Abstract
Sex-specific chromosomes, like the W of most female birds and the Y of male mammals, usually have lost most genes owing to a lack of recombination. We analyze newly available genomes of 17 bird species representing the avian phylogenetic range, and find that more than half of them do not have as fully degenerated W chromosomes as that of chicken. We show that avian sex chromosomes harbor tremendous diversity among species in their composition of pseudoautosomal regions and degree of Z/W differentiation. Punctuated events of shared or lineage-specific recombination suppression have produced a gradient of “evolutionary strata” along the Z chromosome, which initiates from the putative avian sex-determining gene DMRT1 and ends at the pseudoautosomal region. W-linked genes are subject to ongoing functional decay after recombination was suppressed, and the tempo of degeneration slows down in older strata. Overall, we unveil a complex history of avian sex chromosome evolution.
In many species with separate sexes, sex is determined by a pair of heteromorphic sex chromosomes that differ in their size, morphology, and gene content. Mammals have male heterogametic sex chromosomes (XX in female, XY in male), whereas birds have female heterogametic sex chromosomes (ZW in female and ZZ in male) (1). Both sex systems have originated independently from different ancestral autosomes after the two lineages diverged more than 300 million years ago (Ma) (1–4), yet they share many common features with respect to their genomic composition and evolutionary history. Most regions of mammalian Y or avian W chromosomes (chrW) lack recombination except for the pseudoautosomal regions (PARs) that are shared between X and Y, or Z and W, and which encompass only 2% of the human X (5). The nonrecombining region of the Y/W usually has lost almost all functional genes present on the ancestral chromosome except for a few loci, often with sex-specific functions (6), and Y/W chromosomes are often largely composed of repetitive and epigenetically silenced (heterochromatic) DNA (7). Recombination suppression is thought to follow the origination of a sex-determining locus and may initially encompass only a small chromosomal segment surrounding that locus, but then progressively spreads along Y or W chromosomes, possibly owing to the accumulation of sexually antagonistic alleles at PARs that benefit one sex but harm the other (8). Chicken and human sex chromosomes have in parallel evolved recombination suppression in a stepwise manner, manifested as “evolutionary strata” with a gradient of ages. Each stratum is characterized by a distinct range of levels of sequence homology between the remaining XY or ZW gene pairs, reflecting the punctuated evolutionary time points when recombination between the sex chromosomes ceased (9, 10). To date, genomic sequences of 10 mammalian species’ Y chromosomes have been determined (11, 12), and the human Y was found to have five strata (5) probably resulting from Y-linked inversions (13), of which three are shared among all Eutherian mammals (4, 14). No complete avian chrW has been sequenced, but partial annotation of W-linked genes in chicken (Gallus gallus) (15) implied that there are at least three strata (10, 16), and some may have formed independently in different avian lineages (17–20).
A recent investigation of the karyotype of 200 avian species revealed great variation in the size and morphology of chrW, sometimes even between closely related species (21), suggesting a dynamic evolutionary history of bird sex chromosomes. Living birds consist of two sister lineages, the Palaeognathae (flightless ratites and Tinamous) and the Neognathae, which themselves consist of two major lineages, the Galloanserae (land and water fowls) and the Neoaves (all other neognaths). Neoaves are derived from an ancient species radiation and comprise more than 95% of all avian species (22, 23). Karyotyping suggests that most ratite paleognaths, like ostrich (Struthio camelus) and emu (Dromaius novaehollandiae), have homomorphic sex chromosomes with extensive recombining and euchromatic PARs that resemble the ancestral state (24–26). In contrast, their sister group Tinamous shows interspecific variation in sizes and locations of PARs (27). Whether all Neognathae species have an almost completely degenerated chrW similar to that of chicken (28) is unknown. Here, we analyzed female genomes of 17 bird species newly generated by the Avian Genomics Consortium that span the avian taxonomic diversity (23, 29), in order to unveil the broad evolutionary history of bird ZW chromosomes.
Results
Avian sex chromosomes have different lengths of PAR
We collected genome sequences of three Palaeognathae (ostrich, emu, white-throated tinamou), one Galloanserae (Pekin duck) (30), and 13 Neoaves species, including a representative of the sister lineage to all other Neoaves (brown mesite), the Caprimulgiformes (Anna’s hummingbird and chimney swift), core waterbirds (crested ibis to white-tailed tropicbird), and core landbirds (rifleman to woodpecker) (29) (Fig. 1 and table S1). For three species—chicken, emu, and crested ibis—we analyzed sequence data of both sexes (31–33). We additionally improved the ostrich draft genome to a chromosome assembly with optical mapping (34) (N50: 17.7 Mb, table S2). Comparative cytogenetic mapping and genome sequence analysis show that there are very few interchromosomal rearrangements among bird species (35–38); thus, we anchored genome scaffold sequences of the remaining species using the ostrich Z chromosome (chrZ) for all Palaeognathae and chicken Z for all Neognathae (3).
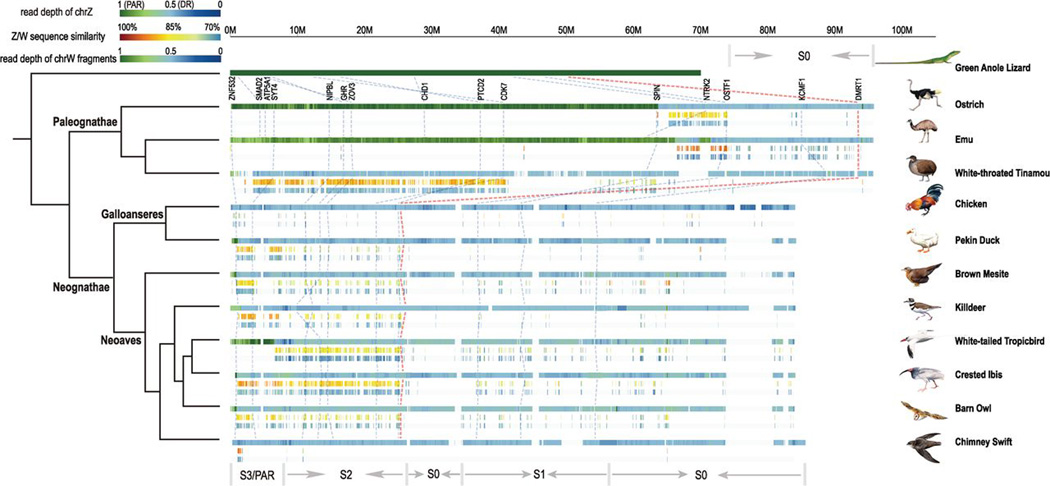
Fig. 1. Evolutionary strata of avian sex chromosomes.
