Abstract
Klebsiella pneumoniae (KP) remains the most prevalent nosocomial pathogen and carries the carbapenemase (KPC) gene which confers resistance towards carbapenem. Thus, it is necessary to discover novel antimicrobials to address the issue of antimicrobial resistance in such pathogens. Natural products such as essential oils are a promising source due to their complex composition. Essential oils have been shown to be effective against pathogens, but the overall mechanisms have yet to be fully explained. Understanding the molecular mechanisms of essential oil towards KPC-KP cells would provide a deeper understanding of their potential use in clinical settings. Therefore, we aimed to investigate the mode of action of essential oil against KPC-KP cells from a proteomic perspective by comparing the overall proteome profile of KPC-KP cells treated with cinnamon bark (Cinnamomum verum J. Presl) essential oil (CBO) at their sub-inhibitory concentration of 0.08% (v/v). A total of 384 proteins were successfully identified from the non-treated cells, whereas only 242 proteins were identified from the CBO-treated cells. Proteins were then categorized based on their biological processes, cellular components and molecular function prior to pathway analysis. Pathway analysis showed that CBO induced oxidative stress in the KPC-KP cells as indicated by the abundance of oxidative stress regulator proteins such as glycyl radical cofactor, catalase peroxidase and DNA mismatch repair protein. Oxidative stress is likely to oxidize and disrupt the bacterial membrane as shown by the loss of major membrane proteins. Several genes selected for qRT-PCR analysis validated the proteomic profile and were congruent with the proteomic abundance profiles. In conclusion, KPC-KP cells exposed to CBO undergo oxidative stress that eventually disrupts the bacterial membrane possibly via interaction with the phospholipid bilayer. Interestingly, several pathways involved in the bacterial membrane repair system were also affected by oxidative stress, contributing to the loss of cells viability.
Introduction
Klebsiella spp. are Gram-negative rod shaped bacteria that cause bacterial pneumonia with a high fatality rate if infection remains untreated in the clinical setting [1]. Globally, the vast majority of Klebsiella infections are hospital-acquired. Nosocomial Klebsiella infections are mainly caused by Klebsiella pneumoniae, the medically most important species of the genus which primarily attacks immune-compromised individuals who are hospitalized and suffer from severe underlying diseases such as diabetes mellitus, chronic pulmonary obstruction or even cancer. It is estimated that Klebsiella spp. cause 8% of all nosocomial bacterial infections in the United States and Europe, with 50.1% of these cases being caused by Klebsiella pneumoniae placing Klebsiella spp. among the eight most important infectious pathogens in hospitals [1]. In 1983, the first report of a plasmid-mediated extended spectrum beta-lactamases (ESBLs) capable of hydrolyzing extended-spectrum cephalosporins was discovered [2,3]. Carbapenems are one of the last lines of antibiotic treatment for severe drug-resistant bacterial infections, and are now the treatment of choice for serious infections caused by pathogens carrying the ESBL gene. This has led to an increased reliance on carbapenems in clinical practice [4]. In tandem with this, the first carbapenemase producing K. pneumoniae isolate was reported in North Carolina in 2001. This enzyme was termed K. pneumoniae carbapenemase (KPC) and conferred resistance to carbapenem antibiotics [5]. KPCs are encoded by the gene blaKPC, whose potential for inter-species and geographic dissemination is largely explained by its location within a Tn3-type transposon, Tn4401 that is capable of inserting itself into diverse plasmids of Gram-negative bacteria. Although K. pneumoniae remains the most prevalent bacterial species carrying KPCs, the enzyme has been identified in several other Gram-negative bacilli such as Escherichia coli, Pseudomonas aeruginosa and Salmonella enterica due to horizontal gene transfer [6]. To worsen this issue, KPC-producing K. pneumoniae (KPC-KP) possesses innate antibiotic resistance in the form of an efflux pump, which generally removes the antibiotics that have penetrated the bacterial membrane, from the cytoplasm into the extracellular environment. Membrane permeability can also be altered in the presence of antibiotics; preventing the access of antibiotics into the cells, which when coupled to the other mechanisms, enables resistance against higher concentrations of antibiotics [7].
In order to address to this particular issue, there had been constant efforts to discover novel antimicrobials for clinical use. Natural products such as essential oil consisting a plethora of chemical compounds, are becoming a popular mainstream platform for researchers in drug discovery [8]. Numerous studies have also demonstrated the efficacy of essential oils from curry plant (Helichrysum italicum (Roth) G. Don fil.), peppermint (Mentha x piperita L.), tea tree (Melaleuca alternifolia (Maiden & Betche) Cheel.) and marjoram (Origanum majorana L.) as promising antimicrobials. Multiple studies have shown the synergistic effects between various essential oils and antibiotics, potentially solving the antibiotic resistance issue in the clinical setting [9–14]. Despite this, only a few studies have been carried out to elucidate the mode of action of several essential oils on different bacteria; most of these studies have postulated that essential oils exert their antimicrobial activities by disrupting bacterial cell membrane and/or their efflux systems through various assays [15–17]. For example, de Souza et al. (2009) postulated that Origanum vulgare L. essential oil affects the membrane permeability of Staphylococcus aureus by studying potassium ion efflux and scanning electron microscopy [15]. Similarly, Silva et al. (2011) hypothesized that coriander essential oil exerts its bactericidal activity towards both Gram-positive and Gram–negative bacteria via membrane damage by measuring their efflux activity, respiratory activity and membrane potential [16]. To further support and understand the antimicrobial activity of essential oils, mass spectrometry-based proteomics analysis has become the tool of choice offering the identification and quantification of the proteome of an organism. There has been a tremendous improvement in instrument performance and the computational tools used in proteomic studies in recent years, which facilitates the understanding of the mechanisms of action of potential antimicrobial agents in the clinical setting. In the most widely used “bottom-up” approach to proteomics, liquid chromatography coupled with mass spectrometry (LC-MS/MS), enables a complex mixture of proteins to be first subjected to enzymatic cleavage; the resulting peptide products are separated based on chemical or physical properties and analyzed using a mass spectrometer. The proteome can then be analyzed, quantified and compared by using third party analytical software such as Progenesis QI (Progenesis Group Sdn. Bhd.) or Perseus (Max Planck Institute of Biochemistry). For instance, Xu et al. (2015) identified the mode of action of paclitaxel as chemotherapeutic drugs in HeLa cells by tampering with the abundance of tumor suppressor PDCD4 via LC-MS/MS proteomic profiling [18]. Similarly, Kawatani et al. (2016) also revealed the role of collismycin A as an iron chelator antagonizing cancer cells, using a proteomic approach [19].
In our previous study, we have successfully shown that cinnamon bark (Cinnamomon verum J. Presl) essential oil (CBO) is, in fact, an effective antimicrobial when used against KPC-KP with an extremely low minimum inhibitory concentration (MIC) of 0.16% (v/v) [11]. Furthermore, combinations of CBO and antibiotic meropenem further reduced the MIC of CBO to 0.08% (v/v) which makes it a putative candidate to be used as an antimicrobial in the clinical setting [11]. Other studies showed that CBO with a low MIC value is effective against a variety of Gram-positive and Gram–negative pathogens. Zamirah and colleagues (2013) demonstrated that CBO is effective against oral pathogens such as Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, Porphyromonas gingivalis, Streptococcus salivarius, S. mitis and S. mutans with a low MIC ranging from 0.02 to 0.06% (v/v) [20]. Kaskatepe et al (2016) also showed that CBO is highly effective against other opportunistic pathogens, including Escherichia coli, Pseudomonas aeruginosa and multidrug-resistant Staphylococcus aureus (MRSA), with a MIC ranging from 0.009% to 0.078% (v/v) [21]. As already mentioned above, preliminary studies had postulated that essential oils affect bacterial membrane and/or their efflux system via numerous assays. Nevertheless, none had explained or gave an overview on the overall changes that the bacterial cells undergo when exposed to essential oils, especially in the perspective of proteomics. Thus, this study was carried out to understand and possibly bridge the missing links among previous studies regarding the mode of action of essential oils as an antimicrobial agent from the proteomic perspective, using CBO and KPC-KP as a model of study.
Materials and methods
CBO and KPC-producing K. pneumoniae
CBO (Cinnamon bark Sri Lanka, lot number: 6488) used throughout the study was purchased from Aroma Trading Ltd. (Milton Keynes, UK). The composition of CBO had been determined in our previous study via GC-MS [10]. The CBO was filter-sterilized using a 0.22 μm PES syringe membrane filter (Bioflow, Malaysia). KPC-producing K. pneumoniae, K. pneumoniae BAA-1705 (KPC-KP) was purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and was cultured on Mueller-Hinton broth (MHB) and agar (MHA) both from Sigma Aldrich, USA.
