Abstract
Heterogeneity in early language development in autism spectrum disorders (ASD) is clinically important and may reflect neurobiologically distinct subtypes. Here we identify a large-scale association between multiple coordinated blood leukocyte gene co-expression modules and multivariate functional neuroimaging (fMRI) response to speech. Gene co-expression modules associated with multivariate fMRI response to speech are different for all pairwise comparisons between typically developing toddlers and toddlers with ASD and either poor versus good early language outcome. Associated co-expression modules are enriched in genes that are broadly expressed in the brain and many other tissues. These co-expression modules are also enriched for ASD, prenatal, human-specific and language-relevant genes. This work highlights distinctive neurobiology in ASD subtypes with different early language outcomes that is present well before such outcomes are known. Associations between neuroimaging measures and gene expression levels in blood leukocytes may offer a unique in-vivo window into identifying brain-relevant molecular mechanisms in ASD.
Autism spectrum disorders (ASD) are heterogeneous at multiple levels (e.g., genetics, cellular and neural systems, cognition, behavior, developmental trajectories, prognosis, response to treatment)1–3. This multi-level heterogeneity presents a significant challenge on the path towards stratified psychiatry and precision medicine4, 5. One dimension of heterogeneity of clinical importance in ASD is early language development and outcome. There is a wide spectrum of variability in early language abilities in the ASD population, from individuals who remain minimally verbal, to those who have difficulties similar to specific language impairment, to those who develop near-typical levels of language function 6, 7. Early language ability is paramount for better understanding a range of clinical phenomena. For example, early language ability is one of the most important predictors of early-intervention response and later life outcomes8–12.
An additional challenge lies in studying relationship between macroscale properties of the brain and the molecular mechanisms at play in early development, and how this relationship may be altered in ASD13. Functional magnetic resonance imaging (fMRI) can be used to gain insight into the macroscale, neural-systems level of organization and its association with cognitive and behavioral functioning. However, the molecular biological underpinnings of this organization are not well understood. Although blood samples are a practical source for assaying atypical gene-expression in early ASD development14, 15, a common question is how relevant they are for understanding atypical neural processes in ASD. The evidence for a genetic basis of ASD is strong16, 17 and genetic variation will likely affect gene expression levels in multiple tissues18, including brain and blood. Thus, identifying associations between the blood leukocyte transcriptome and neuroimaging phenotypes may help shed light on mechanisms affecting early neural systems development in toddlers with ASD as compared to typically-developing toddlers. Such an “in-vivo window” onto the biology of ASD13 may be able to further our understanding of the mechanisms underlying atypical brain development in heterogeneous populations of ASD patients, but may also advance translational work targeted at better monitoring treatment response, predicting prognosis, and in evaluating clinical trials.
Here we ask whether large-scale coordinated gene expression in blood leukocytes is associated with neural responses to speech as measured with fMRI, and whether this association differs between toddlers with ASD and either poor or good early language outcome and typically-developing toddlers. A fundamental question for this work is whether differences in early language outcome are a biologically relevant basis for stratifying ASD. Based on prior work19 suggesting that early language outcome subtypes are underpinned by distinct biology, we predict that early language outcome ASD subtypes will show different profiles of associations between blood leukocyte gene expression and functional neural systems response to speech.
Examining large-scale blood leukocyte transcriptome associations with neuroimaging phenotypes in ASD may also identify novel mechanisms involved in ASD. The omnigenic model20, much like other viewpoints on polygenic risk21, predicts that large numbers of genes are relevant to complex traits like ASD. However, the omnigenic model suggests that these genes do not necessarily need to be specific to brain tissue. Genes that are broadly expressed in one or more tissues, including brain and blood, are predicted to harbor a large amount of the heritability signal and can contribute more to overall risk than the smaller number of tissue-specific genes implicated in a complex trait20. Applied to the current study, we predict that large-scale coordinated transcriptional activity in the blood leukocyte transcriptome could be relevant for explaining neural phenotypes relevant to ASD. The omnigenic model predicts that this large-scale transcriptomic signal would be enriched for genes that are broadly expressed in the brain and many other tissues.
Results
Group-differentiation in superior temporal cortex response to speech and clinical behavioral trajectories over the first 4 years of life
In this study we compared typically-developing toddlers (TD) and age-matched toddlers with ASD, whose language abilities were assessed around 3-4 years of age. Toddlers with ASD were stratified by poor (“ASD Poor”) or good (“ASD Good”) language outcome. ASD Poor is defined by Mullen expressive (EL) and receptive language (RL) T-scores below 1 standard deviation of typical developing age norms (T<40). In contrast, ASD Good is defined by outcome Mullen EL or RL within 1 standard deviation of typical age-norms (T≥40) (see Methods). In prior work we showed that this stratification identifies an ASD subtype with different developmental trajectories and a reduced left-hemisphere superior temporal cortex response to speech, as measured with sleep-fMRI before diagnosis and outcome are known19. The current dataset utilizes a subset of toddlers from the prior paper (n = 19 ASD Poor; n = 24 ASD Good; n = 21 TD), and adds a similar number of new toddlers (n=22 ASD Poor, n=16 ASD Good, and n=16 TD). Thus, we first re-ran longitudinal clinical trajectory and fMRI analyses of this combined dataset.
As previously reported19, all longitudinal clinical measures showed evidence of subtype*age interactions (except for Mullen Fine Motor and ADOS total), indicating that groups differed in the slope of trajectories (see Fig. 1; Supplementary Table 1 for statistics). This difference was generally driven by the ASD Poor group, whose downwards trajectories are indicative of falling further behind age-appropriate norms. All measures also showed main effects of group and were generally due to all groups differing from one another in a hierarchy of ASD Poor as most severe, ASD Good as intermediate, and TD as least severe. With the fMRI data, we also find that the previously reported19 hypoactivation in left hemisphere superior temporal cortex remains stable in this combined sample (Fig. 2 and Supplementary Table 2). Whole-brain between-group analyses did not reveal any regions differentiating the groups. However, the lack of effects in this context are likely to be due to low statistical power for whole-brain between-group comparisons22.
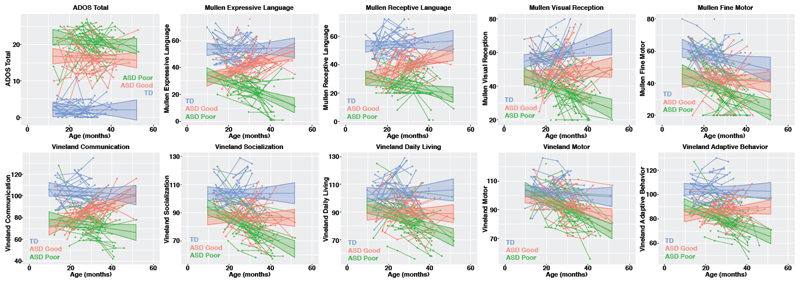
Figure 1. Clinical behavioral trajectories over the first 4 years of life in typically-developing (TD) toddlers and toddlers with ASD and good or poor early language outcome.
This figure shows developmental trajectories over the first 4 years of life for typically-developing (TD) toddlers, toddlers with ASD and good early language outcome (ASD Good) and toddlers with ASD and poor early language outcome (ASD Poor) on clinical behavioral assessment measures such as ADOS total scores, Mullen Scales of Early Learning subscales (Expressive and Receptive Language, Visual Reception, and Fine Motor) and Vineland Adaptive Behavioral Scales (Communication, Socialization, Daily Living Skills, Motor, and Adaptive Behavior). The TD (n=35) group is shown in blue, ASD Good (n=40) in pink, and ASD Poor (n=41) in green. Individual level trajectories are plotted including the group-level trajectory and 95% confidence band.
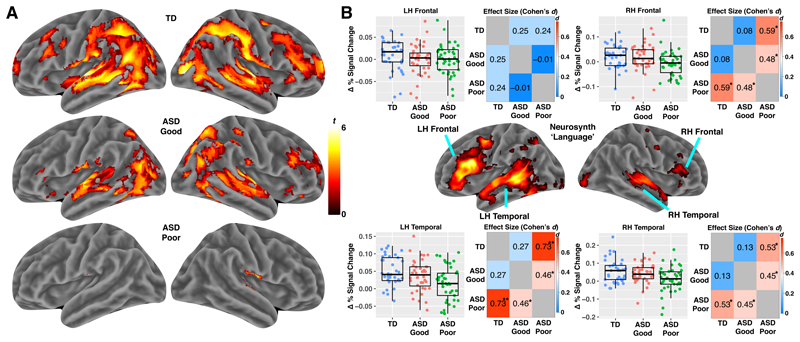
Figure 2. Reduced fMRI response to speech in ASD toddlers with poor early language outcome.