We present evolutionary strata patterns of investigated bird species with partially degenerated chrW, except for chicken and chimney swift as comparison, together with their revised phylogeny (23). A full pattern for all the investigated species is presented in fig. S2 and summarized in fig. S23. chrZs of tinamou and emu are ordered and scaled to the same length as that of ostrich, and all others to that of chicken. Each avian species includes three color-coded tracks from top to bottom: (i) female read depth of chrZ, where PAR is shown in green, differentiated region without recombination between Z/W in blue, and assembly gaps as blanks; (ii) pairwise alignment identities between Z/W plotted from 100% identical (red) to 70% identical (blue); and (iii) female read depth of W-linked sequence fragments, color-coded the same as (i). We labeled regions of each stratum at the bottom: “S0” corresponds to the most ancient evolutionary stratum 0 that is shared by all the species, while the younger strata are abbreviated as S1, S2, etc. Note that different species may not share the same S1, S2, and S3; and part of S0 (25 to 35 Mb in the figure) has been reshuffled because of chromosome inversions that occurred in the ancestor of Neognathae (Fig. 3 and fig. S15). The blank region around 75 to 85 Mb corresponds to a chicken-specific amplification of testis gene clusters (3) (table S7). We also labeled genes previously used for cytological mapping (26) and the reported S0 gene KCMF1 (4) (traced through the phylogeny with blue dotted lines), as well as the putative male sex-determining gene DMRT1(traced with the dotted red line) (39).
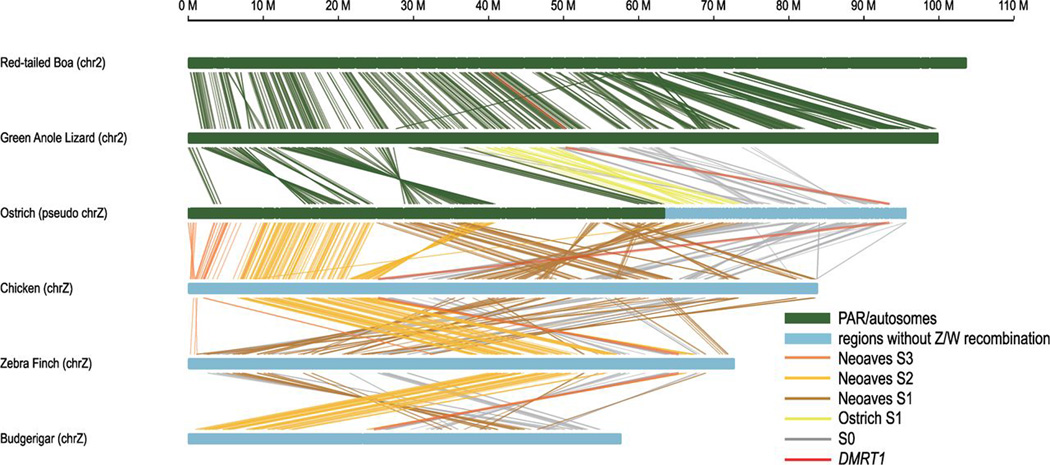
Fig. 3. Gene synteny within avian evolutionary strata.
Mapped is the evolutionary strata gene synteny between sequenced reptile (boa and lizard) and avian species. Each line connects a pair of orthologous genes between species, and different colors of lines represent different strata of genes: S0 (gray), ostrich S1 (yellow), Neognathae S1 (brown), Neoaves S2 (orange), DMRT1 (red). Genomic regions encompassing S0 and S1 have experienced multiple inversions across different species, relocating DMRT1 to the middle of the Z chromosome after the divergence between Neognathae and Palaeognathae. In contrast, genomic regions within Neoaves S2 and S3 show high level of syntenic conservation across species.
To distinguish between the recombining PAR (that has nearly identical sequences between chrZ and chrW) and the region lacking recombination (that has become differentiated between chrZ and chrW) along the sex chromosome, we developed an approach that involves examination of read sequencing depth along chrZ (34). Although most sequences in the female-specific W-linked region may have lost almost all homology with chrZ, some ZW homolog pairs (often called gametologs) may remain. However, W-linked regions are probably too divergent to allow short sequencing reads derived from them [100 base pairs (bp) long in this study] to align with their Z homologs, especially if they are from more ancient strata. Consequently, PARs will show a level of read depth similar to that of autosomes, whereas differentiated regions will have only half the mapped read coverage depth in females (Fig. 1).
Based on this principle, we find evidence of extreme differences in the length of the PAR versus differentiated region on sex chromosomes across species (Fig. 1 and Table 1), in contrast to a lack of such variation in coverage from autosomes (fig. S1). The putative avian male sex-determining geneDMRT1 (39) always resides in the differentiated region in all species, consistent with it being the initial seed for progressive recombination suppression (Fig. 1). However, the size of the PAR varies substantially among species (Table 1). In Palaeognathae, at least two-thirds of chrZ of ostrich and emu are pseudoautosomal and still recombine with chrW, in contrast to only 1% of chrZ of white-throated tinamou (Tinamus guttatus) being PAR (Fig. 1 and Table 1). We confirm that we have identified the true PAR in ostrich, as we find that eight genes that have been cytologically mapped to both chrZ and chrW (26) are appropriately distributed along our assembled ostrich chrZ (Fig. 1). Whereas most Neognathae have restricted recombination between their sex chromosomes into only a very short terminal PAR (Fig. 1 and fig. S2), unexpectedly two Neoavian species, killdeer (Charadrius vociferus) and white-tailed tropicbird (Phaethon lepturus), retain PARs of considerable length, encompassing between 5 and 9% of their chrZ (Table 1). Thus, unlike chicken (40), some Neognathae chrW can pair and recombine with chrZ through a sizable PAR. This also indicates that recombination suppression in the genomic segment adjacent to the PAR has evolved independently in at least some bird lineages (see below).
Table 1.