CBO treatment and protein extraction
KPC-KP cell cultures were divided into two treatment groups, namely control (no treatment) and CBO treated prior to protein extraction. The CBO final concentration used was 0.08% (v/v), as determined by Yang et al. (2017) [11]. Both treatment groups had a final volume of 50 mL, supplemented with Tween 80 at final concentration of 10% to enhance the solubility of CBO and a standard inoculum of 1 × 105 cfu/mL KPC-KP cells. Samples were incubated at 37°C with shaking at 200 rpm for 16 h to obtain sufficient cells for protein extraction. The cells from both treatment groups were pelleted by centrifugation at 9000 rpm for 10 min, washed for at least three times and resuspended in 500 μL cold protein extraction buffer (50 mM ammonium bicarbonate, 10 mM phenylmethylsulfonyl fluoride). Samples were then sonicated on ice at 20 amplitude for 10 cycles to lyse the cells; each cycle consisted of 10 seconds of sonication followed by 20 s of cooling, with a Qsonica Sonicator Q55 (Fischer Scientific, USA). Sonicated samples were then pelleted at 4°C and 10000 rpm for 1 h; supernatants were then collected and quantified via Bradford assay. The protein concentration of each sample was standardized to 1 mg/mL for the subsequent proteomic analysis. Treatment and analysis was standardized in three distinct biological replicates to ensure reproducibility of the experiment.
Peptide digestion
Approximately 100 μg of total protein was resuspended in 100 μL of 50mM ammonium bicarbonate (pH 8.0). RapiGest (Waters Corporation, USA) at final concentration of 0.05% was added to the protein in equal parts. Protein from each sample was then concentrated to 100μL using Vivaspin column (GE Healthcare, USA) with a molecular weight cut-off (MWCO) of 3000 and incubated at 80°C for 15 min. The proteins were reduced in the presence of 5 mM dithiothreitol (DTT) at 60°C for 30 min, and then alkylated in the dark using 10 mM iodoacetamide at room temperature for 45 min. Proteolytic digestion was performed using Trypsin Gold (Promega, USA) at a ratio of 1:200 parts of protein, followed by incubation at 37°C overnight. Tryptic digestion and RapiGest activity were terminated by the addition of 1 μL concentrated trifluoroacetic acid (TFA) followed by the incubation of samples at 37°C for 20 min. The tryptic peptide solution of each sample was centrifuged at 14000 rpm for 20 min, and the resulting supernatants were collected in clean microcentrifuges tube kept at -80°C until subsequent analysis.
Peptide separation and MS analysis
The nanoLC-MS/MS analysis was performed on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific, USA). The samples (2 μL containing 2 μg peptides) were injected and separated on an EASY-nLC 1000 (Dionex, Thermo Scientific, USA) equipped with an Easy-Spray Column Acclaim PepMap C18 100Å (2 μm, 50 μm × 15 cm, Thermo Scientific, USA). Samples were separated at a flow rate of 250 nL/min and using a gradient of 5% to 50% acetonitrile (ACN) in 0.1% formic acid (FA) for 45 min followed by a further gradient to 85% ACN in 0.1% FA for 2 min. The column was equilibrated back to 5% ACN with 0.1% FA over 1 min and maintained at 5% CAN until the next sample injection. Mass spectrometry was conducted in a positive ion mode with a nanospray voltage of 1.5 kV and a source temperature of 250°C. The instrument was operated in a data-dependent acquisition (DDA) mode with an Orbitrap MS (OTMS) survey scan using the following parameters: mass range of m/z 310–1800 with resolving power of 120000, automatic gain control (AGC) of 400000 and a maximum injection time of 50 ms. The Top Speed Mode of 3 seconds was used in the selection of precursors with the monoisotopic charge state of 2 to 7. These precursors were further analysed by MS/MS scanning. All precursors were filtered using a 20 s dynamic exclusion window and intensity threshold of 5000. The MS/MS spectra were analysed using ion trap MS (ITMS) with the following parameters: rapid scan rate with resolving power of 60000, AGC of 100, isolation window of 1.6 m/z and maximum injection time of 250 ms. Precursors were then fragmented by collision-induced dissociation (CID) and high-energy collision dissociation (HCD) at normalized collision energy of 30% to 28%.
Protein identification
Raw data were processed using Thermo Scientific Proteome Discoverer Software v2.1 with the SEQUEST HT search engine. The MS ion intensities were calculated based on the accurate mass and time tag strategy. The accurate alignment of the detected LC retention time and m/z value across different analyses, together with the area under chromatographic elution profiles of the identified peptides could be compared between different samples. For protein identification, data was searched against the Uniprot K. pneumoniae database with 1% strict FDR and 5% relaxed FDR criteria using Percolator. Search parameters were set to 2 miscleavages with fixed modification of carbamidomethylation and variable modification through methionine oxidation or asparagine and glutamine deamidation. A fragment tolerance of 0.6 Da and a precursor tolerance of 10 ppm were used with trypsin as a digestion enzyme. Proteins with at least 2 unique peptides implied a greater confidence of protein identity.
Protein quantification and data analysis
Protein quantification and statistical analyses were performed using Perseus Software v1.6.0.7 (Max Planck Institute of Biochemistry). Each control and treated samples analysis consisted of three biological replicates with three technical replicates, independently injected into the LCMS/MS. The protein file with three technical replicates in txt. format from Proteome Discoverer software were uploaded to Perseus for further comparative analysis between the samples. The data were log2-transformed to stabilise the variance and scale-normalised to the same mean intensity across the technical replicates. The mean values for all three technical replicates of the same biological samples were grouped together in the same matrix and valid values were obtained by filtering with ‘at least 2’, eliminating proteins which only existed in one of the technical replicates. Finally, all biological replicates of the same treatment group were consolidated into the same matrix, with the missing values imputated with the random number derived from a normal distribution. The histograms were plotted to compare the ratio distributions between all samples. Differentially expressed proteins between control and treatment were detected using a T-test, the p-values were also adjusted for multiple-testing using the permutation-based false discovery rate, with a number of randomization of 250. Proteins were considered to be significantly differentially expressed between treatment groups with adjusted p-values of <0.05 and a fold changes of ≤ -1 or ≥ +1.
CBO treatment, RNA extraction and cDNA synthesis
KPC-KP cell culture were treated with CBO or vehicle buffer prior to RNA extraction. The CBO final concentration used was 0.08% (v/v), as determined in our previous work [11]. Both treatment groups had a final volume of 50 mL, and contained Tween 80 at final concentration of 10% to enhance the solubility of CBO. The standard KPC-KP cell inoculum was 1 × 105 cfu/mL. Samples were incubated at 37°C with shaking at 200 rpm for 4 h followed by RNA extraction using the TransZol RNA purification kit (Transgen Biotech, China). RNA (0.5 ng) was subjected to reverse transcription with QuantiNova Reverse Transcription Kit (QIAGEN, Germany) in a 20-μl reaction volume. The synthesized complementary DNA (cDNA) was stored at −20 °C until further use.
Proteomic expression validation through qRT-PCR analysis
The RNA abundance for several upregulated proteins from CBO-treated K. pneumoniae BAA-1705 was determined by qRT-PCR using QuantiNova SYBR Green PCR (QIAGEN, Germany) on CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Inc, USA). The Livak method was employed to assess the relative expression of three upregulated genes, namely cytidine deaminase (cdd), thiamine phosphate synthase (thiE) and uridine phosphorylase (udp), one down regulated gene, namely, 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase (fabA) and two housekeeping genes, namely 16S rRNA and OmpK36 porin. Reactions were performed in triplicate and data were analysed by using the CFX Manage Software (Bio-Rad). The thermal cycling conditions were as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 10 s. In all experiments, no template reactions were used as negative controls.