Panel A shows results of whole-brain analyses (one-tailed t-test) on each group separately (results shown at FDR q<0.05) (TD n = 37; ASD Good n = 40; ASD Poor n = 41). Panel B shows the results of region-of-interest (ROI) analyses testing for subtype differences. ROIs are defined by 4 regions within the Neurosynth ‘Language’ meta-analysis map in left (LH) or right hemisphere (RH) frontal and temporal cortex. ROI data are shown for each individual in the scatter-boxplots (TD, blue, n = 37; ASD Good, pink, n = 40; ASD Poor, green, n = 41). The box in the boxplots indicates the interquartile range (IQR; Q1 indicates the 25th, while Q3 indicates the 75th percentile) and the whiskers indicate Q1-(1.5*IQR) or Q3+(1.5*IQR). The line within the box represents the median. Matrices next to the scatter-boxplots show standardized effect sizes (Cohen’s d) for each pairwise group comparison. Cohen’s d is shown in each cell and also indicated by the color of the cell. Within each cell one star (*) indicates p<0.05, while two stars (**) indicates p<0.005.
Lack of group-differentiation within gene expression data alone
The total sample of n=41 ASD Poor, n=40 ASD Good, and n=37 TD was the largest dataset of toddlers for whom both fMRI and gene expression data were available. We next assessed whether differences in blood leukocyte gene expression might reflect this different neural systems organization between the two ASD language-outcome subtypes. First, we assessed differential expression (DE) between subtypes at the level of individual genes. After correction for multiple comparison, no genes were identified as DE for any pairwise group comparison (Supplementary Table 3). Next, we utilized weighted gene co-expression network analysis (WGCNA) to reduce redundancy between the 14,313 genes, down to 21 discrete co-expression modules. Co-expression modules are summarized by the first principal component score, also known as the module eigengene (ME)23. Similar to the DE analysis at the gene level, there were no ME differences between the two ASD subtypes (Supplementary Table 3). Thus, examining blood leukocyte gene expression data in isolation does not significantly differentiate the groups at the current sample sizes. We next turned to examining associations between gene expression and functional neuroimaging phenotypes.
Large-scale blood leukocyte gene co-expression module association with fMRI response to speech
Multivariate analysis of association between co-expression modules and whole-brain voxel-wise patterns of activation was implemented with partial least squares (PLS) analyses. Of the 63 total latent variable (LV) co-expression-fMRI pairs, PLS identified only one LV pair with a statistically significant association after multiple comparison correction (LV1: d = 65.47, p = 1.99e-4, FDR q = 0.0125; see Supplementary Table 4 for statistics for all PLS LV pairs). LV1 accounts for 20.13% of the covariance between gene expression and fMRI data and is spatially distributed across a number of cortical regions highly relevant to speech, language24, 25 (e.g., primary auditory cortex, superior temporal sulcus, inferior frontal gyrus, ventral premotor cortex, insula), visual and sensorimotor areas (e.g., primary visual cortex, superior parietal cortex, primary somatomotor cortex, premotor cortex), cognitive control (dorsolateral prefrontal cortex), and ‘social brain’ circuitry overlapping with key areas of the default mode network (e.g., posterior cingulate cortex, medial prefrontal cortex, right temporo-parietal junction, superior temporal sulcus) (Fig. 3a). Subcortical regions such as the striatum and thalamus were also implicated and are highly relevant for language processes such as vocal learning25, 26. For example, Area X in song birds is linked to vocal learning and is homologous with human dorsal striatum26.
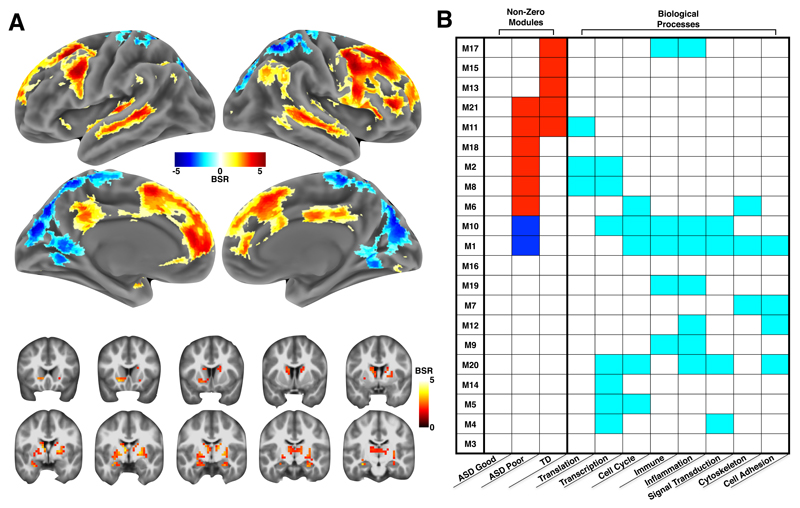
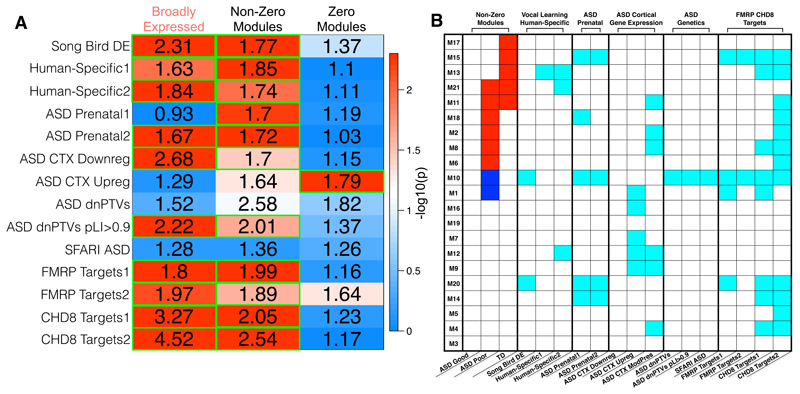
Figure 3. Multivariate gene co-expression-fMRI association in ASD with good or poor early language outcome and typically-developing control toddlers.
Panel A shows the brain regions with the strongest contributions to the multivariate gene co-expression–fMRI association present in the LV1 PLS result. The coloring in each region indicates the bootstrap ratio (BSR) and reflects how important each voxel is to the LV1 PLS result. Areas are shown in panel A if the BSR ≥ 1.96 or BSR ≤ -1.96. Hot colored regions in panel A are interpreted as showing a positive gene co-expression–fMRI correlation — that is, as a module’s eigengene increases, functional activation in response to speech also increases. In contrast, cool colored areas in panel A indicate a negative correlation between a module’s eigengene and functional activation response to speech. The table in panel B describes which modules were the strongest contributors to the LV1 PLS result. Each row indicates one of the 21 co-expression modules used in the PLS analysis. The columns labeled with the heading ‘Non-Zero Modules’ are broken down to indicate gene co-expression–fMRI correlations by group. Cells in these columns are colored red or blue if the gene co-expression–fMRI correlation was non-zero and had 95% confidence intervals (estimated from bootstrapping) that did not include a correlation of 0. These modules are called ‘non-zero’ modules, as they are the strongest contributors or modules of importance to the LV1 PLS result. All other modules with white colored cells are labeled ‘zero’ modules, as the 95% confidence intervals for the gene co-expression–fMRI correlation include 0. Non-zero modules have cells colored in red to indicate a positive gene co-expression–fMRI correlation (i.e. congruent with the interpretation already stated for the hot and cool colored regions in panel A). However, in the case of non-zero modules with cells colored in blue, the previously stated way to interpret the hot and cool colored regions in panel A should reverse (e.g., cool colored regions in panel A reflect positive correlations with a module’s eigengene, while hot colored regions in panel A reflect negative correlations with a module’s eigengene). The remaining columns in panel B with the heading ‘Biological Processes’ annotate each module for enrichments in biological process terms from Metacore GeneGO software. Cyan colored cells indicate modules with enrichments passing FDR q<0.05 for multiple comparison correction. For a complete description of these biological process enrichments, please see Supplementary Table 5.
Extent of non-zero associations across co-expression modules
To better understand the most important co-expression modules for the PLS LV1 result, we first identified what we call ‘non-zero’ association modules. Non-zero modules are defined as gene co-expression modules which have gene co-expression-fMRI correlations with 95% confidence intervals (CI) that exclude a correlation of 0. Non-zero modules comprise about half of all modules analyzed (11/21; 52%) (Fig. 3b; Supplementary Table 4). The remaining modules (10/21; 48%) are referred to as ‘zero’ modules, defined as gene co-expression modules for which a correlation of 0 lies within the 95% CIs. Zero modules contribute little to and/or are unreliable in how they contribute to LV1. Non-zero modules cover a majority (61%) of the transcriptome considered for the WGCNA analysis. This widespread coverage indicates a coordinated and large-scale signal spanning large parts of the blood leukocyte transcriptome associated with macroscale functional neural response to speech measured with fMRI.