Age and gene content of avian evolutionary strata.
| Stratum | Species | Ostrich | Emu | White-throated tinamou |
Pekin duck |
Brown mesite |
Killdeer | White-tailed tropicbird |
Crested ibis |
Barn owl |
|---|---|---|---|---|---|---|---|---|---|---|
| S0 | Age (My)* | 102 | ||||||||
| Z-linked genes | 247 | 224 | 244 | 154 | 137 | 96 | 157 | 182 | 151 | |
| W-linked genes† | 0 | 6/8 | 0 | 0 | 1/1 | 0/0 | 2/3 | 5/4 | 1/2 | |
| W/Z length (Mb) | 0/21.4 | 1.2/19.8 | 0/18.3 | 0.01/32.5 | 0.2/18.4 | 0.1/31.9 | 0.8/20.9 | 0.8/28.5 | 0.3/19.4 | |
| S1‡ | Age (My) | 75 | 23 | 50 | 71–119 | |||||
| Z-linked genes | 108 | 21 | 201 | 333 | 236 | 400 | 246 | 267 | 211 | |
| W-linked genes | 36/19 | 0/0 | 21/9 | 0/4 | 7/3 | 1/1 | 10/10 | 8/5 | 5/4 | |
| W/Z length (Mb) | 5.1/10.6 | 0.3/3.6 | 1.2/17.7 | 0.09/16.9 | 0.08/25.0 | 0.06/20.3 | 0.2/26.7 | 0.5/23.8 | 0.09/26.8 | |
| S2 | Age (My) | PAR | 29 | 92 | 69 | |||||
| Z-linked genes | 461 | 237 | 256 | 279 | 224 | 229 | 203 | |||
| W-linked genes | 94/74 | 14/9 | 23/8 | 27/11 | 37/19 | 33/32 | 12/10 | |||
| W/Z length (Mb) | 16.6/35.1 | 0.5/17.7 | 1.6/19.8 | 2.2/21.2 | 5.0/19.1 | 6.3/20.0 | 1.7/17.8 | |||
| S3 | Age (My) | PAR | PAR | 60 | 47 | 45 | PAR | 53 | 50 | |
| Z-linked genes | 96 | 21 | 24 | 25 | 36 | |||||
| W-linked genes | 13/5 | 7/3 | 3/3 | 2/1 | 5/2 | |||||
| W/Z length (Mb) | 1.3/8.3 | 1.5/3.7 | 2.4/4.7 | 0.6/3.2 | 1.1/6.2 | |||||
| PAR | Z-linked genes | 718 | 619 | 12 | 18 | 19 | 37 | 55 | 8 | 13 |
| Z lengths (Mb) | 63.6 | 65.5 | 0.7 | 1.05 | 0.7 | 4.2 | 6.6 | 0.4 | 0.9 | |
| Total | W/Z genes | 55/1073 | 14/864 | 198/918 | 45/848 | 53/669 | 46/836 | 81/682 | 90/711 | 41/614 |
| W/Z lengths | 5.1/95.6 | 1.6/88.9 | 17.9/71.9 | 1.9/76.5 | 3.4/67.7 | 4.7/82.4 | 6.0/73.2 | 8.3/75.9 | 3.3/71.1 | |
To further quantify Z/W divergence levels within the differentiated region, we searched for assembled W-linked sequences longer than 1 kb, which allow better alignments with chrZ than the raw reads. W gametologs may be embedded within repetitive DNA that cannot be readily assembled into large scaffolds, but unique segments of a species’ chrW are expected to assemble into fragmented scaffold sequences separate from those of chrZ. If these W-segments are only differentiated to a moderate degree, they may have retained sufficient sequence homology to map to the differentiated region along chrZ. Using this approach, we identified candidate W-linked scaffolds up to 25% of the homologous chrZ length with low background mapping to autosomes, indicating effective discrimination from random fragments (34) (fig. S1). We identify abundant W-linked sequences from all three Palaeognathae species, one of the two Galloanserae species and in five of the Neoaves (Table 1 and table S3), and many fewer candidate W-derived fragments homologous to chrZs in chicken and the remaining Neoaves species (Fig. 1 and fig. S2).
Comparison of read depths between the two sexes in chicken, crested ibis, and emu verified that the candidate W-linked scaffolds are specific to females (figs. S3 and S4). Additionally, most of the candidate W-linked scaffolds of chicken exclusively align to W-linked or unmapped (chrUn) fragments of the reference chicken genome derived from Sanger sequencing (fig. S5). These results together confirmed that we have identified truly W-linked scaffolds. We further annotated genes in these W-derived scaffolds, ranging from 24 in the Pekin duck to 55 in ostrich and 198 in the white-throated tinamou (Table 1 and table S4), compared to 26 and 14 W-linked genes previously annotated through transcriptome sequencing in chicken and ostrich, respectively (figs. S6 and 7 and table S5) (15, 41). Some W-linked genes may be missing or fragmented (table S5), if they are highly enriched for transposable elements or too similar to their Z-linked homologs in sequence. Nevertheless, we found the coding sequence lengths of the Z/W gene pairs identified in this work to generally correlate well with those of their chicken Z-linked orthologs (fig. S8). This indicates that, although our current chrW genome assemblies are interrupted by highly repetitive noncoding regions, they contain substantial amounts of coding regions of genes. Overall, we find that avian chrWs show great variation in their degree of degeneration, and more than one-third of the sampled Neognathae species harbor chrWs that are not as fully degenerated as that of chicken (fig. S2).
Evolutionary strata of Palaeognathae species
The identified W-linked fragments are stratified by differences in their occurrences and alignment identities along chrZ (figs. S9 and S10), exhibiting patterns of evolutionary strata. This provides evidence that, similar to those of mammals (4), avian sex chromosomes suppressed recombination through a series of punctuated events (10, 16, 17). The strata present a gradient of ages, reflected by higher levels of sequence homology between chrW and chrZ sequences as one moves toward the PAR (Fig. 1 and figs. S9 to S11). The putative male sex-determining gene of birds, DMRT1 (39), is located within the first stratum (we denote this region as stratum S0) that evolved in the ancestor of all extant birds, around 100 (98 to 108) Ma (23). This is supported by a lack of abundant assembled W-linked fragments due to the ancient origin of S0, and also DMRT1’s close linkage in our new ostrich genome assembly to KCMF1 (Fig. 1), which was also characterized as an S0 gene by an independent study through transcriptome sequencing (4). The boundaries of S0 can still be clearly traced by the nearby stratum in ostrich and emu (Fig. 1), both of which have maintained the most ancestral state of sex chromosome differentiation among the studied birds and share 90% of their Z-linked gene content of S0 (Fig. 2A).
