Abstract
Since 2012, have we in Denmark observed an increase of invasive pneumococcal infections (IPD) due to Streptococcus pneumoniae serotype 24F. We here present epidemiological data on 24F IPD cases, and characterization of 48 24F clinical isolates based on clonal relationship, antimicrobial resistance (AMR) determinants and virulence factors. IPD surveillance data from (1999–2016) were used to calculate the incidence and age-distribution of serotype 24F IPD and the effect of pneumococcal conjugated vaccines (PCV). Characterization of forty-eight 24F isolates (14.7% of all 24F isolates from the period) was based on whole-genome sequencing analysis (WGS). The IPD cases of serotype 24F showed a significant increase (p < 0.05) for all age groups after the PCV-13 introduction in 2010. The majority of tested 24F isolates consisted of two MLST types, i.e. the ST72 and the ST162. Serotype 24F IPD increased in Denmark after the PCV-13 introduction in parallel with an increase of the ST162 clone. The genotypic penicillin binding protein (PBP) profile agreed with the phenotypical penicillin susceptibility. The virulence genes lytA, ply, piaA, piaB, piaC, rspB and the cpsA/wzg were detected in all 24F isolates, while the pspA and zmpC genes were absent.
Introduction
Streptococcus pneumoniae is a ubiquitous bacterium present in the commensal bacterial community in the human nasopharynx. It is responsible for non-invasive infections as well as invasive pneumococcal disease (IPD) with high morbidity and mortality especially among young children and the elderly1. The introduction of the pneumococcal conjugated vaccine (PCV) provided an effective protection against IPD in children. The first PCV on the global market was Prevenar 7 (PCV-7) (Pfizer Vaccines) in 2000 including seven different serotypes (4, 6B, 9V, 14, 18C, 19F and 23F), followed by the 10-valent pneumococcal-conjugate vaccine (PCV-10) (Synflorix, GlaxoSmithKline Biologicals) in 2009 including the serotypes 1, 4, 5, 6B, 7F, 9V, 14, 18C, 19F and 23F, and by the Prevenar 13 vaccine (PCV-13) (Pfizer Vaccines) in 2010 including the serotypes 1, 3, 4, 5, 6A, 6B, 7F, 9V, 14, 18C, 19A, 19F and 23F1. Besides the conjugate vaccines, a pneumococcal polysaccharide vaccine (PPV-23) (Pneumovax®, Merck) based on purified capsular polysaccharides from 23 different serotypes, was introduced in 1996. The PPV-23 vaccine is recommended for patients older than 2 years of age and of high risk for IPD and for the 65+ years age group2,3.
With the introduction of PCV, a significant reduction of IPD cases caused by the included PCV serotypes was seen, but at the same time serotype replacement was observed, with the appearance of new pneumococcal serotypes not included in the vaccines1. One of the non-PCV serotypes that has emerged in recent years is serotype 24F4,5. The non-vaccine serotype 24F is responsible for many penicillin non-susceptible related IPD cases6. Serotype 24F is also linked to erythromycin-clindamycin and tetracycline resistance and is one of the main serotypes that was recovered from young children with IPD between 2011 and 2012 in France7. After the introduction of PCV13 in 2010 in France, serotype 24F, including other non-vaccine serotypes (10A, 12F and 15A), were responsible for approximately 39% of pneumococcal meningitis (PM) cases in children below 5 years of age from 2012 to 20148.
During the last five years, a small increase of IPD cases with serotype 24F has been observed in Denmark1. Because the serotype 24F is an emerging non-pneumococcal vaccine serotype, which to our knowledge is not a part of any pneumococcal vaccine or as part of planned future pneumococcal vaccine. Is it therefore essential to monitor the epidemiology and susceptibility of serotype 24F, and to provide information to the international community that measures against serotype 24F are needed.
The intention of this study was to present epidemiological data based on IPD cases in Denmark in the period from 1999 to 2016. Furthermore, to present a detailed characterization of forty-eight 24F isolates (representing 14.7% of all 24F isolates from the period) using whole-genome sequencing analysis (WGS). Thus, the serotype 24F’s clonal relationship over the years and antimicrobial resistance determinants are presented. Finally, the presence of capsular, toxin and surface related genes are suggested as pneumococcal species.
Material and Methods
Strain collection
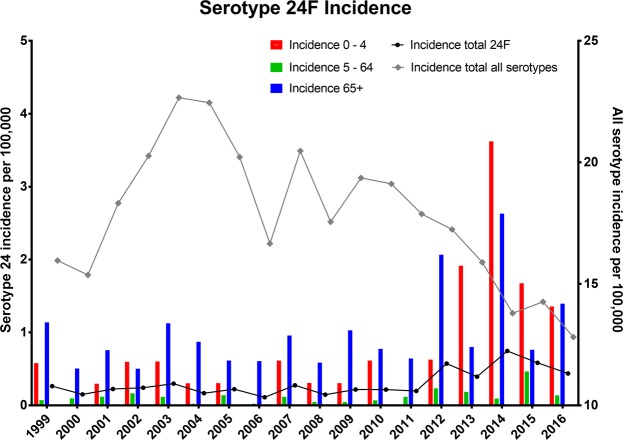
All invasive pneumococcal isolates of serotype 24F from 1999 to 2016 were retrieved from the Danish laboratory surveillance system at the national Neisseria and Streptococcus Reference Laboratory (NSR), Statens Serum Institut (SSI) (Supplementary Table 1). All the IPD serotype 24F cases reported were isolated form patients diagnosed with either bacteraemia (from blood) or meningitis (from cerebrospinal fluid). An IPD case was defined as the occurrence of S. pneumoniae in cerebrospinal fluid, blood or other normally sterile sites9. Incidence and Incidence Rate Ratio (IRR) data from 24F from 1999 to 2014 was previously presented in Slotved et al.1 including information on age, sex, serotype and origin of the pneumococcal isolate (blood, cerebrospinal fluid etc.). Data on the total IPD cases for all age groups in Denmark are presented in Fig. 1 to compare with the serotype 24F. Detailed data on the total IPD cases for all age has previously been presented1,10.