Results and discussion
Comparative proteome profiling of KPC-KP treated with CBO
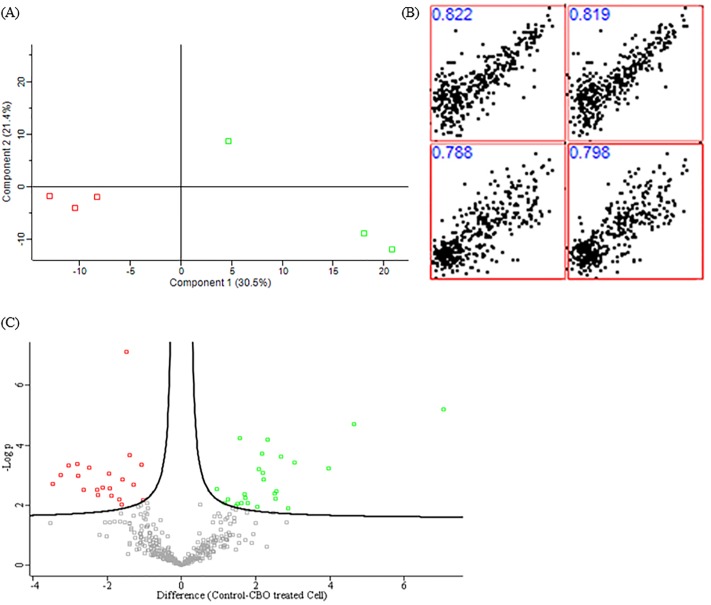
Comparative proteomic analysis was carried out between non-treated and CBO-treated KPC-KP cells in four independent experiments using Perseus Software v1.6.0.7 (Max Planck Institute of Biochemistry). The protein profiles of treated and non-treated cells varied significantly with no outliers from the replicates, as shown in the principle component analysis (PCA) in Fig 1A. In addition, the Pearson correlation values between non-treated and CBO-treated KPC-KP were of high confidence (0.788 to 0.822), indicating that both the compared groups are of the same organism with no contamination within the samples (Fig 1B). The volcano plot (Fig 1C) of the comparative proteome between non-treated and CBO-treated KPC-KP cells showed a total of 46 proteins with significantly different abundance; 25 proteins of which were upregulated, whereas the other 21 proteins were downregulated in response to CBO treatment.
Fig 1. Exploratory analysis output of non-treated and CBO-treated KPC-KP cells using Perseus v1.6.0.7 software.
(A) Principal component analysis (PCA) identifies differences between the non-treated (designated with green square) and CBO-treated (designated with red square) KPC-KP cells. (B) Multi-scatter plot with the Pearson correlation value of one of the profiles from the non-treated and CBO-treated KPC-KP cells. (C) Volcano plot showing up- (designated with green square) and downregulated (designated with red square) proteins from the CBO-treated KPC-KP cells.
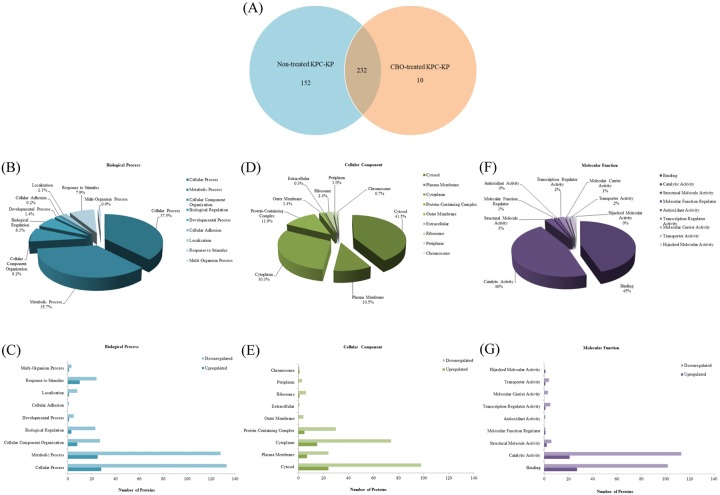
A total of 384 proteins were identified from the non-treated cells, whereas only 242 proteins were identified from the CBO-treated cells, A total of 152 proteins were only expressed in the non-treated cells while 10 proteins were only expressed in the CBO-treated cells, the other 232 proteins were present in both groups of cells (Fig 2A). Proteins that were present in both samples were compared within the Perseus software to measure up- and downregulation of proteins while the rest of the proteins are referred as absent or present from each treatment group (Table 1). Proteins that were differentially present or absent, and up- or downregulated between the two groups of cells were subjected to gene ontology analysis with regards to biological process involvement, cellular component and molecular function (Fig 2). The majority of the proteins identified were linked to cellular and metabolic processes (37.5% and 35.7%), a number were also involved with cellular component organization and response to stimulus (8.2% and 7.9%) (Fig 2B). Cellular component wise, the majority of the proteins identified were categorized under cytosol and cytoplasm (41.5% and 30.3%) followed by macromolecular complex and plasma membrane (11.9% and 10.5%; Fig 2d). The categorization by molecular function identified proteins that were involved in catalytic activity and binding (46% and 45%) in addition to proteins that were structural molecules and or had transcription regulator activity (2.8% and 2.1%) (Fig 2F). Most proteins involved in biological processes, cellular component and molecular function were downregulated (Fig 2C, 2E and 2G).
Fig 2. Comparative proteomic analysis of protein from CBO-treated KPC-KP cells.
(A) Venn diagram of the total protein obtained from non-treated KPC-KP and CBO-treated KPC-KP cells, a total of 394 proteins were identified in non-treated KPC-KPs (384 proteins) and CBO-treated KPC-KPs (242 proteins) together. These total proteins were categorized based on the related biological process (B), cellular component (D) and molecular function (F). The numbers of upregulated and downregulated genes were also compared with respect to the relevant biological process (C), cellular component (E) and molecular function (G). Gene ontology analysis in terms of biological process of protein with significant abundance from CBO-treated KPC-KP cells.
Table 1. Top 100 proteins showing significant abundance difference (together with their accession numbers) in KPC-KP cells treated with CBO.
The list is sorted according to descending positive or negative fold changes of each protein.
| No | Proteins | Gene Name | Uniprot Accession No. | General Function | Fold Change |
|---|---|---|---|---|---|
| Upregulated Proteins | |||||
| 1 | Autonomous glycyl radical cofactor | grcA | A6TCJ1 | Stress response | 7.07 |
| 2 | Beta-galactosidase 2 | lacZ2 | A6TI29 | Carbohydrate metabolism | 4.65 |
| 3 | 50S ribosomal protein L33 | rpmG | B5XTG7 | Protein biosynthesis | 3.97 |
| 4 | Sucrose porin | scrY | P27218 | Transport | 3.05 |
| 5 | ATP-dependent protease ATPase subunit HslU | hslU | B5XZ37 | Stress response | 2.68 |
| 6 | Uridine phosphorylase | udp | P52671 | Pyrimidine biosynthesis | 2.57 |
| 7 | Agmatinase | speB | B5XUB2 | Amine and polyamine biosynthesis | 2.55 |
| 8 | Phosphoribosylaminoimidazole-succinocarboxamide synthase | purC | A6TC99 | Purine biosynthesis | 2.52 |
| 9 | Phosphomethylpyrimidine synthase | thiC | A6TGQ1 | Thiamine biosynthesis | 2.33 |
| 10 | Phosphoribosylformylglycinamidine cyclo-ligase | purM | A6TCB3 | Purine biosynthesis | 2.21 |
| 11 | NAD-dependent malic enzyme | maeA | A6T9K7 | Carbohydrate metabolism | 2.19 |
| 12 | Probable Fe(2+)-trafficking protein | yggX | A6TDX0 | Stress response | 2.17 |
| 13 | Thiamine-phosphate synthase | thiE | A6TGQ0 | Thiamine biosynthesis | 2.08 |
| 14 | Chaperone protein DnaK | dnaK | A6T4F4 | Stress response | 1.73 |
| 15 | Iron-sulfur cluster insertion protein ErpA | erpA | A6T4W0 | Stress response | 1.70 |
| 16 | Cytidine deaminase | cdd | A6TBN1 | Purine biosynthesis | 1.62 |
| 17 | Elongation factor Ts | tsf | A6T4X2 | Protein biosynthesis | 1.58 |
| 18 | Ribosomal protein S12 methylthiotransferase RimO | rimO | A6T6T1 | RNA processing and modification | 1.50 |
| 19 | Dual-specificity RNA methyltransferase RlmN | rlmN | A6TCD6 | RNA processing and modification | 1.26 |
| 20 | Ribosomal RNA small subunit methyltransferase C | rsmC | A6THY6 | RNA processing and modification | 1.16 |
| CBO-treated KPC-KP Exclusive Proteins | |||||
| 21 | Bifunctional protein GlmU | glmU | B5XZM7 | Peptidoglycan biosynthesis | CBO-treated |
| 22 | Carbon storage regulator homolog | csrA | B5XVB9 | Protein biosynthesis | CBO-treated |
| 23 | Cobalamin biosynthesis protein CobD | cobD | A6TDC6 | Cobalamin biosynthesis | CBO-treated |
| 24 | DNA ligase | ligA | A6TC47 | Stress response | CBO-treated |
| 25 | Ferrochelatase | hemH | B5Y0N2 | Porphyrin biosynthesis | CBO-treated |
| 26 | Hydroxyacylglutathione hydrolase | gloB | A6T510 | Secondary metabolite metabolism | CBO-treated |
| 27 | Integration host factor subunit beta | ihfB | A6T704 | DNA processing | CBO-treated |
| 28 | Probable malate:quinone oxidoreductase (Fragment) | mqo | O32719 | Energy synthesis | CBO-treated |
| 29 | tRNA 2-thiocytidine biosynthesis protein TtcA | ttcA | B5XRN3 | RNA processing and modification | CBO-treated |
| Downregulated Proteins | |||||
| 30 | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase | fabA | A6T748 | Fatty acid biosynthesis | -3.46 |
