Abstract
Palmitoylation, the posttranslational thioester linked modification of a 16-carbon saturated fatty acid onto the cysteine residue of a protein, has garnered considerable attention due to its implication in a multitude of disease states. The signature DHHC motif (Asp-His-His-Cys) identifies a family of Protein Acyltransferases (PATs) that catalyze the S-palmitoylation of target proteins via a two-step mechanism. In the first step, autopalmitoylation, palmitate is transferred from palmitoyl-CoA to the PAT, creating a palmitoyl:PAT intermediate and releasing reduced CoA. The palmitoyl moiety is then transferred to a protein substrate in the second step of the reaction. We have developed an in vitro, single-well, fluorescence-based enzyme assay that monitors the first step of the PAT reaction by coupling the production of reduced CoA to the reduction of NAD+ using the α-ketoglutarate dehydrogenase complex. This assay is suitable for determining PAT kinetic parameters, elucidating lipid donor specificity and measuring PAT inhibition by 2-bromopalmitate. Finally, it can be used for High Throughput Screening (HTS) campaigns for modulators of protein palmitoylation.
Keywords: Protein palmitoylation, High Throughput Screening, Autopalmitoylation, post-translational modification, zDHHC proteins
Introductory Statement
The covalent attachment of palmitate to proteins within eukaryote cells was first described over 30 years ago and has been shown to be involved in protein-membrane association, subcellular targeting of proteins between membrane organelles and within micro-domains of individual membrane compartments, protein stability, and protein-protein interactions [1-6]. A diverse and growing list of proteins that undergo palmitoylation include intrinsic and peripherally associated membrane proteins, as well as secreted proteins [7-13]. Although the number and types of proteins that are palmitoylated continually increases, no consensus sequences have been identified.
Deregulation of protein palmitoylation has been linked to a number of diseases. Examples include elevation of zDHHC9 expression levels in colorectal cancer [14] and leukemogenesis [15]; zDHHC3/GODZ is linked to cervical cancer [16]; zDHHC9 and zDHHC15 are associated with X-linked Intellectual Disability [17, 18]; zDHHC8 is a candidate gene connected to schizophrenia [19, 20]; and zDHHC2 mutations have been found in several colorectal cancers [21]. Despite the interest in identifying proteins that are palmitoylated and understanding the mechanism(s) that regulates the addition of palmitate to proteins and the consequences of the modifications, few assay systems have been devised to examine the kinetics of the acylation reaction. As a result, there is a lack of specific inhibitors of protein palmitoylation. Therefore, there is significant need for effective assays that can monitor the protein acyltransferase reaction. Not only will a strong assay give insight into the enzymology of protein palmitoylation, it will provide insights into the biochemical mechanism(s) of substrate recognition and the kinetics of the reaction.
Protein S-palmitoylation proceeds by a two-step mechanism. In the first step, autopalmitoylation, the palmitoyl moiety from palmitoyl-CoA is transferred to the zDHHC cysteine side chain, forming the palmitoyl:enzyme intermediate and producing reduced CoA (CoASH) (Fig. 1A). Once the intermediate is formed, one of two reactions can occur: a) in the presence of the protein substrate, the palmitoyl group of the intermediate can be transferred to a substrate cysteine or b) in the absence of protein substrate, the palmitoyl group of the intermediate can by hydrolyzed, yielding the enzyme and palmitate. The formation of the palmitoyl:enzyme intermediate has been monitored by SDS-PAGE and autoradiography using radiolabeled palmitoyl-CoA as the lipid donor [22-28]. However, there are several limitations to assays that rely on radiolabeled palmitate. First, this assay measures the steady-state amount of palmitoyl:PAT intermediate, not the total amount [25, 26]. Secondly, the use of a radioactive reagent complicates high throughput screening techniques. Finally, sensitivity is a key issue. At the lower limit of the assay's sensitivity, the reaction can accurately detect 5 fmols of transferred palmitate per min, which equates to a signal to noise ratio of approximately 1.5. The main disadvantages of the radioactive assay as it pertains to a high throughput format are the cost of the reagents and the amount of time required to perform each step.
Figure 1. Validation of an in vitro, single well, fluorescence-based coupled assay that measures protein acyl transferase catalyzed autopalmitoylation activity.
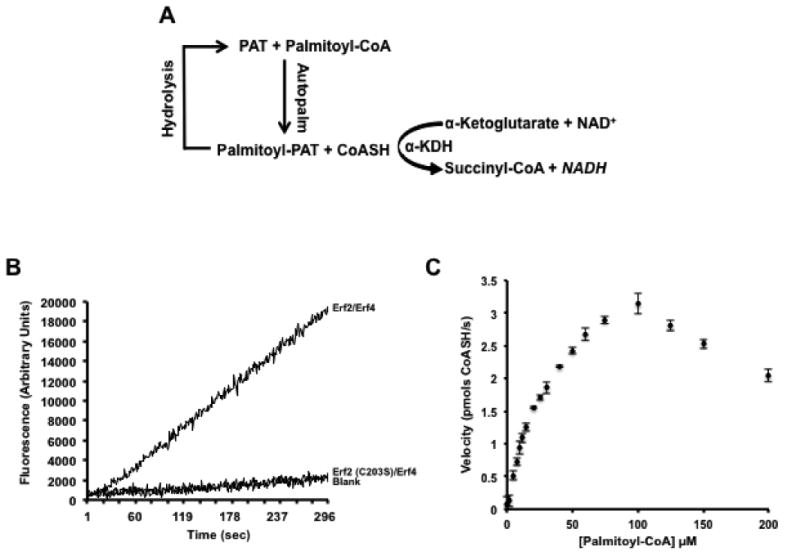
(A) Schematic of the coupled assay reaction. (B) Production of CoASH is dependent on active Erf2-Erf4 complexes. The reaction was performed as described in the Experimental Procedures using 2 μg wild type Erf2-Erf4 complexes, catalytically dead Erf2 (C203S)-Erf4 complexes or boiled Erf2-Erf4 complexes (Blank). Fluorescence was recorded continuously (ex. 340 nm/ em. 460 nm) for 5 min at 30°C. (C) Graphic representation of reaction rates using varying concentrations of palmitoyl-CoA substrate. Reactions were performed in quadruplicate and repeated three times (n=3) at 30°C with intermittent shaking. The fluorescence values (ex. 340 nm/ em. 460 nm) were recorded at 1 min intervals for 10 min. The data was analyzed and fitted using the Michaelis-Menten algorithm of Prism 6 (GraphPad Software, San Diego, CA). The data was corroborated using the Direct Linear Plot method [52, 53]. The amount of CoASH produced was based on a standard curve using known concentrations of CoASH and expressed as pmols/s +/- SD.