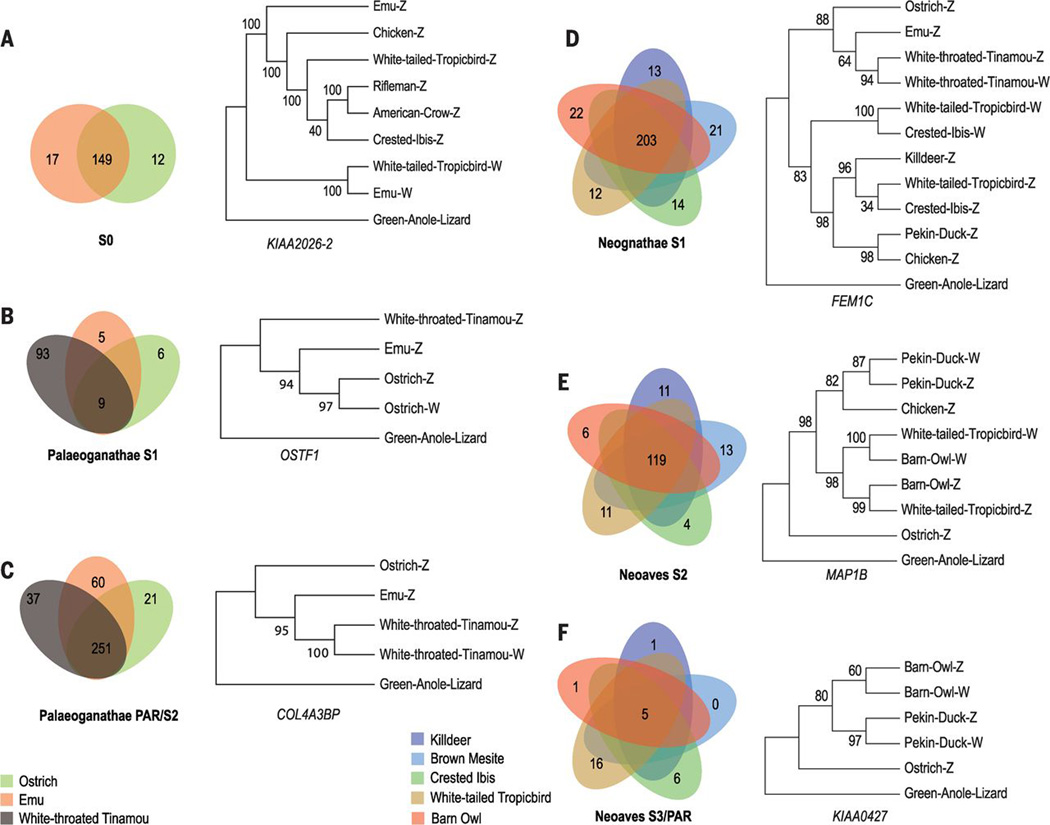
Fig. 2. Emergence of evolutionary strata in different avian lineages.
Shown are Venn diagrams of overlapping gene content between orthologous strata of different Palaeognathae (A to C) and Neognathae (D to F) species. To the right of each diagram is a maximum-likelihood tree for a specific example gene constructed from Z- and W-linked gametologs of that stratum. Additional trees supporting the inference of a certain stratum’s age are shown in the figs. S12, S14, S16, and S17. The tree is constructed with ostrich or green anole lizard as outgroup, with 100 bootstraps. A pattern in which Z gametologs are grouped together by species indicates that Z/W recombination was suppressed before their speciation, whereas clustering of Z and W gametologs of the same species indicates that Z/W recombination was suppressed after the speciation event. “Palaeognathae PAR” shows the number of genes that overlapped between ostrich PAR, emu PAR, and tinamou stratum S2. “Neoaves S3” shows the overlapped gene content between tropicbird PAR versus S3 of four other Neoaves species: killdeer, brown mesite, crested ibis, and barn owl.
Maximum-likelihood trees constructed with multiple S0 genes (including KCMF1) show that Z-linked gametologs of Palaeognathae and Neognathae cluster with each other by species rather than with their W-linked gametologs (fig. S12), confirming that recombination between chrZ and chrW at S0 was abolished in the common ancestor of all birds (17) (Fig. 2A and fig. S12). Consistent with their postulated ancient origin, almost no W-linked genes were identified within the orthologous region of S0 in all the studied species (Table 1), the exception being emu, whose W-linked S0 region has not completely degenerated (23, 31). All residual W-linked fragments within emu S0 show similar levels of divergence from their homologous chrZ sequences (fig. S9), suggesting that they stopped recombining at the same time.
Adjacent to the S0 stratum, we identified one additional stratum (S1) in ostrich and emu, and at least two strata (S1 and S2) in the white-throated tinamou. In contrast to S0, S1 spans different regions in each of the three paleognaths: Each species shares less than 50% of its S1 gene content with the other two (Fig. 2B), and S2 is specific to tinamou, whose orthologous region in the ostrich and emu is PAR (Figs. 1 and 2C). This indicates that these younger strata formed independently in these three anciently diverged species. A small segment of the ostrich PAR is embedded within the inferred emu S1, which suggests a chromosomal rearrangement between these two birds (Fig. 1). Molecular clock dating with Z/W divergence levels at silent sites suggests that stratum S1 of ostrich formed about 75 (34 to 193) Ma and that of emu formed about 23 (11 to 59) Ma, and their sex chromosomes have maintained their current PARs ever since (table S6). White-throated tinamou has a much smaller PAR due to two separate strata that formed about 50 (21 to 95) Ma and 29 (7 to 82) Ma (table S6), after its divergence with ostrich about 58 to 95 Ma (23). Thus, common and lineage-specific events characterize sex chromosome evolution in Palaeognathae birds.
Evolutionary strata of Neognathae species
In comparison, Neognathae species exhibit either three or four strata along their sex chromosomes (Fig. 1). We infer that the oldest Neognathae-specific stratum S1 formed in an ancestor of all Neognathae adjacent to the S0 region described above (Fig. 1). We also recovered W-linked sequence fragments within this stratum by reordering them against the ostrich chrZ as a proxy for the proto-Z (fig. S13). Consistent with its ancestral emergence and previous results in chicken (16), Neognathae S1 harbors almost no identifiable W-linked fragments or genes in Galloanserae, and we find only few W-linked fragments with low sequence identities versus chrZ in Neoaves (figs. S9 and S10). The low density of W fragments within Neognathae S1 hampers us from determining the exact genomic boundaries of this stratum, but phylogenetic trees constructed from Z/W gametologs of S1 confirm that this stratum is shared among all Neognathae (Fig. 2D and fig. S14). Three Z/W gene pairs, SPIN, HINT, and CHD1, which were previously inferred to have stopped recombining before the Neoaves–Galloanserae split (17), are consistently included within the S1 region of Neognathae, despite the shuffling of gene order due to chromosomal rearrangements (fig. S15). By scaling the Z/W divergence level in noncoding regions to that of S2 (see below), we estimate that Neognathae S1 was formed about 71 to 119 Ma, which is consistent with its formation before the inferred split between Neoaves–Galloanserae about 83 to 94 Ma (23) (table S6).