Figure 1.
The IPD incidence in Denmark from 1999 to 2016. The IPD incidence for serotype 24F in all three age groups from 1999 until 2016. Red bars (left Y-axe) represent 0–4 years, green bars (left Y-axe) represent 5–64 years and blue bars (left Y-axe) represent 65+ years. The black curve (left Y-axe) represents the total serotype 24F incidences per year. The gray curve (right Y-axe) represent the total IPD incidences for all serotypes per year.
Identification of pneumococcal isolates
The pneumococci isolates were phenotypically identified by optochin susceptibility and bile solubility tests1,11. All isolates were serotyped either by the Quellung reaction alone or by the Pneumotest Latex kit (SSI-Diagnostica, Copenhagen, Denmark) combined with the Quellung reaction using type-specific pneumococcal rabbit-antisera as previously described (SSIDiagnostica, Copenhagen, Denmark)11.
Characterization of 48 clinical selected isolates
Forty-eight Streptococcus pneumoniae 24F isolates were selected for detailed characterization using whole genome sequencing (Table 1). Forty-seven 24F isolates were isolated from blood and spinal fluid in the period 1999 to 2017, and one historical isolate 24F was cultured from the trachea of an eight-year-old boy in 1943. The 47 isolates were selected to represent 26 children below five years of age and 21 elderly persons above 64 years of age. Otherwise the isolates were randomly selected during the period 1999–2017 to provide a molecular picture of the serotype 24F isolates in Denmark. The isolates represent 14.7% of the total number of 24F IPD cases during the period.
Table 1.
Characteristics of the 47 invasive 24F isolates and one trachea 24F historical isolate from 1943.
| Year | Strain number | Age | Sex | Sample type | Capsular genes (Genotype) | MLST (ST) | PEN* (mg/L) | ERY* (mg/L) | CLI* (mg/L) | ermBd (Tn family) | tetMd (Tn family) | PBP 1a | PBP 2b | PBP 2x |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1943 | 24F-1943 | 8 | Male | Trachea | 24F | 7179 | S (0.032) | S (28.22) | S (25.06) | NR | NR | 0 | 0 | 0 |
| 1999 | 0630-1999 | 0 | Female | Spinal fluid | 24 Group | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2002 | 1030-2002 | 1 | Male | Blood | 24F | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2003 | 0373-2003 | 0 | Female | Blood | 24 Group | (12-19-2-?-6-22-14)b | R (1.0) | R (7.42) | R (6.77) | +(none)c | +(Tn1545) | 17 | 15 | 22 |
| 2004 | 1186-2004 | 0 | Female | Blood | 24F | 72 | S (0.047) | S (27.99) | S (24.08) | NR | NR | 2 | 0 | 0 |
| 2004 | 1073-2004 | 75 | Female | Blood | 24F | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2005 | 0527-2005 | 0 | Male | Spinal fluid | 24F | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2005 | 0741-2005 | 78 | Male | Blood | 24 Group | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2006 | 0194-2006 | 86 | Female | Blood | 24 Group | 72 | S (0.032) | S (27.45) | S (24.47) | NR | NR | 2 | 0 | 0 |
| 2006 | 0579-2006 | 79 | Male | Blood | 24 Group | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2006 | 1119-2006 | 75 | Male | Blood | 24F | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2007 | 0834-2007 | 4 | Female | Blood | 24F | 162 | S (0.047) | S (29.11) | S (24.59) | +(none)c | +(Tn916) | 2 | 0 | 28 |
| 2007 | 0258-2007 | 92 | Female | Blood | 24 Group | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2007 | 1134-2007 | 83 | Female | Blood | 24 Group | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2008 | 0266-2008 | 1 | Female | Spinal fluid | 24F | 11100 | S | S | S | NR | NR | 2 | 18 | 0 |
| 2008 | 1108-2008 | 86 | Female | Blood | 24 Group | 230 | I (0.75)a | R | R | +(none)c | +(Tn1545) | 17 | 15 | 22 |
| 2009 | 0811-2009 | 1 | Male | Blood | 24F | 162 | S (0.032) | S (0.032) | S (25.31) | +(none)c | +(Tn916) | 2 | 0 | 1 |
| 2009 | 0273-2009 | 86 | Female | Blood | 24 Group | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2009 | 1199-2009 | 94 | Male | Blood | 24 Group | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2010 | 0805-2010 | 1 | Male | Spinal fluid | 24F | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2010 | 0510-2010 | 2 | Male | Blood | 24 Group | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2010 | 0993-2010 | 88 | Female | Blood | 24 Group | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2011 | 1853-2011 | 91 | Male | Blood | 24F | 11131 | S | S | S | NR | NR | 0 | 0 | 0 |
| 2012 | 0263-2012 | 1 | Male | Blood | 24F | 162 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2012 | 0272-2012 | 76 | Female | Blood | 24F | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2012 | 0390-2012 | 72 | Male | Blood | 24 Group | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2013 | 0682-2013 | 1 | Male | Blood | 24F | 11100 | S | S | S | NR | NR | 2 | 18 | 0 |
| 2013 | 0699-2013 | 1 | Female | Blood | 24F | 4253 | I (0.5) | R (0.0) | R (0.1) | +(none)c | +(Tn1545) | 17 | 15 | 117 |
| 2013 | 0128-2013 | 75 | Female | Blood | 24 Group | 72 | S (0.032) | S (29.31) | S (24.25) | NR | NR | 2 | 0 | 0 |
| 2013 | 0366-2013 | 76 | Male | Blood | 24 Group | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2014 | 0149-2014 | 1 | Male | Spinal | 24F | 162 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2014 | 0404-2014 | 1 | Male | Blood | 24 Group | 162 | S (0.03) | R (0.1) | R (0.1) | +(none)c | +(Tn916) | 2 | 0 | 28 |
| 2014 | 0669-2014 | 1 | Female | Spinal fluid | 24F | 11100 | S | S | S | NR | NR | 2 | 18 | 0 |
| 2014 | 0747-2014 | 0 | Female | Blood | 24 Group | 162 | S (0.03) | R (0.1) | R (0.1) | +(none)c | +(Tn916) | 2 | 0 | 28 |