In this study, we describe and demonstrate the utility of a single well, fluorescence-based, coupled assay system that monitors the steady-state rate of autopalmitoylation of protein acyl-transferases (PAT) in vitro. This system measures the production of reduced CoA (CoASH), a product of the autopalmitoylation reaction. The CoASH is used in the conversion of α-ketoglutarate to succinyl-CoA with the corresponding reduction of NAD+ to the fluorescent NADH, a reaction that is catalyzed by α-ketoglutarate dehydrogenase complex (α-KDH) [29-33]. Here, we use this assay to demonstrate lipid substrate specificity on the autopalmitoylation activity of the Erf2-Erf4 complex [25, 26]. In addition, we show that the coupled assay can be applied to studying inhibition by 2-bromopalmitate (2-BP), a known inhibitor of protein palmitoylation [34]. Finally, we present evidence that this assay system can be used in a High Throughput Screening format in an attempt to identify modulators of PAT enzymes.
Materials and Methods
Strains, media, and yeast techniques
Yeast and bacteria growth media were prepared as described previously [35]. Cells were grown in synthetic complete (SC) medium or YPD (1% yeast extract, 2% peptone, and 2% glucose) medium [35, 36]. Induction of GAL1, 10 promoters were achieved by adding 4% galactose to SC medium. Yeast transformations were performed using the lithium acetate procedure [37]. The yeast strains used in this study, RJY1842 (MATa/α ade2-1/ade2-1 leu2-3,112/leu2-3,112 ura3-1/ura3-1 trp1-1/trp1-1 his3-11,15/his3-11,15 can1-100/can1-100 erf4Δ::NATr/erf4Δ::NATr GAL+) [25, 26] and RJY1330 (MATα leu2-3,112 ura3-52 ade2Δ ade8Δ lys2-801 ras1::HIS3 ras2::Ras2CS-Ext erf2::KANr <YCp52RAS2>) have been previously described [25, 26, 38].
Plasmid construction
The sequences of the deoxyoligonucleotide primers used to construct zDHHC9 alleles are available upon request. The DNA sequences of all constructs were verified by DNA sequencing (GeneWiz, Plainfield, NJ). The construction of B1302 and B1595 has been described previously [25, 36]. zDHHC9 (accession number: BC006200) and GCP16 (BC001227) were obtained from the Image consortium (Image numbers: 2964425 (zDHHC9) and 3456384 (GCP16)). The zDHHC9 open reading frame was amplified by PCR using primers that would encode a C-terminal 6xHIS epitope as well as an in-frame C-terminal FLAG epitope flanked by sequences homologous to MCS1 of B1192 (pESC-Trp (Stratagene, Santa Clara, CA)) to facilitate ligase independent cloning (LIC), producing B1500 (pESC-Trp zDHHC9). B1856 (pESC-Trp zDHHC9-GCP16) was constructed by inserting the PCR amplified GCP16 possessing the proper flanking sequences into MCS2 of B1500 by LIC.
Protein expression and purification
Erf2-Erf4 enzyme complexes were expressed from the pESC-Leu divergent GAL1,10 promoter in strain RJY1842, which also harbored a second plasmid to enhance galactose induction, pMA210 [39] ([36]. Cultures were grown in synthetic medium containing 2% glucose and lacking leucine and histidine. zDHHC9-GCP16 enzyme complexes were expressed from the pESC-Trp divergent GAL1,10 promoter in strain RJY1842 along with pMA210 and grown overnight at 30°C with shaking (230 RPM) in synthetic medium containing 2% glucose and lacking tryptophan and histidine. Approximately 3×107 cells were inoculated into 50 mls of synthetic medium containing 2% glucose and lacking the appropriate amino acids and the cultures were grown at 30°C with shaking (230 RPM) until the cell density was between 1.6×107 cells/ml (OD600 0.8) and 2.4×107 cells/ml (OD600 1.2). To induce the expression of the enzyme complexes, approximately 50 OD600 of cells (1×109) were seeded into 1 L of YEP containing 2% galactose and the culture was incubated at 30°C with shaking (230 RPM) for 18 h. The cells were harvested by centrifugation at 3000xg for 15 min. The resulting pellet was resuspended in breaking buffer (50 mM Tris-Cl pH 8, 500 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 1x protease inhibitor cocktail (PIC) (1 μM Leupeptin, 2 μM Pepstatin A, and 16 mM Benzamidine), and the cells were lysed using glass beads (400-600 mesh, Sigma-Aldrich, St. Louis, MO) for 40 min with 1 min pulses/1 min cooling (20 cycles) [25, 26, 36]. The resulting extract was spun at 3000xg for 15 min to remove cellular debris and unbroken cells, to yield a whole cell extract (WCE).
The enzyme containing particulate fraction (P19) was isolated from the WCE by centrifugation at 19,000xg for 30 min at 4°C. The soluble portion (S19) of the WCE was decanted and the particulate pellets were solubilized using 0.8% n-Dodecyl-β-D-maltoside (DDM) at 4°C for at least 5 h with gentle agitation [25, 26, 36]. Detergent insoluble particulates were removed from the P19 by centrifugation at 19,000xg for 30 min at 4°C. The soluble P19 samples were then moved to clean tubes. To aid in purification of the 6xHIS:Erf2-Erf4 complexes, urea and imidazole were added to a final concentration of 2 M and 1 mM, respectively (6xHIS:zDHHC9-GCP16 complexes were supplemented with only 1 mM imidazole). The resulting supernatant was incubated with Ni-NTA resin (Five-Prime, Gaithersburg, MD) at 4°C for 1h. The resin was washed once with Solution W (50 mM Tris•HCl, pH 8.5, 0.08% DDM, 5 mM β-ME) containing 300 mM NaCl, and then twice with Solution W containing 150 mM NaCl. The protein was eluted with 50 mM Tris HCl, pH8.5, 150 mM NaCl, 0.08% DDM, 5% glycerol and 250 mM imidazole. Eluates were desalted and the buffer changed to SPB (50 mM Sodium Phosphate Buffer, pH6.8, 10% glycerol) using a column of G-25 resin. Fractions containing 6xHIS:Erf2-Erf4 complexes (or 6xHIS:zDHHC9-GCP16 complexes) were pooled to obtain approximately 0.5 mg of purified Ras PAT per liter of culture. These samples were flash frozen with liquid nitrogen and stored at -80°C for up to 3 years [25].
Coupled Protein Acyltransferase (PAT) Assay
The production of NADH was monitored in 96-well format with a Biotek Mx fluorimeter (Biotek, Winooski, VT) using an excitation of 340 nm and emission of 465 nm [25, 29-31]. The 200 μl reaction contained 2 mM 2-oxoglutarate (α-ketoglutamic acid isolated from pig heart), 0.25 mM NAD+, 0.2 mM thiamine pyrophosphate, 2 μg of purified PAT complex, 1 mM EDTA, 1 mM dithiothreitol, and 32 mU 2-oxoglutarate dehydrogenase (α-ketoglutarate dehydrogenase, Sigma-Aldrich, St. Louis, MO) in 50 mM sodium phosphate buffer, pH6.8. The reaction was initiated by the addition of varying concentrations of palmitoyl-CoA and monitored for 30 min at 30°C. The first 10 min of the reaction was analyzed to determine the initial rates of CoASH release. The PAT specific activity was determined from a standard curve of fluorescence values generated under reaction conditions using different CoASH concentrations. In these reactions, CoASH was added to the standard PAT reaction mixture (without PAT complex) and the reaction was allowed to proceed to equilibrium before fluorescence was measured. To further validate the assay, two types of controls were performed. For the first control, the PAT enzyme was boiled to inactivate the autopalmitoylation activity, and therefore the PAT-dependent formation of NADH, to demonstrate that there is no measurable background hydrolysis of palmitoyl-CoA in the reaction. Boiled enzyme produced significantly less fluorescence (<20%) suggesting no significant contribution from sample heterogeneity or nonspecific fluorescence. Secondly, a catalytically inactive mutant form of the PAT complex (Erf2 (C203S)-Erf4 or zDHHC9 (C169A)-GCP16) was used to determine the baseline activity of the reaction and to show no significant contribution of contaminating activities to the assay results. For reactions that incorporated 2-BP, Erf2-Erf4 complexes were incubated with different concentrations of the inhibitor, 2-BP (dissolved in DMSO), for 20 min at room temperature prior to incorporation into the assay. Assay conditions permitted as much as 6% DMSO without affecting Erf2-Erf4 autopalmitoylation activity (Supplemental Figure S1).