Most non-zero modules can be characterized by a variety of biological processes generally falling within categories such as translation, transcription, cell cycle, immune, inflammation, signal transduction, and cytoskeleton processes. However, enrichments differ substantially depending on the module (Fig. 3b; see Supplementary Table 5 for a complete description of enrichments for each module). For instance, non-zero modules M2, M8, and M11 are primarily translation and transcription modules, while M1 and M10 have enrichments for many of these terms, but not translation and transcription. These biological processes have all been implicated as important in autism. For example, translation processes are affected in many syndromic forms of ASD (e.g., Fragile X Syndrome, Tuberous Sclerosis)27. Many high-confidence ASD-risk genes are known to affect transcription processes (e.g., CHD8)28, 29. Cell cycle processes are involved in aberrant early cell proliferation and increased early brain growth in ASD13, 14. Immune and inflammation processes have been linked to ASD via various lines of evidence30–32. These results supports the idea that ASD-relevant biological processes can be assayed in blood leukocytes and are associated with early developing large-scale functional neuroimaging phenotypes.
Lack of overlap in non-zero modules across ASD subtypes and TD
The majority of non-zero modules are present only in one group (9/11; 81%). In fact, only TD and ASD Poor show evidence of non-zero modules. No non-zero modules are present for the ASD Good subtype. Two (18%) non-zero modules are present in both TD and ASD Poor and are correlated in the same direction. However, the extent of this overlap is not statistically significant (enrichment odds ratio (OR) = 1.67, p = 0.65) (Fig. 3b). This result suggests that different biological mechanisms within each group may underpin the variability observed in macroscale language-relevant fMRI phenotypes. To further test the importance of the ASD language outcome subtype distinction, we next tested whether a simple case–control distinction could enhance sensitivity in detecting gene co-expression–fMRI relationships. This case–control PLS analysis was not able to identify any statistically significant LV pairs at FDR q<0.05 correction for multiple comparisons (Supplementary Table 4). Thus, using early language outcomes as a stratifier in ASD appears to substantially enhance sensitivity for detecting gene co-expression–fMRI relationships.
Non-zero modules are enriched for broadly expressed genes
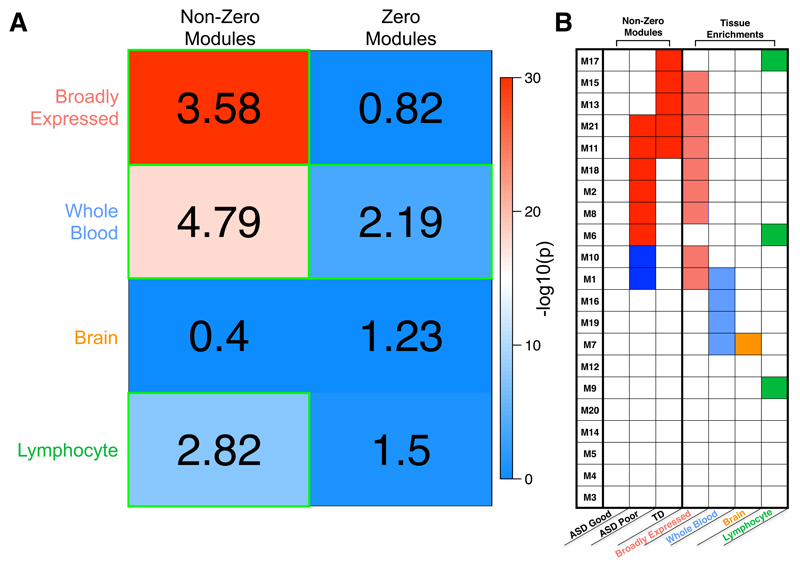
We next examined what class of genes likely heavily contributes to the non-zero modules. Based on ideas from the omnigenic model20, genes that are broadly expressed, i.e. expressed in many tissues including the brain, could also be expressed and measurable in blood leukocytes and, therefore, could be of high relevance for these non-zero modules associated with a functional neuroimaging phenotype. Remarkably, we find that 81% (9/11) of non-zero modules are enriched for broadly expressed genes (OR = 184.5, p = 1.87e-4). All modules enriched for broadly expressed genes were also non-zero modules (Fig. 4b). In contrast, tissue-specific gene lists (e.g., brain, whole blood, lymphocyte) were not heavily enriched in many modules nor over-represented in non-zero modules (brain-specific modules: OR = 0, p = 1; whole blood-specific modules: OR = 0.6, p = 0.96; lymphocyte-specific modules: OR = 4.44, p = 0.53) (Fig. 4b). In addition to running these enrichments at the level of overlap amongst modules, we also ran tests for overlap at the gene level. Around 44% of all broadly expressed genes are present in non-zero modules, amounting to a highly significant enrichment (OR = 3.58, p = 1.48e-93). Whole-blood and lymphocyte-specific genes also showed evidence of enrichment in non-zero modules (blood OR = 4.79, p = 1.57e-18; lymphocyte OR = 2.82, p = 1.94e-8), though whole-blood-specific genes also showed enrichment in zero modules (Fig. 4a). As shown in Fig. 4b, the whole-blood and lymphocyte-specific enrichments are likely driven by genes within 1-2 non-zero modules (e.g., M1, M6, M17). In contrast, the enrichment in broadly expressed genes is driven by genes spread across nearly every single non-zero module.
Figure 4. Tissue class enrichments with sets of non-zero or zero association modules.
Enrichments with different classes of genes taken from the Boyle et al., (2017)20 analysis of tissue-specific or broadly expressed genes from GTEx data. Within panel A, the numbers in each cell represent the enrichment odds ratio, while the coloring represents the –log10 p-value for each hypergeometric test for enrichment. Cells outlined in green pass multiple comparison correction at FDR q<0.05. In panel B, we show all gene co-expression modules (rows) and whether they are enriched for each tissue class (columns). Modules with enrichments passing FDR q<0.05 for multiple comparison correction are indicated as colored cells. The first 3 columns show which modules are those with non-zero associations (colored cells), as shown in Fig. 3b.
Non-zero modules are enriched for differentially expressed genes in a song bird vocal learning model
Given the relationship between expression of genes in non-zero modules and language-relevant functional neuroimaging phenotypes, we next looked to validate whether these genes are brain-relevant and conserved in an animal model of vocal learning. Vocal learning is a language-relevant ability shared between humans and songbirds and has been extensively examined 25, 26, 33. Here we tested whether genes in our non-zero modules show overlap with DE genes from subcortical Area X of singing versus non-singing songbirds. Re-analysis of data from Hilliard et al.33 identified 1,267 DE genes in Area X and of these, 902 overlap with the genes examined in the main PLS analysis. Area X is thought to be homologous with human striatal areas26 (Fig. 1a). Strikingly, 33% of the DE genes in Area X are present in our non-zero modules (OR = 1.77, p = 0.002). In contrast, no such enrichment was present in zero modules (OR = 1.37, p = 0.13) (Fig. 5a). Most of the non-zero enrichment was driven specifically by module M10, as no other non-zero module was specifically enriched in songbird-DE genes (Fig. 5b). These results suggest that a subset of genes in non-zero modules are indeed brain-relevant and conserved between humans and songbirds, which both have an ability for vocal learning.
Figure 5. Vocal learning, human-specific, and ASD-associated enrichments with sets of broadly expressed genes and non-zero or zero association modules.
Panel A shows the results of hyperogeometric tests for enrichment between broadly expressed genes, non-zero, and zero modules (columns) and a variety of different gene lists (rows) relevant to vocal learning, human-specific genes, or genes of relevance to ASD. The numbers in each cell represent the enrichment odds ratio, while the coloring represents the –log10 p-value for each hypergeometric test for enrichment. For details about the gene lists specified in each row, see the Methods section. Cells outlined in green pass multiple comparison correction at FDR q<0.05. Panel B shows a table to indicate which gene co-expression modules (rows) are enriched for a variety of different gene lists (columns). Modules with enrichments passing FDR q<0.05 for multiple comparison correction are indicated as cyan colored cells. The first 3 columns show which modules are those with non-zero associations (colored cells), as shown in Fig. 3b.
Non-zero modules are enriched for transcriptionally human-specific genes
Language requires more than vocal learning and is indeed a uniquely human ability. Therefore, it is possible that components of language ability may be reflected in neural differences between humans and our closest non-human primate relatives (e.g., chimpanzees) which do not possess language34. We therefore investigated whether non-zero modules are enriched for genes that are DE in cortical tissue of humans versus chimpanzees (‘human-specific’ genes). Using two lists of such ‘human-specific’ genes obtained from independent studies34, 35 and which minimally overlap (4.38%), we find that non-zero modules are significantly enriched on both lists, with 33-34% of human-specific genes overlapping with genes from non-zero modules (OR>1.73, p <0.0115) (Fig. 5a). These enrichments are driven by M13, as no other specific modules were enriched across both human-specific gene lists (Fig. 5b). In contrast, no enrichment of human-specific genes is present in zero modules (OR<1.11, p >0.85; Fig. 5a). These results suggest that transcriptional activity of human-specific genes in blood leukocytes is linked to language-relevant fMRI phenotypes measured in TD and ASD toddlers with varying early language abilities.