| 2014 | 0100-2014 | 87 | Female | Blood | 24 Group | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2015 | 0090-2015 | 1 | Female | Blood | 24 Group | 72 | S (0.016)a | S | S | NR | NR | 2 | 0 | 0 |
| 2015 | 0096-2015 | 1 | Male | Blood | 24F | 162 | S (0.032)a | S | S | NR | NR | 2 | 0 | 0 |
| 2015 | 0182-2015 | 1 | Female | Blood | 24 Group | 11100 | S (0.016)a | S | S | NR | NR | 2 | 18 | 0 |
| 2015 | 0233-2015 | 1 | Male | Spinal fluid | 24 Group | 4253 | I (0.5) | R (0.1) | R (0.1) | +(none)c | +(Tn1545) | 17 | 15 | 116 |
| 2015 | 0641-2015 | 1 | Male | Spinal fluid | 24 Group | 162 | S (0.016)a | S | S | NR | NR | 2 | 0 | 0 |
| 2015 | 0120-2015 | 70 | Male | Blood | 24F | 162 | S (0.016)a | S | S | NR | NR | 2 | 0 | 0 |
| 2015 | 0223-2015 | 81 | Female | Blood | 24F | 162 | S (0.03) | R (0.1) | R (0.1) | +(none)c | +(Tn916) | 2 | 0 | 28 |
| 2016 | 0104-2016 | 4 | Female | Blood | 24F | 72 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2016 | 0201-2016 | 2 | Male | Blood | 24 Group | 162 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2016 | 0041-2016 | 71 | Female | Blood | 24 Group | 162 | S | S | S | NR | NR | 2 | 0 | 0 |
| 2016 | 0350-2016 | 68 | Male | Blood | 24 Group | 162 | S (0.03) | R (0.2) | R (0.2) | +(none)c | +(Tn916) | 2 | 0 | 28 |
| 2017 | 0068-2017 | 1 | Female | Blood | 24F | (7-11-10-1-6-?-14)b | S (0.03) | R (0.1) | R (0.2) | +(none)c | +(Tn916) | 2 | 0 | 28 |
| 2017 | 0345-2017 | 1 | Male | Blood | 24 Group | 162 | S (0.03) | R (0.1) | R (0.1) | +(none)c | +(Tn916) | 2 | 0 | 28 |
*Isolates tagged with and S (sensitive) or R (resistance) was only tested with disk diffusion. For penicillin was S/R tagged only based on oxacillin zone diameter, penicillin test was not performed on the isolates. For isolates with an MIC value was the sensititre test performed, except for a few isolates tagged with an “a”.
aPenicillin Etest.
bHas been submitted to the curators of the MLST database.
cDid not harbor any mobile elements of the Tn-family37.
dNR stands for “not relevant” and tag the isolates where ermB and tetM genes were not detected and Tn linkage therefore is not relevant.
Molecular species identification
The isolates were sequenced by paired-end Illumina sequencing. Genomic DNA was extracted using a DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany) and fragment libraries were constructed using a Nextera XT Kit (Illumina, Little Chesterford, UK) followed by 250-bp paired-end sequencing (MiSeqTM; Illumina) according to the manufacturer’s instructions. The paired-end Illumina data were de novo assembled using CLCbio’s Genomics Workbench v.7.5 QIAGEN) reporting only contigs >500 bp using standard settings.
Bioinformatics, including Blast was done using the software the CLC Main Workbench (Version 7.9.1, www.qiagenbioinformatics.com).
All 48 isolates were species identified according to the description by Scholz et al.12. Briefly, the 16S rRNA sequence (accession number: AY485600) identified by Arbique et al.13 was used to identify the nucleotide position at 20312. The S. pneumoniae identification was based on the location of cytosine at the 203 position, while the existence of adenine-residues suggested that the species belonged to another Streptococcus species12.
The presence/absence of a gene was based on a cut-off of 80% coverage and a 95% identity for a positive gene detection in this study14.
The genomic sequence data for the 48 isolates are deposited in the Genbank (https://www.ebi.ac.uk/ena) (ENA project no. PRJEB31691).
The species identification was confirmed by Multilocus Sequence Analysis (MLSA) analysis using the 9 housekeeping genes (ddl, gdh, rpoB, sodA, map, pfl, ppaC, pyk, tuf) as described by Bishop et al.15 and Killian et al.16.
Molecular characterization of their capsular genes
Sequences from all 48 isolates were checked for the 92 capsular polysaccharide genes (CPS genes) by BLAST. The FASTA files for the capsular locus sequences were retrieved from the NCBI database, using the accession numbers CR931632-CR931722, JF911515.1 and HV580364.117,18.
Identification of serogroup/type 24F was performed according to the presence/absence of genes using a cut-off of 80% coverage and a 95% identity as described by Sheppard et al.14 and Kapatai et al.18. However, differentiating further into a specific serotype within the group 24 can be difficult18. We therefore only presented genotypes, where the coverage and identity clearly indicated a specific genotype.
Multilocus sequence typing (MLST)
MLST was performed using the PubMLST DataBase (https://pubmlst.org/spneumoniae/) to identify the sequence type (ST) for each of the 24F S. pneumoniae strains. Analysis of the STs and assignment to CC was performed using PHYLOViZ 2.0 programme (http://phyloviz.readthedocs.io/en/latest/#). The STs that shared at least six of seven allelic variants composed a CC (clonal complex)19.
A phylogenetic tree based on single nucleotide polymorphisms (SNP’s) analysis of the core genome was performed on the 48 isolates. Identification of SNP’s’ was performed using BWA-mem for mapping and GATK with filtering set to remove positions with less than 10-fold depth and 90% unambiguous variant calls as implemented in NASP20 against isolate 0100-2014’s chromosome, which was used as a reference strain in the SNP alignment after removal of duplicated regions using NUCmer. The resulting SNP matrix was purged for recombination using Gubbins21. FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/, accessed 25-09-2018) was used to visualize the phylogenetic tree.