| 31 | ATP-dependent protease subunit HslV | hslV | B5XZ36 | Stress response | -3.25 |
| 32 | HTH-type transcriptional regulator IscR | iscR | A6TCF2 | Stress response | -3.05 |
| 33 | Ribosome maturation factor RimP | rimP | A6TEI9 | Ribosome biogenesis | -2.80 |
| 34 | 50S ribosomal protein L32 | rpmF | A6T7E9 | Translation | -2.78 |
| 35 | Recombination-associated protein RdgC | rdgC | B5Y109 | DNA processing | -2.48 |
| 36 | Peptide methionine sulfoxide reductase MsrB | msrB | A6T7R0 | Stress response | -2.27 |
| 37 | 6-phosphogluconate dehydrogenase, decarboxylating | gnd | P41576 | Carbohydrate metabolism | -2.24 |
| 38 | 50S ribosomal protein L17 | rplQ | A6TEU7 | Protein biosynthesis | -2.13 |
| 39 | Cysteine desulfurase IscS | iscS | A6TCF1 | Stress response | -1.95 |
| 40 | 50S ribosomal protein L5 | rplE | A6TEW0 | Protein biosynthesis | -1.94 |
| 41 | Uridine kinase | udk | A6TBG6 | Pyrimidine biosynthesis | -1.89 |
| 42 | Probable septum site-determining protein MinC | minC | A6TAX3 | Cell division | -1.67 |
| 43 | UDP-4-amino-4-deoxy-L-arabinose—oxoglutarate aminotransferase | arnB | A6TFA0 | Lipopolysaccharide biosynthesis | -1.62 |
| 44 | S-adenosylmethionine synthase | metK | A6TDV1 | Protein biosynthesis | -1.58 |
| 45 | Glutamate—tRNA ligase | gltX | A6TC43 | Protein biosynthesis | -1.49 |
| 46 | Arginine—tRNA ligase | argS | A6TB43 | Protein biosynthesis | -1.39 |
| 47 | HTH-type transcriptional regulator MalT | malT | A6TF41 | Carbohydrate metabolism | -1.28 |
| 48 | Protein TolB | tolB | B5XZC1 | Cell division | -1.10 |
| 49 | Acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha | accA | A6T4Y7 | Lipid metabolism | -1.08 |
| 50 | 30S ribosomal protein S3 | rpsC | B5XNA0 | Protein biosynthesis | -1.04 |
| Control KPC-KP Exclusive Proteins | |||||
| 51 | 10 kDa chaperonin | grosA | B5Y369 | Stress response | Control |
| 52 | 1-deoxy-D-xylulose-5-phosphate synthase | dxs | A6T5F3 | Thiamine biosynthesis | Control |
| 53 | 3-dehydroquinate synthase | aroB | A6TF13 | Chorismate biosynthesis | Control |
| 54 | 3-methyl-2-oxobutanoate hydroxymethyltransferase | panB | A6T4T0 | Pantothenate biosynthesis | Control |
| 55 | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase | ubiD | A6TGM1 | Ubiquinone biosynthesis | Control |
| 56 | 3-phosphoshikimate 1-carboxyvinyltransferase | aroA | B5XY87 | Chorismate biosynthesis | Control |
| 57 | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | ispH | A6T4G3 | Isoprenoid biosynthesis | Control |
| 58 | 50S ribosomal protein L34 | rpmH | A6TG05 | Protein biosynthesis | Control |
| 59 | 6-phosphogluconolactonase | pgl | A6T6J6 | Carbohydrate metabolism | Control |
| 60 | Adenine phosphoribosyltransferase | apt | A6T5N2 | Purine metabolism | Control |
| 61 | Aminomethyltransferase | gcvT | A6TDR7 | Protein biosynthesis | Control |
| 62 | Argininosuccinate synthase | argG | A6TEJ0 | Protein biosynthesis | Control |
| 63 | ATP synthase epsilon chain | atpC | A6TG35 | Energy synthesis | Control |
| 64 | ATP-dependent RNA helicase RhlB | rhlB | A6TGG9 | RNA processing and modification | Control |
| 65 | Biosynthetic arginine decarboxylase | speA | B5XUB1 | Amine and polyamine biosynthesis | Control |
| 66 | Catalase-peroxidase | katG | A6T9H9 | Stress response | Control |
| 67 | Cell division protein ZipA | zipA | A6TC48 | Cell division | Control |
| 68 | Cell division topological specificity factor | minE | A6TAX5 | Cell division | Control |
| 69 | Chorismate synthase | aroC | A6TC15 | Chorismate biosynthesis | Control |
| 70 | Chromosomal replication initiator protein DnaA | dnaA | B5XT51 | DNA processing | Control |
| 71 | D-amino acid dehydrogenase | dadA | A6TAW4 | Protein biosynthesis | Control |
| 72 | Dihydroorotase | pyrC | A6T7D6 | Pyrimidine biosynthesis | Control |
| 73 | Dihydroorotate dehydrogenase (quinone) | pyrD | A6T739 | Pyrimidine biosynthesis | Control |
| 74 | Dihydroxy-acid dehydratase | ilvD | A6TGF8 | Protein biosynthesis | Control |
| 75 | DNA gyrase inhibitor YacG | yacG | A6T4P3 | DNA processing | Control |
| 76 | DNA mismatch repair protein MutS | mutS | A6TD24 | Stress response | Control |
| 77 | DnaA initiator-associating protein DiaA | diaA | A6TEG8 | DNA processing | Control |
| 78 | Electron transport complex subunit C | rnfC | B5XWP9 | Energy synthesis | Control |
| 79 | Elongation factor P—(R)-beta-lysine ligase | epmA | A6TH74 | Protein biosynthesis | Control |
| 80 | Endonuclease V | nfi | A6TGQ5 | Stress response | Control |
| 81 | Exodeoxyribonuclease 7 small subunit | xseB | B5Y0W9 | DNA processing | Control |
| 82 | Formate-dependent phosphoribosylglycinamide formyltransferase | purT | B5XQ21 | Purine metabolism | Control |
| 83 | Fructose-6-phosphate aldolase | fsa | A6TGD7 | Carbohydrate metabolism | Control |
| 84 | Gamma-glutamyl phosphate reductase | proA | A6T561 | Protein biosynthesis | Control |
| 85 | Glucans biosynthesis protein D | mdoD | B5XWS2 | Glycan metabolism | Control |
| 86 | Glucokinase | glk | A6TC33 | Energy synthesis | Control |
| 87 | Glucosamine-6-phosphate deaminase | nagB | A6T6C1 | Carbohydrate metabolism | Control |
| 88 | Glutamate 5-kinase | proB | A6T562 | Protein biosynthesis | Control |
| 89 | Glycerol-3-phosphate acyltransferase | plsB | A6TGV0 | Phospholipid biosynthesis | Control |
| 90 | Histidine biosynthesis bifunctional protein HisB | hisB | A6TBC5 | Protein biosynthesis | Control |
| 91 | Holliday junction ATP-dependent DNA helicase RuvA | ruvA | B5XQ04 | Stress response | Control |
| 92 | HTH-type transcriptional regulator CysB | cysB | P45600 | Protein biosynthesis | Control |
| 93 | HTH-type transcriptional repressor PurR | purR | A6TA06 | Purine metabolism | Control |
| 94 | Hydroxyethylthiazole kinase | thiM | A6TBJ8 | Thiamine biosynthesis | Control |
| 95 | Imidazole glycerol phosphate synthase subunit HisF | hisF | A6TBC8 | Protein biosynthesis | Control |
| 96 | Iron-binding protein IscA | iscA | A6TCE9 | Stress response | Control |
| 97 | Ketol-acid reductoisomerase (NADP(+)) | ilvC | A6TGG1 | Protein biosynthesis | Control |
| 98 | Large-conductance mechanosensitive channel | mscL | B5XNC0 | Transport | Control |
| 99 | L-lactate dehydrogenase | lldD | A6TFK0 | Energy synthesis | Control |
| 100 | L-threonine 3-dehydrogenase | tdh | A6TFL2 | Protein biosynthesis | Control |
Of the overlapping 232 proteins identified in both treatment groups, only 41 proteins showed significant differences, in terms of fold change between the groups. The majority of the proteins which showed significant abundance difference were downregulated (51.2%) when compared to the non-treated KPC-KP cells, whereas the other 48.8% of the proteins were upregulated as shown in Table 1. Proteins that were absent or present in each treatment group were also listed in Table 1, with proteins that were only present in the CBO-treated KPC-KP cells listed under the upregulated proteins whereas proteins which were only present in the non-treated KPC-KP cells listed under the downregulated protein section. These proteins were then subjected to KEGG pathway analysis to elucidate mechanism involved in the action of CBO on KPC-KP cells.