Non-zero modules are enriched for highly active prenatal co-expression modules associated with ASD
Several lines of evidence point towards ASD pathophysiology having key impact on prenatal brain development13, 36–38. We therefore examined whether non-zero modules are enriched for genes that are members of co-expression modules that show high levels of prenatal expression and that possess a number of highly-penetrant ASD-associated genes. Using lists from two independent studies of the BrainSpan atlas39 examining either cortical-only37 or cortical and subcortical regions36, we find that approximately 32% of genes in prenatal and ASD-associated co-expression modules also appear in non-zero modules (OR>1.7, p <0.0056) (Fig. 5a), whereas only 15-17% are present in zero modules (OR<1.19, p>0.74). Non-zero modules M15 and M10 drove the enrichment, as no other non-zero modules showed evidence of enrichment for genes in either ASD-associated prenatal gene lists (Fig. 5b). Overall, this evidence supports the idea some of the genes present in non-zero modules are also genes that are members of prenatally active and ASD-associated co-expression modules.
Non-zero modules are enriched with genes from ASD-downregulated co-expression modules from frontal and temporal cortex tissue
While establishing that non-zero modules overlap with prenatally relevant co-expression modules that harbor ASD-relevant genes, a caveat to this result is that those prenatal, ASD-associated co-expression modules were identified from the BrainSpan dataset39, which for obvious reasons does not contain prenatal tissue from ASD donors. Thus, to more directly connect non-zero modules with cortical gene expression in diagnosed ASD patients, we used gene-expression data from post-mortem frontal and temporal cortical tissue of ASD patients40. Non-zero modules are enriched for genes that are members of ASD-downregulated frontal and temporal cortex co-expression modules (OR = 1.70, p = 0.03). Enrichments at trend levels were also seen for genes from ASD-upregulated co-expression modules (OR = 1.64, p = 0.0502, FDR q = 0.0586) (Fig. 5a). However, no specific non-zero modules seemed to drive this enrichment (Fig. 5b). While zero modules were not enriched in genes from ASD-downregulated modules (OR = 1.15, p = 0.75), zero modules were enriched for genes from ASD-upregulated modules (OR = 1.79, p = 2.80e-5) (Fig. 5a). These results further point towards the ASD- and brain-relevance of genes identified via their non-zero association between expression in blood leukocytes and language-relevant functional neuroimaging phenotypes.
Non-zero modules with preservation of network structure between ASD blood and cortical tissue
Utilizing the same gene-expression dataset from post-mortem cortical tissue from ASD patients40, we next examined whether co-expression network structure of non-zero modules identified in blood might be preserved in ASD frontal and temporal cortical tissue. This is important, as it highlights specific modules where co-expression network connectivity patterns are similar between blood leukocytes and brain tissue. While non-zero modules M8 and M11 showed moderate evidence of preservation (2< Zsummary <6), the non-zero M2 module was the highest ranking of all modules with evidence of high-moderate preservation (Zsummary = 8.1) (Supplementary Fig. 1). M2 is highly enriched for the term ‘translation in mitochondria’ (Supplementary Table 5) and many of M2’s hub genes encode proteins that are localized to mitochondria (e.g., MRPS12, NDUFS3, NDUFB8, HINT2, MRPL14) (Supplementary Table 6). This evidence could be relevant in light of possible mitochondrial dysfunction in autism41. Other notable M2 hub genes are DGCR6 and BOLA2. Both are located within prominent ASD-associated CNV regions of 22q11.21 (DGCR6) and 16p11.2 (BOLA2)42. Interestingly with regard to evolutionarily accelerated human-specific genes, BOLA2 is known for human-specific duplications and shows upregulated expression in human versus chimpanzee induced pluripotent stem cells (iPSCs)43, 44. In patients with 16p11.2 CNVs, 96% of breakpoints include human-specific duplications of BOLA244. Deletions and duplications of 16p11.2 are linked to language and its associated neural circuitry45–47. Thus, the evidence here could suggest that BOLA2 is an important ASD-relevant 16p11.2 locus, but also is more generally relevant for the human-specific capacity to develop language and the neural systems supporting that development.
Non-zero modules are enriched for ASD de novo protein-truncating variants and cortically ASD-downregulated co-expression modules
We next tested non-zero modules for enrichments with different classes of genetic variants associated with ASD. We first examined enrichment with high-penetrance rare de novo protein truncating variants (dnPTVs). Amongst the genes highlighted by Kosmicki et al.,48 with ≥2 dnPTVs in ASD, 43% are also present in non-zero modules, resulting in an enrichment at trend level significance (OR = 2.58, p = 0.08, FDR q = 0.0915). The lack of significant enrichment may be due to the limited number of known dnPTVs that overlap with the subset of genes considered in our analysis (i.e. 28). When we relax the criterion to ≥1 dnPTVs in ASD but add the constraint that the gene should also have a probability of loss-of-function intolerance (pLI)≥0.949, this enabled us to study a larger set (155) of putative ASD-relevant dnPTVs. Under this criterion, we find a significant enrichment of these ASD risk genes in non-zero modules (OR = 2.01, p = 0.02) (Fig. 5a), including ADNP, ANKRD11, DYRK1A, ILF2, KDM5B, KDM6B, MED13L, PHF2, PTEN, SPAST, SUV420H1, TRIP12, WDFY3, ZC3H4. Non-zero module M10 is the primary driver behind this enrichment (Fig. 5b) and includes ADNP, ANKRD11, DYRK1A, KDM5B, TRIP12, and ZC3H4. Of these notable M10 genes, ADNP is within the top 20 hub genes (Supplementary Table 6). In contrast, zero modules were not enriched for these ASD risk genes, either amongst the criteria of ≥2 dnPTVs or with ≥1 dnPTVs and pLI ≥0.9 (OR < 1.82, p > 0.24) (Fig. 5a). In addition, and contrary to the enrichment with ASD-associated dnPTVs, we could not find any enrichment amongst the 543 ASD-associated genes annotated on SFARI Gene (https://gene.sfari.org)50 for non-zero (OR = 1.36, p = 0.66) or zero modules (OR = 1.26, p = 0.44) (Fig. 5a). This evidence suggests that some high-penetrance ASD-associated genes are detectable within blood leukocyte gene expression data and show strong association to in-vivo functional neuroimaging phenotypes relevant for early language heterogeneity in ASD.
Non-zero modules are enriched for FMRP and CHD8 targets
While non-zero modules do not contain some of the most well-known and highly-penetrant ASD-associated genes, such as FMR1 and CHD8, non-zero modules may nevertheless overlap with the molecular networks linked to these genes. One way to examine this hypothesis is through testing non-zero modules for enrichment with downstream targets of these highly important genes. Non-zero modules are highly enriched for both FMRP and CHD8 targets across two different target lists (OR>1.89, p<0.0269) (Fig. 5a). Numerous modules drive these enrichments, such as M10 and M15 for FMRP targets and M10, M8, M13, and M15 for CHD8 targets (Fig. 5b). In contrast, zero modules were not enriched for target genes of either FMRP or CHD8 (Fig. 5a). These results suggest that non-zero modules also contain genes that are members of FMRP and CHD8-related networks.
Broadly expressed genes are a prominent source of signal driving enrichments
Finally, given the prominent overlap between broadly expressed genes and non-zero modules, we tested whether many of the other enrichments with non-zero modules were driven by broadly expressed genes. We first examined the enrichment of broadly expressed genes with all of the gene lists already tested. Remarkably, we found that nearly all gene lists enriched in non-zero modules are also highly enriched in broadly expressed genes (Fig. 5a). Furthermore, once broadly expressed genes are removed from these lists, the enrichments with non-zero modules largely disappear (Supplementary Fig. 2). This suggests that broadly expressed genes drive the enrichments of these lists in non-zero modules.
Discussion
Here we find one large-scale association between coordinated gene co-expression modules in blood leukocytes with multivariate fMRI response to speech. Highlighting the distinctiveness of ASD language outcome subtypes, we find that blood leukocyte co-expression modules associated with multivariate fMRI response to speech are different for all pairwise comparisons between groups of TD toddlers or toddlers with ASD and either poor or good language outcome. Given the early ages when blood samples and fMRI data were collected, it is clear that this association manifests well before stable diagnoses and final language outcomes are known. Co-expression modules of importance in TD but not ASD may signal normative biological processes associated with the development of language-related neural circuitry. These normative processes may be affected in ASD. In addition, modules that diverge between ASD subtypes may indicate risk or protective mechanisms that push different ASD individuals towards different early developmental language outcomes. Thus, in contrast to the idea that ASD is a uniform condition with similar underlying biological mechanisms in all diagnosed individuals, these results indicate that a behavioral stratifier such as early language outcome holds important information to help understand how the underlying biology may be differentially linked to the way macroscale neural systems develop.