Antibiotic susceptibility testing (phenotypic tests)
Screening of antibiotic susceptibility was performed on all 48 isolates by disk diffusion using Mueller–Hinton 5% blood agar with NAD (Oxoid, Denmark) incubated in ambient air with 5% CO2 at 35 °C and oxacillin, penicillin, erythromycin and clindamycin discs (Oxoid, Denmark). Some isolates were also tested using the E-test randomly, however not on a regularly basis. Isolates showing non-susceptibility were tested using a Microbroth dilution test (Sensititre, Streptococcus species MIC Plate, STP6F, Trek Diagnostic System, USA).
MIC determination of penicillin G was done by using either a gradient test (Etest; bioMérieux), before 2010, or a broth microdilution method (Sensititre, Streptococcus species MIC Plate, STP6F, Trek Diagnostic System, USA), after 2010. S. pneumoniae ATCC 49619 was used as a quality control strain. Interpretation of susceptibility was done according to the breakpoints described in EUCAST (http://www.eucast.org/clinical_breakpoints/).
We do not routinely perform phenotypical screening of tetracycline on the isolates.
Genotypic antibiotic resistance profile
Penicillin (PEN) susceptibility in pneumococci is associated with penicillin-binding proteins (PBP) which in penicillin non-susceptible strains of pneumococci are modified to low-binding-affinity versions of the native PBP1A, PBP2B and PBP2X. The 48 isolates were analyzed for their PBP signature, based on a genotyping proposal and algorithm described for PBP1A, PBP2B and PBP2X22, where the combination of the three PBP signatures determines the level of beta-lactam resistance. The 48 isolates were tested by BLAST with the published types of predictive mutations vs. resistance levels of PBP1A, PBP2B and PBP2X proteins as described in Li et al.22 (Table 1).
Erythromycin (ERY), clindamycin (CLI) and tetracycline (TET) genotypic profile
The 48 S. pneumoniae genomes were analyzed for the genes ermB (NCBI, FJ667782), mefA (NCBI, KU739790), mefE (NCBI, NC_003098.1 (R6), and Tn916 gene tet(M) (FR671418)23. ResFinder 2.1 (https://cge.cbs.dtu.dk/services/ResFinder/) (80% ID threshold and 60% minimum length settings)24 was used to confirm the presence of the three genes.
Virulence gene profile
Presence of capsular, toxin and surface related genes used for pneumococcal virulence characterization25 and presence of genes with potential species discrimination:
The presence of the virulence genes lytA and ply26 for the identification of S. pneumoniae were tested as recommended by Centers for Disease Control (CDC)12,27.
Genes piaA, piaB and piaC (GenBank: AF338658.1) coding for membrane proteins and ATP-binding proteins as previously described14,27.
Genes zmpB28 and zmpC (AE005672:75858-76420)29, which are paralogous zinc metalloproteases30.
Genes for the pneumococcal surface proteins (pspA) (Genbank: AF516671)31 and psrp32.
The partial capsular gene cpsA, also known as the wzg gene, was tested for its presence/absence (Genbank: AF057294:2134–2473)33.
Gene rpsB encoding for ribosomal protein S229.
Data analysis
Data were analyzed using Graph Pad Prism version 7 (GraphPad Software) for descriptive statistical analysis. RStudio version 1.1.447 and R version 3.5.0 for Windows was used for all calculations of incidence rate, incidence rate ratio (IRR) and confidence interval (CI) (http://www.r-project.org/). Two tailed Fisher’s Exact Test in R was used to calculate P-values. P < 0.05 was considered significant. The raw data for calculation of the incidence rate are presented in the Supplementary Table 1.
Ethical considerations
The study was a retrospective, population-based study based on national laboratory surveillance data on isolates from patients with IPD. Since data and samples from patients were collected routinely for national surveillance purposes, no ethical approval or informed consent from patients or guardians were required. The study was approved by the Danish Data Protection Agency (record number 2007-41-0229).
Results
Prevalence/incidence of invasive pneumococcal disease in 1999–2016 due to S. pneumoniae serotype 24F
Data from the incidence rates of serotype 24F IPD cases from 1999 until 2016 are presented in Fig. 1 and Table 2. The incidence of serotype 24F IPD cases was low in the period from 1999–2007 (0.22 per 100.000, CI: 0.17–0.26) and 2008–2010 (0.19 per 100.000, CI: 0.09–0.30), while an increase in incidence (0.49 per 100.000, CI: 0.29–0.69) was observed in all age groups from 2011 to 2016. The introduction of PCV-7 in 2007 did not affect the 24F incidence in any of the age groups, while the introduction of PCV-13 in 2010 showed a significant increase in prevalence, two years after the introduction. The increase in the IRR of serotype 24F IPD varied with an IRR of 3.69 (CI: 1.30-10.53) in infants (P = 0.0083), 3.78 (CI: 1.73-8.32) in the age group from 5 to 64 years ((P < 0.001) and 1.74 (1.74 CI: 1.08–2.80) in the age group +65 (P = 0.025), although the general mean incidence was very low.
Table 2.
The mean incidence rates and 95% confidence intervals are presented for serotype 24F for all three age groups and in total. Incidence Rate Ratios (IRRs) are calculated by dividing a mean incidence from one period by a mean incidence from another period. IRR values were statistically significant (P < 0.01) calculated using a two tailed Fisher’s Exact Test.
| Age | 1999–2007 (Before introduction of PCV-7) |
2008–2010 (After introduction of PCV-7 and before introduction of PCV-13) |
2011–2016 (After introduction of PCV13) |
1999–2007 versus 2008–2010 |
2008–2010 versus 2011–2016 |
|---|---|---|---|---|---|
| Mean incidence of serotype 24F per 100,000 95% confidence interval of the mean incidence (lower CI; upper CI) |
IRR, 95% confidence interval of the mean incidence (lower CI; upper CI) Two tailed Fisher’s Exact Test |
||||
| 0–4 years | 0.37 (0.17–0.56) | 0.41 (−0.03–0.85) | 1.53 (0.22–2.84) | 1.11 (0.35–3.50) P = 0.77 |
3.69 (1.30–10.53) P = 0.0083* |
| 5–64 years | 0.10 (0.06–0.13) | 0.05 (0.02–0.09) | 0.21 (0.06–0.35) | 0.56 (0.25–1.26) P = 0.17 |
3.78 (1.72–8.32) P = 0.0002* |
| 65 + years | 0.79 (0.60–0.98) | 0.80 (0.25–1.35) | 1.38 (0.53–2.23) | 1.01 (0.61–1.67) P = 1 |
1.74 (1.08–2.80) P = 0.025* |
| Total all age group | 0.22 (0.17–0.26) | 0.19 (0.09–0.30) | 0.49 (0.29–0.69) | 0.89 (0.60–1.33) P = 0.63 |
2.53 (1.73–3.69) P < 0.00001* |
The general IPD incidence for all age groups and serotypes in Denmark was reduced since the introduction of PCV-7 (Fig. 1).
