Abstract
The Radical SAM Superfamily contains over 100,000 homologous enzymes that catalyze a remarkably broad range of reactions required for life, including metabolism, nucleic acid modification, and biogenesis of cofactors. While the highly conserved SAM-binding motif responsible for formation of the key 5’-deoxyadenosyl radical intermediate is a key structural feature that simplifies identification of superfamily members, our understanding their structure-function relationships is complicated by the modular nature of their structures, which exhibit varied and complex domain architectures. To gain new insight about these relationships, we classified the entire set of sequences into similarity-based subgroups that could be visualized using sequence similarity networks. This superfamily-wide analysis reveals important features that had not previously been appreciated from studies focused on one or a few members. Functional information mapped to the networks indicates which members have been experimentally or structurally characterized, their known reaction types, and their phylogenentic distribution. Despite the biological importance of radical SAM chemistry, the vast majority of superfamily members have never been experimentally characterized in any way, suggesting that many new reactions remain to be discovered. In addition to 20 subgroups with at least one known function, we identified additional subgroups made up entirely of sequences of unknown function. Importantly, our results indicate that even general reaction types fail to track well with our sequence similarity-based subgroupings, raising major challenges for function prediction for currently identified and new members that continue to be discovered. Interactive similarity networks and other data from this analysis are available from the Structure-Function Linkage Database.
Keywords: Radical SAM superfamily census; classification of Radical SAM enzymes by sequence similarity; subgroups and families in the Radical SAM superfamily; multiple domain structures of Radical SAM superfamily enzymes; structure-function mapping, phylogenetic representation; sequence similarity networks
1. INTRODUCTION: OVERVIEW OF THE RADICAL SAM SUPERFAMILY
The widely studied Radical S-adenosylmethionine (SAM) Superfamily (RSS) was originally defined using bioinformatics techniques to survey the 650 RSS members available at that time. It described a homologous group of enzymes united by their utilization of SAM in a radical mechanism (Sofia, Chen, Hetzler, Reyes-Spindola, & Miller, 2001). The original sequence set came from 126 species representing all Kingdoms of life and included many of the first RSS enzymes to be characterized: lysine 2,3-aminomutase (LAM), (Moss & Frey, 1987), biotin synthase (BioB) (Lotierzo, Tse Sum Bui, Florentin, Escalettes, & Marquet, 2005; Reyda, Dippold, Dotson, & Jarrett, 2008) lipoyl synthase (LipA) (Cicchillo, Iwig, et al., 2004; Miller et al., 2000), pyruvate-formate lyase activase (PflA) (Knappe & Sawers, 1990; Vey et al., 2008), and anaerobic ribonucleoside-triphosphate reductase activase (NrdG) (Padovani, Thomas, Trautwein, Mulliez, & Fontecave, 2001). From a chemical perspective, to be considered a member of the RSS, Sofia defined three characteristics that were minimally required:
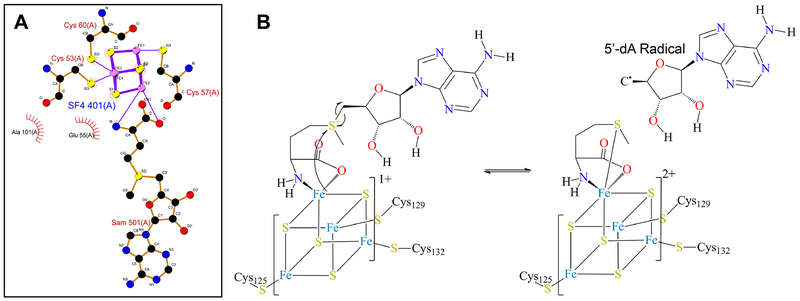
A unique three-cysteine motif that binds the [Fe4S4] cluster, leaving the apical iron free to bind the SAM moiety in a bidentate manner. This canonical motif, [CX3CX2C] is associated with a single domain and largely conserved in known structures (Fig. 1A). The vast majority of RSS proteins (more than 90 %) contain this motif and the rest exhibit several different variations that have either cysteine residues one and two or two and three separated by two or three other residues. The number of residues between alternate cysteine pairs (cysteines one and two or cysteines two and three) can vary from three to 22.
A common activation step involved in formation of the 5’-deoxyadenosyl (5’-dA) radical and methionine (Met) (Fig. 1B).1
A requirement for an external electron donor to catalyze the initial reduction of the [Fe4S4] cluster.
Fig. 1.
(A) [Fe4-S4] binding motif from biotin synthase (PDB: 1R30). Image created using LigPlot+ (Laskowski & Swindells, 2011). (B) The common activation step associated with the canonical RSS.
In this work, we refer to this superfamily as the “canonical” RSS to distinguish it from several other unrelated superfamilies that use SAM in a radical-like reaction.
The original set of 650 sequences has now grown over 150-fold (not counting sequences from the large compilations of metagenomic data now coming available). Known functions reveal that RSS enzymes catalyze a dizzying array of disparate and essential chemistries that range from the formation of complex metal cofactors (Dinis, Wieckowski, & Roach, 2016) (e.g. the formation of the [FeFe]-hydrogenase metallocofactor in HydG (Pilet et al., 2009) and HydE (Nicolet et al., 2008)) to the formation of more than half of the over two dozen known organic cofactors (for example, biotin (Lotierzo, Tse Sum Bui, Florentin, Escalettes, & Marquet, 2005; Reyda, Dippold, Dotson, & Jarrett, 2008), lipoic acid (Cicchillo, Lee, et al., 2004; Miller et al., 2000), menaquinone (vitamin K) (Hiratsuka et al., 2008) and pyrroloquinonoline quinone (PQQ) (Barr et al., 2016; Puehringer, Metlitzky, & Schwarzenbacher, 2008)). They are also involved in the modification of nucleic acids, often via methylation of aromatic carbon centers, repair of DNA dimers (as in spore photoproduct lyase (SPL) (Benjdia, Heil, Barends, Carell, & Schlichting, 2012; Yang & Li, 2015)), the formation of the wybutosine base on tRNA (Young & Bandarian, 2011), and the formation of complex natural products such as antibiotics (Mahanta, Hudson, & Mitchell, 2017a, 2017b) (for example nosiheptide (LaMattina et al., 2017; Yu et al., 2009) and bleomycin (Tao et al., 2007)). Despite many excellent reviews (see (Broderick, Duffus, Duschene, & Shepard, 2014; Dowling, Vey, Croft, & Drennan, 2012; Grell, Goldman, & Drennan, 2015; Lanz & Booker, 2012; Vey & Drennan, 2011; Wang et al., 2014) for some recent examples) and an ever growing corpus of work that addresses mechanistic and structural features of individual or small groups of RSS enzymes, relatively little is known about the structure-function relationships across all of the members of this complex and fascinating superfamily because the huge proportion of RSS enzymes remain experimentally uncharacterized.
As with the chemical perspective, the RSS is difficult to define from a sequence and structural perspective due to the varied multi-domain architectures (MDAs) in which the core superfamily motif is embedded. For the global analysis described here, we started with the sequence sets that represent the superfamily domain containing the SAM binding motif available from the widely used Pfam (Finn et al., 2016) and InterPro (Finn et al., 2017) resources, then expanded and curated the set to obtain over 100,000 non-redundant sequences representing the RSS superfamily(see Methods).
Besides the canonical RSS, we note that at least three other homologous sets of other fold types catalyze RSS-like reactions using mechanisms that differ in key ways from those of the RSS described here. We have provisionally defined these groups as superfamilies because the members within each are sufficiently diverse that we expect they may catalyze additional reactions besides those for which they are named. We were unable to find sequence or structural relationships between the canonical RSS and any of these other superfamilies. As a result, we do not include any of the members of these other superfamilies in the canonical RSS.
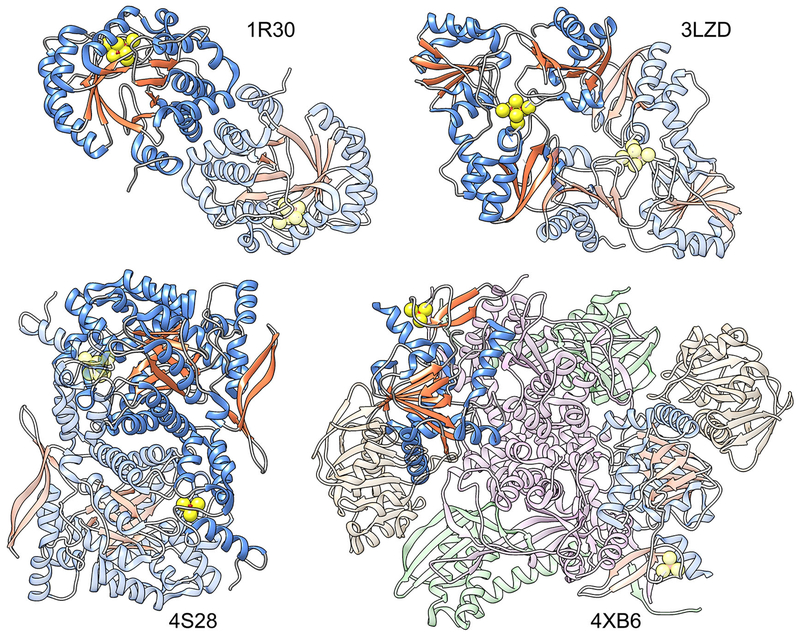
These superfamilies include the radical SAM phosphonate metabolism superfamily (Kamat, Williams, & Raushel, 2011) that catalyzes the demethylation and cyclization of a phosphine by alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase (PhnJ), the radical SAM phosphomethylpyrimidine synthase superfamily (Coquille et al., 2013) that catalyzes the phosphomethylpyrimidine synthesis reaction, and the radical SAM 3-amino-3-carboxypropyl radical forming superfamily involved in diphthamide biosynthesis (Zhang et al., 2010). For the latter, both the radical formation and structural origin of the SAM binding motif show differences from the canonical RSS, i.e., the initial radical formed is a 3-amino-3-carboxypropyl rather than the 5’-deoxyadenosyl radical, while the iron-sulfur cluster is bound by a three-cysteine motif using cysteine residues from three different domains (rather than from a single domain as in the canonical RSS). Although these superfamilies are not discussed further in this report, additional information, including sequence similarity networks and other data about these non-canonical superfamilies is available from the Structure-Function Linkage Database (SFLD) (Akiva et al., 2014). Fig. 2 shows a comparison of a canonical RSS structure with those of these three noncanonical superfamilies. An additional superfamily has been described in the literature that produces a 5’-dA radical but differs in other important details, such as in the source of the radical. For example, methylmalonyl-coenzyme A mutase (Mancia et al., 1996) is a member of a class of enzymes that uses coenzyme B12 (adenosylcobalamin) as a cofactor to generate the 5’-dA radical).