Adjacent to S1, we identify a younger Neognathae stratum S2 that spans a similar region in all Neognathae (Fig. 1), but Z/W gametologs within this region cluster by lineage rather than by chromosomes (Fig. 2E and fig. S16), suggesting the Galliformes (chicken), Anseriformes (Pekin duck), and Neoaves may have each independently formed stratum S2, after the shared origination of S1. On the basis of either the proposed divergence times between these lineages (23) or from Z/W sequence divergence level in noncoding regions, we estimate that the duck stratum S2 arose 68 (18 to 127) Ma and that of Neoaves arose 69 (66 to 72) Ma (table S6), whereas the chicken S2 was previously estimated to have arisen 34 to 54 Ma (16).
One Neoaves species, the white-tailed tropicbird, seems to have experienced no further recombination suppressions after stratum S2, and its current PAR appears to be comparable to the Neoaves ancestor PAR (Fig. 1). By contrast, orthologs of tropicbird PAR genes are located in differentiated regions of all the other Neoaves, including brown mesite (Mesitornis unicolor), suggesting that the youngest stratum S3 evolved independently in different lineages. Indeed, only five genes overlap between the different S3 regions (Fig. 2F), and Z-linked genes from S3 cluster with W-linked gametologs from the same species and not with Z-linked orthologs of other species (Fig. 2F and fig. S17). Two Z/W gene pairs within the S3 region, ATP5A1 and UBAP2, were previously shown to have suppressed recombination independently among different bird species (17). Together, these results indicate that recombination in S3 ceased in some lineages after the Neoaves species radiation, and we estimate that S3 emerged at different time points in different lineages (between 45 and 53 Ma; Table 1 and table S6), by scaling its Z/W divergence level at noncoding regions to that of Neoaves S2.
Putative Z- and W-linked inversions causing recombination suppression
Recombination suppression at evolutionary strata may have been mediated by chromosomal inversions, as has been suggested for mammals (9, 13). Genomic comparison of four avian species with de novo assembled genomes against the reptile outgroups green anole lizard (Anolis carolinensis) (42) and boa snake (Boa constrictor) (43) (Fig. 3 and fig. S18) allow us to infer the presence of inversions on chrZ and further inform on the formation paths of evolutionary strata in birds. Both cytogenetic mapping (44) and our genomic comparison have shown that ostrich has maintained the most ancestral gene synteny relative to other avian species with only a few intrachromosomal rearrangements (Fig. 3). We specifically compared gene synteny spanning the ostrich S0 with the assembled genomes of green anole lizard and boa snake and to cytogenetic data from three other reptiles (rat snake, gecko lizard, Chinese soft-shelled turtle) (45), and identified a chromosomal inversion on ostrich chrZ relative to reptiles, which are all collinear within their orthologous regions on the corresponding autosome (fig. S18). This suggests that an inversion on the proto-Z abolished recombination between chrZ and chrW at S0 in an ancestor of all birds. Gene content and synteny within the ostrich S1 region is largely conserved between the green anole lizard and ostrich, suggesting that a putative inversion creating the ostrich-specific S1 stratum must have occurred on chrW (Fig. 3).
Multiple genomic rearrangements within Neognathae S0 and S1 regions among Neognathae species preclude us from inferring the origin of Neognathae S1 (figs. S13 and S15). However, both cytogenetic (35) and genomic mapping (Fig. 3) place DMRT1 close to the telomere of Palaeognathae chrZ but in the middle of chrZ for most other Neognathae species. This suggests that at least one Z-linked inversion, which may have been associated with the formation of Neognathae S1, has relocated DMRT1 in the ancestor of all Neognathae. This putative inversion has shuffled part of the oldest stratum S0 containingDMRT1 in between the younger strata Neoaves S2 and S1 in chicken (Fig. 3 and fig. S15), which explains why DMRT1 was placed in a more recent stratum in a previous study using chicken Z chromosome coordinates as reference (fig. S15) (16). Gene synteny is to a large extent conserved across species within both S2 and S3 (Fig. 3 and figs. S18 and S19). This suggests that if inversions are responsible for recombination suppression between chrZ and chrW at S2 and S3, they originated on chrW.
Tempo of W degeneration in birds
Once recombination is suppressed between chrW and chrZ, the functions of W-linked genes are expected to decay (6). Y chromosome genome and transcriptome sequencing of 15 mammals has shown a loss of more than 95% of all genes hypothesized to have been present on the ancestral Y of each lineage (4, 12). In contrast, we find extensive heterogeneity across birds in levels of W degeneration, as measured by the proportion of residual W-linked genes or sequences relative to their Z-linked homologs (Table 1). Heterogeneity in W degeneration is found not only between species groups whose strata evolved independently (e.g., Paleognathae versus Neognathae), but also within homologous strata dating back to a common ancestor, and which therefore have carried the same ancestral gene content and underwent degeneration for the same length of time (Fig. 1 and Table 1). This is exemplified by the W-linked region of Neoaves stratum S2 having become almost completely differentiated and lost nearly all genes in 8 out of 13 Neoaves species (fig. S2), whereas tropicbird and crested ibis have retained at least 20% of their ancestral W-linked genes relative to the homologous Z-linked regions (Table 1). Within each stratum, we estimate that chrWs of different species have lost 52 to 95% of their ancestral gene content (Fig. 4, A to G), which may be an overestimate owing to difficulties in assembling W gametologs within highly repetitive regions (figs. S6 and S7). Functional decay is also apparent at assembled W gametologs that are present in the nonrecombining region of chrW (46): We find that about 26 to 50% of W-linked genes within each stratum have likely become pseudogenes, owing to the accumulation of premature stop codons and/or frame-shift mutations (Fig. 4, A to G). In addition, W-linked genes in all species consistently show significantly higher rates of protein evolution than their Z-linked gametologs (comparing pairwise rates versus chicken; Wilcoxon test: P < 0.05, fig. S20), indicating that they experience less efficient purifying selection on the nonrecombining chrW than the Z gametologs (46, 47). Disabling mutations may have also accumulated in regulatory regions: W-linked gametologs within the differentiated region of ostrich, emu, and duck are expressed at a significantly lower level than both their Z-linked gametologs and lizard orthologs (Wilcoxon test, P < 0.05, Fig. 4H and fig. S21) in different tissues, and W gametologs with disrupted open reading frames are down-regulated even more (fig. S22).