The rate of the coupled reaction (α-ketoglutarate dehydrogenase) must have a first-order dependence on the concentration of CoASH, a product of autopalmitoylation. However, this assumes a VMAX rate of the autopalmitoylation reaction (the first reaction) and therefore the concentration of palmitoyl-CoA would need to be at saturation conditions. This is not realistic for this reaction given the interference in the activity observed at higher concentrations of palmitoyl-CoA (Figure 1C). Instead, for measurements of the rate of the coupled reaction (at sub-VMAX concentrations of CoASH) to provide accurate information concerning the initial rate of autopalmitoylation, a steady-state concentration of CoASH must be reached before the rate of autopalmitoylation diminishes noticeably from its initial value. Using the method of Storer and Cornish-Bowden [40], we calculated the time needed to reach a steady-state concentration of CoASH (334pM) as approximately 2 seconds, where the rate of autopalmitoylation is about 3% of the limiting rate of the coupled reaction and where the ratio of the CoASH dependent rate of the coupled reaction to the rate of autopalmitoylation is 0.99. Therefore, in practice, there is no appreciable time lag while the system reaches equilibrium. Therefore, the accumulation of NADH adequately represents the rate of autopalmitoylation.
Radioactivity-based 2-BP labeling assay
Covalent modification of Erf2-Erf4 by 3H-2-BP was performed by combining 8 μg (approximately 29 pmols of Erf2-Erf4 per time point) of native Erf2-Erf4 complex (or boiled Erf2-Erf4) and 100 μM 2-BP (specific activity, 0.4 Ci/mmol) [9,10-3H] 2-BP (American Radiolabeled Chemicals, St. Louis, MO) in 200 μL of 50 mM sodium phosphate buffer, pH 7.4 and 0.5 mM dithiothreitol. The reaction was incubated at 30°C and 50 μL samples removed to tubes containing 12 μL of 5x reducing protein loading buffer to stop the reactions at the indicated times. Samples were boiled for 3 min, centrifuged for 3 min at 13,000xg to removed insoluble particulates and separated by SDS-PAGE (10%). To visualize the radioactivity, after separation, the gel was soaked in En3Hance (Perkin-Elmer, Waltham, MA), as per the manufacturer's instructions and dried under a vacuum. Typical film exposure times ranged from 2-4 days. To determine the number of dpm comprising each labeled Erf2 band, the bands corresponding to Erf2 were excised from the dried gel using the exposed film as a template and soaked in 500 μL Soluene S-350 (Perkin-Elmer, Waltham, MA) and 81 μL of a 50% (w/v) solution of hydroxylamine (1 M; final) (Sigma/Aldrich, St. Louis, MO) for 18 h at 37°C in scintillation vials to dissolve the acrylamide and release the radioactivity. Scintillation cocktail (5 ml) was added directly to the Soluene S-350 dissolved samples in the vials. The samples were allowed to equilibrate for 2 h and the number of cpm determined by scintillation counting (Beckman-Coulter, LS6500, Indianapolis, IN). The counting efficiency of the tritium was determined by adding a known amount of tritium standard (dpm) (American Radio Chemicals, St. Louis, MO) to the vials containing samples and the vials recounted. Counting efficiency was calculated as the difference between the cpm from the combination of standard plus sample and the sample alone divided by the amount of dpm of the standard (approximately 90%). To determine the efficiency with which the tritium was released from the acrylamide, native Erf2-(FLAG)Erf4 bound with anti-FLAG conjugated to agarose beads (Sigma-Aldrich, St. Louis, MO) was incubated with 3H-2-BP for 30 min at 30°C. The free 3H-2-BP label was washed away extensively (5x 1 ml) with buffer. The Erf2-(FLAG)Erf4 complexes were removed from the beads with 0.1 M glycine, transferred to a fresh tube and neutralized with 1 M Tris·Cl, pH 7.5. One fifth of the sample was submitted for scintillation counting (with 500 μL Soluene S-350 added) and the remainder of the sample was separated by SDS-PAGE (10%). The radioactivity was isolated and released under the same conditions as the time point samples. Comparison of the dpm obtained directly from the control complexes and those obtained from the gel slices showed the procedure to be approximately 35% efficient.
Results and Discussion
Development of an in vitro fluorescence-based coupled enzymatic assay to monitor autopalmitoylation
The first step in the palmitoylation reaction, autopalmitoylation, can be monitored by the production of CoASH. The amount of CoASH produced in turn can be coupled to the reduction of NAD+ to NADH by α-ketoglutarate dehydrogenase (Fig. 1A). NADH has fluorescence excitation and emission wavelengths of 340 nm and 465 nm, respectively [29]. The α-KDH reaction is essentially irreversible (-ΔGo (pH 7.0) of approximately 10 kcal/mole) and has a Km value for CoASH of less the 0.1 μM [30]. When NAD+ and α-ketoglutarate are present in molar excess compared to CoASH, the reaction proceeds to completion with approximately zero-order kinetics. The source of α-ketoglutarate dehydrogenase is pig heart, which can be inhibited to a slight degree by NADH and succinyl-CoA. This inhibition is abolished if the NAD+ concentration is in great excess relative to the concentration of CoASH being assayed [31].
As proof of principle, we performed a pilot study measuring yeast Erf2-Erf4 autopalmitoylation activity. The production of NADH was monitored fluorometrically in a single cuvette using a PTI Double-Double QuantaMaster 3 Fluorometer (340 nm excitation/ 465 nm emission). Although 2 ug of PAT enzyme are used in the reaction, increases in NADH fluorescence can be detected using PAT enzyme amounts as low as 0.5 ug (data not shown). The reaction proceeded linearly throughout the 5 min monitoring phase (Fig. 1B) and maintained its linearity for more than 25 min (data not shown). Denaturing Erf2-Erf4 by boiling abolishes the formation of NADH demonstrating that there is no measurable background hydrolysis of palmitoyl-CoA (Fig. 1B). In addition, an inactive mutant of Erf2, Erf2 C203S, that was purified in parallel with wild type Erf2-Erf4, does not produce NADH above that of the boiled wild type Erf2, demonstrating the increase in fluorescence is not due to a contaminating moiety from the enzyme preparation protocol. These controls demonstrate that even though there is a small amount of drift of the fluorescence signal, the signal is specific for active, wild type Erf2-Erf4. Similar results were obtained using the mammalian ortholog of Erf2-Erf4, zDHHC9-GCP16 (data not shown). A standard curve of known amounts of exogenously added CoASH, ranging from 0-3000 pmol CoASH, was used to quantify the amount of steady-state autopalmitoylation, as the formation of the intermediate and production of CoASH are in a 1:1 stoichiometry.