These findings may be of high translational importance. Both neuroimaging methods and blood sampling to quantify the leukocyte transcriptome with high-throughput techniques are feasible to collect from ASD patients with different levels of impairment and at early ages. In-vivo examination of the molecular mechanisms and their associations with higher-level macroscale neural systems and heterogeneity in clinical phenotypes will be important for furthering progress towards precision medicine13. Endeavors such as evaluating early-age treatment response, monitoring clinical trials, developing prediction tools for diagnosis and prognosis can all be facilitated with this approach to understanding links between gene expression, macroscale neural systems, and behavioral levels of analysis. Future work will be necessary to determine whether similar associations are present in older children and adults with ASD. Given the inability to directly and non-invasively assay gene expression from brain tissue in living patients, the current approach offers a novel in-vivo window into how molecular mechanisms are associated with ongoing and dynamic macroscale neural systems development across the lifespan in ASD.
Another striking feature of these results is the large-scale nature of the association that covers a majority of the blood leukocyte transcriptome considered by the co-expression analysis. This feature matches predictions from the omnigenic model20. The omnigenic model suggests that for any complex trait or disorder (e.g., ASD), the majority of heritability signal is spread widely throughout most of the genome. The omnigenic model also suggests that the numerous widespread ‘peripheral’ genes of small effect likely interact within gene regulatory networks with a smaller set of ‘core’ genes with much larger effect. Here we find evidence that higher-impact rare dnPTVs in ASD that are intolerant to loss of function mutations are enriched amongst non-zero modules. Furthermore, we also find that many targets of FMRP and CHD8 are enriched in non-zero modules. Thus, the massive number of genes present within non-zero modules may point to a large peripheral background of small risk common variants that could work en masse and interact in important ways with genes that can be higher-impact core mechanisms.
The omnigenic model makes another key prediction, namely that the such associations can be detectable in many tissue types other than the brain, such as blood leukocytes. The omnigenic model suggests that a large percentage of the genes associated with a complex trait are likely to be broadly expressed genes. Here we find evidence of large overlap between broadly expressed genes and non-zero modules - around 44% of all broadly expressed genes exist within non-zero association modules. In contrast to the fact that nearly all non-zero modules (e.g., 81%) were enriched with broadly expressed genes, only 2 non-zero modules (e.g., 18%) were enriched in lymphocyte-specific genes. Thus, this large-scale gene co-expression-fMRI association is largely driven by genes broadly expressed in the brain and many other tissues rather than lymphocyte-specific genes. While we observed enrichments between non-zero modules and genes implicated in vocal learning, human-specific genes, ASD-associated prenatal co-expression modules, cortically ASD-downregulated co-expression modules, ASD dnPTVs, and FMRP and CHD8 targets, most of these enrichments likely emerged because each gene list is heavily enriched in broadly expressed genes. Removing broadly expressed genes from these lists results in elimination of nearly all significant enrichments with non-zero modules. Overall, these results highlight the importance of broadly expressed genes as a novel class of mechanisms for further study in ASD.
There are some limitations and caveats to keep in mind. First, the number of genes investigated in the final co-expression and PLS analyses are a subset of the total number of genes in the entire genome that could be considered. Therefore, while non-zero modules do cover a large proportion of the genes examined in the analysis, they do not cover a large majority of the entire genome. The extent of coverage of non-zero modules is certainly compatible with ideas about polygenic architecture behind complex neural phenotypes21. However, the coverage of non-zero modules cannot be interpreted with respect to the omnigenic model in terms of sheer size. The current study does however evaluate predictions from the omnigenic model, particularly with respect to the importance of broadly expressed genes. However, this result can also be consistent with polygenic viewpoints, particularly if most of the polygenic associations reside within broadly expressed genes. Second, because the expression data is measured from a non-neural tissue, many brain-specific genes are not considered in the analyses. Thus, the current dataset cannot say anything about the importance or lack thereof with regard to brain-specific genes, nor can we make comparisons about the relative importance of broadly expressed genes versus brain-specific genes.
To summarize, we identify a large-scale association between multiple coordinated blood leukocyte gene co-expression modules and multivariate fMRI response to speech. Associated co-expression modules are different for all pairwise comparisons between TD toddlers and toddlers with ASD and good versus poor early language outcome. The associated co-expression modules are highly enriched in broadly expressed genes as well as ASD, prenatal, human-specific, and language-relevant genes. These results are congruent with predictions from polygenic and omnigenic models and suggest that gene expression in peripheral cells like blood leukocytes are associated with in-vivo functional neural response to language that differentiates ASD toddlers with poor versus good early language outcomes. The study showcases a novel in-vivo approach that could be used in future work towards precision medicine goals.
Methods
Participants
This study was approved by the Institutional Review Board at University of California, San Diego. Parents provided written informed consent according to the Declaration of Helsinki and were paid for their participation. Identical to the approach used in our earlier studies14, 15, 19, 51, 52, toddlers were recruited through two mechanisms: community referrals (e.g., website) or a general population-based screening method called the 1-Year Well-Baby Check-Up Approach53 that allowed for the prospective study of ASD beginning at 12 months based on a toddler’s failure of the CSBS-DP Infant-Toddler Checklist54, 55. All toddlers were tracked from an intake assessment around 12 months and followed roughly every 12 months until 3–4 years of age. All toddlers, including normal control subjects, participated in a series of tests collected longitudinally across all visits, including the Autism Diagnostic Observation Schedule (ADOS; Module T, 1, or 2)56, the Mullen Scales of Early Learning57, and the Vineland Adaptive Behavior Scales58. All testing occurred at the University of California, San Diego Autism Center of Excellence (ACE). No randomization procedures were implemented as part of the data collection process. Data collection and analyses were not performed blind to the conditions of the experiment.
A total of n=118 toddlers were scanned with fMRI and had available gene expression data. No statistical methods were used to pre-determine sample sizes, but our sample sizes are currently amongst the largest of any fMRI study to date on ASD at very early ages in toddlerhood. From these 118 toddlers, n=81 ASD individuals were examined and were split into 2 language outcome subtypes. n=41 individuals with ASD (34 male, 7 female) were classified as ‘poor’ language outcome (ASD Poor), based on the criteria of having both Mullen EL and RL T-scores more than 1 standard deviation below the norm of 50 (i.e. T<40) at the final testing time-point (mean age at fMRI scan = 29.53 months, SD at fMRI scan = 8.04, range = 12-46 months). Another n=40 individuals with ASD (30 male, 10 female) were classified as ‘good’ language outcome (ASD Good), based on having either Mullen EL or RL T-scores greater than or equal to 40 (i.e. T ≥ 40) at the final testing time-point (mean age at fMRI scan = 29.73 months, SD at fMRI scan = 8.51, range = 12-45 months). The usage of the term ‘Good’ here is not used to refer to ability level in absolute terms, but more reflects ability relative to the ASD Poor subgroup. These ASD subtypes were compared to n=37 typically-developing toddlers (21 male, 16 female; mean age at fMRI scan = 26.19 months, SD at fMRI scan = 10.20, range = 12-45 months). ASD subtypes and TD did not statistically differ in age at the time of scanning (F(2,115) = 1.87, p = 0.15). For more demographic and phenotypic information, please see Supplementary Table 7.
Blood Sample Collection, RNA extraction, quality control and samples preparation
Four to six milliliters of blood was collected into EDTA-coated tubes from toddlers on visits when they had no fever, cold, flu, infections or other illnesses, or use of medications for illnesses 72 hours prior blood draw. Blood samples were passed over a LeukoLOCK™ filter (Ambion, Austin, TX, USA) to capture and stabilize leukocytes and immediately placed in a 20°C freezer. Total RNA was extracted following standard procedures and manufacturer’s instructions (Ambion, Austin, TX, USA). LeukoLOCK disks (Ambion Cat #1933) were freed from RNA-later and Tri-reagent (Ambion Cat #9738) was used to flush out the captured lymphocyte and lyse the cells. RNA was subsequently precipitated with ethanol and purified though washing and cartridge-based steps. The quality of mRNA samples was quantified by the RNA Integrity Number (RIN), values of 7.0 or greater were considered acceptable59, and all processed RNA samples passed RIN quality control. Quantification of RNA was performed using Nanodrop (Thermo Scientific, Wilmington, DE, USA). Samples were prep in 96-well plates at the concentration of 25 ng/µl.
Gene expression and data processing
RNA was assayed at Scripps Genomic Medicine (La Jolla, CA, USA) for labeling, hybridization, and scanning using the Illumina BeadChips pipeline (Illumina, San Diego, CA, USA) per the manufacturer’s instruction. All arrays were scanned with the Illumina BeadArray Reader and read into Illumina GenomeStudio software (version 1.1.1). Raw data was exported from Illumina GenomeStudio, and data pre-processing was performed using the lumi package60 for R (http://www.R-project.org) and Bioconductor (http://www.bioconductor.org)61. Raw and normalized data are part of larger sets deposited in the Gene Expression Omnibus database (GSE42133; GSE111175).