Fig. 2.
Comparison of a canonical RSS structure with structures from unrelated superfamilies of other fold types whose members catalyze RSS-like chemistry. Chains containing the [Fe4S4] cluster are colored by secondary structure, with helices in blue and strands in orange; one copy of the chain per structure is highlighted. The sulfur atoms from the [Fe4S4] clusters and from their adjacent cysteine residues are shown as spheres. Left to right top row: canonical RSS, biotin synthase, PDB: 1R30; Radical SAM 3-amino-3-carboxypropyl Radical Forming Superfamily, diphthamide synthetase, PDB: 3LZD; bottom row: Radical SAM Phosphomethylpyrimidine Synthase Superfamily, phosphomethylpyrimidine synthase, PDB: 4S28; and Radical SAM Phosphonate Metabolism Superfamily, PDB: 4XB6. The physiological unit of all of these structures is a homo-2-mer except for 4XB6, which is a hetero-8-mer.
This paper reports new findings regarding sequence-structure-function relationships in the canonical RSS that can be uniquely accessed from large-scale comparisons of their sequences. Sequence similarity networks (SSNs) annotated with different types of functional and other information (Atkinson, Morris, Ferrin, & Babbitt, 2009) are used to summarize these relationships across the entire superfamily. Guided by the subgroupings emerging from these comparisons, we generated a classification of the RSS that includes known functions (knowns) along with the majority of members that are of unknown function (unknowns). For sequences sufficiently similar to characterized knowns, the large-scale context that resulted may provide clues useful for predicting some of their functional features. This global context can also be used to guide identification of informative targets for biochemical and structural characterization of unknowns.
The results of this work, along with additional data and information about the RSS, are available from the SFLD at http://sfld.rbvi.ucsf.edu/. SSNs for the subgroups and families curated in the SFLD, and other information, can be downloaded and interactively visualized and studied using the freely available Cytoscape software (Shannon et al., 2003).
2. RESULTS AND DISCUSSION
In this study, we first describe the RSS from a structural perspective, including the known variations across MDAs that typify the superfamily. Next, we provide a global view of sequence similarity relationships among the RSS using SSNs to illustrate the subgroups into which we partitioned these sequences to establish a comprehensive classification of the entire superfamily based on sequence similarity (in contrast to the majority of previously published RSS classifications that are based on various descriptions of reaction similarity). Mapping the SSNs with functional information describing the coverage of known functions and solved structures reveals how little we know about the breadth of chemical and structural variation across the RSS, while mapping the types of life in which members are found supports previous suggestions that the RSS is of ancient origin. Finally, we describe differences between similarity-based and reaction-based classifications of the RSS and discuss the implications of these differences for function prediction of unknowns.
2.1. STRUCTURAL OVERVIEW OF THE RSS
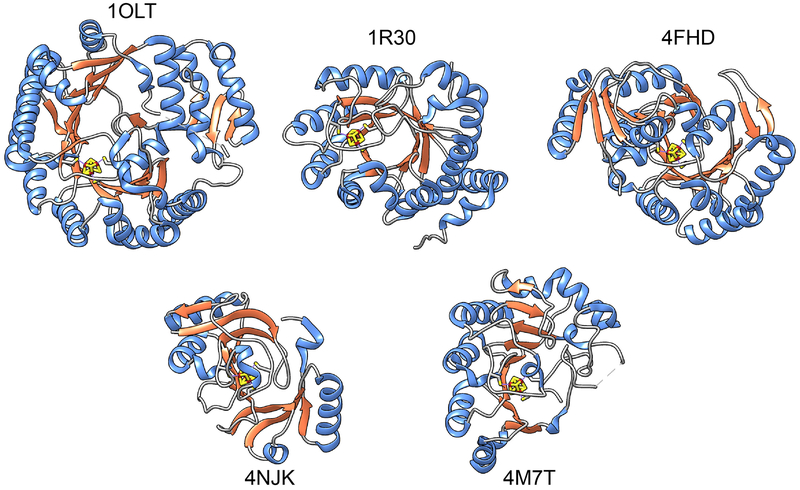
Until recently, it was thought that only two folds were represented in the canonical RSS superfamily, a full triose phosphate isomerase (TIM) barrel type ((β/α)8, CATH (Dawson, Sillitoe, Lees, Lam, & Orengo, 2017) topology 3.20.20.70) and a variant three-quarter barrel ((β/α)6, CATH domain 3.80.30.20). As the number of characterized structures has grown, greater structural diversity has become apparent, with topologies ranging from the full TIM barrel to several additional variants of pruned-down barrels. Some examples are given in Fig. 3.
Fig. 3.
Structural examples of some full-length RSS members of varied architectures. Structures are aligned to show the [Fe4S4] clusters in a similar orientation. For structures with multiple chains, only chain A is shown. Secondary structure coloring is the same as in Fig 2. Top row: 1OLT, coproporphyringen III oxidase, (β/α)6; 1R30, biotin synthase, (β/α)8; 4FHD, spore product lyase, (β/α)6; Bottom row: 4NJK, 7-carboxy-7deazaguanine synthase, (β6/α3); 4M7T, 2-deoxy-scillo-inosamine dehydrogenase (β5/α4).
Much previous work has provided in-depth information about some of the varied domain types that make up these proteins (for example (Dowling, Vey, Croft, & Drennan, 2012; Grell, Goldman, & Drennan, 2015; Vey & Drennan, 2011)). A superfamily-wide description of RSS component domains and their structural roles as they are defined and discussed in this paper is provided in Box 1.
Box 1: Definitions of RSS structural domains used in this work.
The canonical RSS described in this work is defined based on sequence and structural similarity rather than by chemical similarity. Each protein includes several structural elements or domains that make up its multiple domain architecture.
Superfamily domain:
Structural domain that contains the core structural motif shown in Fig. 1A that functions in binding the iron-sulfur cluster involved in activation of S-adenosyl methionine. It is conserved throughout the canonical superfamily and represents the minimum structural motif required for superfamily membership (with at least one known exception; see Footnote 1). It ranges from 46–250 residues in length. The great majority of RSS members have at least one of these motifs; some members have two or three of them.
Functional domain:
The superfamily domain plus accessory domain(s) and other inserts or extensions required for tailoring RS functionality for specific roles. These accessory domains may be conserved in a subset of RSS enzymes (such as the SPASM/Twitch domain-containing enzymes) but are not conserved across the entire superfamily. Their lengths range from 46–1449 residues. In some RSS members, the superfamily and functional domains are the same.
Full-length RSS protein:
This represents the complete amino acid sequence, and may include domains with other functions than those performed by the RSS functional domain. These “extra” domains may be conserved within a subgroup or family but are not conserved across the entire RSS. They may be fused with the RSS functional domain in the full-length RSS polypeptide; alternatively, they can be found as separate enzymes in some organisms.
Full-length RSS proteins range from 46 to just over 2,000 residues. They can be composed of only a superfamily or a functional domain, or as a longer fusion protein, as described above.
The shortest full-length RSS enzyme that is not a fragment is 46 residues in length, while the longest is over 2,000 residues (UniProt 2017). Much of this large variation in size can be ascribed to the large proportion of RSS enzymes represented by multi-domain structures, many of them made up of accessory domains required for RSS function or that are involved in tailoring function in some full-length proteins. Both types of accessory domains are unrelated to the core superfamily domain and are homologous to each other only within specific subgroup(s) or families. For example, the B12-dependent enzymes require Vitamin B12, and so include a cobalamin-binding domain fused to the radical SAM domain. Our analysis, along with that of the InterPro resource (data not shown) suggests that most RSS proteins are on average composed of two or more domains.
The variations in MDA structures represented in the RSS can be estimated from Pfam data, which describe proteins in terms of their constituent domains. Although the PFAM model (PF04055) itself represents only the radical SAM superfamily domain, it is accompanied by a prediction of the additional domain types found in the full-length sequences of the canonical RSS. Table 1 reports on the number of MDAs they predict. The highest confidence MDA predictions are those that are found in multiple sequences, rather than the large number of predicted MDAs that are represented by only one superfamily sequence.
Table 1.
Number of MDAs represented by 1 or more proteins in InterPro
| # Predicted MDAs | Minimum # of proteins/MDA |
|---|---|
| 20 | 1,000 |
| 46 | 200 |
| 60 | 100 |
| 73 | 50 |
| 89 | 25 |
| 831 | 1 |
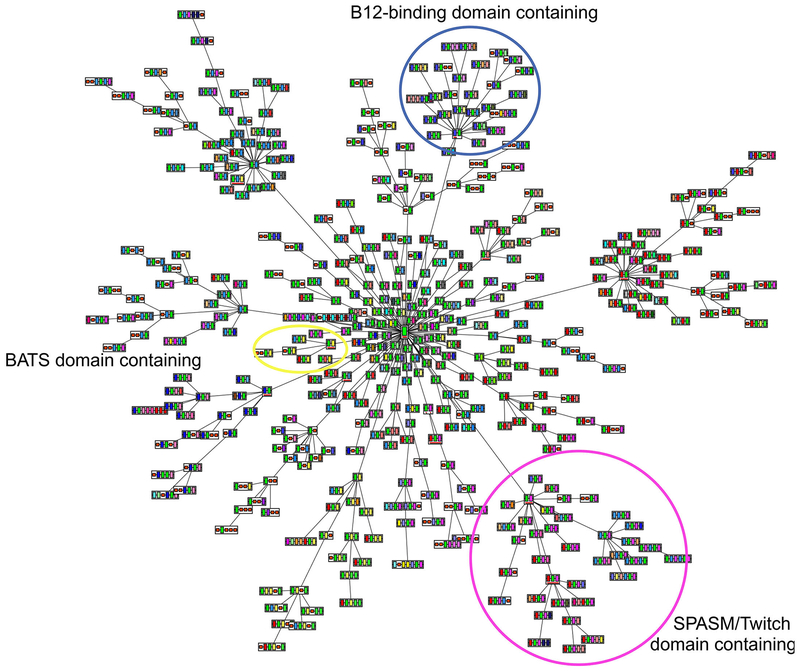
Fig. 4 shows the primary MDAs predicted by Pfam as a network which is centered on the core radical SAM superfamily domain, along with the MDA variants to which it links. As highlighted in Fig. 4, some of these MDAs have multiple domain patterns that cluster together based on the domains they have in common.