Determination of the kinetic parameters for Erf2-Erf4 autopalmitoylation using a 96-well format
The coupled fluorescence assay was developed for a 96-well plate format, thus allowing a number of samples to be run simultaneously. As an initial evaluation of the assay, varying concentrations of palmitoyl-CoA were used to assess the activity of the enzyme over a 1 h period (Fig. 1C). The palmitoyl-CoA concentrations varied from 0-200 μM. Plotting the initial change in fluorescence versus the concentration of palmitoyl-CoA, we observed a curve that approached saturation until approximately 100 μM of palmitoyl-CoA, and then showed an apparent decrease in the activity for the concentrations above 100 μM. The observed decrease could be a result of inhibition of α-KDH by high concentrations of palmitoyl-CoA [31], or more likely the result of formation of palmitoyl-CoA micelles reducing the effective concentration of palmitoyl-CoA. The approximate critical micelle concentration for palmitoyl-CoA is in the range of 80-100 μM. Accordingly, we limited the palmitoyl-CoA concentrations to below 100 μM and determined the VMAX, 43 pmols CoASH produced/min/μg, and KM of the reaction to be 43 μM for palmitoyl-CoA, which agreed with the values we calculated using a single cuvette reaction in a PTI Double-Double QuantaMaster 3 Fluorometer.
The effect of detergents on the autopalmitoylation reaction
All zDHHC protein acyltransferases so far identified are integral membrane proteins with four or more transmembrane domains [41-44]. Therefore, isolation and characterization of zDHHC PATS requires detergent solubilization. To determine the effect of different detergents on the autopalmitoylation reaction, we screened for changes in autopalmitoylation activity using a 72 detergent collection (Hampton Research, Aliso Viejo, CA). The detergent was added to the reaction prior to the Erf2-Erf4 enzyme at a final concentration equal to the critical micelle concentration (CMC) for the respective detergent. The ratio of autopalmitoylation in the presence of detergent to the autopalmitoylation in the “no detergent added” control for each detergent is shown in TABLE 1. Over 50% of the detergents examined provided at least 70% of the autopalmitoylation activity when compared to the control. Although there was no noticeable trend amongst the non-ionic and zwitter-ionic detergents effect on autopalmitoylation, ionic detergents had the most consistently detrimental effect on activity.
Table 1.
Effect of detergents on the enzymatic autopalmitoylation activity of the Erf2-Erf4 Complex. NA (Not Applicable), NI (Non-Ionic), Z (Zwitter-Ionic) and I (Ionic).
| Detergent | Detergent Type | Percent of Control |
|---|---|---|
| No Detergent | NA | 100 |
| Anapoe® 35 | NI | 96 |
| n-Octyl-β-D-thioglucoside | NI | 90 |
| IPTG | NI | 87 |
| ZWITTERGENT® 3-12 | Z | 87 |
| C12E9 | NI | 86 |
| DDMAB | Z | 85 |
| LDAO | NI | 85 |
| C12E8 | NI | 85 |
| CYMAL® -1 | NI | 84 |
| CYPFOS-3 | NI | 83 |
| MEGA® -9 | NI | 83 |
| ZWITTERGENT® 3-14 | Z | 82 |
| n-Tetradecyl-β-D-maltoside | NI | 82 |
| n-Dodecyl-β-D-maltoside | NI | 82 |
| FOS-Choline® -12 | Z | 82 |
| 1-s-Nonyl-β-D-thioglucoside | NI | 81 |
| n-Undecyl-β-D-maltoside | NI | 80 |
| n-Dodecyl-N,N-dimethylglycine | Z | 80 |
| Pluronic® F-68 | NI | 79 |
| DDAO | NI | 79 |
| Sucrose monolaurate | NI | 79 |
| n-Tridecyl-β-D-maltoside | NI | 78 |
| CYMAL®–6 | NI | 76 |
| n-Nonyl-β-D-maltoside | NI | 75 |
| ZWITTERGENT® 3-08 | Z | 75 |
| n-Decyl-β-D-maltoside | NI | 75 |
| TRITON® X–100 | NI | 75 |
| FOS-Choline® -10 | Z | 75 |
| FOS-Choline® -9 | Z | 74 |
| n-Hexadecyl-β-D-maltoside | NI | 74 |
| n-Decyl-β-D-thiomaltoside | NI | 74 |
| n-Nonyl-β-D-maltoside | NI | 74 |
| n-Decanoylsucrose | NI | 72 |
| CYMAL®–5 | NI | 72 |
| n-Dodecyl-β-D-maltotrioside | NI | 72 |
| HEGA-10 | NI | 71 |
| n-Nonyl-β-D-glucoside | NI | 66 |
| Big CHAP, Deoxy | NI | 64 |
| CYMAL® -4 | NI | 63 |
| n-Octyl-β-D-thiomaltoside | NI | 62 |
| n-Octanoylsucrose | NI | 62 |
| n-Octyl-β-D-glucoside | NI | 61 |
| MEGA®8 | NI | 60 |
| HECAMEG | NI | 60 |
| Anapoe® X-405 | NI | 58 |
| Anapoe® X-305 | NI | 58 |
| CYMAL® -2 | NI | 57 |
| C8E5 | NI | 57 |
| 1-s-Heptyl-β-D-thioglucoside | NI | 54 |
| ZWITTERGENT®3-10 | Z | 52 |
| n-Heptyl-β-D-thioglucopyranoside | NI | 51 |
| Anapoe® 20 | NI | 50 |
| Thesit® | NI | 50 |
| OPOE | NI | 47 |
| CHAPS | Z | 46 |
| Anapoe® 80 | NI | 44 |
| CYMAL®3 | NI | 44 |
| CHAPSO | Z | 43 |
| C-HEGA-11 | NI | 39 |
| FOS-Choline® -8 | Z | 39 |
| Anapoe® 58 | NI | 37 |
| n-Hexyl-β-D-glucopyranoside | NI | 36 |
| Anapoe® X-114 | NI | 34 |
| Anapoe® C10E9 | NI | 31 |
| Anapoe® C13E8 | NI | 31 |
| Anapoe® C10E6 | NI | 31 |
| HEGA-9 | NI | 30 |
| HEGA-8 | NI | 25 |
| C-HEGA®10 | NI | 24 |
| CTAB | I | 23 |
| C-HEGA-9 | NI | 20 |
| Anapoe® C12E10 | NI | 14 |
| BAM | I | 0 |
| No Enzyme | NA | 0 |
The effect of acyl-CoA chain length on autoacylation activity
The availability of acyl-CoAs of varying chain lengths and the non-radioactive nature of the coupled assay allowed us to test directly the specificity of the chain length of the acyl donor for Erf2-Erf4. We determined the VMAX and KM kinetic parameters for acyl-CoAs of carbon lengths between 8 and 20 (C8:0 – C20:0), as well as palmitoleoyl CoA (C16:1 n7), cis-vaccenoyl CoA (C18:1 n7) and oleoyl CoA (C18:1 n9). The mean KM values ranged between 9 and 90 μM for all the hydrocarbon chains examined (Fig. 2; inset). We also observed a similar phenomenon as for palmitoyl-CoA, where the velocity of the reaction decreased at higher concentrations of substrate, suggesting that perhaps, at higher concentrations of the longer chain lengths, the hydrocarbon was not as soluble. However, by lowering the concentrations of the acyl-CoA substrates, we were able to overcome the insolubility issues and obtain highly reproducible data to calculate the parameters.
