Abstract
We present version 10 of OrthoMaM, a database of orthologous mammalian markers. OrthoMaM is already 11 years old and since the outset it has kept on improving, providing alignments and phylogenetic trees of high-quality computed with state-of-the-art methods on up-to-date data. The main contribution of this version is the increase in the number of taxa: 116 mammalian genomes for 14,509 one-to-one orthologous genes. This has been made possible by the combination of genomic data deposited in Ensembl complemented by additional good-quality genomes only available in NCBI. Version 10 users will benefit from pipeline improvements and a completely redesigned web-interface.
Keywords: orthologous sequences, mammals, coding sequences, phylogenomics, comparative genomics
OrthoMaM is a database of high-quality orthologous sequence alignments and phylogenetic trees from mammalian genomes. It has been, for instance, used for developing new molecular markers, inferring mammalian phylogenies, simulating sequences for testing alignment filtering methods, and studying the evolution of base composition in protein-coding sequences. Previous versions of our database only included mammalian genomes from Ensembl (Ranwez et al. 2007; Douzery et al. 2014). With the progress of sequencing techniques, the number of genomes included in NCBI, but not yet in Ensembl, grows each year. This motivated us to totally rethink our database to include the annotated mammalian genomes available in NCBI only.
To identify the core set of orthologous sequences to be used, we rely on the OrthoMaM v8 pipeline described in Douzery et al. (2014). Briefly, we isolate 1-to-1 orthologous genes among pairs of our pillar placental species (Homo-Mus, Homo-Canis, and Mus-Canis) by using Ensembl v91 annotations (Zerbino et al. 2018). These clusters are then enriched by adding sequences of 69 additional mammals annotated in Ensembl v91 as 1-to-1 orthologues to the human gene, and turned into clusters of 1-to-1 orthologous CDSs by selecting the longest transcript of each gene. These proto-clusters are aligned using MAFFT at the amino acid level (Katoh and Standley 2013).
The CDSs of the 47 annotated mammal genomes available in NCBI (Rigden and Fernández 2017) but not in Ensembl as of March 2018 are then used to enrich the proto-alignments. For each proto-alignment, an HMM profile is created via hmmbuild using default parameters of the HMMER toolkit (Eddy 2011). Additionally, all HMM profiles are concatenated and summarized using hmmpress to construct an HMM database. Then, for each NCBI CDS Ci, hmmscan is used on the HMM database to get the best hits among the proto-alignments—denoted bestAl(Ci). For each proto-alignment Aj, the most similar sequences for each species S—denoted bestSeq(S, Aj)—are detected via hmmsearch. Outputs from hmmsearch and hmmscan are discarded if the first hit score is not substantially better than the second (hit2 < 0.9 hit1) and are combined in a best-reciprocal-hit fashion: Suppose that we are given a NCBI CDS sequence Ci for species Sk; then denoting Aj the best proto-alignment for Ci, if the first sequence of bestSeq(Sk, bestAl(Ci)) equals Ci and none of the other sequences Cz≠ Ci in bestSeq(Aj) is such that bestAl(Sk, Cz) =Aj, then Ci is considered to belong to Aj. This ensures our orthology predictions for the NCBI CDSs to be robust. The enriched orthologous clusters have been thoughtfully aligned and filtered via the OMM_MACSE pipeline to construct high-quality codon alignments relying on MACSE v2 (Ranwez et al. 2018), MAFFT (Katoh and Standley 2013), and HMMcleaner (Philippe et al. 2017). Phylogenetic trees for each CDS alignment are constructed under maximum likelihood (ML) with RAxML (Stamatakis 2014) under the GTR+Γ model (Yang 1994).
OrthoMaM v10 also includes an ad hoc nonorthologous sequence detection method. For a given marker, the distance between the most recent common ancestor (MRCA) of placental sequences and any sequence of the marker, denoted here Si, depends mostly on two factors: the considered marker (some genes evolve faster) and the genome to which Si belongs (some species evolve faster). We use linear regression to explain the patristic distance on inferred ML trees between a sequence Si and the placental MRCA based on those two factors. When the linear regression prediction departs from the observed patristic distance (standardized residual >3), we consider the sequence as spurious and remove it.