Fig. 4:
Predicted domain architectures created using ArchSchema (Tamuri & Laskowski, 2010). Shown are 435 major architecture types of the more than 1,500 representative domain architectures predicted for the 63,785 representative RSS protein sequences in Pfam version 27. These architectures represent 171 distinct domains. The central green rectangle underlined in red in the figure represents the core superfamily domain shared by all members of the canonical RSS, which is repeated in each MDA image shown. Edges connecting individual domains distinguish each complete MDA. The domain (rectangle) connecting each cluster to the larger MDA network is also underlined in red. The circled clusters represent the SPASM/Twitch-like domain (magenta), BATS-like domain (yellow) and B12-binding-like domain (blue) clusters.
A wide literature has addressed the notion that functional diversity within enzyme superfamilies can be achieved through many routes (see, for example, (Brown & Babbitt, 2014; Furnham et al., 2012)). The analysis reported here identifies sequence and structural features that unite the RSS proteins on the scale of the superfamily and describes key types of variation known to confer structural and functional specificity. These findings are consistent with and extend similar conclusions made by others for smaller groups of RSS members. Viewed from this global perspective, the key structure-function relationship of the RSS reveals a conserved “plug-and-play” superfamily domain embedded in many different MDAs. Together with the conserved core motif delivered by the superfamily domain, additional MDA domains enable and distinguish the catalytic and specificity features of the varied reactions of the superfamily. Although this model is undoubtedly an over-simplification with respect to the refined mechanistic understanding of the interplay of structural and chemical features for several unique RSS reactions, it provides a useful foundation for creation of a global classification of the superfamily based on sequence similarity, further refined using available structural and functional information.
2.2. A NEW CLASSIFICATION OF THE RSS BASED ON SEQUENCE SIMILARITY SUBGROUPINGS
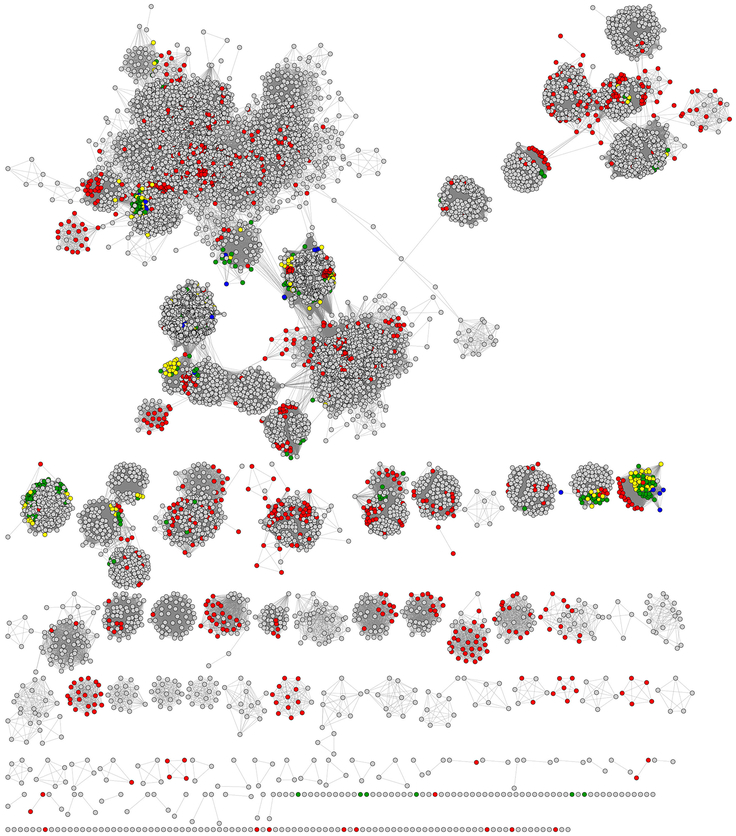
As of December 2017, the RSS curated in the SFLD contains 113,776 unique sequences. To guide our mapping of structure-function relationships across the superfamily, we generated a representative SSN (see Methods for details), followed by a “divide and conquer” strategy to define subgroups of similarity-based clusters. One or more biochemically or structurally characterized enzymes can be associated with 20 of these subgroups; 22 additional subgroups containing no known reactions or structures were also identified and classified in the SFLD as “uncharacterized subgroups.” The SSN shown in Fig. 5 provides a summary view of these Level 1 subgroups of our RSS classification. Together, they form the basis for the new classification of the RSS described in this work. Sequences that were too diverse to be confidently assigned to either named or uncharacterized subgroups were not analyzed further.
Fig. 5.
Representative SSN for the RSS showing major Level 1 subgroups. The SSN was generated from the 113,776 full length RSS sequences in the SFLD. Sequences that share >50% pairwise identical were binned into 10,741 representative nodes (circles). Edges (lines between representative nodes) were drawn between representative nodes if the mean of the BLAST (Altschul et al., 1997) E-values (used as scores) between any pair of sequences in that node was at least 1 × 10−20. At this E-value, the network has 13,591,858 representative edges with a mean sequence identity of 26 % across a mean alignment length of 300 residues The networks are laid out using the prefuse force directed layout in Cytoscape. Twenty subgroups are denoted by distinct colors and numbered according to Supplemental Table 1. Colored nodes are further specified by size and shape: Large nodes are colored if they include at least one sequence assigned to a numbered subgroup. Diamond-shaped nodes specify that at least one of the sequences in that node has been experimentally characterized (but not structurally characterized). Nodes shaped like a downward arrow have at least one protein that has been structurally characterized. Small circular colored nodes have been assigned to a subgroup but are comprised entirely of sequences of unknown function. Small gray circular nodes have not been assigned to a subgroup and are comprised entirely of sequences of unknown function. The 22 largest of these entirely gray clusters are curated in the SFLD as “Uncharacterized Radical SAM Subgroups.” Some small colored clusters and singletons randomly laid out at the bottom of the image are not labeled with a number because they belong to a larger numbered cluster of the same color but fail to meet the E-value cutoff for drawing edges connecting them to that cluster. (The largest subgroup, subgroup 17, provides an example. In this visualization, both the large cluster at the top left and the smaller clusters and singletons that are colored magenta near the bottom of the image belong to subgroup 17, but these nodes are too diverse to be connected to the main subgroup because their similarities fall below the E-value threshold (1 × 10−20) used for drawing edges to the main subgroup. Note that each representative node may contain many individual sequences (see section 2.3).
The SFLD defines subgroups at different levels of detail (e.g., Level 1, Level 2, Level 3 subgroups) as sets of homologous sequences for which the similarities among all members in each subgroup are greater within that subgroup than they are to any sequence in another subgroup. Other information, such as structural variations, MDA organization, and detailed variations in subgroup-specific SAM binding motifs add support for these subgroup boundaries. In particular, as the iron-sulfur cluster binding motifs are highly conserved in the RSS, they are easy to identify so that small differences within each motif and its flanking sequences help to distinguish subgroups. As warranted by available information, more detailed examination of primary (Level 1) subgroups allows curation of additional subgroup levels within it, each representing subsets of sequences that share more detailed sequence, structural, or functional features than are shared across the parent subgroup (see Supplementary Table 1).
At the most granular level of the classification, the SFLD defines “reaction families” that represent sets of sequences within a superfamily or subgroup for which good evidence suggests that all family members catalyze the same reaction using a similar mechanism (Holliday et al., 2017). Each reaction family is curated using one or more “founder” enzymes that have been biochemically (and often, structurally) characterized. Along with the founder enzyme(s), expert curators may assign to these families uncharacterized sequences that are sufficiently similar to the founder sequence and that conserve functionally distinguishing residues (see for example (Brown, Gerlt, Seffernick, & Babbitt, 2006; Holliday et al., 2017)).
Although the use of computational annotation transfer expands the set of sequences that can be functionally assigned, our confidence in these assignments is still limited by the lack of direct experimental evidence of the annotated activity. ,To aid users in evaluating the confidence of these assignments, these sequences are annotated in the SFLD with the IEA (Inferred from Electronic Annotation) evidence code to differentiate them from sequences with the IDA (Inferred from Direct Assay) evidence code. Even using this electronic annotation transfer protocol, more than 50,000 RSS sequences fail to meet our criteria for assigning them to reaction families.
The 20 colored and numbered subgroups shown in Fig. 5 are designated in this work as Level 1 subgroups. Within some of these, an additional 17 subgroups have been classified in the SFLD as Lever 2 or Level 3 subgroups that are children of a Level 1 subgroup. Our classification also includes 101 reaction families for which 85 chemical reactions that have been described in the primary literature. Accompanying this report, Supplementary Table S1 provides a list of the RSS reaction families curated in the SFLD along with their known overall chemical transformations. Other features provided by this table include images of the overall reactions, member accession numbers from several public sequence and structure databases, conserved residues and motifs, and the known types of life to which the majority of members of each subgroup belongs.