Bacterial membrane disruption
The Gram negative bacterial cell outer barrier consists of three separate component, the outer membrane, the peptidoglycan and the plasma membrane [22]. Both the outer membrane and the plasma membrane are made up of a phospholipid bilayer embedded with membrane proteins. The biochemical feature which differentiates the layers is the presence of lipopolysaccharides uniquely in the outer membrane. Of the identified proteins from the KPC-KP cells, 10.5% were located at the bacterial membrane (Fig 2D and 2E). Following exposure to CBO, our proteomic profiling showed 5 outer membrane exclusive proteins and 26 plasma membrane exclusive proteins were completely lost after the exposure to CBO (Table 2). Similarly, Wu and colleagues (2016) found that 3-p-trans-coumaroyl-2-hydroxyquinic acid, a phenolic compound isolated from Himalayan cedar essential oil caused bacterial membrane damage and the loss of membrane proteins due to specific interactions between the compound and the lipid and proteins within the bacterial membrane [23]. This disrupted the membrane integrity of the bacteria which caused the loss of plasma membrane protein. The downregulation of outer membrane integrity regulators such as the TolB porin-interacting protein, the large-conductance mechanosensitive channel and outer membrane protein assembly factor BamA in KPC-KP cells exposed to CBO also indicated the loss of membrane integrity in KPC-KP cells [24]. In addition, proteins involved in energy generation, such as the ATP synthase, the electron transport complex and the NADH-quinone oxidoreductases which are often embedded within the bacterial membrane were lost completely. This is yet another indicator for disrupted bacterial membrane integrity that may contribute to bacterial cell killing through the shutdown ofenergy production in CBO-treated KPC-KP cells. With compromised plasma membrane integrity, intracellular proteins easily escape into the extracellular environment as suggested by Ukuku et al. (2007) [25].
Table 2. List of proteins belonging to the outer membrane and plasma membrane of KPC-KP cells and their relative status in proteomic profiling.
| Uniprot Accession No. | Protein | Status |
|---|---|---|
| Outer membrane protein | ||
| A6TGU6 | Maltoporin 2 | Lost |
| P40786 | Nucleoside-specific channel-forming protein tsx | Lost |
| P24017 | Outer membrane protein A | Upregulated by 2.04 folds |
| A6T4X9 | Outer membrane protein assembly factor BamA | Lost |
| A6T7G4 | Penicillin-binding protein activator LpoB | Lost |
| P27218 | Sucrose porin | Lost |
| Plasma membrane protein | ||
| A6TGM1 | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase | Lost |
| A6TG35 | ATP synthase epsilon chain | Lost |
| B5XZ37 | ATP-dependent protease ATPase subunit HslU | Lost |
| B5XUB1 | Biosynthetic arginine decarboxylase | Lost |
| A6TC48 | Cell division protein ZipA | Lost |
| A6TAX5 | Cell division topological specificity factor | Lost |
| A6T4F4 | Chaperone protein DnaK | Upregulated by 1.73 folds |
| B5XT51 | Chromosomal replication initiator protein DnaA | Lost |
| A6TDC6 | Cobalamin biosynthesis protein CobD | Upregulated* |
| A6TAW4 | D-amino acid dehydrogenase | Lost |
| A6T739 | Dihydroorotate dehydrogenase (quinone) | Lost |
| B5XWP9 | Electron transport complex subunit C | Lost |
| B5XWS2 | Glucans biosynthesis protein D | Lost |
| A6TGV0 | Glycerol-3-phosphate acyltransferase | Lost |
| A6TDR6 | Glycine cleavage system H protein | Lost |
| B5XNC0 | Large-conductance mechanosensitive channel | Lost |
| A6TFK0 | L-lactate dehydrogenase | Lost |
| A6TEQ3 | Malate dehydrogenase | Lost |
| B5XXP0 | NAD(P)H dehydrogenase (quinone) | Lost |
| A6TBX3 | NADH-quinone oxidoreductase subunit B | Lost |
| A6TBX2 | NADH-quinone oxidoreductase subunit C/D | Lost |
| A6TC99 | Phosphoribosylaminoimidazole-succinocarboxamide synthase | Upregulated by 2.52 folds |
| Q07411 | Polyphosphate kinase | Lost |
| O32719 | Probable malate:quinone oxidoreductase (Fragment) | Upregulated* |
| A6TDG4 | Prolipoprotein diacylglyceryl transferase | Lost |
| P27219 | PTS system sucrose-specific EIIBC component | Lost |
| A6TC94 | Succinyl-diaminopimelate desuccinylase | Lost |
| A6THZ7 | Thymidine phosphorylase | Lost |
| A6TGL3 | Ubiquinone/menaquinone biosynthesis C-methyltransferase UbiE | Lost |
| A6T4N3 | UDP-N-acetylglucosamine—N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase | Lost |
*refers to the proteins that were only present in CBO-treated KPC-KP cells.
Oxidative stress
CBO contains a plethora of different chemical compounds which are dominated by a large group of oxygenated terpenes and terpenoids [10]. These compounds are hypothesized to be responsible for CBO’s action on the bacterial membrane. From our proteomic data, upregulation of proteins such as autonomous glycyl radical cofactor and catalase peroxidase in the CBO-treated KPC-KP cells suggest the presence of significant oxidative stress. Autonomous glycyl radical cofactor is upregulated by 7.07 fold, and acts as a radical domain which protects pyruvate formate lyase from oxidative stress. Wagner et al. (2001) and Shisler et al. (2014) found that upregulation of glycyl radical cofactor indicates oxidative stress which affects the pyruvate formate lyase that is involved in glucose metabolism [26, 27]. In addition, catalase peroxidase, an enzyme which alleviates oxidative stress from reactive oxygen species (ROS), was also induced when KPC-KP cells were exposed to CBO. Under condition of oxidative stress, high abundance ROS cause oxidative damage to nucleic acids [28]. The detection of the DNA mismatch repair protein MutS and the DNA ligase following the exposure to CBO showed that genetic materials of KPC-KP had been damaged, and that elevated expression of these proteins could alleviate some of the oxidative DNA damage. The work of Vogel et al. (2011) showed that oxidative stress leads to the degradation of ribosomal protein [29] and this appears to be in line with our findings in CBO-treated KPC-KP cells where 14 ribosomal proteins and ribosome related proteins showed decreased abundance. Specifically, 30S ribosomal protein S3; 50S ribosomal proteins L17, L32, L34 and L5; ribosomal protein L11 methyltransferase; ribosomal RNA large subunit methyltransferase E, F and I; ribosomal RNA small subunit methyltransferase B and G; ribosome maturation factor M and P, and ribosome binding factor A were all reduced in abundance. As one of the key proteins in the maturation of 30S ribosomal subunit, ribosome maturation factor RimP protein is crucial in the process of protein translation by allowing correct pairing of the mRNA and the corresponding anticodon of the tRNA [30, 31]. In agreement with this finding is the decrease in abundance of the other three 50S ribosomal subunit fragments namely, L5, L17 and L32. This further supports the idea that induction of oxidative stress by CBO affects the abundance of ribosomal subunit fragments, as a result of the denaturation of protein fragments or the interruption of protein synthesis in KPC-KP cells. Cosentino et al. (2014) found that bergamot essential oil induced the production of ROS in polymorphonuclear leukocytes, contributing to enhanced eradication of infection [32]. Another study by Yoo et al (2005) also suggest that eugenol, one of the main constituents of essential oils such as from nutmeg and cinnamon bark induces the production of ROS in leukemia cells, eventually killing the cells by initiating apoptosis [33]. The induction of oxidative stress by essential oils seems to be in conflict with the perception that essential oils contain high concentration of antioxidants. We previously found that the CBO used in this study comprised 13 compounds, of which nine were antioxidants whereas the other four were not [10, 34–38]. These non-antioxidant compounds may be responsible for inducing the oxidative stress observed, by generating ROS or inhibiting anti-oxidizing systems [39]. Mimica-Dukić et al. (2016) suggested that essential oils may act both as antioxidants and also as pro-oxidant due to their complex chemistry [40]. Additionally, a single compound might exhibit dual antioxidant and prooxidant effects. For instance, Bezerra et al. (2017) found that eugenol had such dual effects, acting as an antioxidant in free-radical scavenging while inducing DNA damage via ROS generation [41]. So, the prooxidant activity within CBO may contribute to the oxidative stress, leading to lipid peroxidation in the cell membrane, while inhibiting the antioxidant activity, as indicated in the proteomic profile.