Figure 2. The effect of acyl CoA substrate specificity on Erf2-Erf4 autoacylation activity.
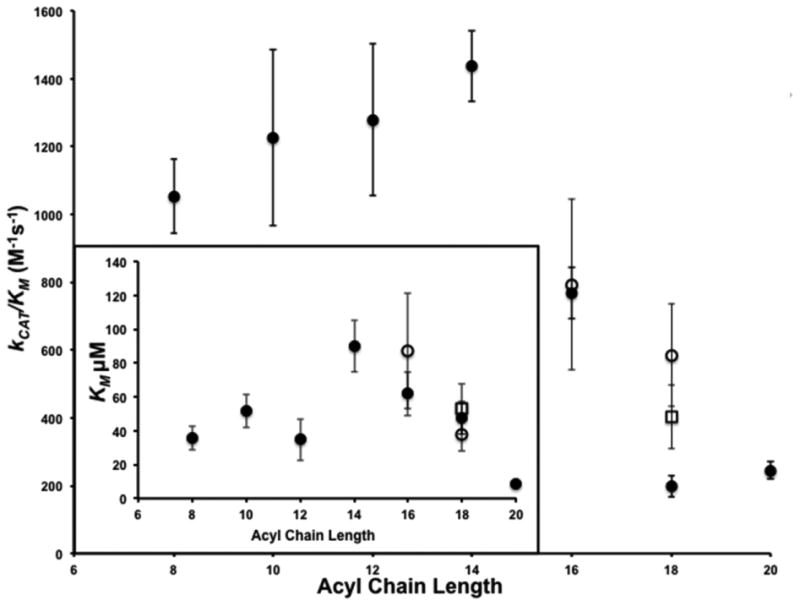
Graphical representation of specificity constants (kCAT/KM) for acyl CoAs of varying saturated hydrocarbon chain lengths (C8:0-C20:0; closed circles), unsaturated lengths (palmitoleoyl CoA (C16:1 n7), cis-vaccenoyl CoA (C18:1 n7) (open circles) and oleoyl CoA (C18:1 n9) open square) for the Erf2-Erf4 steady-state autoacylation activity. The assays were performed in triplicate and repeated three times (n=3). (Inset) Graphical representation of the KM values used to calculate the kCAT/KM values (above). The saturated acyl hydrocarbon chains are denoted by closed circles, palmitoleoyl and cis-vaccenoyl chains are denoted by open circles and the oleoyl chain denoted by an open square.
We used the specificity constant (kCAT/KM) to compare the relative rates of reaction of each of the substrates and plotted these values as a function of acyl chain length (Fig. 2). Acyl chain lengths between 8 and 14 carbons produced the greatest activity followed by acyl chain lengths of 16 carbons. Saturated chain lengths of 18 and 20 carbons produced the lowest activities. De-saturating the 18 carbon chain length (cis-vaccenoyl-CoA) increased the activity between 2- and 3-fold. These data demonstrate that protein acyltransferases may be able to utilize acyl-CoAs of various chain lengths with varying degrees of saturation as substrates for acylation and could serve as a possible explanation for the heterogeneity observed with acyl modifications to proteins [27, 45].
2-bromopalmitate covalently modifies and inhibits the autopalmitoylation activity of Erf2-Erf4
2-Bromopalmitate has been used as a lipid metabolism inhibitor for over 50 years [46, 47]. Initially, we observed that the Erf2-Erf4 activity inhibition by 2-BP could not be diluted or removed from the reaction (data not shown) suggesting the inhibition of palmitoylation was irreversible. In addition, 2-BP covalently modifies native Erf2 at a rate of approximately 0.1 pmol/min (Fig. 3A; squares) as measured by incorporation of 3H-2-BP into Erf2-Erf4 complexes (Fig. 3A, lower panel). On a molar ratio basis, 2-BP was in 172-fold excess compared to the amount of Erf2-Erf4 complexes. A calculation of the amount of Erf2-Erf4 complexes labeled reveals 5 pmols compared to the 5 nmols of 2-BP added to the reaction. This results in one molecule of 2-BP reacting with an Erf2-Erf4 complex for every 1000 molecules of 2-BP. As bromide at the 2 position is a good leaving group and is most likely the site of covalent bond formation between Erf2 and 2-BP, the reaction product is not hydroxylamine labile (data not shown). Interestingly, the incorporation of the tritium signal is not detectable if the Erf2-Erf4 complex is denatured before addition of 3H-2-BP (Fig. 3A; diamonds) implying the native structure of the Erf2-Erf4 complex is required for labeling by 2-BP.
Figure 3. Covalent modification and inhibition of Erf2-Erf4 autopalmitoylation activity by 2-Bromopalmitate.