To examine the contribution of the highly conserved RSS superfamily domain to the topology of the SSN generated from the full-length sequences shown in Fig. 5, we generated an SSN using only this typically shorter domain. The resulting network (not shown) is substantially similar to that of Fig. 5, indicating that the conserved RSS superfamily domain dominates the sequence signal captured by the all-by-all BLAST comparisons. This interpretation is consistent with other work suggesting that the superfamily domain provides the foundational structural machinery required for radical SAM chemistry while decorations/domain additions to the superfamily and functional domains enable the wide variation in function for which the superfamily is known, for example, (Dowling, Vey, Croft, & Drennan, 2012; Vey & Drennan, 2011).
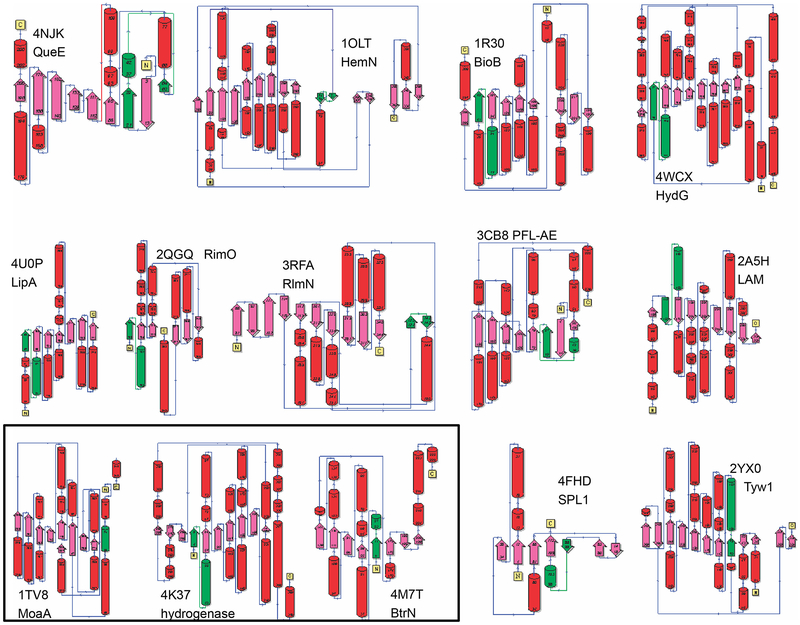
Fig. 6 shows secondary structure diagrams from several subgroups for the core radical SAM superfamily domains. The [Fe4-S4]-AdoMet binding motif is highlighted in each. This view indicates that not only are the overall topologies of these proteins varied, but the position of the motif in these sequences varies as well, illustrating yet another way in which nature has expanded the use of this plug and play motif to support broadly different reactions. Although the topologies shown are associated with different subgroups, due to the still poor structural coverage of the superfamily, we cannot assert that each typifies the majority of the structures of its associated subgroup.
Fig. 6.
Secondary structure topologies of representative radical SAM superfamily domains. Images for several RSS subgroups created using the PDBSum website (de Beer, Berka, Thornton, & Laskowski, 2014). Red: helices, pink: beta strands, green: [Fe4-S4]-AdoMet binding motif. The common abbreviations of the enzyme names and their PDB identifiers are shown on the figure.
For each image shown in Fig. 6, the full names, common names (in parentheses), PDB identifiers, and Level 1 subgroup numbers (in square brackets) are: 7-carboxy-7-deazaguanine synthase (QueE), 4NJK, [1]; oxygen-independent coproporphyrinogen-III oxidase 1 (HemN) 1OLT, [2]; biotin synthase (BioB) 1R30, [6]; [Fe] hydrogenase maturase (HydG) 4WCX, [6]; lipoyl synthase (LipA) 4U0P, [11]; ribosomal protein S12 (aspartate89-C3)-methylthiotransferase (RimO) 2QGQ, [12]; 23S rRNA (adenine2503-C2)-methyltransferase (RlmN), 3RFA, [13]; pyruvate formate-lyase activase (PFL-AE) 3C8F, [15]; L-lysine 2,3-aminomutase (LAM) 2AH5 [16]; cyclic pyranopterin phosphate synthase (MoaA) 1TV8, [17, Level 3]; anaerobic Cys-type sulfatase-maturating enzyme (AnSME) 4K36, [17, Level 3]; 2-deoxy-scyllo-inosamine dehydrogenase (BtrN) 4M7T, [17, Level 2]; spore photoproduct lyase 1 (SPL1) 4FHD, [19]; tRNA 4-demethylwyosine synthase (Tyw1) 2YX0 [20]. These and other data, expanded to include all RSS subgroups of our classification, are provided in Supplemental Table 1.
2.3. A LARGE SCALE VIEW OF RSS SEQUENCE-STRUCTURE-FUNCTION RELATIONSHIPS REVEALS HOW LITTLE WE KNOW
The representative network shown in Fig. 5 provides an estimate of the breadth of experimental coverage for RSS subgroups. Although this visualization may appear to infer that several of these numbered Level 1 subgroups include many characterized functions and structures, this first-pass interpretation could lead to the incorrect inference that a significant proportion of RSS members are knowns. This results because the figure was intended to highlight how broadly and in which subgroups experimentally characterized enzymes sample the RSS sequence space, rather than to convey the proportion of that sequence space that has been experimentally characterized. Table 2 provides a direct count of the number of representative nodes and the total number of sequences in each of the 20 numbered Level 1 subgroups classified in the SFLD. There are 9,137 representative nodes in the 20 numbered subgroups shown in Fig. 5 containing a total of 97,276 sequences. As detailed below, the overwhelming majority of these sequences have not been experimentally characterized in any way. (The number of sequences presented in Table 2 includes 97,276 sequences of the 113,776 sequences in the RSS, as the remainder are sufficiently diverse that they could not be confidently assigned to a named subgroup or even to one of the “uncharacterized” subgroups in the SFLD. None of these remainder sequences have characterized functions.)
Table 2.
Number of representative nodes and sequences in each Level 1 subgroup curated in the SFLD
| Level 1 Subgroup # | Subgroup Name | # Rep Nodes | # Sequences |
|---|---|---|---|
| 1 | 7-carboxy-7-deazaguanine synthase like | 220 | 4,126 |
| 2 | Anaerobic coproporphyrinogen-III oxidase like | 1,098 | 12,939 |
| 3 | Antiviral proteins | 57 | 312 |
| 4 | AviX12-like | 3 | 7 |
| 5 | B12-binding domain containing | 1,615 | 6,858 |
| 6 | BATS domain containing | 242 | 8,239 |
| 7 | DesII-like | 1 | 12 |
| 8 | Elongater protein-like | 267 | 2,626 |
| 9 | F420, menaquinone cofactor biosynthesis | 154 | 3,271 |
| 10 | FeMo cofactor biosynthesis protein | 68 | 813 |
| 11 | Lipoyl synthase like | 113 | 5,416 |
| 12 | Methylthiotransferase | 828 | 14,884 |
| 13 | Methyltransferase (Class A) | 315 | 6,632 |
| 14 | Methyltransferase (Class D) | 64 | 572 |
| 15 | Organic radical-activating enzymes | 587 | 7,847 |
| 16 | PLP-dependent | 162 | 2,679 |
| 17 | SPASM/twitch domain containing | 3,251 | 18,807 |
| 18 | Spectinomycin biosynthesis (SpcY-like) | 5 | 6 |
| 19 | Spore photoproduct lyase like | 14 | 437 |
| 20 | tRNA wybutosine-synthesizing | 73 | 793 |
Another indicator of the proportion of RSS sequences that are unknowns is the large amount of gray space in Fig 5. There are 8,254 small gray representative nodes (out of 10,741 total nodes in the figure) that are entirely made up of unknowns. Although these gray nodes are not included in Table 2, the number of sequences in each of these 22 “uncharacterized” subgroups in the SFLD range from 44 to 2,635.
Especially daunting for achieving a realistic estimate of the functional and structural variation in the RSS, the proportion of characterized structures and molecular functions reported in the primary literature also remains remarkably small. Fewer than 4,000 RSS members (less than 4% of the total number of sequences in the RSS) are described as experimentally characterized in the reviewed section of the UniProt Knowledge Base (UniProt 2017), Swiss -Prot. (Swiss-Prot offers the largest high quality compilation of known protein functions currently available.) Thus, the experimentally determined proportion of RSS enzymes is likely to remain much smaller than that of the unknowns. Likewise, the structural coverage of the RSS is also extraordinarily low, with less than 100 structures representing even fewer unique proteins in the world-wide PDB (Berman, Henrick, & Nakamura, 2003).
Our knowledge of the extent of structural variation in the canonical RSS is also small due to the still poor structural sampling of its sequence space. Our attempts to use modelling of RSS to ascertain independently the range of fold variation that comprise the RSS functional domain failed to identify additional fold types (data not shown). As with other investigations of this superfamily, this analysis was limited by the poor availability of structurally characterized RSS enzymes for use as modelling templates.
2.4. THE ANCIENT LINEAGE OF THE RSS
It has been previously suggested that the RSS is of ancient origin, based in part on its ubiquity across the biosphere (see, for example, (Holliday et al., 2007; Vey & Drennan, 2011)). The global analysis of the RSS reported here reveals the broad extent of RSS members in all three Kingdoms of life, providing additional support for this notion. The SSN shown in Fig. 7 indicates that the vast majority of RSS members are bacterial, while archaeal sequences represent the next largest proportion. Although a small proportion compared to bacteria, archaeal sequences are found globally across the network, likely indicating evolutionary emergence of RSS enzymes and divergence into major subtypes prior to the archaeal-eubacterial split. Few members are found in eukaryotic organisms and these are largely grouped together in a small number of subgroups. Some of these eukaryotic enzymes have been associated with disease-causing mutations in humans, for example, the MoaA (Hanzelmann & Schindelin, 2004) and LipA (Baker et al., 2014) families of the cyclic pyranopterin phosphate Level 3 subgroup of the SPASM/Twitch Level 1 subgroup and the Level 1 lipoyl synthase subgroup, respectively.
Fig. 7.
RSS SSNs mapped with type of life. The same representative network shown in Fig. 5 except that node coloring is by type of life as defined in the SFLD. Representative nodes are colored by the dominant type of life in each. Bacteria: gray, Archaea: red, Invertebrates: yellow, Vertebrates: blue, Plants, green.