KEGG pathway analysis
When analysing significant pathways via the Panther classification system we found that the main pathways associated with the proteins identified included the lipid, cell wall and lipopolysaccharide biosynthesis pathways.
Lipid biosynthesis
Lipids have critical functions in the bacterial cytoplasmic and outer membranes in separating the bacterial cytoplasm from the external environment [42]. KEGG pathway analysis identified 4 proteins which are involved in the lipid biosynthesis pathway that were affected by CBO treatment. Two of these proteins were downregulated in CBO-treated KPC-KP cells: 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase (-3.46 fold) and acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha (-1.08 fold). While two other proteins were undetectable following CBO treatment: glycerol-3-phosphate acyltransferase and large-conductance mechanosensitive channel protein. The former two proteins are key components in the fatty acid biosynthesis pathway of Gram-negative bacteria. Acetyl-coenzyme A carboxylase carboxyl transferase is one of the major enzymes in the synthesis of malonyl-CoA, a substrate required in the synthesis of fatty acids [43]. 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase functions as an essential mediator in the synthesis of phospholipids in bacteria [44, 45]. As shown in Table 1, glycerol-3-phosphate acyltransferase (G3PAT) was undetectable in the CBO-treated KPC-KP cells. G3PAT, a rate determining enzyme belonging to the glycerolphospholipid metabolism pathway catalyzes the synthesis of phosphatidic acid from glycerol-3-phosphate and long-chain acyl-CoA, is an essential precursor in the synthesis of the bacterial phospholipid bilayer [46, 47]. The absence of G3PAT indicates a perturbed bacterial membrane repair system. This further indicates a disruption in the integrity of the phospholipid membrane in CBO-treated KPC-KP cells. The absence of the large-conductance mechanosensitive channel protein in the treated group also indicated a disrupted membrane structure as this channel regulates membrane stretching and stability under osmotic stress [48, 49]. Upon exposure to CBO, oxidative stress may promote protein denaturation and so affect bacterial cell membrane stretch capacity under osmotic pressure, eventually leading to increased CBO influx and bacterial cell killing.
Cell wall biosynthesis
Peptidoglycan in the cell wall is a major structural component in prokaryotic cells, forming a stable layer which protects the bacteria from lysis under osmotic stress [50]. The proteomic profile showed that three essential proteins in bacterial cell wall synthesis were lost upon exposure to CBO. This may indicate that CBO inhibited the expression of these proteins or that CBO-induced oxidative stress resulted in degradation of these proteins, so preventing cell wall synthesis and repair, and eventually causing cell death. These proteins included UDP-N-acetylglucosamine—N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase (murG); penicillin-binding protein activator LpoB and succinyl-diaminopimelate desuccinylase. The MurG protein is involved in the biosynthesis of the N-acetylmuramic acid-N-acetylglucosamine intermediate, which form a single unit of peptidoglycan cell wall. In the absence of murG, no N-acetylmuramic acid would not be linked to N-acetylglucosamine, preventing the formation of peptidoglycan bilayer linkage and eventually killing the cell due to osmotic pressure and oxidative stress [50]. Additionally, succinyl-diaminopimelate desuccinylase also play a major role in maintaining the structure and integrity of the peptidoglycan, as this protein is essential in the synthesis of meso-diaminopimelic acid which is one of the penta-peptides found linked to N-acetylmuramic acid, and is crucial for the cross-linkage with the opposite layer of N-acetylglucosamine. Interestingly, penicillin-binding protein activator LpoB which stimulates the biosynthesis of peptidoglycan molecules was not detected in CBO treated KPC-KP cells. These findings may indicate that the peptidoglycan biosynthesis and repairing had been completely shut down [51].
Lipopolysaccharide biosynthesis
Lipopolysaccharide (LPS) is a highly acylated saccharolipid located on the outer layer of the outer membrane of Gram-negative bacteria. LPS is crucial in the maintenance of membrane integrity and has a barrier function which prevents the passive diffusion of hydrophobic solutes, such as antibiotics and detergents into the cell [52]. Many studies have postulated that CBO exerts its bactericidal activity through damage to the outer membrane which allows other bactericidal molecules to enter the cell, eventually killing the cells [11, 15, 16]. LPS consist of a few components including lipid A; core oligosaccharide and O-antigen [53]. We identified three proteins that are involved in LPS biosynthesis that became undetectable upon CBO exposure. These were O-antigen export system ATP-binding protein (RfbB); UDP-4-amino-4-deoxy-L-arabinose—oxoglutarate aminotransferase (arnB) and UDP-3-O-acyl-N-acetylglucosamine deacetylase (lpxC). In order to achieve resistance towards antibiotics such as polymyxin, the 4-amino-4-deoxy-L-arabinose moiety must be added to the lipid A [54]. This reaction is catalyzed by the arnB protein which is not detected in the KPC-KP cells exposed to CBO. Furthermore, lpxC protein, a critical enzyme which catalyzes the synthesis of lipid A was also undetectable in CBO-treated KPC-KP cells. 90% of the bacterial outer membrane consists of lipid A, which is crucial for the attachment of core oligosaccharide as well as o-antigen, granting antibiotic resistance to bacteria while maintaining the membrane integrity. In the absence of the lpxC protein, the outer membrane integrity cannot be maintained, compromising cellular resistance against antibiotics. Due to the importance of this protein in lipid A biosynthesis, lpxC has been a target for the development of novel antimicrobial drugs [55, 56]. The O-antigen export system ATP-binding protein, which facilitates the export of O-antigen into the outer membrane was also not detected following exposure to CBO. Thus, CBO may contain compounds that inhibit the expression of these important membrane proteins and sensitize bacteria to antibiotics and changes in osmotic pressure.
qRT-PCR analysis of differentially expressed proteins
Standard curve for qRT-PCR
We selected four genes for further validation of our proteomic data using qRT-PCR together with two housekeeping genes, namely 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase (fabA), cytidine deaminase (cdd), thiamine phosphate synthase (thiE), uridine phosphorylase (udp), 16s rRNA and OmpK36 porin. Out of these, three were significantly higher in abundance and one was less abundant in the proteomic profile following exposure to CBO. The designed primers for each selected genes were listed in S1 Table. Their efficiency ranged within 90 and 100%.
Relative expression level of selected genes
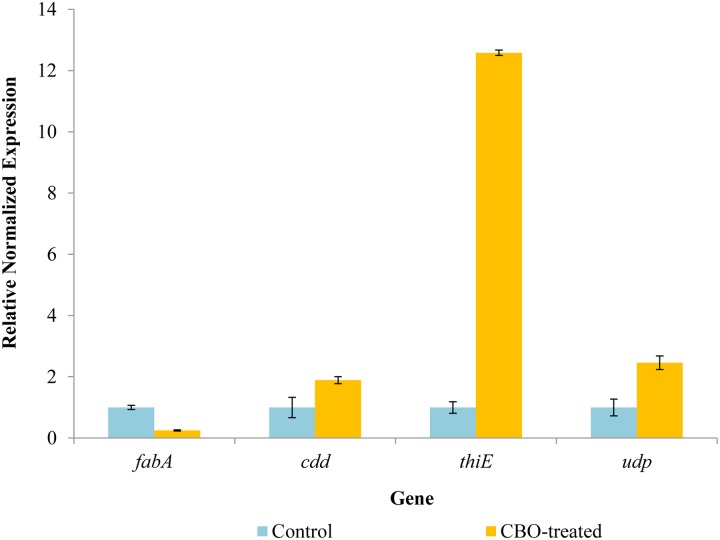
The relative abundance of mRNA for the differentially expressed proteins investigated was measured by qRT-PCR, comparing the untreated cells with CBO-treated KPC-KP cells (Fig 3). Proteomic profiling found that fabA was downregulated by -3.46 fold, whereas cdd, thiC and udp were upregulated by 1.62, 2.08 and 2.57 fold respectively (Table 1). The qRT-PCR analysis of mRNA abundance found that fabA was downregulated by 0.8 fold whereas cdd, thiC and udp were upregulated by 0.8, 11 and 1.4 fold respectively. The pattern in mRNA abundance change following CBO treatment mirrors that of the protein abundance for these four gene products, indicating that at least part of the change in the abundance for these four proteins is due to changes in gene transcription. The differences in the magnitude of the fold changes between the protein abundance and mRNA transcript abundance may be a consequence of oxidative stress-induced protein degradation or different stabilities for the mRNA species. Nevertheless, the trends in expression changes shown in the protein profiles and mRNA transcription levels are consistent with each other and validate our results obtained from proteomic profiling.