Identification of the capsular genes for serotype 24F
While all isolates were confirmed phenotypically to be serotype 24F, it was only possible to correctly identify the genotype to group level for 20/48 serotype 24F isolates. For the 28 remaining isolates the 24F capsular genes were correctly identified (Table 1).
Phylogenetic analysis by MLST
The ST72 belonging to Clonal complex (CC) 72 prevailed among the others STs and included 23 isolates. This was followed by ST162 belonging to CC156 and consisting of 14 isolates.
The CC230 consisted of our isolates (1 ST230, 2 ST4253 and 1 ST(12-19-2-?-6-22-14). Three different singletons ST7179 (1 isolate), ST11100 (4 isolates) and ST4253 (2 isolates) were detected (Table 3).
Table 3.
Virulence genes detected in the S. pneumoniae isolates. These parameters were used for positive gene detection: Cut-off of overlap as 80% and 95% identity. LytA26, ply29, piaA/piaB/piaC (AF338658), rpsB (MF375925), cpsA/wzg (AF057294:2134–2473) were detected in all 48 isolates. PspA (AF516671) and zmpC (AE005672:75858–76420)29 were not detected in any of the isolates.
| Year | Strain number | MLST (ST)/Clonal Complex | psrP (30) | zmpB (26) |
|---|---|---|---|---|
| 1943 | 24F-1943 | 7179/singleton | + | Negative |
| 1999 | 0630-1999 | 72/72 | + | Negative |
| 2002 | 1030-2002 | 72/72 | + | Negative |
| 2003 | 0373-2003 | (12-19-2-?-6-22-14)/230 | Negative | Negative |
| 2004 | 1186-2004 | 72/72 | + | Negative |
| 2004 | 1073-2004 | 72/72 | + | Negative |
| 2005 | 0527-2005 | 72/72 | + | Negative |
| 2005 | 0741-2005 | 72/72 | + | Negative |
| 2006 | 0194-2006 | 72/72 | + | Negative |
| 2006 | 0579-2006 | 72/72 | + | Negative |
| 2006 | 1119-2006 | 72/72 | + | Negative |
| 2007 | 0834-2007 | 162/156 | Negative | + |
| 2007 | 0258-2007 | 72/72 | + | Negative |
| 2007 | 1134-2007 | 72/72 | + | Negative |
| 2008 | 0266-2008 | 11100/singleton | Negative | Negative |
| 2008 | 1108-2008 | 230/230 | Negative | Negative |
| 2009 | 0811-2009 | 162/156 | Negative | + |
| 2009 | 0273-2009 | 72/72 | + | Negative |
| 2009 | 1199-2009 | 72/72 | + | Negative |
| 2010 | 0805-2010 | 72/72 | + | Negative |
| 2010 | 0510-2010 | 72/72 | + | Negative |
| 2010 | 0993-2010 | 72/72 | + | Negative |
| 2011 | 1853-2011 | 11131/singleton | + | Negative |
| 2012 | 0263-2012 | 162/156 | Negative | + |
| 2012 | 0272-2012 | 72/72 | + | Negative |
| 2012 | 0390-2012 | 72/72 | + | Negative |
| 2013 | 0682-2013 | 11100/singleton | Negative | Negative |
| 2013 | 0699-2013 | 4253/230 | Negative | Negative |
| 2013 | 0128-2013 | 72/72 | + | Negative |
| 2013 | 0366-2013 | 72/72 | + | Negative |
| 2014 | 0149-2014 | 162/156 | Negative | + |
| 2014 | 0404-2014 | 162/156 | Negative | + |
| 2014 | 0669-2014 | 11100/singleton | Negative | Negative |
| 2014 | 0747-2014 | 162/156 | Negative | + |
| 2014 | 0100-2014 | 72/72 | + | Negative |
| 2015 | 0090-2015 | 72/72 | + | Negative |
| 2015 | 0096-2015 | 162/156 | Negative | + |
| 2015 | 0182-2015 | 11100/singleton | Negative | Negative |
| 2015 | 0233-2015 | 4253/230 | Negative | + |
| 2015 | 0641-2015 | 162/156 | Negative | + |
| 2015 | 0120-2015 | 162/156 | Negative | + |
| 2015 | 0223-2015 | 162/156 | Negative | Negative |
| 2016 | 0104-2016 | 72/72 | + | Negative |
| 2016 | 0201-2016 | 162/156 | Negative | + |
| 2016 | 0041-2016 | 162/156 | Negative | + |
| 2016 | 0350-2016 | 162/156 | Negative | + |
| 2017 | 0068-2017 | (7-11-10-1-6-?-14)/156 | Negative | + |
| 2017 | 0345-2017 | 162/156 | Negative | + |
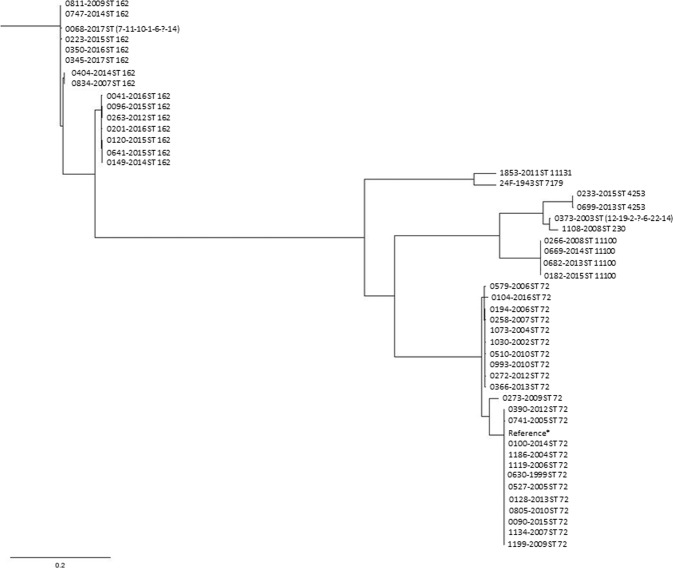
MLST sequence types correlated to clade relationships depicted in the core SNP phylogeny (Fig. 2) except for isolate 1186–2004.