Fig. 4. Evolution of avian W-linked genes.
(A to G) For each stratum of the example species, we show a pie chart of gene composition on the W with regards to whether their open reading frames (ORFs) are predicted to be intact or not. The x axis is the estimated age, and y axis is the Z/W divergence level of each stratum. Within each pie chart, we show W-linked genes that cannot be found within the assembled sequences (gray), genes with disrupted ORFs that contain premature stop codons or frameshift mutations (blue), and genes with intact ORFs (red). Note that the number of missing genes may be overestimated, owing to the difficulty of assembling W-linked genes surrounded by repetitive sequences. (H) Comparison of gene expression between Z and W linked gametologs in ostrich brain and liver samples, relative to their autosomal lizard orthologs. We show significance level of Wilcoxon test comparing the gametolog expression versus that of lizard (**P < 0.01). W gametologs show a significant down-regulation of gene expression. (I) Correlation between gene loss rate and age of stratum. Each data point represents a stratum of a certain bird species (dot) in Fig. 1 or human (triangle). The data for the human Y chromosome are derived from (49, 51).
Older strata generally have lost more W-linked genes (Fig. 4, A to G), yet we find a significant negative correlation between the rate of gene loss (measured as percent of gene loss per million years) versus the age of stratum in birds (R2 = 0.87, Pearson’s test P-value = 2.87 × 10−12, Fig. 4I), similar to those seen in mammalian and chicken strata (12, 16, 49). Thus, older strata appear to lose genes at a lower rate in both clades, regardless of their different types of sex chromosome inheritance. This matches theoretical predictions that interference effects between linked loci (Hill-Robertson effects), which are ultimately driving Y/W degradation, diminish over evolutionary time owing to the decrease in the number of functional genes that are targets for selection in older strata (49).
Discussion
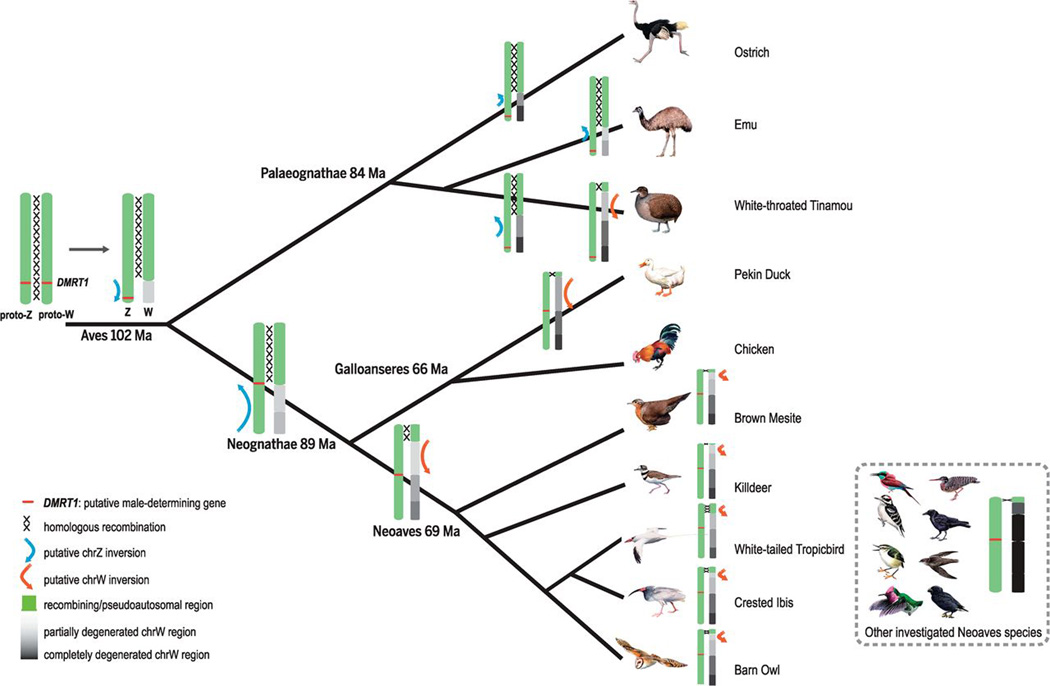
Our results (Fig. 1, summarized in fig. S23) allow us to reconstruct a likely evolutionary formation trajectory of avian sex chromosome strata (detailed in Fig. 5). We hypothesize that genetic sex determination involving DMRT1 originated in an ancestor of all birds, and a Z-linked inversion suppressing recombination between chrZ and chrW may have been selected by the accumulation of sexually antagonistic alleles nearby DMRT1, forming S0, the only stratum shared among all bird species. This event was followed by numerous independent inversions along the proto–sex chromosome pair accompanying various speciation events. Each event resulted in spreading recombination suppression across genomic regions adjacent to the non-recombining region and decreasing the size of the PAR. Separate evolutionary strata emerged among the different Palaeognathae species. We propose that Neognathae stratum S1 emerged in the ancestor of Neognathae, possibly involving the translocation of the Z-linked DMRT1 gene toward the center of the Z chromosome. This was followed by the independent formation of stratum S2 in Galliformes, Anseriformes, and the ancestor of Neoaves. Finally some, but not all, Neognathae lineages formed independent S3 strata.
Fig. 5. Hypothesized formation scenarios of avian evolutionary strata.
We designate DMRT1 on the chromosome with the red bar, and homologous recombination between proto–sex chromosomes at PAR (in green) with intersecting lines. Gray or black regions on the W chromosome represent the female-specific region lacking recombination, with darker color representing higher level of differentiation between Z/W. We also show inferred Z- or W-linked inversions that have led to the formation of certain evolutionary strata on the tree.