Fig 3. Expression patterns of fabA, cdd, thiE and udp genes in KPC-KP cells subjected to CBO treatment.
Results are presented as differential relative transcript abundance.
CBO exerts antimicrobial activity on KPC-KP cells through membrane disruption, and proteomic profiling shows that the membrane damage induced was due to oxidative stress. This is supported by the increased in abundance of oxidative stress regulators when KPC-KP cells were exposed to CBO. A review by Itri et al. (2014) consolidated information from studies involving oxidative stress and membrane damage. The study concluded that oxidative stress would reduce the permeability and integrity of the plasma membrane, leading to leakage of intracellular contents and eventually killing the bacterial cells [57]. The membrane disruptive effects of CBO could also be deduced from our proteomic profiles, and numerous proteins showed decreased abundance or were lost following CBO exposure. Interestingly, CBO treatment also interfered with the biosynthesis of the plasma membrane, cell wall and outer membrane, disabling the structural repair system. The proteomic profiles were validated using qRT-PCR analysis, where proteomic changes were mirrored by changes in corresponding mRNA abundance. Together with the evidence from our previous study on the antimicrobial potential and the mode of action of CBO against KPC-KP cells [11], we showed that the antibacterial activity of CBO derives from its ability to induce oxidative stress in bacterial cells. The resulting oxidation disrupts the bacterial membrane, eventually enabling the influx of ROS into the cells and, at the same time, leads to intracellular content leakage. ROS induce genetic damage and impair DNA and membrane repair systems. Our results demonstrate that CBO causes oxidative stress, damage to bacterial membranes, cellular leakage and cell killing.
Supporting information
(ZIP)
(XLSX)
(DOCX)
Acknowledgments
The authors like to thank all members of the Floral Biotechnology Laboratory, Universiti Putra Malaysia. All authors read and approved the final manuscript. Additionally, the authors would also like to thank Malaysia Genome Institute (MGI) for providing the proteomic facilities throughout this study.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The research received funding UPM Internal Grant (GP-IPS/2016/9505800), Malaysian Medical Association (MMA), and Fundamental Research Grant Scheme (FRGS/1/2018/SKK11/PERDANA/02/1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Durdu B, Hakyemez IN, Bolukcu S, Okay G, Gultepe B, Aslan T. Mortality markers in nosocomial Klebsiella pneumoniae bloodstream infection. Springerplus. 2016;5(1): 1892 10.1186/s40064-016-3580-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4): 657–686. 10.1128/CMR.18.4.657-686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ur Rahman S, Ali T, Ali I, Khan NA, Han B, Gao J. The Growing genetic and functional diversity of extended spectrum beta-lactamases. Biomed Res Int. 2018;2018:9519718 10.1155/2018/9519718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhomberg PR, Jones RN. Summary trends for the Meropenem Yearly Susceptibility Test Information Collection Program: a 10-year experience in the United States (1999–2008). Diagn Microbiol Infect Dis. 2009;65(4): 414–426. 10.1016/j.diagmicrobio.2009.08.020 [DOI] [PubMed] [Google Scholar]
- 5.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4): 1151–1161. 10.1128/AAC.45.4.1151-1161.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 2009;53(8): 3365–3370. 10.1128/AAC.00126-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang SK, Low LY, Yap PSX, Yusoff, Mai CW, Lai KS, et al. Plant-derived antimicrobials: insights into mitigation of antimicrobial resistance. Rec Nat Prod. 2018;12(4): 295–316. [Google Scholar]
- 8.Yap PSX, Yiap BC, Ping HC, Lim SHE. Essential oils, a new horizon in combating bacterial antibiotic resistance. Open Microbiol J. 2014;8: 6–14. 10.2174/1874285801408010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenzi V, Muselli A, Bernardini AF, Berti L, Pages JM, Amaral L, et al. Geraniol restores antibiotic activities against multidrug-resistant isolates from gram-negative species. Antimicrob Agents Chemother. 2009;53(5): 2209–2211. 10.1128/AAC.00919-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yap PSX, Krishnan T, Chan KG, Lim SHE. Antibacterial mode of action of Cinnamomum verum bark essential oil, alone and in combination with piperacillin, against a multidrug-resistant Escherichia coli strain. J Microbiol Biotechnol. 2015;25(8): 1299–1306. 10.4014/jmb.1407.07054 [DOI] [PubMed] [Google Scholar]
- 11.Yang SK, Yusoff K, Mai CW, Lim WM, Yap WS, Lim SHE, et al. Additivity vs synergism: investigation of the additive interaction of cinnamon bark oil and meropenem in combinatory therapy. Molecules. 2017;22(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yap PSX, Lim SHE, Hu CP, Yiap BC. Combination of essential oils and antibiotics reduce antibiotic resistance in plasmid-conferred multidrug resistant bacteria. Phytomedicine. 2013;20(8–9): 710–713. 10.1016/j.phymed.2013.02.013 [DOI] [PubMed] [Google Scholar]
- 13.Yap PSX, Yang SK, Lai KS, Lim SHE. Essential oils: the ultimate solution to antimicrobial resistance in Escherichia coli. 2017.
- 14.Yang SK, Yap PSX, Krishnan T, Yusoff K, Chan KG, Yap WS, et al. Mode of action: synergistic interaction of peppermint (Mentha x piperita L. Carl) essential oil and meropenem against plasmid-mediated resistant E. coli. Rec Nat Prod. 2018;12(6): 13. [Google Scholar]
- 15.de Souza EL, de Barros JC, de Oliveira CE, da Conceicao ML. Influence of Origanum vulgare L. essential oil on enterotoxin production, membrane permeability and surface characteristics of Staphylococcus aureus. Int J Food Microbiol. 2010;137(2–3): 308–311. 10.1016/j.ijfoodmicro.2009.11.025 [DOI] [PubMed] [Google Scholar]
- 16.Silva F, Ferreira S, Queiroz JA, Domingues FC. Coriander (Coriandrum sativum L.) essential oil: its antibacterial activity and mode of action evaluated by flow cytometry. J Med Microbiol. 2011;60: 1479–1486. 10.1099/jmm.0.034157-0 [DOI] [PubMed] [Google Scholar]
- 17.Turgis M, Han J, Caillet S, Lacroix M. Antimicrobial activity of mustard essential oil against Escherichia coli O157:H7 and Salmonella typhi. Food Control. 2009;20(12): 1073–1079. [Google Scholar]
- 18.Xu H, Dephoure N, Sun H, Zhang H, Fan F, Liu J, et al. Proteomic profiling of paclitaxel treated cells identifies a novel mechanism of drug resistance mediated by PDCD4. J Proteome Res. 2015;14(6): 2480–2491. 10.1021/acs.jproteome.5b00004 [DOI] [PubMed] [Google Scholar]
- 19.Kawatani M, Muroi M, Wada A, Inoue G, Futamura Y, Aono H, et al. Proteomic profiling reveals that collismycin A is an iron chelator. Scientific Reports. 2016;6: 38385 10.1038/srep38385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zainal-Abidin Z, Mohd-Said S, Adibah F, Majid A, Mustapha WAW, Jantan I. Anti-bacterial activity of cinnamon oil on oral pathogens. The Open Conference Proceedings Journal. 2013;4: 5. [Google Scholar]
- 21.Kaskatepe B, Kiymaci ME, Simsek D, Erol HB, Erdem SA. Comparison of the contents and antimicrobial activities of commercial and natural cinnamon oils. Indian J Pharm Sci. 2016;78(4): 8.27168676 [Google Scholar]
- 22.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2(5): a000414 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, Bai J, Zhong K, Huang Y, Qi H, Jiang Y, et al. Antibacterial activity and membrane-disruptive mechanism of 3-p-trans-coumaroyl-2-hydroxyquinic acid, a novel phenolic compound from pine needles of Cedrus deodara, against Staphylococcus aureus. Molecules. 2016;21(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63(4): 1008–1025. 10.1111/j.1365-2958.2006.05571.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ukuku DO, Geveke DJ, Cooke P, Zhang HQ. Membrane damage and viability loss of Escherichia coli K-12 in apple juice treated with radio frequency electric field. J Food Prot. 2008;71(4): 684–690. [DOI] [PubMed] [Google Scholar]