A larger primary dataset of blood leukocyte gene expression was available from 383 samples from 314 toddlers with the age range of 1-to-4 years old. The samples were assayed using the Illumina microarray platform on three batches. The datasets were combined by matching the Illumina Probe ID and probe nucleotide sequences. The final set included a total of 20,194 gene probes. Quality control analysis was performed to identify and remove 23 outlier samples from the dataset. Samples were marked as outlier if they showed low signal intensity (average signal two standard deviations lower than the overall mean), deviant pairwise correlations, deviant cumulative distributions, deviant multi-dimensional scaling plots, or poor hierarchical clustering, as described elsewhere14. The high-quality dataset included 360 samples from 299 toddlers. High reproducibility was observed across technical replicates (mean Spearman correlation of 0.97 and median of 0.98). Thus, we randomly removed one of each of two technical replicates from the primary dataset. From the subjects in the larger primary dataset, n=118 also had task-fMRI data and thus a total of n=105 from the Illumina HT12 platform along with n=13 from the Illumina WG6 platform were used in this study. Batch was not asymmetrically distributed across one subgroup more than another, as chi-square analyses on the contingency table between subgroup and batch show no effect (χ2(4) = 4.772, p = 0.3115). ASD subtypes and TD toddlers also did not statistically differ in age at the time of blood sampling (F(2,115) = 1.74, p = 0.17). The 20,194 probes were then collapsed to 14,313 genes based on picking the probe with maximal mean expression across samples. Data were quantile normalized and then adjusted for batch effects, sex, and RIN. This batch, sex, and RIN adjusted data were utilized in all further downstream analyses. We also checked for differences in proportion estimates of different leukocyte cell types (i.e. neutrophils, B cells, T cells, NK cells, and monocytes) using the CellCODE deconvolution method62, but found no evidence of differences across groups for any cell type (see Supplementary Table 8). In addition to the primary analyses using WGCNA, differential expression analysis at the level of individual genes was also conducted using limma63, and DE genes were identified if they passed Storey FDR q<0.0564. Data distributions were assumed to be normal but this was not formally tested for each gene. Further enrichment tests were used annotate which co-expression modules are enriched for such DE genes.
Weighted Gene Co-Expression Network Analysis
We reduced the number of features in the gene expression dataset from 14,313 genes down to 21 modules of tightly co-expressed genes. This data reduction step was achieved using weighted gene co-expression network analysis (WGCNA), implemented within the WGCNA library in R23. Correlation matrices estimated with the robust correlation measure of biweight midcorrelation were computed and then converted into adjacency matrices that retain the sign of the correlation. These adjacency matrices were then raised to a soft power of 16 (see Supplementary Fig. 3a). This soft power was chosen by finding the first soft power where a measure of R2 scale-free topology model fit saturates at least above R2 > 0.865 and where the slope was between -1 and -266. The soft power thresholded adjacency matrix was then converted into a topological overlap matrix (TOM) and then a TOM dissimilarity matrix (e.g., 1-TOM). The TOM dissimilarity matrix was then input into agglomerative hierarchical clustering using the average linkage method. Gene modules were defined from the resulting clustering tree, and branches were cut using a hybrid dynamic tree cutting algorithm (deepSplit parameter = 4) (see Supplementary Fig. 3b). Modules were merged at a cut height of 0.2, and the minimum module size was set to 100. Only genes with a module membership was r > 0.3 were retained within modules. For each gene module, a summary measure called the module eigengene (ME) was computed as the first principal component of the scaled (standardized) module expression profiles. We also computed module membership for each gene and module. Module membership indicates the correlation between each gene and the module eigengene (see Supplementary Table 6). Genes that could not be clustered into any specific module are left within the M0 module, and this module was not considered in any further analyses. Analysis of group differences in MEs were also conducted using linear models and correction for multiple comparisons at FDR q<0.05 (see Supplementary Table 3; Supplementary Fig. 4). Data distributions were assumed to be normal but this was not formally tested for each module. Further WGCNA analyses were run separately within each group in order to check for preservation of detected modules across groups at a soft power threshold of 20. These analyses all indicated high levels of preservation (Zsummary>10)67 across nearly all detected modules for each pairwise group comparison (see Supplementary Fig. 5).
fMRI Data Acquisition and Task Design
The fMRI task was identical to that used in our previously published studies19, 68–70 and consisted of three types of speech stimuli (complex forward speech, simple forward speech, and backward speech) as well as rest blocks interspersed between task blocks to forestall possible habituation across blocks. Blocks were 20 seconds in duration. All speech conditions were created using the same female speaker. Two contrasts of interest were analyzed in this study: all speech conditions versus rest and forward (simple + complex) versus backward speech. At early language learning ages, when neonates, infants, and toddlers are not yet experts at language, forward and backward speech both activate language-relevant temporal areas; thus, specific comparisons between them tend to be non-significant70, 71. Therefore, forward and backward speech stimuli both appear to be effective in stimulating language-sensitive cortices, by perhaps both being treated as potentially language-relevant by the language-inexperienced infant and toddler brain. Thus, although we have specifically analyzed both contrasts, because of this age-related caveat for forward versus backward speech, our main contrast of interest was all speech versus rest.
Imaging data were collected on a 1.5 Tesla General Electric MRI scanner during natural sleep at night; no sedation was used. High-resolution T1-weighted anatomical scans were collected for warping fMRI data into standard atlas space. Blood oxygenation level-dependent (BOLD) signal was measured across the whole brain with echoplanar imaging during the language paradigm (echo time = 30 ms, repetition time = 2,500 ms, flip angle = 90 degrees, bandwidth = 70 kHz, field of view = 25.6 cm, in-plane resolution = 4 x 4 mm, slice thickness = 4 mm, 31 slices).
Analysis of head motion via framewise displacement (FD) and DVARS indicated that head motion was minimal (mean FD<0.25) for nearly all subjects in all groups (ASD Good mean = 0.11 mm, sd = 0.23; ASD Poor mean = 0.07 mm, sd = 0.08; TD mean = 0.07 mm, sd = 0.03) and that groups did not differ in either mean FD (F(2,115) = 1.12, p = 0.33) or mean DVARS (F(2,115) = 1.93, p = 0.15; ASD Good mean = 8.81, sd = 2.85; ASD Poor mean = 8.61, sd = 2.57; TD mean = 7.75, sd = 2.01).
fMRI Data Analyses
Preprocessing of functional imaging data was implemented within the Analysis of Functional NeuroImages (AFNI) software package. The preprocessing pipeline was comprised of motion correction, normalization to Talairach space, and smoothing (8mm full-width at half-maximum (FWHM) Gaussian kernel). First-level and second-level mass-univariate whole-brain activation analyses were modeled with the general linear model (GLM) in SPM8 (http://www.fil.ion.ucl.ac.uk/spm/). Events in first-level models were modeled using the canonical hemodynamic response function and its temporal derivative. All first-level GLMs included motion parameters as covariates of no interest. High-pass temporal filtering was applied with a cutoff of 0.0078 Hz (1/128 seconds) in order to remove low frequency drift in the time series. For whole-brain analyses, the distributions were assumed to be normal but this was not formally tested for every voxel.
Group-level analysis were implemented using the general linear model in SPM8. We ran whole-brain analyses for the contrast of All Speech vs Rest within and between-groups and thresholded at a voxelwise FDR q<0.0572. For between-group region of interest (ROI) analysis we used meta-analytic ROIs from the Neurosynth term ‘language’73 of frontal and temporal cortex areas in both hemispheres, identical to those used in a prior paper19. We computed the difference in percent signal change for All Speech vs Rest and used this as the dependent variable in a linear model that tests subtype membership as the main independent variable of interest, while covarying for sex. Data are plotted in Fig. 2b for each individual to show the distribution of the data. No group showed heavy deviations from normality and all regions showed evidence of homogeneity of variance between groups. Follow-up tests for pairwise group comparisons used Welch’s t-test.
fMRI-Gene Expression Association Analysis
To assess multivariate fMRI-gene expression relationships we used partial least squares (PLS) analysis74, 75. PLS is widely used in the neuroimaging literature, particularly when explaining multivariate neural responses in terms of multivariate behavioral patterns of variation or a design matrix. Given that the current dataset is massively multivariate both in terms of fMRI and gene expression datasets, we used PLS to elucidate how variation in neural response to speech across large-scale neural systems covaries with gene expression as measured by module eigengene values of co-expression modules. PLS allows for identifying such relationships by finding latent fMRI-gene expression variable pairs (LV) that maximally explain covariation in the dataset and which are uncorrelated with other latent fMRI-gene expression variable pairs. The strength of such covariation is denoted by the singular value (d) for each brain-behavior LV, and hypothesis tests can be made via using permutation tests on the singular values. Furthermore, identifying brain regions that most strongly contribute to each LV pair is made via bootstrapping, whereby a bootstrap ratio is created for each voxel, and represents the reliability of that voxel for contributing strongly to the LV pattern identified. The bootstrap ratio is roughly equivalent to a Z statistic and can be used to threshold data to find voxels that reliably contribute to an LV pair.
