detected by PCR in 5 of 28 (18%) SCCs from UV-exposed sites.21 Again, this may reflect differences in precise sites of the UV-exposed SCCs used in the two studies. Alternatively, it is possible that PCR has higher sensitivity and therefore can detect smaller quantities of RNA than the ISH techniques used in the present study. The failure to detect FcaPV-2 RNA in any of the UV-exposed SCCs in the present study suggests that FcaPV-2 is unlikely to be a common cause of SCCs from the pinnae or eyelid of cats.
In addition to using ISH to determine whether or not FcaPV-2 nucleic acid was present within neoplastic cells, the localization of E6/E7 hybridization signals were also determined because evidence from studies of human cervical cancer have shown papillomavirus hybridization signals change as lesions progress from hyperplasia to neoplasia.6 In the cats, areas of hyperplastic epithelium surrounding the SCCs consistently contained intense ISH nuclear hybridization signals in the upper epithelial layers and dot-like cytoplasmic and nuclear hybridization signals restricted to the basal and suprabasal keratinocytes. A similar hybridization pattern has been reported for the earliest precursor lesion of cervical cancer and is thought to indicate the presence of a productive papillomavirus infection.6 The presence of a similar hybridization pattern in the feline samples therefore suggests that the hyperplastic epidermis surrounding SCCs contains a productive papillomavirus infection. This is supported by the high copy numbers of FcaPV-2 which have been previously reported in some feline SCCs.21
Within the feline SCCs that contained hybridization signals, the hybridization pattern appeared as intense dot-like signals throughout all layers of the epidermis or with progressive loss of nuclear hybridization signals in the upper layers of the epidermis. In humans, this hybridization pattern is seen as pre-neoplastic lesions progress to true neoplasms.6 This is thought to be due to the deregulation of the viral genome leading to increased E6/E7 transcripts throughout all layers of the epithelium.5 Normally, in a productive infection, E6/E7 transcription is restricted to basal and suprabasal keratinocytes. Due to the deregulation of normal viral gene expression, such neoplastic lesions contain no or reduced viral replication. Due to the loss of normal regulation of E6/E7 gene transcription, these lesions would be expected to contain abundant E6/E7 transcripts using RT-qPCR, which has been reported in a proportion of feline SCCs.21
One potential disadvantage of RNAscope ISH is the possibility that both RNA and DNA could be detected by the hybridization. Therefore, we assessed hybridization patterns after a pretreatment with either DNase or RNAse. The intense diffuse nuclear signals represent co-detection of RNA and DNA, likely due to the presence of single-stranded forms of viral DNA that occur during upregulated episome synthesis when the DNA load is high, as in cells in the productive phase of infection.6 The majority of punctate signals within the hyperplastic regions and SCC are consistent with RNA, as these were largely removed with pretreatment with RNase and not with DNase.
In humans, the distinction between an early pre-neoplastic lesion and a lesion that has undergone malignant transformation is vital in determining the appropriate treatment and prognosis.1 To help with this differentiation when histology alone cannot provide a clear distinction, evaluation of the RNA expression patterns using RNAscope ISH can be used as a biomarker for transformation.6 In the present study using cats, there was a clear distinction in the hybridization pattern between hyperplastic lesions and SCC, with the latter having a hybridization pattern consistent with a transformative infection. Identification of feline transformative phase lesions could help to determine treatment strategies; however, in veterinary medicine, the high cost of RNAscope ISH precludes its use, at least at this time, as a diagnostic aid. However, it is interesting that the pattern of E6/E7 hybridization signals within the feline SCCs and the human cervical lesions were so similar. This suggests that, as in cervical cancers, the papillomaviral gene transcription changes as lesions progress from pre-neoplastic to neoplastic. The altered gene transcription within the SCCs provides further evidence that papillomaviruses are not present simply as an incidental infection within the neoplasms.
In conclusion, FcaPV-2 E6/E7 mRNA was detected within 5 of 18 feline cutaneous SCCs, including 5 of 9 SCCs from UV-protected skin. The presence of FcaPV-2 E6/E7 transcription within the neoplasms provides additional evidence that papillomaviruses can influence the development of these common cancers in cats. The hybridization patterns of E6/E7 within the feline lesions were similar to that observed in human papillomavirus-induced cervical cancers.
Supplementary Material
Acknowledgements
This project was supported by research funds provided by North Carolina State University, College of Veterinary Medicine.