- 26.Shisler KA, Broderick JB. Glycyl radical activating enzymes: structure, mechanism, and substrate interactions. Arch Biochem Biophys. 2014;546: 64–71. 10.1016/j.abb.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner AF, Schultz S, Bomke J, Pils T, Lehmann WD, Knappe J. YfiD of Escherichia coli and Y06I of bacteriophage T4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochem Biophys Res Commun. 2001;285(2): 456–462. 10.1006/bbrc.2001.5186 [DOI] [PubMed] [Google Scholar]
- 28.Willi J, Kupfer P, Evequoz D, Fernandez G, Katz A, Leumann C, et al. Oxidative stress damages rRNA inside the ribosome and differentially affects the catalytic center. Nucleic Acids Res. 2018;46(4): 1945–1957. 10.1093/nar/gkx1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogel C, Silva GM, Marcotte EM. Protein expression regulation under oxidative stress. Mol Cell Proteomics. 2011;10(12): M111 009217 10.1074/mcp.M111.009217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunner AE, Nord S, Wikstrom PM, Williamson JR. The effect of ribosome assembly cofactors on in vitro 30S subunit reconstitution. J Mol Biol. 2010;398(1): 1–7. 10.1016/j.jmb.2010.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nord S, Bylund GO, Lovgren JM, Wikstrom PM. The RimP protein is important for maturation of the 30S ribosomal subunit. J Mol Biol. 2009;386(3): 742–753. 10.1016/j.jmb.2008.12.076 [DOI] [PubMed] [Google Scholar]
- 32.Cosentino M, Luini A, Bombelli R, Corasaniti MT, Bagetta G, Marino F. The essential oil of bergamot stimulates reactive oxygen species production in human polymorphonuclear leukocytes. Phytother Res. 2014;28(8): 1232–1239. 10.1002/ptr.5121 [DOI] [PubMed] [Google Scholar]
- 33.Yoo CB, Han KT, Cho KS, Ha J, Park HJ, Nam JH, et al. Eugenol isolated from the essential oil of Eugenia caryophyllata induces a reactive oxygen species-mediated apoptosis in HL-60 human promyelocytic leukemia cells. Cancer Lett. 2005;225(1): 41–52. 10.1016/j.canlet.2004.11.018 [DOI] [PubMed] [Google Scholar]
- 34.Carvalho RL, Cabral MF, Germano TA, de Carvalho WM, Brasil IM, Gallão MI, et al. Chitosan coating with trans-cinnamaldehyde improves structural integrity and antioxidant metabolism of fresh-cut melon. Postharvest Biol Tech. 2016;113: 29–39. [Google Scholar]
- 35.Ciftci O, Ozdemir I, Tanyildizi S, Yildiz S, Oguzturk H. Antioxidative effects of curcumin, beta-myrcene and 1,8-cineole against 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced oxidative stress in rats liver. Toxicol Ind Health. 2011;27(5): 447–453. 10.1177/0748233710388452 [DOI] [PubMed] [Google Scholar]
- 36.Bubols GB, Vianna Dda R, Medina-Remon A, von Poser G, Lamuela-Raventos RM, Eifler-Lima VL, et al. The antioxidant activity of coumarins and flavonoids. Mini Rev Med Chem. 2013;13(3): 318–334. [DOI] [PubMed] [Google Scholar]
- 37.Dahham SS, Tabana YM, Iqbal MA, Ahamed MB, Ezzat MO, Majid AS, et al. The anticancer, antioxidant and antimicrobial properties of the sesquiterpene beta-caryophyllene from the essential oil of Aquilaria crassna. Molecules. 2015;20(7): 11808–11829. 10.3390/molecules200711808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavan B, Dalpiaz A, Marani L, Beggiato S, Ferraro L, Canistro D, et al. Geraniol pharmacokinetics, bioavailability and its multiple effects on the liver antioxidant and xenobiotic-metabolizing enzymes. Front Pharmacol. 2018;9: 18 10.3389/fphar.2018.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahal A, Kumar A, Singh V, Yadav B, Tiwari R, Chakraborty S, et al. Oxidative stress, prooxidants, and antioxidants: the interplay. Biomed Res Int. 2014;2014: 761264 10.1155/2014/761264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mimica-Dukić N, Orčić D, Lesjak M, Šibul F. Essential oils as powerful antioxidants: misconception or scientific fact? Medicinal and aromatic crops: production, phytochemistry, and utilization ACS Symposium Series. 1218: American Chemical Society; 2016. p. 187–208. [Google Scholar]
- 41.Bezerra DP, Militao GCG, de Morais MC, de Sousa DP. The dual antioxidant/prooxidant effect of eugenol and its action in cancer development and treatment. Nutrients. 2017;9(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cronan JE, Thomas J. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 2009;459: 395–433. 10.1016/S0076-6879(09)04617-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broussard TC, Price AE, Laborde SM, Waldrop GL. Complex formation and regulation of Escherichia coli acetyl-CoA carboxylase. Biochemistry. 2013;52(19): 3346–3357. 10.1021/bi4000707 [DOI] [PubMed] [Google Scholar]
- 44.Emiola A, Andrews SS, Heller C, George J. Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc Natl Acad Sci USA. 2016;113(11): 3108–3113. 10.1073/pnas.1521168113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moynié L, Leckie SM, McMahon SA, Duthie FG, Koehnke A, Taylor JW, et al. Structural insights into the mechanism and inhibition of the β-hydroxydecanoyl-acyl carrier protein dehydratase from Pseudomonas aeruginosa. J Mol Biol. 2013;425(2): 365–377. 10.1016/j.jmb.2012.11.017 [DOI] [PubMed] [Google Scholar]
- 46.Wendel AA, Lewin TM, Coleman RA. Glycerol-3-phosphate acyltransferases: rate limiting enzymes of triacylglycerol biosynthesis. Biochim Biophys Acta. 2009;1791(6): 501–506. 10.1016/j.bbalip.2008.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao J, Rock CO. Phosphatidic acid synthesis in bacteria. Biochim Biophys Acta. 2013;1831(3): 495–502. 10.1016/j.bbalip.2012.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Birkner JP, Poolman B, Kocer A. Hydrophobic gating of mechanosensitive channel of large conductance evidenced by single-subunit resolution. Proc Natl Acad Sci USA. 2012;109(32): 12944–12949. 10.1073/pnas.1205270109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blount P, Iscla I, Moe PC, Li Y. MscL: The bacterial mechanosensitive channel of large conductance Curr Top Membr. 58: Academic Press; 2007. p. 201–33. [Google Scholar]
- 50.Lovering AL, Safadi SS, Strynadka NC. Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem. 2012;81: 451–478. 10.1146/annurev-biochem-061809-112742 [DOI] [PubMed] [Google Scholar]
- 51.Egan AJ, Jean NL, Koumoutsi A, Bougault CM, Biboy J, Sassine J, et al. Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B. Proc Natl Acad Sci USA. 2014;111(22): 8197–8202. 10.1073/pnas.1400376111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang G, Meredith TC, Kahne D. On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr Opin Microbiol. 2013;16(6): 779–785. 10.1016/j.mib.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenfeld Y, Shai Y. Lipopolysaccharide (endotoxin)-host defense antibacterial peptides interactions: role in bacterial resistance and prevention of sepsis. Biochim Biophys Acta. 2006;1758(9): 1513–1522. 10.1016/j.bbamem.2006.05.017 [DOI] [PubMed] [Google Scholar]
- 54.Breazeale SD, Ribeiro AA, Raetz CR. Origin of lipid A species modified with 4-amino-4-deoxy-L-arabinose in polymyxin-resistant mutants of Escherichia coli. An aminotransferase (ArnB) that generates UDP-4-deoxyl-L-arabinose. J Biol Chem. 2003;278(27): 24731–24739. 10.1074/jbc.M304043200 [DOI] [PubMed] [Google Scholar]
- 55.Lee CJ, Liang X, Gopalaswamy R, Najeeb J, Ark ED, Toone EJ, et al. Structural basis of the promiscuous inhibitor susceptibility of Escherichia coli LpxC. ACS Chem Biol. 2014;9(1): 237–246. 10.1021/cb400067g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barb AW, Zhou P. Mechanism and inhibition of LpxC: an essential zinc-dependent deacetylase of bacterial lipid A synthesis. Curr Pharm Biotechnol. 2008;9(1): 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Itri R, Junqueira HC, Mertins O, Baptista MS. Membrane changes under oxidative stress: the impact of oxidized lipids. Biophys Rev. 2014;6(1): 47–61. 10.1007/s12551-013-0128-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(ZIP)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.