The PLS analyses reported here were implemented within the plsgui Matlab toolbox (www.rotman-baycrest.on.ca/pls/). Here we input first-level all speech versus rest contrast images into the PLS. For gene expression data, we input module eigengene values for all 21 co-expression modules. For statistical inference on identified fMRI-gene expression LV pairs, a permutation test was run with 10,000 permutations. To identify reliably contributing voxels for fMRI-gene expression LVs and to compute 95% confidence intervals (CIs) on fMRI-gene expression correlations, bootstrapping was used with 10,000 resamples. To show voxels that most reliably contribute to significant fMRI-gene expression LVs, we thresholded data for visualization at a bootstrap ratio (BSR) of 1.96 and -1.96. The strength of fMRI-gene expression correlations for significant LVs was displayed as a bar graph with 95% bootstrap CIs as error bars. Gene co-expression modules whereby 95% CIs do not encompass 0 are denoted as ‘non-zero’ association modules. All other modules where 95% CIs include 0 are denoted as ‘zero’ modules.
From the PLS results we tested whether non-zero associations across modules were common across ASD subtypes or common across ASD subtypes and TD. To test this question we counted the overlap amongst non-zero association modules in each group and ran hypergeometric tests that explicitly test for statistically significant overlap or commonality of non-zero associations across groups.
Enrichment Tests
Tests for functional (process-level) enrichment across all modules were implemented using the MetaCore GeneGO software platform. Further gene set enrichment tests (hypergeometric tests and enrichment odds ratio) were done on tissue-specific gene lists. First, we annotated each co-expression module by enrichment with 4 types of gene classes of relevance as defined by GTEx data reported from Boyle et al.20 These classes were 1) broadly expressed genes, 2) brain-specific genes, 3) whole-blood specific genes, and 4) lymphocyte-specific genes. The background pool number for these hypergeometric tests was 14,313. Next, we tested whether non-zero modules were heavily enriched with modules from one or more of these gene classes. The background total for these tests was set to the total number of co-expression modules (e.g., 21).
Further enrichment tests were done across a wider range of gene lists of theoretical importance. Song birds are often used as animal models relevant for the vocal learning component of language25, 26, 33. We investigated enrichments with differentially expressed genes taken from a microarray dataset of Area X of song birds33. To identify differentially expressed (DE) genes between singing versus non-singing birds, we re-analyzed this dataset (GEO Accession ID: GSE34819) using limma63, and DE genes were identified if they passed Storey FDR q<0.0564. Given the uniquely human nature of language, we also tested hypotheses regarding enrichments amongst genes that are transcriptionally different in the cortical tissue between humans and chimpanzees (i.e. human-specific genes). These tests were done across gene lists from two independent investigations on human-specific gene expression differences, where the common overlap amongst the two lists is small (4.38%)34, 35. Ample evidence suggests that prenatal brain developmental periods are critical for ASD13, 36–38. To test enrichment with prenatal ASD-associated co-expression modules, we utilized co-expression modules from two independent studies that analyzed the Allen Institute BrainSpan dataset39 – 1) Eising et al., analyzed data from both subcortical and cortical regions and identified modules M3, M9, and M12 as ASD-associated and prenatally active36; 2) Parikshak et al., analyzed only cortical regions and identified M2 and M3 as ASD-associated and prenatally active37. There is 23% overlap between these two gene lists. We also tested enrichments with gene lists known to be associated with ASD, either via genetic evidence or evidence from cortical transcriptomic dysregulation. In particular, we examined de novo protein-truncating variants (dnPTV) associated with ASD48, ASD-associated genes from the SFARI Gene (https://gene.sfari.org)50, and differentially expressed cortical co-expression modules measured from ASD post-mortem frontal and temporal cortex tissue40. For ASD-associated dnPTVs we used a list of 38 genes from Kosmicki et al.,48 with ≥2 dnPTVs in ASD and which also showed 0 dnPTVs in the normative ExAC database49. We additionally used a more relaxed criteria of ≥1 dnPTVs in ASD and 0 dnPTVs in ExAC combined with a probability of loss-of-function intolerance (pLI) ≥0.949, which resulted in 211 genes. Finally, we tested for enrichments with known downstream targets of highly penetrant mutations known to be associated with ASD – FMRP and CHD8. For each, we had lists of downstream targets for two independent studies76–79, where the overlap for FMRP targets was 3.71% and 27.61% for CHD8 targets. FDR q<0.05 was used to identify significant enrichments after multiple comparison correction.
Co-expression Network Preservation Across ASD Brain and Blood Datasets
We also wanted to understand whether co-expression modules detected in blood leukocytes showed preservation of co-expression network patterns in ASD post-mortem cortical tissue from frontal and temporal cortex. To achieve this aim, we utilized ASD post-mortem frontal and temporal cortex RNA-seq data from Parikshak et al.,40. Using the same preprocessed data as Parikshak et al., we computed Zsummary module preservation statistics and evaluated which modules detected from ASD blood leukocyte datasets are preserved in ASD cortical frontal and temporal cortical tissue sampled from similar sites as those detected in the PLS LV1 map. Zsummary > 10 indicates strong preservation, while Zsummary between 2 and 10 indicates moderate preservation67.
Supplementary Material
Acknowledgments
This research was supported by grants to EC and KP (NIMH R01-MH080134 (KP), NIMH R01-MH104446 (KP), NFAR grant (KP), NIMH Autism Center of Excellence grant P50-MH081755 (EC, KP), NIMH R01-MH036840 (EC), NIMH R01-MH110558 (EC, NEL), NIMH U01-MH108898 (EC), NIDCD R01-DC016385 (EC, KP, MVL), CDMRP AR130409 (EC), Simons Foundation 176540 (EC)). The work was additionally supported by an ERC Starting Grant (ERC-2017-STG; 755816) to MVL, and a grant from the Brain & Behavior Research Foundation (NARSAD) to TP. We thank Richard Znamirowski, Clelia Ahrens-Barbeau, Stephanie Solso, Kathleen Campbell, Maisi Mayo, and Julia Young for help with data collection, Stuart Spendlove and Melanie Weinfeld for assistance with clinical characterization of subjects.
Footnotes
Accession codes
Raw blood leukocyte gene expression data is available via Gene Expression Omnibus (GEO) with the following accession codes: GSE42133; GSE111175. Song bird Area X gene expression data is available on GEO (GSE34819).
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Data availability
The raw data that support the findings from this study are publicly available from the NIH National Database for Autism Research (NDAR). Raw blood leukocyte gene expression data is publicly available via Gene Expression Omnibus (GEO) (GSE42133; GSE111175). Song bird Area X gene expression data is publicly available on GEO (GSE34819). GTEx data is publicly available at https://gtexportal.org. ASD post-mortem cortical gene expression can be found at https://github.com/dhglab/Genome-wide-changes-in-lncRNA-alternative-splicing-and-cortical-patterning-in-autism.
Code availability
Code for implementing all analyses can be found at https://github.com/mvlombardo/asdlangoutcomebloodgexfmripls.
Author Contributions
E.C., K.P., L.E., M.V.L., and T.P. conceived the idea and designed the study. M. V. L. conceived and performed all analyses. T.P., V.G., V.W., R.A.I.B., and N.E.L. aided data analyses. E.C., K.P., L.E., L.L., and C.C.B., collected data. E.C., K.P., L.E., N.E.L., T.P., and M.V.L., obtained grant funding. M.V.L. and E.C. wrote the manuscript. All authors contributed to editing the manuscript.
Competing Interests
The authors declare no competing interests.