As in the previous versions of our database, we use exon positions in CDS alignments to infer exon orthology and alignments (see Douzery et al. 2014 for details). Phylogenetic trees for each exon alignment are reconstructed as for CDSs.
OrthoMaM v10 database is available at http://www.orthomam.univ-montp2.fr/orthomam_v10/. For each CDS and exon marker, we provide gene level information (gene name, GO annotation), full sequence traceability information (sequence identifier in Ensembl/NCBI, filtering details), nucleotide and amino acid alignments, phylogenetic trees, as well as several evolutionary indicators such as relative evolutionary rate and G + C content. The improved web-interface (see fig. 1) provides a user-friendly way to query the database based on alignment content (number of sequences, number of species that should be represented, and alignment length), evolutionary indicators, or sequence similarity using blast.
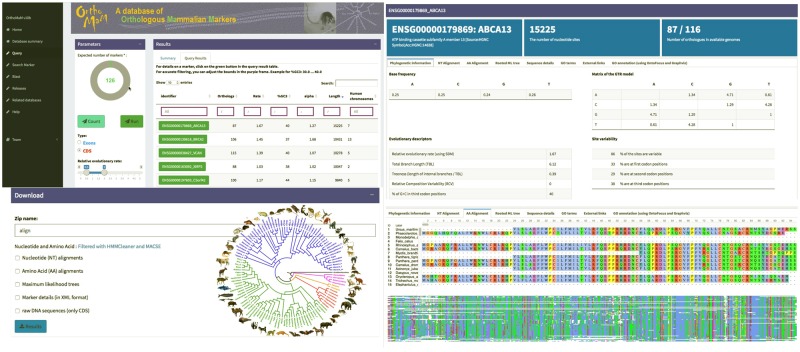
Fig. 1.
Screenshots from the OrthoMaM website version 10. Here, the database is queried for CDSs with a relative evolutionary rate between 0.5 and 2, a GC3 between 23% and 45%, a parameter of the gamma distribution ranging from 1 to 2, and an alignment length >1,000 characters (top left). The result returns 126 target CDSs, for which information can be downloaded (bottom left). The longest CDS (ABCA13) is visualized with the corresponding phylogenetic information (top right) and AA alignment (bottom right).
This new reactive web site, which has been completely redesigned to improve the user experience, the triplication of the number of species, the major update of the pipeline for orthology prediction and sequence filtering should make OrthoMaM v10 a central resource for anyone interested in mammalian comparative genomics.
Acknowledgments
Our project largely benefited from the Montpellier Bioinformatics Biodiversity cluster computing platform. This work was supported by the Agence Nationale de la Recherche “Investissements d’avenir/Bioinformatique” (ANR-10-BINF-01-02 project Ancestrome), Labex CeMEB “Centre Méditerranéen de l’Environnement et de la Biodiversité” (ANR-10-LABX-0004), and the European Research Council (ERC-2015-CoG-683257 project ConvergeAnt). This is contribution ISEM 2018-292 of the Institut des Sciences de l’Evolution de Montpellier.
References
- Douzery EJP, Scornavacca C, Romiguier J, Belkhir K, Galtier N, Delsuc F, Ranwez V.. 2014. OrthoMaM v8: a database of orthologous exons and coding sequences for comparative genomics in mammals. Mol Biol Evol. 31(7):1923–1928. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, de Vienne DM, Ranwez V, Roure B, Baurain D, Delsuc F.. 2017. Pitfalls in supermatrix phylogenomics. Eur J Taxon. 283:1–25. [Google Scholar]
- Ranwez V, Delsuc F, Ranwez S, Belkhir K, Tilak MK, Douzery EJP.. 2007. OrthoMaM: a database of orthologous genomic markers for placental mammal phylogenetics. BMC Evol Biol. 7(1):241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwez V, Douzery EJP, Cambon C, Chantret N, Delsuc F.. 2018. MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol Biol Evol. 35(10):2582–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden D. J., Fernández X. M.. 2017. The 2018 Nucleic Acids Research database issue and the online molecular biology database collection. Nucleic acids research. 46(D1):D1–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 39(3):306–314. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, et al. 2018. Ensembl 2018. Nucleic Acids Res. 46(D1):D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]