As another indication of the ancient origins of the superfamily, many of the reactions RSS enzymes are known to catalyze are fundamental to all types of life. For example, the RSS is involved in the biosynthesis of over half of the known organic cofactors. Table 3 shows some of the Level 1 subgroups and their constituent families involved in the biosynthesis of some of these cofactors, along with the types of life in which these enzymes are found.
Table 3.
RSS enzymes involved in biosynthesis of organic cofactors
| L1 Subgroup Name and (#) | Family | Cofactor | Type of Lifea |
|---|---|---|---|
| anaerobic coproporphyrinogen-III oxidase (2) | heme degradation proteins (HutW/ChuW) | Heme (degradation) | B |
| oxygen-independent coproporphyrinogen-III oxidase 1 (HemN) | Heme (biosynthesis) | BPVI | |
| oxygen-independent coproporphyrinogen-III oxidase 2 (HemZ) | B | ||
| B12-binding domain (5) | anaerobic magnesium-protoporphyrin-IX monomethyl ester cyclase | Bacteriochlorophyll/anaerobicly synthesized chlorophyll | B |
| BATS domain containing (6) | 2-iminoacetate synthase (ThiH) | Thiamine | ABPI |
| [Fe] hydrogenase maturase (HydG) | [Fe] Hydrogenase metallocofactor assembly | BPI | |
| biotin synthase | Biotin | ABPVI | |
| HMD cofactor maturase (HmdB) | 5,10-Methenyltetrahydromethanopterin hydrogenase cofactor | AB | |
| [FeFe] hydrogenase maturase (HydE) | [FeFe] hydrogenase metallocofactor assembly | BPI | |
| F420, menaquinone cofactor biosynthesis (9) | ((2,3,4,5-tetrahydroxypentyl)amino)dihydropyrimidine-2,4-dione synthase (CofH) | 8-hydroxy-5-deazaflavin (Coenzyme F420) | ABP |
| 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase (CofG) | ABPV | ||
| aminofutalosine synthase (mqnE) | Menaquinone via futalosine (two steps along the same pathway) | AB | |
| cyclic dehypoxanthine futalosine synthase () | ABPV | ||
| FeMo cofactor biosynthesis protein (10) | FeMo cofactor biosynthesis protein (nifB) | FeMo cofactor | AB |
| lipoyl synthase (11) | lipoyl synthase | Lipoyl | ABPVI |
| SPASM/twitch domain containing (17) | coenzyme PQQ synthesis protein E (PqqE) | Pyrroloquinoline quinone (PQQ) | B |
| cyclic pyranopterin phosphate synthase (MoaA) | Molybdenum cofactor | ABPVI | |
| alternative heme biosynthesis C (AhbC) | Heme B (two steps along the same pathway) | AB | |
| alternative heme biosynthesis D (AhbD) | |||
| C-terminal tyrosine decarboxylase (MftC) | Mycofactocin peptide maturase | B | |
| heme D1 biosynthesis (NirJ) | Heme d1 | B | |
| tungsten cofactor oxidoreducase radical SAM maturase | Tungsten containing cofactors | AB |
Type of life: B: bacteria, P: plants, V: vertebrates, I: invertebrates, A: archaea
The RSS also contains a large number of families responsible for the modification and repair of DNA and RNA, another fundamental and ancient requirement for life. For example, the methylthiotransferase subgroup 12, MTTases, catalyze a C-H to C-S bond conversion in the methylthiolation of tRNA. Four families can be assigned to this subgroup: MiaB-like, CDK5RAP1, RimO, and MtaB (Anantharaman, Koonin, & Aravind, 2001). MTTases appear to be made up of an N-terminal MTTase domain, a central radical-generating fold and a TRAM domain (an acronym representing two named domains, TRM2 and the MiaB), found at the C-terminal end. The TRAM domain can bind to an RNA substrate and appears to be important for substrate recognition (Lee et al., 2009; UniProt 2017). In addition to the radical-generating [Fe4-S4] cluster domain found in the middle of the protein, the N-terminal MTTase domain contains three cysteines that bind a second [Fe4-S4] cluster, which could be involved in the thiolation reaction. Within the RSS, the TRAM domain is unique to the MTTase family and is not found in any other known superfamily members. This family is found in all types of life, reflecting the critical role of the reaction it catalyzes.
Other families in the RSS that have representation across all domains of life include the viperin family (antiviral proteins subgroup 3), known to be involved in antiviral activity (although the molecular function is unknown), the elongator protein-like family from the subgroup of the same name, thought to catalyze the tRNA wobble uridine modification at C5 of tRNA, and the Class A methyltransferase families of subgroup 13, adenosine C2 methyltransferase (RlmN-like) and adenosine C8 methyltransferase (Cfr-like). These subgroup 13 proteins utilize two SAM molecules, one as a methyl donor and one as the source of the 5’-dA radical.
Together, these observations lend additional support to previous suggestions that RSS ancestors evolved early in the history of life due to their ability to provide a catalytically simple but powerful mechanism for activating atoms that are typically unreactive in biological organisms. This fundamental synthetic power, their activity in the biosynthesis, degradation, modification and repair of molecules essential to life, along with the range and breadth of their coverage in the biosphere confirms on the global scale of the superfamily the notion that the lineage of the RSS is indeed ancient.
2.5. CLASSIFICATION OF THE RSS BASED ON SEQUENCE SIMILARITY DIFFERS FROM ITS CLASSIFICATION BASED ON CHEMISTRY
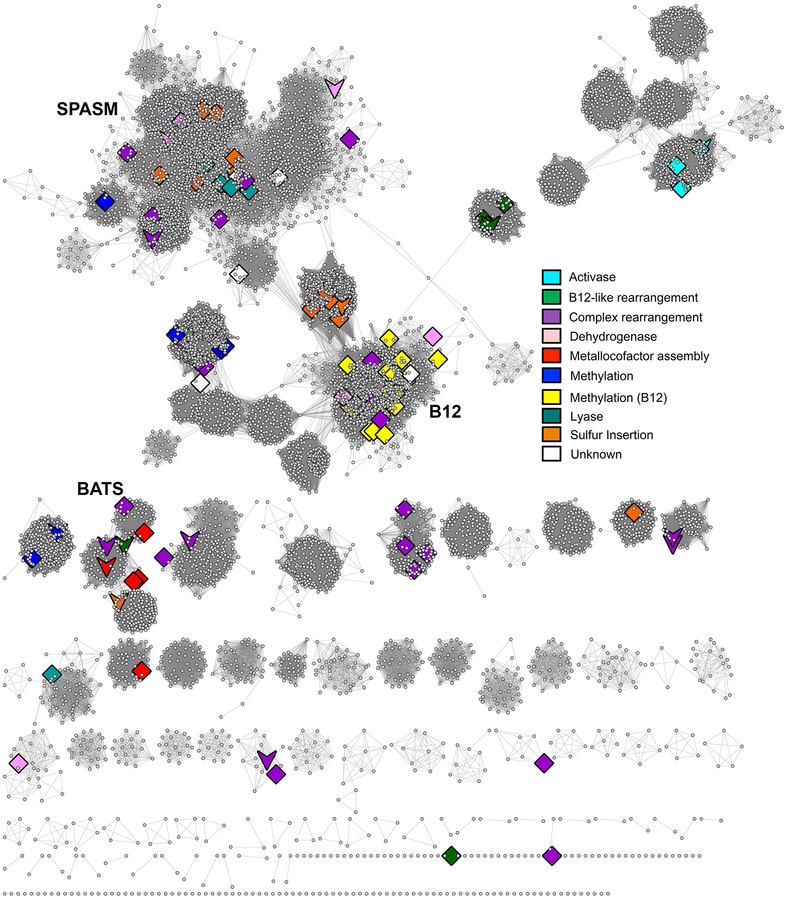
Divergence of ancestral proteins that use the conserved mechanistic machinery of the RSS superfamily domain has led to the wide variety of overall chemical transformations that can be broadly categorized into several general types of chemistry, for example, the insertion of a sulfur atom, complex rearrangements, creation of a glycyl-radical, or methylation (see Supplementary Table S1 for details). Fig. 8 illustrates how some of these major reaction types map to the SSN. As indicated by the figure, these general reaction types do not cluster discretely with specific subgroups. Even for the knowns of the B12-binding methylthiotransferase-like reaction type (yellow diamonds), which appear to map discretely to the main cluster of subgroup 5, several other reaction types map to this subgroup as well. Supplemental Table 1 describes 27 known chemical reactions in this subgroup to which only a total of 1307 sequences have been assigned to a family in the SFLD. Another 5551 sequences in subgroup 5 remain classified only at the subgroup level. These disconnects between sequence-similarity derived clustering and the types of reactions found in those clusters suggest that not even these very general reaction types can be assigned to unknowns based only on sequence similarity (although accurate annotation transfer of reaction type from characterized enzymes to closely related sequences in the same SFLD family can be done with some confidence, especially if key specificity-determining residues are conserved).
Fig. 8.
SSN of RSS mapped with general types of RSS chemistry. The same representative network shown in Fig. 5 except that the highlighted nodes and coloring are by general reaction type as indicated in the key. Diamonds: large representative nodes include at least one functionally characterized member colored by dominant reaction type as shown in the accompanying key. Downward arrows: representative nodes include at least one structurally characterized member.
Other types of chemical classifications fail to track well with our sequence similarity-based classification as well (not shown). For example, the stoichiometry and utilization of SAM differs widely across the RSS. This view identifies three classes that differ with respect to the fate of SAM (Booker, 2009). Class I enzymes utilize SAM catalytically. Class II represents the glycyl radical activating class, i.e. those reactions that abstract a hydrogen atom from a glycine residue in a protein substrate. Class III enzymes, by far the largest set of RSS enzymes currently known, utilize SAM stoichiometrically. However, our results indicate that SAM is utilized both stoichiometrically and catalytically within a subgroup. Even for two enzymes in the Level 2 subgroup of the Level 1 BATS domain-containing subgroup, PylB utilizes SAM as a true cofactor, whereas HydE utilizes it stoichiometrically.