References
- 1.Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Happe F, Ronald A, Plomin R. Time to give up on a single explanation for autism. Nat Neurosci. 2006;9:1218–1220. doi: 10.1038/nn1770. [DOI] [PubMed] [Google Scholar]
- 3.Lai MC, Lombardo MV, Chakrabarti B, Baron-Cohen S. Subgrouping the autism "spectrum": reflections on DSM-5. PLoS Biol. 2013;11:e1001544. doi: 10.1371/journal.pbio.1001544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapur S, Phillips AG, Insel TR. Why has it taken so long for biological psychiatry to develop clinical tests and what to do about it? Mol Psychiatry. 2012;17:1174–1179. doi: 10.1038/mp.2012.105. [DOI] [PubMed] [Google Scholar]
- 5.Lombardo MV, Lai MC, Baron-Cohen S. Big data approaches to decomposing heterogeneity across the autism spectrum. bioRxiv. 2018 doi: 10.1101/278788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kjelgaard MM, Tager-Flusberg H. An Investigation of Language Impairment in Autism: Implications for Genetic Subgroups. Lan Cogn Process. 2001;16:287–308. doi: 10.1080/01690960042000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tager-Flusberg H, Kasari C. Minimally verbal school-aged children with autism spectrum disorder: the neglected end of the spectrum. Autism Res. 2013;6:468–478. doi: 10.1002/aur.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howlin P. Outcome in high-functioning adults with autism with and without early language delays: implications for the differentiation between autism and Asperger syndrome. J Autism Dev Disord. 2003;33:3–13. doi: 10.1023/a:1022270118899. [DOI] [PubMed] [Google Scholar]
- 9.Perry A, et al. Predictors of outcome for children receiving intensive behavioral intervention in a large, community-based program. Res Autism Spectr Disord. 2011;5:592–603. [Google Scholar]
- 10.Gotham K, Pickles A, Lord C. Trajectories of autism severity in children using standardized ADOS scores. Pediatrics. 2012;130:e1278–1284. doi: 10.1542/peds.2011-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ventner A, Lord C, Schopler E. A follow-up study of high-functioning autistic children. J Child Psychol Psychiatry. 1992;33:489–507. doi: 10.1111/j.1469-7610.1992.tb00887.x. [DOI] [PubMed] [Google Scholar]
- 12.Szatmari P, et al. Similar developmental trajectories in autism and Asperger syndrome: from early childhood to adolescence. J Child Psychol Psychiatry. 2009;50:1459–1467. doi: 10.1111/j.1469-7610.2009.02123.x. [DOI] [PubMed] [Google Scholar]
- 13.Courchesne E, et al. The ASD living biology: From cell proliferation to clinical phenotype. Mol Psychiatry. 2018 doi: 10.1038/s41380-018-0056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pramparo T, et al. Cell cycle networks link gene expression dysregulation, mutation, and brain maldevelopment in autistic toddlers. Mol Syst Biol. 2015;11:841. doi: 10.15252/msb.20156108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pramparo T, et al. Prediction of autism by translation and immune/inflammation coexpressed genes in toddlers from pediatric community practices. JAMA Psychiatry. 2015;72:386–394. doi: 10.1001/jamapsychiatry.2014.3008. [DOI] [PubMed] [Google Scholar]
- 16.Geschwind DH, State MW. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 2015;14:1109–1120. doi: 10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandin S, et al. The Heritability of Autism Spectrum Disorder. JAMA. 2017;318:1182–1184. doi: 10.1001/jama.2017.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.GTEx Consortium et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombardo MV, et al. Different functional neural substrates for good and poor language outcome in autism. Neuron. 2015;86:567–577. doi: 10.1016/j.neuron.2015.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyle EA, Li YI, Pritchard JK. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wray NR, Wijmenga C, Sullivan PF, Yang J, Visscher PM. Common Disease Is More Complex Than Implied by the Core Gene Omnigenic Model. Cell. 2018;173:1573–1580. doi: 10.1016/j.cell.2018.05.051. [DOI] [PubMed] [Google Scholar]
- 22.Cremers HR, Wager TD, Yarkoni T. The relation between statistical power and inference in fMRI. PLoS One. 2017;12:e0184923. doi: 10.1371/journal.pone.0184923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hickok G, Poeppel D. The cortical organization of speech processing. Nature reviews. Neuroscience. 2007;8:393–402. doi: 10.1038/nrn2113. [DOI] [PubMed] [Google Scholar]
- 25.Konopka G, Roberts TF. Insights into the Neural and Genetic Basis of Vocal Communication. Cell. 2016;164:1269–1276. doi: 10.1016/j.cell.2016.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfenning AR, et al. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science. 2014;346 doi: 10.1126/science.1256846. 1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 28.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernier R, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan JT, et al. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol Psychiatry. 2010;68:368–376. doi: 10.1016/j.biopsych.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 31.Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- 32.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hilliard AT, Miller JE, Fraley ER, Horvath S, White SA. Molecular microcircuitry underlies functional specification in a basal ganglia circuit dedicated to vocal learning. Neuron. 2012;73:537–552. doi: 10.1016/j.neuron.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konopka G, et al. Human-specific transcriptional networks in the brain. Neuron. 2012;75:601–617. doi: 10.1016/j.neuron.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, et al. Disruption of an Evolutionarily Novel Synaptic Expression Pattern in Autism. PLoS Biol. 2016;14:e1002558. doi: 10.1371/journal.pbio.1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eising E, et al. A set of regulatory genes co-expressed in embryonic human brain is implicated in disrupted speech development. Mol Psychiatry. 2018 doi: 10.1038/s41380-018-0020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parikshak NN, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willsey AJ, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parikshak NN, et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540:423–427. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17:290–314. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders SJ, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchetto MCN, et al. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503:525–529. doi: 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nuttle X, et al. Emergence of a Homo sapiens-specific gene family and chromosome 16p11.2 CNV susceptibility. Nature. 2016;536:205–209. doi: 10.1038/nature19075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hippolyte L, et al. The Number of Genomic Copies at the 16p11.2 Locus Modulates Language, Verbal Memory, and Inhibition. Biol Psychiatry. 2016;80:129–139. doi: 10.1016/j.biopsych.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 46.Demopoulos C, et al. Abnormal Speech Motor Control in Individuals with 16p11.2 Deletions. Sci Rep. 2018;8 doi: 10.1038/s41598-018-19751-x. 1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berman JI, et al. Abnormal auditory and language pathways in children with 16p11.2 deletion. Neuroimage Clin. 2015;9:50–57. doi: 10.1016/j.nicl.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kosmicki JA, et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet. 2017;49:504–510. doi: 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abrahams BS, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pierce K, Conant D, Hazin R, Stoner R, Desmond J. Preference for geometric patterns early in life as a risk factor for autism. Arch Gen Psychiatry. 2011;68:101–109. doi: 10.1001/archgenpsychiatry.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pierce K, et al. Eye Tracking Reveals Abnormal Visual Preference for Geometric Images as an Early Biomarker of an Autism Spectrum Disorder Subtype Associated With Increased Symptom Severity. Biol Psychiatry. 2016;79:657–666. doi: 10.1016/j.biopsych.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pierce K, et al. Detecting, studying, and treating autism early: the one-year well-baby check-up approach. J Pediatr. 2011;159:458–465 e451-456. doi: 10.1016/j.jpeds.2011.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wetherby A, Prizant B. Communication and Symbolic Behavior Scales Developmental Profile, First Normed Edition. Paul H. Brookes; Baltimore: 2002. [Google Scholar]
- 55.Wetherby AM, Brosnan-Maddox S, Peace V, Newton L. Validation of the Infant-Toddler Checklist as a broadband screener for autism spectrum disorders from 9 to 24 months of age. Autism. 2008;12:487–511. doi: 10.1177/1362361308094501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lord C, et al. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 2000;30:205–223. [PubMed] [Google Scholar]
- 57.Mullen EM. Mullen scales of early learning. American Guidance Service, Inc; Circle Pine, MN: 1995. [Google Scholar]
- 58.Sparrow S, Cicchetti D, Balla D. Vineland-II scales of adaptive behavior: survey form manual. American Guidance Service Inc; Circle Pines, MN: 2005. [Google Scholar]
- 59.Schroeder A, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 61.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chikina M, Zaslavsky E, Sealfon SC. CellCODE: a robust latent variable approach to differential expression analysis for heterogeneous cell populations. Bioinformatics. 2015;31:1584–1591. doi: 10.1093/bioinformatics/btv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society, Series B. 2002;64:479–498. [Google Scholar]
- 65.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 66.Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eyler LT, Pierce K, Courchesne E. A failure of left temporal cortex to specialize for language is an early emerging and fundamental property of autism. Brain. 2012;135:949–960. doi: 10.1093/brain/awr364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Redcay E, Courchesne E. Deviant functional magnetic resonance imaging patterns of brain activity to speech in 2-3-year-old children with autism spectrum disorder. Biol Psychiatry. 2008;64:589–598. doi: 10.1016/j.biopsych.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Redcay E, Haist F, Courchesne E. Functional neuroimaging of speech perception during a pivotal period in language acquisition. Dev Sci. 2008;11:237–252. doi: 10.1111/j.1467-7687.2008.00674.x. [DOI] [PubMed] [Google Scholar]
- 71.Dehaene-Lambertz G, Dehaene S, Hertz-Pannier L. Functional neuroimaging of speech perception in infants. Science (New York, N.Y. 2002;298:2013–2015. doi: 10.1126/science.1077066. [DOI] [PubMed] [Google Scholar]
- 72.Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- 73.Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 2011;8:665–670. doi: 10.1038/nmeth.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krishnan A, Williams LJ, McIntosh AR, Abdi H. Partial Least Squares (PLS) methods for neuroimaging: a tutorial and review. Neuroimage. 2011;56:455–475. doi: 10.1016/j.neuroimage.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 75.McIntosh AR, Lobaugh NJ. Partial least squares analysis of neuroimaging data: applications and advances. Neuroimage. 2004;23(Suppl 1):S250–263. doi: 10.1016/j.neuroimage.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 76.Cotney J, et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6 doi: 10.1038/ncomms7404. 6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sugathan A, et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci USA. 2014;111:E4468–4477. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Greenblatt EJ, Spradling AC. Fragile X mental retardation 1 gene enhances the translation of large autism-related proteins. Science. 2018;361:709–721. doi: 10.1126/science.aas9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.