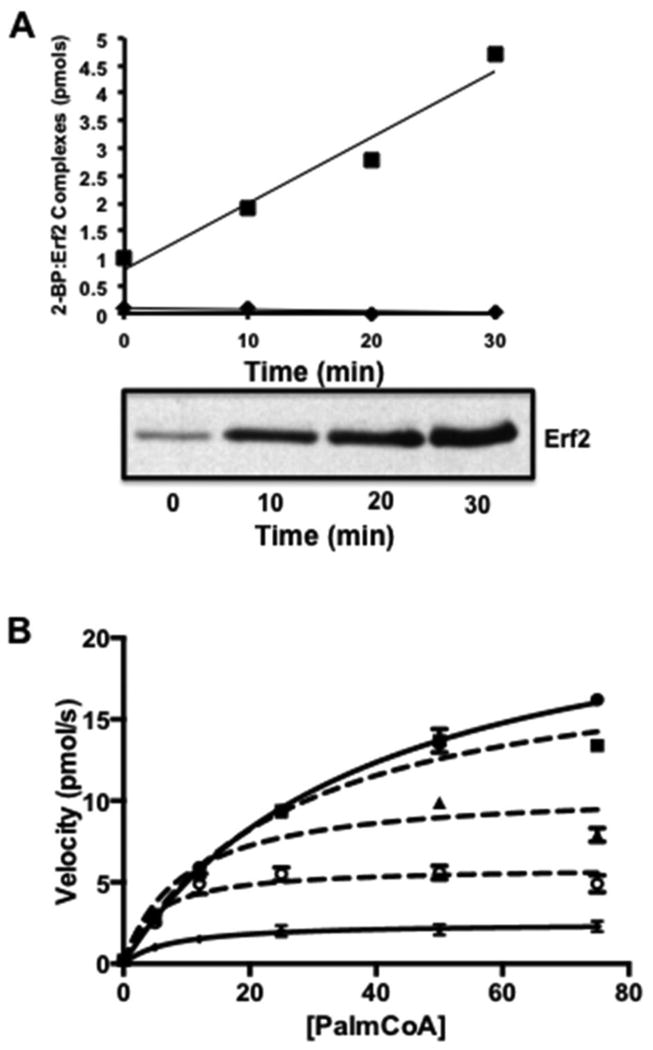
(A) Native Erf2-Erf4 complexes and boiled Erf2-Erf4 were incubated with 3H-2-BP for the times shown. The products of the reactions were separated by SDS-PAGE and subjected to autoradiography (bottom panel). The intensities of the signals corresponding to palmitoyl-Erf2 were determined by cutting the bands from the gels, dissolving the gel slices in Soluene S-350 to release the tritium and the amount of radioactivity determined by scintillation counting [22]. Tritium-based counts were converted to pmols and plotted versus time (top panel). Wild type Erf2-Erf4 complexes are denoted by closed circles and boiled Erf2-Erf4 denoted by closed diamonds. (B) Reaction rates of Erf2-Erf4 autopalmitoylation activity with increasing concentrations of palmitoyl-CoA in the presence of 2-BP. Samples include Erf2-Erf4 (closed circles), Erf2-Erf4 plus 50 μM 2-BP (closed squares), Erf2-Erf4 plus 100 μM 2-BP (closed triangles), Erf2-Erf4 plus 150 μM 2-BP (open circles) and Erf2 (C203S)-Erf4 (Xs). Reactions were performed in triplicate (n=3) at 30°C. The fluorescence values (ex. 340 nm/ em. 460 nm) were recorded at 1 min intervals for 10 min. The data was analyzed and fitted using Prism 6 (Michaelis-Menten algorithm) (GraphPad Software, San Diego, CA). The values for VMAX and KM were corroborated using the Direct Linear Plot method of Cornish-Bowden [52, 53].
The effect of 2-BP on autopalmitoylation was examined using the coupled assay. The initial rate of autopalmitoylation with increasing concentrations of palmitoyl-CoA was measured under conditions where the Erf2-Erf4 enzyme complex had been pre-incubated with 2-BP. When no 2-BP was present, a curve of velocity versus palmitoyl-CoA concentration was observed similar to previous curves for the wild type enzyme. However, when 2-BP is pre-incubated with the enzyme, the velocities and the VMAX are decreased in a dose dependent manner. Three concentrations of 2-BP (50 μM, 100 μM and 150 μM) were compared, resulting in increasing levels of inhibition with an IC50 value of approximately 100 μM. Intriguingly, the amount of inhibition due to 2-BP was never as much as observed for the catalytically inactive Erf2 (C203S)-Erf4 mutant (approximately 80% of the catalytically dead enzyme control). Although it was previously reported that the Ki for 2-BP was 10 μM [24], a concentration of at least 100 μM is required to reduce the Vmax of Erf2-Erf4 complex by 50% in this assay. The difference may be due to 2-BP's effect on other steps required for palmitoylation in addition to covalent modification of the enzyme [48].
In the past, the method of choice was radiolabeled palmitate molecules, which had its drawbacks. First, steady state levels of the palmitoylated species can be monitored, but the turnover of the enzyme cannot be easily quantified. Second, visualization can require an inordinate amount of time. Finally, the radiolabel is cost prohibitive. The price of 3H-palmitoyl-CoA is approximately $33-50 per assay. For a library the size of 40,000 compounds, the cost of the 3H-palmitoyl-CoA from commercial sources is therefore prohibitive (>$1M). The cost can be reduced by synthesizing 3H-palmitoyl-CoA from 3H-palmitate, CoASH and acyl CoA synthetase (data not shown), lowering the cost to approximately $0.30 per assay ($12,000 for a 40,000 compound screen), but detecting the radioactive signal is still problematic. By comparison, the cost of performing the single well, fluorescence-based coupled assay is approximately $0.07 per assay ($2800 for a 40,000 compound library). Included in this amount is the cost of expressing and purifying the PAT enzyme from the galactose-inducible S. cerevisiae over-production system.
Protein palmitoylation plays a role in a variety of cellular processes involved in cancer, cardiovascular disease, infectious disease, and neurological disorders [49] and is required for membrane association and neoplasm promoting activity of H- and N-Ras [50]. Palmitoylation has also been shown to play an important role in other cancer targets. For example, estrogen signaling in breast cancer involves palmitoylation [51]. Despite the central role of palmitoylation in the subcellular localization and function of numerous signaling proteins involved in cancer and several neurological disorders, no specific, high affinity, palmitoylation inhibitors exist and a detailed understanding of the relationship between zDHHC enzyme and substrates is lacking. Therefore, a potential application of this technology is to use high throughput screening procedures to identify inhibitors of protein palmitoylation. The autopalmitoylation/coupled assay is reproducible and robust (TABLE 2). For the Erf2-Erf4 complex, the assay has a Z′-value of 0.87, a Coefficient of Variance of 6.2 and a signal-to-noise ratio of 33. In addition, the enzyme complex can be isolated in relatively large quantities from yeast and is stable for greater than 24 h at 4°C. The activity of the human ortholog of the Erf2-Erf4 complex, zDHHC9-GCP16, is similarly robust and reproducible with a Z′-value of 0.77, a Coefficient of Variance of 2.6 and a signal-to-noise ratio of 21.
Table 2. Kinetic and High Throughput Screening Parameters for zDHHC9-GCP16 and Erf2-Erf4 Complexes.
| Parameter | zDHHC9-GCP16 | Erf2-Erf4 |
|---|---|---|
| Autopalmitoylation Activity (pmol/min/μg) | 47.0 +/- 2.5 | 43.0 +/- 3.0 |
| KM (μM) | 45 +/- 4 | 43 +/- 8 |
| Yield (mg/L) | 1-3 | 2-4 |
| Z′-Valuea | 0.77 | 0.87 |
| Coefficient of Variancea | 2.6 | 6.2 |
| Signal-to-Noise Ratioa | 21 | 33 |
| Stability (4°C) | >24H | >24H |
Kinetic and screening parameters: zDHHC9-GCP16 and Erf2-Erf4
Values were determined on 3 successive days using the National Center for Advancing Translational Sciences High Throughput Screening Criteria.
In the present study, we have developed a fluorescence-based assay that can measure the turnover activity of the protein acyltransferase and to determine kinetic parameters. We have demonstrated the usefulness of this assay to measure the autoacylation activity of Erf2-Erf4 using several acyl-CoAs of varying chain lengths. Finally, we have shown that this assay can be used to examine protein acyl transferase inhibition in a manner that is reproducible and robust, and therefore applicable to HTS applications.