Figure 2.
SNP alignment of all 48 isolates based on their SNP site location. *Isolate 0100-2014 was used as a reference strain in the SNP alignment. Strain number and MLST (ST) number have been added to each strain. The observed clusters of isolates contain mainly a cluster of susceptible ST72 and a cluster with cotrimoxazole-resistant ST162 isolates.
Comparison of phenotypic and genotypic antibiotic resistance profiles
BLAST results of the PBPs protein types and the 48 genomes showed that the majority of the isolates showed combinations of the three PBPs types as previously described22, but several new PBP protein type combinations were also observed. The new PBPs combinations were 2-0-28, 2-18-0, 2-0-1, 0-0-0 (all penicillin susceptible) and 17-15-116/117 (both penicillin resistant). Comparison of the PBP combination with the phenotypical penicillin susceptibility (Table 1), showed an agreement with the prediction in the MM model22. All the penicillin non-susceptible isolates had the 17 (PBP1a) −15 (PBP2b) -X (The PBP2x showed variable numbers) signatures.
For erythromycin-clindamycin, the BLAST results in general agreed with the phenotypic antibiotic susceptibility result. The presence of the ermB gene was linked to the resistance of erythromycin and clindamycin. Only two isolates (0834-2007 and 0811-2009) showed different genotypic and phenotypic results. The ResFinder 2.1 search confirmed the above results (Table 1). The twelve isolates harbouring the ermB gene also harboured the tet(M) gene according to ResFinder 2.1. Based on the information from the resistance gene accession number provided by the ResFinder 2.1, none of the isolates harbouring the ermB gene were found positive for the presence of mobile elements of Tn-family, while both Tn917 (8 isolates) and Tn1545 (4 isolates) was found in the isolates harbouring the tet(M) gene (Table 1).
Characterization of pneumococcal virulence genes
The virulence genes lytA and ply, the combined genes piaA, piaB, and piaC for membrane and ATP-binding proteins, the rpsB gene encoding for ribosomal protein S2, and the partial capsular cpsA (wzg) gene were detected in all 48 isolates.
The zinc metalloprotease related gene zmpB was only detected in isolates from 2007 and forwards.
The surface protein related gene psrp was most commonly found in isolates before 2012. Regarding the presence of zmpB and psrp, forty-one isolates harbored only one of the genes while seven isolates did not show any of the genes, and none harbored both genes.
The pneumococcal surface protein A gene (pspA) and the zinc metalloprotease zmpC gene were absent from all isolates.
Discussion
With the introduction of PCV-7 in 2007 in Denmark in the children’s vaccination program, a reduction in IPD with PCV-included serotypes was observed. However, as noted in other countries, non-vaccine serotypes have emerged, and it is therefore important to monitor the appearance of replacement serotypes1,5,7,34. Serotype 24F has been observed to be one of the emerging non-PCV serotypes5,7,28. Although serotype 24F is only among the 20 most common cause of IPD in Denmark, it is one of the serotypes which have increased most after the PCV-13 introduction in Denmark1. Serotype 24F is described as a type with a high potential for invasive disease5, and it is therefore an important serotype to keep under surveillance. Figure 1 shows that the total incidence of 24F has been relatively steady until 2012, when a significant increase in serotype 24F IPD cases were observed with a peak in 2014 (Table 2 and Fig. 1). In general, the total IPD in Denmark has been reduced since the introduction of the PCV (Fig. 1)1,10.
The analyzed MLST data show, that the majority of the isolates belong to ST72 and ST162. According to the MLST database, ST72 is a well-known clone of serotype 24F. The distribution between the previously described susceptible ST72 and resistant ST1627 was also observed in this study (Table 1). ST72 belonged to the penicillin susceptible lineages of CC72, while ST162 (belonging to CC156) also penicillin-susceptible was observed to be cotrimoxazole-resistant and found also among 9 V isolates7. The CC156 has emerged after the PCV-13 era7, which is also in accordance with our observation of the increase in ST162 (Table 1). The ST162/CC156 are furthermore described as the PMEN3 clone (Spain9V-156), particular related to the PCV included serotype 9V and serotype 1435. It has been suggested that the appearance of PMEN3 clone of serotype 14 might be due to capsular switching35, which also might be the situation with the serotype 24F ST162/CC156 isolates observed in this study.
The ST162 lineage is described as a more successful lineage than the CC72 and the multidrug resistant lineage CC2307 (Janoir et al. 2016).
Four of the 48 isolates were CC230, one ST230, two ST4253 and a new ST profile (isolate 0373-2003). As described by Janoir et al.7, CC230 is known as a highly resistant clonal complex, which we can confirm in this study (Table 1). The CC230 has previously been described in relation to a Danish penicillin resistant serotype 14, and are referred to as Denmark14-32 PMEN clone. It has also been suggested that the CC230 serotype 24F might be due to capsular switching from a serotype 14 (https://www.pneumogen.net/pmen/, accessed 24th September 2018)36. Figure 2 shows that the clustering of isolates based on the SNP site location corresponded well with the MLST.