Several general patterns also emerge from comparing the evolutionary history of avian and mammalian sex chromosomes. Unlike in mammals, no sex-autosome fusions contributed to the formation of ancestral avian evolutionary strata, but recombination became abolished in a similar stepwise manner in both birds and mammals. The observation that recombination suppression gradually spreads outwards from the region containing the putative sex-determining gene DMRT1 (Fig. 1 and fig. S9) in birds mimics patterns of strata formation in mammals, where the oldest stratum contains the sex-determining SRY gene. We infer that if inversions caused recombination suppression in birds, they must have occurred on both the Z and W chromosomes (Fig. 5). In contrast, all the inversions suppressing recombination in mammals probably occurred on the Y (13).
This different pattern is consistent with a model in which sexual antagonism drives the evolution of suppressed recombination between sex chromosomes (8, 50). Assuming that most sexual antagonism is caused by sexual selection in males, alleles that are beneficial to males but detrimental to females may be more common than female-beneficial male-detrimental mutations (8, 50). In XY systems, Y-linked inversions that trap such male-beneficial female-detrimental alleles into the male-limited region along the Y will be immediately selected for. In ZW systems, however, neither Z- nor W-linked inversions can restrict such alleles to males, but either can be favored by preventing male-beneficial sexual antagonistic alleles from recombining onto the female-specific W chromosome. In the chicken, male function genes are accumulating on the Z chromosome, including a large array of testis-expressed genes termed “Z amplicons” (3), providing empirical support for the accumulation of sex-specific genes along the sex chromosomes (table S7).
In conclusion, our results provide a detailed view of the evolutionary history of avian sex chromosomes, involving both shared and lineage-specific recombination suppression events. These events differ by their emergence times, the genomic regions spanned, and the tempo of Z/W differentiation within the affected region, which together underlie the diversity of avian sex chromosome composition. Even within homologous strata of the same age and size, we find marked variation in the extent to which they have lost their W-linked genes in different lineages. It is not entirely clear why rates of avian sex chromosome evolution should differ so dramatically, and whether, or to what extent, this can be attributed to sexual selection, which is widely manifested in birds. Surprisingly, several Neognathae species have retained a large number of genes within the nonrecombining segment of their W chromosome (Table 1), contrary to the general assumption that ancient sex-linked regions invariably degenerate almost completely.
Supplementary Material
Acknowledgments
We thank B. Vicoso, R. Bowie, M. Sayres, I. Nanda, and D. Charlesworth for insightful comments and discussions; A. Thais for sharing the green anole lizard photo; O. Ryder and W. Wang for providing the ostrich sample; and the teams of N. Xiao and Z. Bin at OpGen Inc. for performing the optical mapping and technical assistance. We also thank five anonymous reviewers for constructive comments and suggestion. This work was supported by the National Key Basic Research Program of China (973 Program: 2013CB945200) to G.Z. Q.Z. is supported by grants from the National Institutes of Health (GM076007 and GM093182) and a Packard Fellowship to D.B. Raw sequence reads and assembled genomes and gene annotation are deposited at http://phybirds.genomics.org.cn/genome.jsp.
REFERENCE AND NOTES
- 1.Bull JJ. Evolution of Sex Determining Mechanisms. Menlo Park, CA: Benjamin/Cummings; 1983. [Google Scholar]
- 2.Fridolfsson AK, et al. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8147–8152. doi: 10.1073/pnas.95.14.8147. pmid: 9653155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellott DW, et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature. 2010;466:612–616. doi: 10.1038/nature09172. pmid: 20622855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortez D, et al. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508:488–493. doi: 10.1038/nature13151. pmid: 24759410. [DOI] [PubMed] [Google Scholar]
- 5.Ross MT, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. pmid: 15772651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. pmid: 11127901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. pmid: 12815422. [DOI] [PubMed] [Google Scholar]
- 8.Rice WR. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution. 1987;41:911–914. doi: 10.1111/j.1558-5646.1987.tb05864.x. [DOI] [PubMed] [Google Scholar]
- 9.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. pmid: 10542153. [DOI] [PubMed] [Google Scholar]
- 10.Nam K, Ellegren H. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics. 2008;180:1131–1136. doi: 10.1534/genetics.108.090324. pmid: 18791248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G, et al. Comparative analysis of mammalian Y chromosomes illuminates ancestral structure and lineagespecific evolution. Genome Res. 2013;23:1486–1495. doi: 10.1101/gr.154286.112. pmid: 23788650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellott DW, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. pmid: 24759411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemaitre C, et al. Footprints of inversions at present and past pseudoautosomal boundaries in human sex chromosomes. Genome Biol. Evol. 2009;1:56–66. doi: 10.1093/gbe/evp006. pmid: 20333177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandstedt SA, Tucker PK. Evolutionary strata on the mouse X chromosome correspond to strata on the human X chromosome. Genome Res. 2004;14:267–272. doi: 10.1101/gr.1796204. pmid: 14762062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayers KL, et al. RNA sequencing reveals sexually dimorphic gene expression before gonadal differentiation in chicken and allows comprehensive annotation of the W-chromosome. Genome Biol. 2013;14:R26. doi: 10.1186/gb-2013-14-3-r26. pmid: 23531366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright AE, Moghadam HK, Mank JE. Trade-off between selection for dosage compensation and masculinization on the avian Z chromosome. Genetics. 2012;192:1433–1445. doi: 10.1534/genetics.112.145102. pmid: 22997237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handley LJ, Ceplitis H, Ellegren H. Evolutionary strata on the chicken Z chromosome: Implications for sex chromosome evolution. Genetics. 2004;167:367–376. doi: 10.1534/genetics.167.1.367. pmid: 15166161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellegren H, Carmichael A. Multiple and independent cessation of recombination between avian sex chromosomes. Genetics. 2001;158:325–331. doi: 10.1093/genetics/158.1.325. pmid: 11333240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Kloet RS, de Kloet SR. Evolution of the spindlin gene in birds: Independent cessation of the recombination of sex chromosomes at the spindlin locus in neognathous birds and tinamous, a palaeognathous avian family. Genetica. 2003;119:333–342. doi: 10.1023/b:gene.0000003842.72339.df. pmid: 14686612. [DOI] [PubMed] [Google Scholar]