Supplementary Material
Figure S1 Erf2-Erf4 autopalmitoylation activity tolerates relatively high concentrations of dimethylsulfoxide (DMSO). Erf2-Erf4 catalyzed autopalmitoylation reactions containing varying percentages of DMSO were performed as described in Materials and Methods using 2 μg wild type Erf2-Erf4 complex. The reaction proceeded for 5 min at 30°C while continuously monitoring the fluorescence intensity (ex. 340 nm/ em. 460 nm). The reactions were performed eight times (n=8) and are expressed as the mean +/- SD. The P-values are given on the figure. The P-value for 0 and 6 percent DMSO is 0.7.
Footnotes
This work was supported in part by National Institutes of Health Grants GM073977 and CA050211 (to RJD) and the Fred Wright Cancer Endowment. We would like to thank Krishna Reddy, Jeremy Baker and Laura Pendleton for critical reading of the manuscript and many helpful and insightful discussions.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Milligan G, Parenti M, Magee AI. The dynamic role of palmitoylation in signal transduction. Trends in biochemical sciences. 1995;20:181–187. doi: 10.1016/s0968-0004(00)89004-0. [DOI] [PubMed] [Google Scholar]
- 2.Dunphy JT, Linder ME. Signalling functions of protein palmitoylation. Biochimica et biophysica acta. 1998;1436:245–261. doi: 10.1016/s0005-2760(98)00130-1. [DOI] [PubMed] [Google Scholar]
- 3.Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochimica et biophysica acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 4.El-Husseini Ael D, Bredt DS. Protein palmitoylation: a regulator of neuronal development and function. Nature reviews. Neuroscience. 2002;3:791–802. doi: 10.1038/nrn940. [DOI] [PubMed] [Google Scholar]
- 5.Silvius JR. Mechanisms of Ras protein targeting in mammalian cells. J Membr Biol. 2002;190:83–92. doi: 10.1007/s00232-002-1026-4. [DOI] [PubMed] [Google Scholar]
- 6.Bijlmakers MJ, Marsh M. The on-off story of protein palmitoylation. Trends Cell Biol. 2003;13:32–42. doi: 10.1016/s0962-8924(02)00008-9. [DOI] [PubMed] [Google Scholar]
- 7.Zevian S, Winterwood NE, Stipp CS. Structure-function analysis of tetraspanin CD151 reveals distinct requirements for tumor cell behaviors mediated by alpha3beta1 versus alpha6beta4 integrin. J Biol Chem. 2011;286:7496–7506. doi: 10.1074/jbc.M110.173583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delandre C, Penabaz TR, Passarelli AL, Chapes SK, Clem RJ. Mutation of juxtamembrane cysteines in the tetraspanin CD81 affects palmitoylation and alters interaction with other proteins at the cell surface. Experimental cell research. 2009;315:1953–1963. doi: 10.1016/j.yexcr.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou B, Liu L, Reddivari M, Zhang XA. The palmitoylation of metastasis suppressor KAI1/CD82 is important for its motility- and invasiveness-inhibitory activity. Cancer research. 2004;64:7455–7463. doi: 10.1158/0008-5472.CAN-04-1574. [DOI] [PubMed] [Google Scholar]
- 10.Scheffer KD, Gawlitza A, Spoden GA, Zhang XA, Lambert C, Berditchevski F, Florin L. Tetraspanin CD151 mediates papillomavirus type 16 endocytosis. Journal of virology. 2013;87:3435–3446. doi: 10.1128/JVI.02906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma C, Rabinovitz I, Hemler ME. Palmitoylation by DHHC3 is critical for the function, expression, and stability of integrin alpha6beta4. Cellular and molecular life sciences : CMLS. 2012;69:2233–2244. doi: 10.1007/s00018-012-0924-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagnoux-Palacios L, Dans M, van't Hof W, Mariotti A, Pepe A, Meneguzzi G, Resh MD, Giancotti FG. Compartmentalization of integrin alpha6beta4 signaling in lipid rafts. J Cell Biol. 2003;162:1189–1196. doi: 10.1083/jcb.200305006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Kovalenko OV, Tang W, Claas C, Stipp CS, Hemler ME. Palmitoylation supports assembly and function of integrin-tetraspanin complexes. J Cell Biol. 2004;167:1231–1240. doi: 10.1083/jcb.200404100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansilla F, Birkenkamp-Demtroder K, Kruhoffer M, Sorensen FB, Andersen CL, Laiho P, Aaltonen LA, Verspaget HW, Orntoft TF. Differential expression of DHHC9 in microsatellite stable and instable human colorectal cancer subgroups. British journal of cancer. 2007;96:1896–1903. doi: 10.1038/sj.bjc.6603818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuiffo B, Ren R. Palmitoylation of oncogenic NRAS is essential for leukemogenesis. Blood. 2010:3598–3605. doi: 10.1182/blood-2009-03-213876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cha SH, Kim C, Choi BK, Kim HJ, Baek SY. C-arm assessment of cervical pedicle screw: screw coaxial fluoroscopy and oblique view. Spine (Phila Pa 1976) 2007;32:1721–1727. doi: 10.1097/BRS.0b013e3180b9f29b. [DOI] [PubMed] [Google Scholar]
- 17.Raymond FL, Tarpey PS, Edkins S, Tofts C, O'Meara S, Teague J, Butler A, Stevens C, Barthorpe S, Buck G, Cole J, Dicks E, Gray K, Halliday K, Hills K, Hinton J, Jones D, Menzies A, Perry J, Raine K, Shepherd R, Small A, Varian J, Widaa S, Mallya U, Moon J, Luo Y, Shaw M, Boyle J, Kerr B, Turner G, Quarrell O, Cole T, Easton DF, Wooster R, Bobrow M, Schwartz CE, Gecz J, Stratton MR, Futreal PA. Mutations in ZDHHC9, which encodes a palmitoyltransferase of NRAS and HRAS, cause X-linked mental retardation associated with a Marfanoid habitus. Am J Hum Genet. 2007;80:982–987. doi: 10.1086/513609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mansouri MR, Marklund L, Gustavsson P, Davey E, Carlsson B, Larsson C, White I, Gustavson KH, Dahl N. Loss of ZDHHC15 expression in a woman with a balanced translocation t(X;15)(q13.3;cen) and severe mental retardation. Eur J Hum Genet. 2005;13:970–977. doi: 10.1038/sj.ejhg.5201445. [DOI] [PubMed] [Google Scholar]
- 19.Chen WY, Shi YY, Zheng YL, Zhao XZ, Zhang GJ, Chen SQ, Yang PD, He L. Case-control study and transmission disequilibrium test provide consistent evidence for association between schizophrenia and genetic variation in the 22q11 gene ZDHHC8. Human molecular genetics. 2004;13:2991–2995. doi: 10.1093/hmg/ddh322. [DOI] [PubMed] [Google Scholar]
- 20.Mukai J, Liu H, Burt RA, Swor DE, Lai WS, Karayiorgou M, Gogos JA. Evidence that the gene encoding ZDHHC8 contributes to the risk of schizophrenia. Nat Genet. 2004;36:725–731. doi: 10.1038/ng1375. [DOI] [PubMed] [Google Scholar]