The historical trachea isolate 24F-1943 was found to harbor a new PBP signature (0-0-0)22. The isolate did not show any clonal relationship with the other forty-seven 24F isolates from the period 1999–2017, and it only showed some relation to one other single isolate (isolate 1853–2011) based on the SNP site location (Fig. 2). It is well-known that capsular switching occurs regularly among pneumococcal isolates36, and it is therefore a possible explanation that the 24F isolates we see today in Denmark are due to capsular switching in other serotypes7,36.
In the study by Li et al.22, penicillin susceptibility could be predicted by the signature of the three penicillin-binding proteins PBP1a, PBP2b, and PBP2x. We also found an excellent correlation between the signature of the three PBPs and the phenotypical penicillin susceptibility. Interestingly, the four penicillin non-susceptible isolates all had the 17-15-x signature, while all susceptible isolates had low-numbered PBP signatures.
Phenotypic resistance to erythromycin/clindamycin was in general agreement with the genotypic resistance and presence of relevant genes (Table 1). The presence of the ermB gene was linked to resistance toward erythromycin and clindamycin and not to the mefA and mefE genes. The 12 isolates which were positive for the ermB gene, also harbored the tetM gene. Of note, two isolates harboring both the ermB gene and the tetM gene were erythromycin susceptible, which has been seen by others37.
The selection of virulence genes in this study was inspired by other studies12,14,26–33. A more comprehensive list of various pneumococcal genes can be found in the study by Gámez et al.25. The virulence genes (lytA, ply, the combined genes piaA, piaB and piaC, and the cpsA (wzg)), were detected in all 48 isolates, thus confirming their common use for identification of S. pneumoniae species using molecular tests14,26,29,38 (Table 3).
An interesting observation was made regarding the two genes pdrp and zmpB (Table 3). None of the 48 isolates harbored both the psrp gene and the zmpB gene; most of the isolates had only one of the genes, while for seven isolates the two genes were absent. Interestingly, psrp was detected in the isolates before 2012. The psrp gene has been described as a gene found primarily in antibiotic susceptible strains36, which we also observed. Only the MLST ST7179 and the well-known susceptible MLST ST72 strain harbored the psrp gene (Table 3). Although the zinc metalloprotease related gene zmpB is described as being widespread in S. pneumoniae30, we only detected the gene in Danish isolates from 2007 and onwards. The zmpB gene was only detected in the non-susceptible strains, particularly in ST162 (Table 3). We have not been able to find any studies showing that the zmpB gene is linked to antibiotic susceptibility. A 24F isolate with the zmpB gene was first detected in 2007 and became more common in the 24F isolates with the increase in the 24F incidence in 2013 (Fig. 1).
Conclusion
We have seen an increase in serotype 24F IPD in Denmark after the introduction of the PCV-13 vaccine in 2010. It was not significantly associated with an increase in antibiotic resistance or virulence determinants, but was observed in parallel with an increase of the ST162 clone.
Supplementary information
Acknowledgements
Monja Hammer, Lasse Jessen Schwartz, and Kirsten Burmeister are acknowledged for their skilled laboratory work and input to this study. We acknowledge the Danish Departments of Clinical Microbiology for submitting invasive pneumococcal isolates for national surveillance throughout the study period. We thank Eramus+for the Erasmus placement stipend for Ioanna Drakaki Kavalari, Department of Biology, National and Kapodistrian University of Athens, Greece.
Author Contributions
I.D.K. and H.C.S. designed the study, analysed the data and drafted the manuscript. K.A.K. and K.F. analysed and reviewed the data, contributed to the manuscript and critically revised the manuscript. All authors have approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41983-8.
References
- 1.Slotved H, Dalby T, Hoffmann S. The effect of pneumococcal conjugate vaccines on the incidence of invasive pneumococcal disease caused by ten non-vaccine serotypes in Denmark. Vaccine. 2016;34:769–774. doi: 10.1016/j.vaccine.2015.12.056. [DOI] [PubMed] [Google Scholar]
- 2.Slotved HC. Other age groups than children need to be considered as carriers of Streptococcal pneumoniae serotypes. Human Vaccines and Immunotherapeutics. 2016;12:2670–2674. doi: 10.1080/21645515.2016.1197451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borrow R, Heath PT, Siegrist C-A. Use of pneumococcal polysaccharide vaccine in children: what is the evidence? Curr. Opin. Infect. Dis. 2012;25:292–303. doi: 10.1097/QCO.0b013e3283531b0f. [DOI] [PubMed] [Google Scholar]
- 4.Camilli R, et al. Impact of pneumococcal conjugate vaccine (PCV7 and PCV13) on pneumococcal invasive diseases in Italian children and insight into evolution of pneumococcal population structure. Vaccine. 2017;35:4587–4593. doi: 10.1016/j.vaccine.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Balsells E, Guillot L, Nair H, Kyaw MH. Serotype distribution of Streptococcus pneumoniae causing invasive disease in children in the post-PCV era: A systematic review and meta-analysis. PLoS One. 2017;12:e0177113. doi: 10.1371/journal.pone.0177113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muñoz-Almagro C, et al. Serotypes and clones causing invasive pneumococcal disease before the use of new conjugate vaccines in Catalonia, Spain. J. Infect. 2011;63:151–162. doi: 10.1016/j.jinf.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Janoir C, Lepoutre A, Gutmann L, Varon E. Insight into resistance phenotypes of emergent non 13-valent pneumococcal conjugate vaccine type pneumococci isolated from invasive disease after 13-valent Pneumococcal conjugate vaccine implementation in France. Open Forum Infect. Dis. 2016;3:1–9. doi: 10.1093/ofid/ofw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alari A, et al. Impact of pneumococcal conjugate vaccines on pneumococcal meningitis cases in France between 2001 and 2014: A time series analysis. BMC Med. 2016;14:1–11. doi: 10.1186/s12916-016-0755-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harboe ZB, et al. Temporal Trends in Invasive Pneumococcal Disease and Pneumococcal Serotypes over 7 Decades. Clin. Infect. Dis. 2010;50:329–337. doi: 10.1086/649872. [DOI] [PubMed] [Google Scholar]