- 20.García-Moreno J, Mindell DP. Rooting a phylogeny with homologous genes on opposite sex chromosomes (gametologs): A case study using avian CHD. Mol. Biol. Evol. 2000;17:1826–1832. doi: 10.1093/oxfordjournals.molbev.a026283. pmid: 11110898. [DOI] [PubMed] [Google Scholar]
- 21.Rutkowska J, Lagisz M, Nakagawa S. The long and the short of avian W chromosomes: No evidence for gradual W shortening. Biol. Lett. 2012;8:636–638. doi: 10.1098/rsbl.2012.0083. pmid: 22417794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hackett SJ, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. pmid: 18583609. [DOI] [PubMed] [Google Scholar]
- 23.Jarvis ED, et al. Whole genome analyses resolve the early branches to the tree of life of modern birds. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa A, Murata K, Mizuno S. The location of Z- and W-linked marker genes and sequence on the homomorphic sex chromosomes of the ostrich and the emu. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4415–4418. doi: 10.1073/pnas.95.8.4415. pmid: 9539751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishida-Umehara C, et al. Differentiation of Z and W chromosomes revealed by replication banding and FISH mapping of sex-chromosome-linked DNA markers in the cassowary (Aves, Ratitae) Chromosome Res. 1999;7:635–640. doi: 10.1023/a:1009236103013. pmid: 10628664. [DOI] [PubMed] [Google Scholar]
- 26.Tsuda Y, Nishida-Umehara C, Ishijima J, Yamada K, Matsuda Y. Comparison of the Z and W sex chromosomal architectures in elegant crested tinamou (Eudromia elegans) and ostrich (Struthio camelus) and the process of sex chromosome differentiation in palaeognathous birds. Chromosoma. 2007;116:159–173. doi: 10.1007/s00412-006-0088-y. pmid: 17219176. [DOI] [PubMed] [Google Scholar]
- 27.Pigozzi MI. Diverse stages of sex-chromosome differentiation in tinamid birds: Evidence from crossover analysis in Eudromia elegans and Crypturellus tataupa. Genetica. 2011;139:771–777. doi: 10.1007/s10709-011-9581-1. pmid: 21567220. [DOI] [PubMed] [Google Scholar]
- 28.Pigozzi MI. Origin and evolution of the sex chromosomes in birds. Biocell. 1999;23:79–95. pmid: 10904535. [PubMed] [Google Scholar]
- 29.Zhang G, et al. Comparative genomics across modern bird species reveal insights into avian genome evolution and adaptation. Science. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, et al. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nat. Genet. 2013;45:776–783. doi: 10.1038/ng.2657. pmid: 23749191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vicoso B, Kaiser VB, Bachtrog D. Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc. Natl. Acad. Sci. U.S.A. 2013;110:6453–6458. doi: 10.1073/pnas.1217027110. pmid: 23547111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye L, et al. A vertebrate case study of the quality of assemblies derived from next-generation sequences. Genome Biol. 2011;12:R31. doi: 10.1186/gb-2011-12-3-r31. pmid: 21453517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan WL, et al. Genome-wide patterns of genetic variation in two domestic chickens. Genome Biol. Evol. 2013;5:1376–1392. doi: 10.1093/gbe/evt097. pmid: 23814129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Materials and methods are available as supplementary material on Science Online.
- 35.Nanda I, Schlegelmilch K, Haaf T, Schartl M, Schmid M. Synteny conservation of the Z chromosome in 14 avian species (11 families) supports a role for Z dosage in avian sex determination. Cytogenet. Genome Res. 2008;122:150–156. doi: 10.1159/000163092. pmid: 19096210. [DOI] [PubMed] [Google Scholar]
- 36.Dalloul RA, et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): Genome assembly and analysis. PLOS Biol. 2010;8:e1000475. doi: 10.1371/journal.pbio.1000475. pmid: 20838655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warren WC, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. pmid: 20360741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellegren H, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491:756–760. doi: 10.1038/nature11584. pmid: 23103876. [DOI] [PubMed] [Google Scholar]
- 39.Smith CA, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. pmid: 19710650. [DOI] [PubMed] [Google Scholar]
- 40.Guioli S, Lovell-Badge R, Turner JM. Error-prone ZW pairing and no evidence for meiotic sex chromosome inactivation in the chicken germ line. PLOS Genet. 2012;8:e1002560. doi: 10.1371/journal.pgen.1002560. pmid: 22412389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yazdi HP, Ellegren H. Old but not (so) degenerated—slow evolution of largely homomorphic sex chromosomes in ratites. Mol. Biol. Evol. 2014;31:1444–1453. doi: 10.1093/molbev/msu101. pmid: 24618361. [DOI] [PubMed] [Google Scholar]
- 42.Alföldi J, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477:587–591. doi: 10.1038/nature10390. pmid: 21881562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bradnam KR, et al. Assemblathon 2: Evaluating de novo methods of genome assembly in three vertebrate species. Gigascience. 2013;2:10. doi: 10.1186/2047-217X-2-10. pmid: 23870653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawai A, et al. Different origins of bird and reptile sex chromosomes inferred from comparative mapping of chicken Z-linked genes. Cytogenet. Genome Res. 2007;117:92–102. doi: 10.1159/000103169. pmid: 17675849. [DOI] [PubMed] [Google Scholar]
- 45.Kawai A, et al. The ZW sex chromosomes of Gekko hokouensis (Gekkonidae, Squamata) represent highly conserved homology with those of avian species. Chromosoma. 2009;118:43–51. doi: 10.1007/s00412-008-0176-2. pmid: 18685858. [DOI] [PubMed] [Google Scholar]
- 46.Berlin S, Ellegren H. Fast accumulation of nonsynonymous mutations on the female-specific W chromosome in birds. J. Mol. Evol. 2006;62:66–72. doi: 10.1007/s00239-005-0067-6. pmid: 16320115. [DOI] [PubMed] [Google Scholar]
- 47.Itoh Y, Kampf K, Arnold AP. Disruption of FEM1C-W gene in zebra finch: Evolutionary insights on avian ZW genes. Chromosoma. 2009;118:323–334. doi: 10.1007/s00412-008-0199-8. pmid: 19139913. [DOI] [PubMed] [Google Scholar]
- 48.Hughes JF, et al. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature. 2012;483:82–86. doi: 10.1038/nature10843. pmid: 22367542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bachtrog D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics. 2008;179:1513–1525. doi: 10.1534/genetics.107.084012. pmid: 18562655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charlesworth D, Charlesworth B. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet. Res. 1980;35:205–214. doi: 10.1017/s0016672300014051. pmid: 6930353. [DOI] [PubMed] [Google Scholar]
- 51.Wilson Sayres MA, Makova KD. Gene survival and death on the human Y chromosome. Mol. Biol. Evol. 2013;30:781–787. doi: 10.1093/molbev/mss267. pmid: 23223713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.