Another way to map between reaction type and similarity groupings is provided by the Gene Ontology (Holliday, Davidson, Akiva, & Babbitt, 2017) which is used in many large resources to describe similarities in enzyme function. Here, enzyme reactions can be defined using the Enzyme Nomenclature Commission classification (Tipton, 1994), which defines enzyme reactions by EC number. Our automated comparison of known overall chemical reactions of the RSS using EC-BLAST (Rahman, Cuesta, Furnham, Holliday, & Thornton, 2014), confirms that similarity in EC classification also fails to track with sequence similarity. Issues with annotating unknowns with the EC number of the most similar characterized enzyme has been raised previously for other enzyme superfamilies (Babbitt, 2003).
The general disconnect between chemical classifications of RSS reactions and the similarity-based classification presented in this work suggests that functional and mechanistic prediction of newly discovered RSS sequences may not be asserted with confidence without biochemical characterization. One way to address this challenge is for experiment and bioinformatics to work together to achieve breakthroughs in resolving these limitations. For example, many recent studies have used multiple types of orthogonal information to obtain functional clues about unknowns (Radivojac et al., 2013). The context provided by SSNs along with information such as genome context, structural models and in silico docking have been especially powerful in suggesting functional properties for many proteins (for example (Calhoun et al., 2018; Hermann et al., 2007; Kalyanaraman et al., 2008; Mashiyama et al., 2014; Zhao et al., 2013)). In addition, the extensive variations in MDAs for the majority of the RSS proteins provides a somewhat unique advantage for functional inference for this and other superfamilies with complex MDAs, such as the haloacid dehalogenase superfamily ((Burroughs, Allen, Dunaway-Mariano, & Aravind, 2006). As fusion proteins can be used to provide key evidence of functional association, (Marcotte et al., 1999), a superfamily-wide strategy focusing on broad-scale mining of the MDA data such as shown in Fig. 4, especially if used conjunction with other orthogonal data (Gerlt, Babbitt, Jacobson, & Almo, 2012), may aid in forming useful functional hypotheses for unknowns that share conserved aspects of their MDAs.
2.6. A LOOK TO THE FUTURE: TARGETING UNKNOWNS FOR EXPERIMENTAL CHARACTERIZATION
As the number of RSS sequences discovered in genome and metagenome projects continues to grow, the proportion of the RSS that can be experimentally characterized will continue to diminish. This scarcity of experimental data has significant consequences for predicting functions of unknowns. Functional annotation in public databases now depends on computational annotation transfer and is still heavily based on the assumption that sequence similarity between unknowns and the most similar protein(s) that have been experimentally characterized provides adequate support for annotation transfer. While this usually works well for closely related sequences, it can also lead to high levels of misannotation, particularly in functionally diverse enzyme superfamilies such as the RSS (Schnoes, Brown, Dodevski, & Babbitt, 2009). Likewise, the even greater scarcity of characterized structures limits our knowledge of unexplored regions of the RSS sequence space (Fig. 5). Our attempts to use modelling of RSS sequences to ascertain the range of fold variants that comprise the superfamily failed, as noted in section 2.3, as the current scarcity of structures and their uneven breadth of coverage available for use as modelling templates diminishes our ability to even ask this question.
Conservation of functionally important residues, including active site features and associated mechanistic knowledge, has long provided important clues about specific functional features of unknowns in many superfamilies and families (for example, (Brown, Gerlt, Seffernick, & Babbitt, 2006; Holliday et al., 2017)). Table 2 and Fig. 5 allow estimation of the current state of experimental coverage for the RSS, showing that some subgroups (such as the B12-binding domain subgroup #5 in Fig. 5) are broadly covered relative to other subgroups. Many other numbered subgroups have far fewer characterized enzymes; the 22 uncharacterized subgroups have none at all. As variations in active site conservation patterns, genome context, structural features or biological phenotypes are available to inform experimental design, it may be possible to expand the coverage of these neglected subgroups. Especially relevant for the RSS, the rich variation among its MDAs could support a global analysis of accessory domains to gain new functional clues about RSS unknowns.
Comparison of conservation patterns have aided in the discrimination of RSS subgroups and families as well. For example, of the more than 5,000 proteins that can be assigned to the BioB-like Level 2 subgroup (of the BATs domain subgroup 6), only about 3,200 can be annotated with that reaction based on the presence of functionally important active site residues. The remainder are missing one or more of the residues considered critical for performing the canonical biotin synthase reaction (Betz et al., 2015; Holliday, Davidson, Akiva, & Babbitt, 2017), leading to the assertion that a large proportion of enzymes annotated with the BioB function in the Gene Ontology database (Ashburner et al., 2000) have been found to be misannotated (Holliday, Davidson, Akiva, & Babbitt, 2017).
Broadening functional knowledge across the RSS requires as a necessary first step identifying unknowns that are likely to represent fruitful targets. While superfamily members associated with important biological phenotypes or that are of particular interest for other reasons will remain key motivations for characterization, additional strategies to investigate unknowns throughout the broader sequence space are needed. The subgroup classification presented here identifies the least characterized subgroups of the RSS; examination of differences between their conservation patterns and those of better characterized subgroups will be useful in choosing targets likely to represent new reactions.
Many new strategies are now available for deeper evaluation of unknowns. As noted earlier, large-scale in silico modelling can lead to functional hypotheses that can be validated and studied in detail using targeted biochemical and structural characterization, while in silico docking of metabolites and genome context predictions can be evaluated using many types of experimental approaches. Our subgroup classification, along with the interactive versions of subgroup and family SSNs available for download from the SFLD, were created in part to provide a global context for target selection in the RSS and to facilitate application of the varied strategies now available to assign their functions.
2.7. METHODS
2.7.1. Collection of RSS sequences
To initiate populating the RSS for the SFLD, we collected the full-length sequences in September 2012 associated with Pfam model PF04055 and InterPro family IPR007197, removed duplicate sequences and resolved other differences. This set was last updated using the SFLD automated update protocol on 7/9/14. Functional domains of this representative sequence set superfamily were last updated on 11/22/17.
2.7.2. Representative networks
The full set of 113,776, sequences was clustered using CD-HIT (Li & Godzik, 2006) at 50% pairwise identity, resulting in 10,741 representative nodes. Edges were drawn between representative nodes if the BLAST E-value (used as a score) was ≤ 1 × 10−20. The representative network shown in Figs 5, 7 and 8 was calculated following the Pythoscape infrastructure (Barber & Babbitt, 2012) where similarity between each pair of representative nodes was calculated as the mean of the all-by-all comparison scores between the sequences in each representative node pair. The networks were visualized using the Prefuse force directed layout (mean edge E-value) in Cytoscape (Smoot, Ono, Ruscheinski, Wang, & Ideker, 2011). As the calculated network represents a multidimensional set of n-1 pairwise comparisons where n is the number of sequences, these data must be compressed for visualization in two dimensions. Other work has indicated that two-dimensional visualization provides an acceptable estimate of the similarity relationships in thresholded SSNs (see (Atkinson, Morris, Ferrin, & Babbitt, 2009), Supplementary Figure 1 statistics).
2.7.3. Determining subgroups and families
RSS subgroups were defined semi-automatically using as a guide representative SSNs annotated with a list of RSS reactions currently known in the primary literature. These initial subgroup boundaries were refined based on manual inspection of the network across a range of E-value thresholds and further informed from MDA data and available structural information. The aim was to choose an E-value score threshold for drawing edges that resulted in sequence clusters that minimally split nodes with similar reactions while maximally splitting those of different reactions. The final mean E-value score shown in Fig. 5 and used to visualize the named subgroups curated in the SFLD is 1 × 10−20. Nodes of the same color that ended up in different clusters in Fig. 5 reflect the extent to which this could not be achieved and likely resulted from several issues that include uneven rates of evolution of subgroups. For example, SFLD curation of the highly divergent Level 1 subgroup 17, which represents proteins with a SPASM/Twitch domain, required the creation of Level 2 and Level 3 subgroupings as the entire Level 1 subgroup could not be united at a statistically significant score using a single hidden Markov Model (HMM) (Eddy, 2011). This diversity is also reflected in Fig. 5: the E-value chosen to distinguish the majority of RSS subgroups was unable to cluster all of the SPASM/Twitch domain subgroup members together. In contrast, at this E-value threshold, all of the representative nodes of subgroup 1 cluster together due to the greater similarity among members of this subgroup relative to those of subgroup 17.
In theory, the assignment of families in the SFLD is relatively simple, requiring at minimum, at least one member that has been biochemically characterized, with at least some residues and features identified that are known to be functionally important in that reaction. However, transfer of function is problematic for representative nodes, each of which may contain substantially divergent sequences that may catalyze reactions that are different from that catalyzed by the characterized “founder” enzyme. To address this issue, we computed and examined for each subgroup (at the most detailed subgroup level available) SSNs in which each node represents a single sequence (data not shown). This allowed a detailed comparison of all sequences in a subgroup for conservation as well as differences among key residues known to be functionally important in the characterized member(s). Family assignments were then made guided by these data.
2.7.4. Annotation of the RSS in the sfld
Annotation of subgroups and families is supported by several types of information, including multiple sequence alignments (MSAs) created using Clustal Omega (Sievers et al., 2011) as the sizes of their sequence sets allowed. Hidden Markov models (HMMs) used in curation of subgroups and families were created using HMMER 3. Each family is annotated with at least one reference from the primary literature and its overall chemical transformation (where known). Where possible, the conserved functional residues are reported (in the majority of cases, these are associated with the [Fe4S4] binding motif).
Detailed annotation of the RSS is provided in Supplemental Table 1 of this work. Key data and information is also provided for download from the SFLD (see http://sfld.rbvi.ucsf.edu). These include MSAs and interactive SSNs at the subgroup and family levels that can be visualized and manipulated using Cytoscape. Sequence sets, chemical reactions, and other data are also available.