- 21.Oyama T, Miyoshi Y, Koyama K, Nakagawa H, Yamori T, Ito T, Matsuda H, Arakawa H, Nakamura Y. Isolation of a novel gene on 8p21.3-22 whose expression is reduced significantly in human colorectal cancers with liver metastasis. Genes Chromosomes Cancer. 2000;29:9–15. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1001>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 22.Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- 23.Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 24.Jennings BC, Nadolski MJ, Ling Y, Baker MB, Harrison ML, Deschenes RJ, Linder ME. 2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. Journal of lipid research. 2009;50:233–242. doi: 10.1194/jlr.M800270-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell DA, Mitchell G, Ling Y, Budde C, Deschenes RJ. Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J Biol Chem. 2010;285:38104–38114. doi: 10.1074/jbc.M110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell DA, Hamel LD, Ishizuka K, Mitchell G, Schaefer LM, Deschenes RJ. The Erf4 subunit of the yeast Ras palmitoyl acyltransferase is required for stability of the Acyl-Erf2 intermediate and palmitoyl transfer to a Ras2 substrate. J Biol Chem. 2012;287:34337–34348. doi: 10.1074/jbc.M112.379297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jennings BC, Linder ME. DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J Biol Chem. 2012;287:7236–7245. doi: 10.1074/jbc.M111.337246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linder ME, Jennings BC. Mechanism and function of DHHC S-acyltransferases. Biochemical Society transactions. 2013;41:29–34. doi: 10.1042/BST20120328. [DOI] [PubMed] [Google Scholar]
- 29.Shepherd D, Yates DW, Garland PB. The relationship between the rates of conversion of palmitate into citrate or acetoacetate and the acetyl-coenzyme A content of rat-liver mitochondria. The Biochemical journal. 1965;97:38C–40C. doi: 10.1042/bj0970038c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massey V. The composition of the ketoglutarate dehydrogenase complex. Biochimica et biophysica acta. 1960;38:447–460. doi: 10.1016/0006-3002(60)91280-4. [DOI] [PubMed] [Google Scholar]
- 31.Garland PB, Shepherd D, Yates DW. Steady-state concentrations of coenzyme A, acetyl-coenzyme A and long-chain fatty acyl-coenzyme A in rat-liver mitochondria oxidizing palmitate. The Biochemical journal. 1965;97:587–594. doi: 10.1042/bj0970587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerbey AL, Radcliffe PM, Randle PJ. Diabetes and the control of pyruvate dehydrogenase in rat heart mitochondria by concentration ratios of adenosine triphosphate/adenosine diphosphate, of reduced/oxidized nicotinamide-adenine dinucleotide and of acetyl-coenzyme A/coenzyme A. The Biochemical journal. 1977;164:509–519. doi: 10.1042/bj1640509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molnos J, Gardiner R, Dale GE, Lange R. A continuous coupled enzyme assay for bacterial malonyl-CoA:acyl carrier protein transacylase (FabD) Analytical biochemistry. 2003;319:171–176. doi: 10.1016/s0003-2697(03)00327-0. [DOI] [PubMed] [Google Scholar]
- 34.Webb Y, Hermida-Matsumoto L, Resh MD. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J Biol Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- 35.Sherman F, Fink GR, Hicks JB. Laboratory course manual: Methods in yeast genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1986. [Google Scholar]
- 36.Budde C, Schoenfish MJ, Linder ME, Deschenes RJ. Purification and characterization of recombinant protein acyltransferases. Methods. 2006;40:143–150. doi: 10.1016/j.ymeth.2006.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartels DJ, Mitchell DA, Dong X, Deschenes RJ. Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6775–6787. doi: 10.1128/mcb.19.10.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 40.Storer AC, Cornish-Bowden A. The kinetics of coupled enzyme reactions. Applications to the assay of glucokinase, with glucose 6-phosphate dehydrogenase as coupling enzyme. The Biochemical journal. 1974;141:205–209. doi: 10.1042/bj1410205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, 3rd, Davis NG. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohno Y, Kihara A, Sano T, Igarashi Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochimica et biophysica acta. 2006;1761:474–483. doi: 10.1016/j.bbalip.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 43.Draper JM, Smith CD. Palmitoyl acyltransferase assays and inhibitors (Review) Mol Membr Biol. 2009;26:5–13. doi: 10.1080/09687680802683839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukata Y, Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nature reviews. Neuroscience. 2010;11:161–175. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- 45.Liang X, Nazarian A, Erdjument-Bromage H, Bornmann W, Tempst P, Resh MD. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J Biol Chem. 2001;276:30987–30994. doi: 10.1074/jbc.M104018200. [DOI] [PubMed] [Google Scholar]
- 46.Burges RA, Butt WD, Baggaley A. Some effects of alpha-bromopalmitate, an inhibitor of fatty acid oxidation, on carbohydrate metabolism in the rat. The Biochemical journal. 1968;109:38P–39P. doi: 10.1042/bj1090038pb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chase JF, Tubbs PK. Specific inhibition of mitochondrial fatty acid oxidation by 2-bromopalmitate and its coenzyme A and carnitine esters. The Biochemical journal. 1972;129:55–65. doi: 10.1042/bj1290055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davda D, El Azzouny MA, Tom CT, Hernandez JL, Majmudar JD, Kennedy RT, Martin BR. Profiling targets of the irreversible palmitoylation inhibitor 2-bromopalmitate. ACS chemical biology. 2013;8:1912–1917. doi: 10.1021/cb400380s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsutsumi R, Fukata Y, Fukata M. Discovery of protein-palmitoylating enzymes. Pflugers Archiv : European journal of physiology. 2008;456:1199–1206. doi: 10.1007/s00424-008-0465-x. [DOI] [PubMed] [Google Scholar]
- 50.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 51.Pedram A, Razandi M, Deschenes RJ, Levin ER. DHHC-7 and -21 are palmitoylacyltransferases for sex steroid receptors. Mol Biol Cell. 2012;23:188–199. doi: 10.1091/mbc.E11-07-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eisenthal R, Cornish-Bowden A. The direct linear plot. A new graphical procedure for estimating enzyme kinetic parameters. The Biochemical journal. 1974;139:715–720. doi: 10.1042/bj1390715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cornish-Bowden A, Eisenthal R. Estimation of Michaelis constant and maximum velocity from the direct linear plot. Biochimica et biophysica acta. 1978;523:268–272. doi: 10.1016/0005-2744(78)90030-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Erf2-Erf4 autopalmitoylation activity tolerates relatively high concentrations of dimethylsulfoxide (DMSO). Erf2-Erf4 catalyzed autopalmitoylation reactions containing varying percentages of DMSO were performed as described in Materials and Methods using 2 μg wild type Erf2-Erf4 complex. The reaction proceeded for 5 min at 30°C while continuously monitoring the fluorescence intensity (ex. 340 nm/ em. 460 nm). The reactions were performed eight times (n=8) and are expressed as the mean +/- SD. The P-values are given on the figure. The P-value for 0 and 6 percent DMSO is 0.7.