References
- 1.WHO Guidelines for Screening and Treatment of Precancerous Lesions for Cervical Cancer Prevention Geneva; 2013. [PubMed] [Google Scholar]
- 2.Altamura G, Corteggio A, Pacini L, Conte A, Pierantoni GM, Tommasino M, et al. Transforming properties of Felis catus papillomavirus type 2 E6 and E7 putative oncogenes in vitro and their transcriptional activity in feline squamous cell carcinoma in vivo. Virology 2016;496:1–8. [DOI] [PubMed] [Google Scholar]
- 3.Boscolo-Rizzo P, Pawlita M, Holzinger D. From HPV-positive towards HPV-driven oropharyngeal squamous cell carcinomas. Cancer Treat Rev 2015. [DOI] [PubMed]
- 4.Connolly K, Manders P, Earls P, Epstein RJ. Papillomavirus-associated squamous skin cancers following transplant immunosuppression: one Notch closer to control. Cancer treatment reviews 2014;40(2):205–214. [DOI] [PubMed] [Google Scholar]
- 5.Doorbar J The papillomavirus life cycle. J Clin Virol 2005;32 Suppl 1:S7–15. [DOI] [PubMed] [Google Scholar]
- 6.Evans MF, Peng Z, Clark KM, Adamson CS, Ma XJ, Wu X, et al. HPV E6/E7 RNA in situ hybridization signal patterns as biomarkers of three-tier cervical intraepithelial neoplasia grade. PLoS One 2014;9(3):e91142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross TLIP, Walder EJ, et al. : Skin Diseases of the Dog and Cat: Clinical and Histopathologic Diagnosis Oxford: Blackwell Science, 2005. [Google Scholar]
- 8.Luff JA, Affolter VK, Yeargan B, Moore PF. Detection of six novel papillomavirus sequences within canine pigmented plaques. J Vet Diagn Invest 2012;24(3):576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munday JS. Papillomaviruses in felids. Vet J 2014;199(3):340–347. [DOI] [PubMed] [Google Scholar]
- 10.Munday JS, Aberdein D. Loss of retinoblastoma protein, but not p53, is associated with the presence of papillomaviral DNA in feline viral plaques, Bowenoid in situ carcinomas, and squamous cell carcinomas. Vet Pathol 2012;49(3):538–545. [DOI] [PubMed] [Google Scholar]
- 11.Munday JS, Dunowska M, De Grey S. Detection of two different papillomaviruses within a feline cutaneous squamous cell carcinoma: case report and review of the literature. N Z Vet J 2009;57(4):248–251. [DOI] [PubMed] [Google Scholar]
- 12.Munday JS, French AF, Gibson IR, Knight CG. The presence of p16 CDKN2A protein immunostaining within feline nasal planum squamous cell carcinomas is associated with an increased survival time and the presence of papillomaviral DNA. Vet Pathol 2013;50(2):269–273. [DOI] [PubMed] [Google Scholar]
- 13.Munday JS, French AF, Peters-Kennedy J, Orbell GM, Gwynne K. Increased p16CDKN2A protein within feline cutaneous viral plaques, bowenoid in situ carcinomas, and a subset of invasive squamous cell carcinomas. Vet Pathol 2011;48(2):460–465. [DOI] [PubMed] [Google Scholar]
- 14.Munday JS, Gibson I, French AF. Papillomaviral DNA and increased p16CDKN2A protein are frequently present within feline cutaneous squamous cell carcinomas in ultraviolet-protected skin. Vet Dermatol 2011;22(4):360–366. [DOI] [PubMed] [Google Scholar]
- 15.Munday JS, Kiupel M, French AF, Howe L. Amplification of papillomaviral DNA sequences from a high proportion of feline cutaneous in situ and invasive squamous cell carcinomas using a nested polymerase chain reaction. Vet Dermatol 2008;19(5):259–263. [DOI] [PubMed] [Google Scholar]
- 16.Munday JS, Kiupel M, French AF, Howe L, Squires RA. Detection of papillomaviral sequences in feline Bowenoid in situ carcinoma using consensus primers. Vet Dermatol 2007;18(4):241–245. [DOI] [PubMed] [Google Scholar]
- 17.Munday JS, Peters-Kennedy J. Consistent detection of Felis domesticus papillomavirus 2 DNA sequences within feline viral plaques. J Vet Diagn Invest 2010;22(6):946–949. [DOI] [PubMed] [Google Scholar]
- 18.Munday JS, Witham AI. Frequent detection of papillomavirus DNA in clinically normal skin of cats infected and noninfected with feline immunodeficiency virus. Vet Dermatol 2010;21(3):307–310. [DOI] [PubMed] [Google Scholar]
- 19.Murphy S Cutaneous squamous cell carcinoma in the cat: current understanding and treatment approaches. J Feline Med Surg 2013;15(5):401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rector A, Van Ranst M. Animal papillomaviruses. Virology 2013;445(1–2):213–223. [DOI] [PubMed] [Google Scholar]
- 21.Thomson NA, Munday JS, Dittmer KE. Frequent detection of transcriptionally active Felis catus papillomavirus 2 in feline cutaneous squamous cell carcinomas. J Gen Virol 2016;97(5):1189–1197. [DOI] [PubMed] [Google Scholar]
- 22.Wieland U, Kreuter A, Pfister H. Human papillomavirus and immunosuppression. Current problems in dermatology 2014;45:154–165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