Supplementary Material
ACKNOWLEDGEMENTS
Support for this work acknowledges NIH R01 GM-60595 (P. Babbitt), NIH R01 GM-122595 (S. Booker), NSF DBI-1356193 (P. Babbitt and G. Holliday), and NIH U54-GM093342 (P Babbitt). Some of the results described in this paper were initially developed as part of a workshop on the RSS sponsored by the Enzyme Function Initiative. The authors thank David Mischel and Benjamin Polacco for technical support of the SFLD and Kathy Clement for help with generating graphics images of SSNs.
ABBREVIATIONS
- SAM
S-Adenosylmethionine
- RSS
Radical SAM Superfamily
- LAM
lysine 2,3-aminomutase
- BioB
Biotin Synthase
- LipA
Lipoyl Synthase
- PflA
Pyruvate formate-lyase activase
- NrdG
Anaerobic ribonucleotide reductase activase
- 5’-dA
5’-deoxyadenosyl
- Met
Methionine
- PQQ
Pyrroloquinonoline quinone
- SPL
Spore product lyase
- MDA
Multi-domain architecture
- PhnJ
Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase
- SFLD
Structure-Function Linkage Database
- SSN
sequence similarity network
- Knowns
Sequences of known function
- Unknowns
Sequences of unknown function
- TIM
Triose phosphate isomerase
- HMM
hidden Markov Model
- MSA
multiple sequence alignment
Footnotes
Although it has long been assumed that the only chemical feature that all RSS members have in common is the formation of the 5’-dA radical from SAM by the structurally conserved iron-sulfur cluster-binding motif, even this most fundamental and “obligate” descriptor of the canonical RSS no longer holds. At least one exception to this rule has now been identified. Although it is clearly homologous to other canonical RSS sequences, a tryptophan methyltransferase TsrM involved in the biosynthesis of the thiopeptide antibiotic thiostrepton, does not catalyze formation of a 5’-dA radical (Blaszczyk et al., 2016; Pierre et al., 2012). A few other similar as yet uncharacterized cobalamin-dependent methylases that methylate sp2-hybridized carbon centers may represent additional exceptions.
COMPETING FINANCIAL INTERESTS STATEMENT
None
REFERENCES
- Akiva E, Brown S, Almonacid DE, Barber AE 2nd, Custer AF, Hicks MA, … Babbitt PC (2014). The Structure-Function Linkage Database. Nucleic Acids Res, 42(Database issue), D521–530. doi: 10.1093/nar/gkt1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, & Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res, 25(17), 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, & Aravind L (2001). TRAM, a predicted RNA-binding domain, common to tRNA uracil methylation and adenine thiolation enzymes. FEMS Microbiol Lett, 197(2), 215–221. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, … Sherlock G (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet, 25(1), 25–29. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson HJ, Morris JH, Ferrin TE, & Babbitt PC (2009). Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One, 4(2), e4345. doi: 10.1371/journal.pone.0004345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbitt PC (2003). Definitions of enzyme function for the structural genomics era. Curr Opin Chem Biol, 7(2), 230–237. [DOI] [PubMed] [Google Scholar]
- Baker PR 2nd, Friederich MW, Swanson MA, Shaikh T, Bhattacharya K, Scharer GH, … Van Hove JL (2014). Variant non ketotic hyperglycinemia is caused by mutations in LIAS, BOLA3 and the novel gene GLRX5. Brain, 137(Pt 2), 366–379. doi: 10.1093/brain/awt328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber AE 2nd, & Babbitt PC (2012). Pythoscape: A framework for generation of large protein similarity networks. Bioinformatics. doi: 10.1093/bioinformatics/bts532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr I, Latham JA, Iavarone AT, Chantarojsiri T, Hwang JD, & Klinman JP (2016). Demonstration That the Radical S-Adenosylmethionine (SAM) Enzyme PqqE Catalyzes de Novo Carbon-Carbon Cross-linking within a Peptide Substrate PqqA in the Presence of the Peptide Chaperone PqqD. J Biol Chem, 291(17), 8877–8884. doi: 10.1074/jbc.C115.699918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjdia A, Heil K, Barends TR, Carell T, & Schlichting I (2012). Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme. Nucleic Acids Res, 40(18), 9308–9318. doi: 10.1093/nar/gks603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H, Henrick K, & Nakamura H (2003). Announcing the worldwide Protein Data Bank. Nat Struct Biol, 10(12), 980. doi: 10.1038/nsb1203-980 [DOI] [PubMed] [Google Scholar]
- Betz JN, Boswell NW, Fugate CJ, Holliday GL, Akiva E, Scott AG, … Broderick JB (2015). [FeFe]-hydrogenase maturation: insights into the role HydE plays in dithiomethylamine biosynthesis. Biochemistry, 54(9), 1807–1818. doi: 10.1021/bi501205e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszczyk AJ, Silakov A, Zhang B, Maiocco SJ, Lanz ND, Kelly WL, … Booker SJ (2016). Spectroscopic and Electrochemical Characterization of the Iron-Sulfur and Cobalamin Cofactors of TsrM, an Unusual Radical S-Adenosylmethionine Methylase. J Am Chem Soc, 138(10), 3416–3426. doi: 10.1021/jacs.5b12592 [DOI] [PubMed] [Google Scholar]
- Booker SJ (2009). Anaerobic functionalization of unactivated C-H bonds. Curr Opin Chem Biol, 13(1), 58–73. doi: 10.1016/j.cbpa.2009.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick JB, Duffus BR, Duschene KS, & Shepard EM (2014). Radical S-adenosylmethionine enzymes. Chem Rev, 114(8), 4229–4317. doi: 10.1021/cr4004709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, & Babbitt PC (2014). New insights about enzyme evolution from large scale studies of sequence and structure relationships. J Biol Chem, 289(44), 30221–30228. doi: 10.1074/jbc.R114.569350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Gerlt JA, Seffernick JL, & Babbitt PC (2006). A gold standard set of mechanistically diverse enzyme superfamilies. Genome Biol, 7(1), R8. doi: 10.1186/gb-2006-7-1-r8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Allen KN, Dunaway-Mariano D, & Aravind L (2006). Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol, 361(5), 1003–1034. doi: S0022–2836(06)00777–7 [pii] 10.1016/j.jmb.2006.06.049 [DOI] [PubMed] [Google Scholar]
- Calhoun S, Korczynska M, Wichelecki DJ, San Francisco B, Zhao S, Rodionov DA, … Sali A (2018). Prediction of enzymatic pathways by integrative pathway mapping. Elife, 7. doi: 10.7554/eLife.31097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchillo RM, Iwig DF, Jones AD, Nesbitt NM, Baleanu-Gogonea C, Souder MG, … Booker SJ (2004). Lipoyl synthase requires two equivalents of S-adenosyl-L-methionine to synthesize one equivalent of lipoic acid. Biochemistry, 43(21), 6378–6386. doi: 10.1021/bi049528x [DOI] [PubMed] [Google Scholar]
- Cicchillo RM, Lee KH, Baleanu-Gogonea C, Nesbitt NM, Krebs C, & Booker SJ (2004). Escherichia coli lipoyl synthase binds two distinct [4Fe-4S] clusters per polypeptide. Biochemistry, 43(37), 11770–11781. doi: 10.1021/bi0488505 [DOI] [PubMed] [Google Scholar]
- Coquille S, Roux C, Mehta A, Begley TP, Fitzpatrick TB, & Thore S (2013). High-resolution crystal structure of the eukaryotic HMP-P synthase (THIC) from Arabidopsis thaliana. J Struct Biol, 184(3), 438–444. doi: 10.1016/j.jsb.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson NL, Sillitoe I, Lees JG, Lam SD, & Orengo CA (2017). CATH-Gene3D: Generation of the Resource and Its Use in Obtaining Structural and Functional Annotations for Protein Sequences. Methods Mol Biol, 1558, 79–110. doi: 10.1007/978-1-4939-6783-4_4 [DOI] [PubMed] [Google Scholar]
- de Beer TA, Berka K, Thornton JM, & Laskowski RA (2014). PDBsum additions. Nucleic Acids Res, 42(Database issue), D292–296. doi: 10.1093/nar/gkt940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinis P, Wieckowski BM, & Roach PL (2016). Metallocofactor assembly for [FeFe]-hydrogenases. Curr Opin Struct Biol, 41, 90–97. doi: 10.1016/j.sbi.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Dowling DP, Vey JL, Croft AK, & Drennan CL (2012). Structural diversity in the AdoMet radical enzyme superfamily. Biochim Biophys Acta, 1824(11), 1178–1195. doi: 10.1016/j.bbapap.2012.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR (2011). Accelerated Profile HMM Searches. PLoS Comput Biol, 7(10), e1002195. doi: 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, … Mitchell AL (2017). InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res, 45(D1), D190–D199. doi: 10.1093/nar/gkw1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, … Bateman A (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res, 44(D1), D279–285. doi: 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnham N, Sillitoe I, Holliday GL, Cuff AL, Laskowski RA, Orengo CA, & Thornton JM (2012). Exploring the evolution of novel enzyme functions within structurally defined protein superfamilies. PLoS Comput Biol, 8(3), e1002403. doi: 10.1371/journal.pcbi.1002403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC, Jacobson MP, & Almo SC (2012). Divergent evolution in enolase superfamily: strategies for assigning functions. J Biol Chem, 287(1), 29–34. doi: 10.1074/jbc.R111.240945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell TA, Goldman PJ, & Drennan CL (2015). SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes. J Biol Chem, 290(7), 3964–3971. doi: 10.1074/jbc.R114.581249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann P, & Schindelin H (2004). Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc Natl Acad Sci U S A, 101(35), 12870–12875. doi: 10.1073/pnas.0404624101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, Shoichet BK, & Raushel FM (2007). Structure-based activity prediction for an enzyme of unknown function. Nature, 448(7155), 775–779. doi: 10.1038/nature05981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka T, Furihata K, Ishikawa J, Yamashita H, Itoh N, Seto H, & Dairi T (2008). An alternative menaquinone biosynthetic pathway operating in microorganisms. Science, 321(5896), 1670–1673. doi: 10.1126/science.1160446 [DOI] [PubMed] [Google Scholar]