- 10.Harboe ZB, et al. Impact of 13-valent pneumococcal conjugate vaccination in invasive pneumococcal disease incidence and mortality. Clin. Infect. Dis. 2014;59:1066–1073. doi: 10.1093/cid/ciu524. [DOI] [PubMed] [Google Scholar]
- 11.Slotved H-C, et al. External Quality Assurance for Laboratory Identification and Capsular Typing of Streptococcus pneumoniae. Sci. Rep. 2017;7:13280. doi: 10.1038/s41598-017-13605-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholz CFP, Poulsen K, Kilian M. Novel molecular method for identification of Streptococcus pneumoniae applicable to clinical microbiology and 16S rRNA sequence-based microbiome studies. J. Clin. Microbiol. 2012;50:1968–73. doi: 10.1128/JCM.00365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arbique JC, et al. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J. Clin. Microbiol. 2004;42:4686–4696. doi: 10.1128/JCM.42.10.4686-4696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheppard CL, et al. Clinical streptococcal isolates, distinct from Streptococcus pneumoniae, but containing the β-glucosyltransferasettsgene and expressing serotype 37 capsular polysaccharide. PeerJ. 2017;5:e3571. doi: 10.7717/peerj.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bishop CJ, et al. Assigning strains to bacterial species via the internet. BMC Biol. 2009;7:3. doi: 10.1186/1741-7007-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilian M, et al. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One. 2008;3:e2683. doi: 10.1371/journal.pone.0002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentley SD, et al. Genetic Analysis of the Capsular Biosynthetic Locus from All 90 Pneumococcal Serotypes. PLoS Genet. 2006;2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapatai G, et al. Whole genome sequencing of Streptococcus pneumoniae: development, evaluation and verification of targets for serogroup and serotype prediction using an automated pipeline. PeerJ. 2016;4:e2477. doi: 10.7717/peerj.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slotved H-C, Dalby T, Hoffmann S. Multilocus sequence types of invasive pneumococcal isolates from Danish infants (0–90 days) 2003–2013. BMC Res. Notes. 2015;8:563. doi: 10.1186/s13104-015-1540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahl JW, et al. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb. genomics. 2016;2:e000074. doi: 10.1099/mgen.0.000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, et al. Penicillin-binding protein transpeptidase signatures for tracking and predicting β-lactam resistance levels in Streptococcus pneumoniae. MBio. 2016;7:1–9. doi: 10.3391/mbi.2016.7.1.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science (80-.). 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gámez G, et al. The variome of pneumococcal virulence factors and regulators. BMC Genomics. 2018;19:1–18. doi: 10.1186/s12864-017-4376-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvalho MDGS, et al. Evaluation and Improvement of Real-Time PCR Assays Targeting lytA, ply, and psaA Genes for Detection of Pneumococcal DNA. J. Clin. Microbiol. 2007;45:2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyllie AL, et al. Molecular surveillance of nasopharyngeal carriage of Streptococcus pneumoniae in children vaccinated with conjugated polysaccharide pneumococcal vaccines. Sci. Rep. 2016;6:23809. doi: 10.1038/srep23809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camilli R, et al. Zinc metalloproteinase genes in clinical isolates of Streptococcus pneumoniae: Association of the full array with a clonal cluster comprising serotypes 8 and 11A. Microbiology. 2006;152:313–321. doi: 10.1099/mic.0.28417-0. [DOI] [PubMed] [Google Scholar]
- 29.Wyllie AL, et al. Sequencing of the variable region of rpsB to discriminate between Streptococcus pneumoniae and other streptococcal species. Open Biol. 2017;7:170074. doi: 10.1098/rsob.170074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bek-Thomsen, M., Poulsen, K. & Kilian, M. Occurrence and Evolution of the Paralogous Zinc Metalloproteases IgA1 Protease, ZmpB, ZmpC, and ZmpD in Streptococcus pneumoniae and Related Commensal Species. MBio3, e00303-12-e00303-12 (2012). [DOI] [PMC free article] [PubMed]
- 31.Abeyta M, Hardy GG, Yother J. Genetic Alteration of Capsule Type but Not PspA Type Affects Accessibility of Surface-Bound Complement and Surface Antigens of Streptococcus pneumoniae. Infect. Immun. 2003;71:218–225. doi: 10.1128/IAI.71.1.218-225.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blumental S, et al. Virulence factors of Streptococcus pneumoniae. Comparison between African and French invasive isolates and implication for future vaccines. PLoS One. 2015;10:1–17. doi: 10.1371/journal.pone.0133885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pai R, Gertz RE, Beall B. Sequential Multiplex PCR Approach for Determining Capsular Serotypes of Streptococcus pneumoniae Isolates. J. Clin. Microbiol. 2006;44:124–131. doi: 10.1128/JCM.44.1.124-131.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waight PA, et al. Effect of the 13-valent pneumococcal conjugate vaccine on invasive pneumococcal disease in England and Wales 4 years after its introduction: an observational cohort study. Lancet. Infect. Dis. 2015;15:535–43. doi: 10.1016/S1473-3099(15)70044-7. [DOI] [PubMed] [Google Scholar]
- 35.Càmara, J. et al. Evolution of the β-lactam-resistant Streptococcus pneumoniae PMEN3 clone over a 30 year period in Barcelona, Spain. J. Antimicrob. Chemother. 1–11, 10.1093/jac/dky305 (2018). [DOI] [PubMed]
- 36.Pantosti A, et al. A Novel, Multiple Drug–Resistant, Serotype 24F Strain of Streptococcus pneumoniae That Caused Meningitis in Patients in Naples, Italy. Clin. Infect. Dis. 2002;35:205–208. doi: 10.1086/341250. [DOI] [PubMed] [Google Scholar]
- 37.Varaldo PE, Montanari MP, Giovanetti E. Genetic Elements Responsible for Erythromycin Resistance in Streptococci. Antimicrob. Agents Chemother. 2009;53:343–353. doi: 10.1128/AAC.00781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varghese R, Jayaraman R, Veeraraghavan B. Current challenges in the accurate identification of Streptococcus pneumoniae and its serogroups/serotypes in the vaccine era. J. Microbiol. Methods. 2017;141:48–54. doi: 10.1016/j.mimet.2017.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.