- Holliday GL, Brown SD, Akiva E, Mischel D, Hicks MA, Morris JH, … Babbitt PC (2017). Biocuration in the structure-function linkage database: the anatomy of a superfamily. Database (Oxford), 2017. doi: 10.1093/database/bax045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday GL, Davidson R, Akiva E, & Babbitt PC (2017). Evaluating Functional Annotations of Enzymes Using the Gene Ontology. Methods Mol Biol, 1446, 111–132. doi: 10.1007/978-1-4939-3743-1_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday GL, Thornton JM, Marquet A, Smith AG, Rebeille F, Mendel R, … Warren MJ (2007). Evolution of enzymes and pathways for the biosynthesis of cofactors. Nat Prod Rep, 24(5), 972–987. doi: 10.1039/b703107f [DOI] [PubMed] [Google Scholar]
- Kalyanaraman C, Imker HJ, Fedorov AA, Fedorov EV, Glasner ME, Babbitt PC, … Jacobson MP (2008). Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening. Structure, 16(11), 1668–1677. doi: 10.1016/j.str.2008.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat SS, Williams HJ, & Raushel FM (2011). Intermediates in the transformation of phosphonates to phosphate by bacteria. Nature, 480(7378), 570–573. doi: 10.1038/nature10622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe J, & Sawers G (1990). A radical-chemical route to acetyl-CoA: the anaerobically induced pyruvate formate-lyase system of Escherichia coli. FEMS Microbiol Rev, 6(4), 383–398. [DOI] [PubMed] [Google Scholar]
- LaMattina JW, Wang B, Badding ED, Gadsby LK, Grove TL, & Booker SJ (2017). NosN, a Radical S-Adenosylmethionine Methylase, Catalyzes Both C1 Transfer and Formation of the Ester Linkage of the Side-Ring System during the Biosynthesis of Nosiheptide. J Am Chem Soc, 139(48), 17438–17445. doi: 10.1021/jacs.7b08492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz ND, & Booker SJ (2012). Identification and function of auxiliary iron-sulfur clusters in radical SAM enzymes. Biochim Biophys Acta, 1824(11), 1196–1212. doi: 10.1016/j.bbapap.2012.07.009 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, & Swindells MB (2011). LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model, 51(10), 2778–2786. doi: 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- Lee KH, Saleh L, Anton BP, Madinger CL, Benner JS, Iwig DF, … Booker SJ (2009). Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily. Biochemistry, 48(42), 10162–10174. doi: 10.1021/bi900939w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, & Godzik A (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics, 22(13), 1658–1659. doi: 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- Lotierzo M, Tse Sum Bui B, Florentin D, Escalettes F, & Marquet A (2005). Biotin synthase mechanism: an overview. Biochem Soc Trans, 33(Pt 4), 820–823. doi: 10.1042/BST0330820 [DOI] [PubMed] [Google Scholar]
- Mahanta N, Hudson GA, & Mitchell DA (2017a). Correction to Radical S-Adenosylmethionine Enzymes Involved in RiPP Biosynthesis. Biochemistry, 56(45), 6072. doi: 10.1021/acs.biochem.7b01056 [DOI] [PubMed] [Google Scholar]
- Mahanta N, Hudson GA, & Mitchell DA (2017b). Radical S-Adenosylmethionine Enzymes Involved in RiPP Biosynthesis. Biochemistry, 56(40), 5229–5244. doi: 10.1021/acs.biochem.7b00771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancia F, Keep NH, Nakagawa A, Leadlay PF, McSweeney S, Rasmussen B, … Evans PR (1996). How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 A resolution. Structure, 4(3), 339–350. [DOI] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, & Eisenberg D (1999). Detecting protein function and protein-protein interactions from genome sequences. Science, 285(5428), 751–753. [DOI] [PubMed] [Google Scholar]
- Mashiyama ST, Malabanan MM, Akiva E, Bhosle R, Branch MC, Hillerich B, … Babbitt PC (2014). Large-scale determination of sequence, structure, and function relationships in cytosolic glutathione transferases across the biosphere. PLoS Biol, 12(4), e1001843. doi: 10.1371/journal.pbio.1001843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Busby RW, Jordan SW, Cheek J, Henshaw TF, Ashley GW, … Marletta MA (2000). Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry, 39(49), 15166–15178. [DOI] [PubMed] [Google Scholar]
- Moss M, & Frey PA (1987). The role of S-adenosylmethionine in the lysine 2,3-aminomutase reaction. J Biol Chem, 262(31), 14859–14862. [PubMed] [Google Scholar]
- Nicolet Y, Rubach JK, Posewitz MC, Amara P, Mathevon C, Atta M, … Fontecilla-Camps JC (2008). X-ray structure of the [FeFe]-hydrogenase maturase HydE from Thermotoga maritima. J Biol Chem, 283(27), 18861–18872. doi: 10.1074/jbc.M801161200 [DOI] [PubMed] [Google Scholar]
- Padovani D, Thomas F, Trautwein AX, Mulliez E, & Fontecave M (2001). Activation of class III ribonucleotide reductase from E. coli. The electron transfer from the iron-sulfur center to S-adenosylmethionine. Biochemistry, 40(23), 6713–6719. [DOI] [PubMed] [Google Scholar]
- Pierre S, Guillot A, Benjdia A, Sandstrom C, Langella P, & Berteau O (2012). Thiostrepton tryptophan methyltransferase expands the chemistry of radical SAM enzymes. Nat Chem Biol, 8(12), 957–959. doi: 10.1038/nchembio.1091 [DOI] [PubMed] [Google Scholar]
- Pilet E, Nicolet Y, Mathevon C, Douki T, Fontecilla-Camps JC, & Fontecave M (2009). The role of the maturase HydG in [FeFe]-hydrogenase active site synthesis and assembly. FEBS Lett, 583(3), 506–511. doi: 10.1016/j.febslet.2009.01.004 [DOI] [PubMed] [Google Scholar]
- Puehringer S, Metlitzky M, & Schwarzenbacher R (2008). The pyrroloquinoline quinone biosynthesis pathway revisited: a structural approach. BMC Biochem, 9, 8. doi: 10.1186/1471-2091-9-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Clark WT, Oron TR, Schnoes AM, Wittkop T, Sokolov A, … Friedberg I (2013). A large-scale evaluation of computational protein function prediction. Nat Methods, 10(3), 221–227. doi: 10.1038/nmeth.2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman SA, Cuesta SM, Furnham N, Holliday GL, & Thornton JM (2014). EC-BLAST: a tool to automatically search and compare enzyme reactions. Nat Methods, 11(2), 171–174. doi: 10.1038/nmeth.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyda MR, Dippold R, Dotson ME, & Jarrett JT (2008). Loss of iron-sulfur clusters from biotin synthase as a result of catalysis promotes unfolding and degradation. Arch Biochem Biophys, 471(1), 32–41. doi: 10.1016/j.abb.2007.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoes AM, Brown SD, Dodevski I, & Babbitt PC (2009). Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol, 5(12), e1000605. doi: 10.1371/journal.pcbi.1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, … Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 13(11), 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, … Higgins DG (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol, 7, 539. doi: 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, & Ideker T (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics, 27(3), 431–432. doi: 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, & Miller NE (2001). Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res, 29(5), 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamuri AU, & Laskowski RA (2010). ArchSchema: a tool for interactive graphing of related Pfam domain architectures. Bioinformatics, 26(9), 1260–1261. doi: 10.1093/bioinformatics/btq119 [DOI] [PubMed] [Google Scholar]
- Tao M, Wang L, Wendt-Pienkowski E, George NP, Galm U, Zhang G, … Shen B (2007). The tallysomycin biosynthetic gene cluster from Streptoalloteichus hindustanus E465–94 ATCC 31158 unveiling new insights into the biosynthesis of the bleomycin family of antitumor antibiotics. Mol Biosyst, 3(1), 60–74. doi: 10.1039/b615284h [DOI] [PubMed] [Google Scholar]
- Tipton KF (1994). Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB). Enzyme nomenclature. Recommendations 1992. Supplement: corrections and additions. Eur J Biochem, 223(1), 1–5. [DOI] [PubMed] [Google Scholar]
- Consortium UniProt. (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res, 45(D1), D158–D169. doi: 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vey JL, & Drennan CL (2011). Structural insights into radical generation by the radical SAM superfamily. Chem Rev, 111(4), 2487–2506. doi: 10.1021/cr9002616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vey JL, Yang J, Li M, Broderick WE, Broderick JB, & Drennan CL (2008). Structural basis for glycyl radical formation by pyruvate formate-lyase activating enzyme. Proc Natl Acad Sci U S A, 105(42), 16137–16141. doi: 10.1073/pnas.0806640105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Woldring RP, Roman-Melendez GD, McClain AM, Alzua BR, & Marsh EN (2014). Recent Advances in Radical SAM Enzymology: New Structures and Mechanisms. ACS Chem Biol. doi: 10.1021/cb5004674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, & Li L (2015). Spore photoproduct lyase: the known, the controversial, and the unknown. J Biol Chem, 290(7), 4003–4009. doi: 10.1074/jbc.R114.573675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AP, & Bandarian V (2011). Pyruvate is the source of the two carbons that are required for formation of the imidazoline ring of 4-demethylwyosine. Biochemistry, 50(49), 10573–10575. doi: 10.1021/bi2015053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Duan L, Zhang Q, Liao R, Ding Y, Pan H, … Liu W (2009). Nosiheptide biosynthesis featuring a unique indole side ring formation on the characteristic thiopeptide framework. ACS Chem Biol, 4(10), 855–864. doi: 10.1021/cb900133x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, … Lin H (2010). Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature, 465(7300), 891–896. doi: 10.1038/nature09138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Kumar R, Sakai A, Vetting MW, Wood BM, Brown S, … Jacobson MP (2013). Discovery of new enzymes and metabolic pathways by using structure and genome context. Nature, 502(7473), 698–702. doi: 10.1038/nature12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.