Abstract
Members of the neurexin gene family, neurexin 1 (NRXN1), neurexin 2 (NRXN2), and neurexin 3 (NRXN3) encode important components of synaptic function implicated in autism and other neurodevelopmental/neuropsychiatric disorders. Loss of function variants have been reported predominantly in NRXN1, with fewer such variants detected in NRXN2 and NRXN3. Evidence for segregating NRNX3 variants has particularly been lacking. Here, we report identification by chromosomal microarray analysis (CMA) of a rare exonic deletion affecting the NRXN3 alpha isoform in a three-generation Chinese family. The proband, a 7-year-old boy, presented with motor and language delay and met the clinical diagnostic criteria for autism. He also presented with moderate intellectual disability, attention deficit hyperactivity disorder (ADHD) and facial dysmorphic features. The mother and maternal grandfather, both deletion carriers, presented with variable degrees of language and communication difficulties, as well as neuropsychiatric problems such as schizophrenia and temper tantrums. A compilation of sporadic cases with deletions involving part or all of NRXN3 revealed that 9 of 23 individuals (39%) displayed features of autism. The evidence for co-segregation in our family further supports a role for NRXN3 in autism and neurodevelopmental/neuropsychiatric disorders but demonstrates intra-family variable expressivity due to this NRXN3 deletion, with schizophrenia and facial dysmorphism being potential novel features of NRXN3 haploinsufficiency.
Keywords: NRXN3, intragenic deletion, autism, intra-family expressivity
Introduction
Neurexins are presynaptic cell-adhesion molecules encoded by paralogous genes, NRXN1, NRXN2 and NRXN3. Each of these encodes alpha and beta isoforms [Sudhof, 2008]. Neurexins are cell-surface receptors that bind neuroligins to form a Ca2+-dependent neurexin/neuroligin complex at synapses in the central nervous system; they are required for efficient neurotransmission and play key roles in synaptic contacts and function [Tabuchi and Sudhof, 2002; Reissner et al., 2008]. All three genes had been implicated in autism spectrum disorders (ASD) [Feng et al., 2006; Autism Genome Project Consortium et al., 2007; Kim et al., 2008; Glessner et al., 2009; Rujescu et al., 2009; Ching et al., 2010; Gauthier et al., 2011; Liu et al., 2012; Gjørlund et al., 2012; Stessman et al., 2017; Mohrmann et al., 2011; Dachtler et al., 2014; Born et al., 2015; Boyle et al., 2015; Vaags et al., 2012].
Loss of function variants had been mostly identified in NRXN1 among patients with neurodevelopmental or neuropsychiatric disorders, supporting NRXN1 as a susceptibility gene for ASD, schizophrenia and intellectual disability [Feng et al., 2006; Szatmari et al., 2007; Kim et al., 2008; Glessner et al., 2009; Rujescu et al., 2009; Ching et al., 2010; Gauthier et al., 2011; Liu et al., 2012; Gjørlund et al., 2012; Stessman et al., 2017]. Variable expressivity and incomplete penetrance frequently were observed [Woodbury-Smith et al., 2017; Lowther et al., 2017]. Variants involving NRXN2 and NRXN3 are much rarer but reports are accumulating [Gauthier et al., 2011; Mohrmann et al., 2011; Dachtler et al., 2014; Born et al., 2015; Boyle et al., 2015; Vaags et al., 2012]. Several findings support a role for NRXN2 in autism, but the contribution of NRXN2 to brain function might be less than other NRXN genes since NRXN2 deletion does not seem to impair cognitive competence [Gauthier et al., 2011; Mohrmann et al., 2011; Dachtler et al., 2014; Born et al., 2015; Boyle et al., 2015].
Loss of function sequence variants in NRXN3 gene are very rare: only one stop gain variant was detected among 60,706 individuals in the Exon Aggregation Consortium (ExAC) database. While sporadic deletion cases have been reported in databases and publications, these presented limited segregation evidence to support a causal relationship between NRXN3 deletion and autism [Schlade-Bartusiak et al., 2008; Cingoz et al., 2011; Vaags et al., 2012; Griswold et al., 2012; Riegel et al., 2014; Faheem et al., 2015; Nicita et al., 2015; https://decipher.sanger.ac.uk/]. Familial exonic NRXN3 deletions were reported previously in autistic children from four unrelated pedigrees of Western origin. Among the four pedigrees, one deletion was inherited from a carrier mother without ASD diagnosis, another from an unaffected father. The third deletion was inherited from an affected father with subclinical autism and the fourth deletion was de novo. Thus, the four pedigrees provided limited evidence for co-segregation of the deletion with a neurodevelopmental phenotype [Vaags et al., 2012]. NRXN3 has also been implicated in neuropsychiatric disorders such as schizophrenia [Hu et al., 2013], but overall, studies of NRXN3 in autism and other neuropsychiatric disorders are quite limited. Here, we describe a three-generation Chinese family carrying a deletion at 14q24.3–31.1 involving the NRXN3 alpha isoform (Fig.1). This pedigree provides important supporting evidence for the association between NRXN3 and neurodevelopmental/neuropsychiatric conditions, including autism.
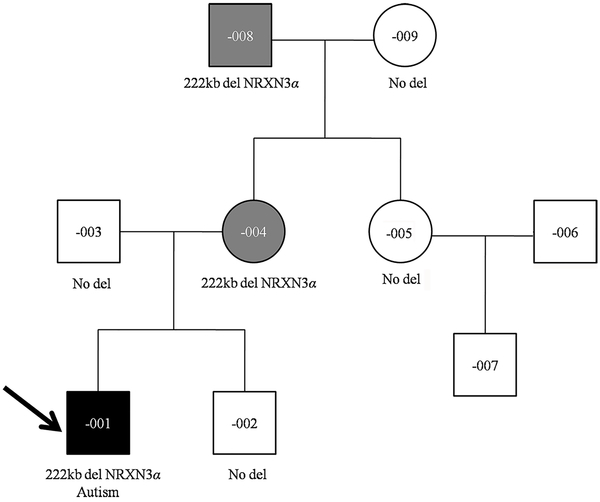
Fig. 1:
Pedigree of our large Chinese family with deletion at the NRXN3 locus
Black-filled symbol represents ASD affected individual, gray-filled symbols represent affected individuals, and unfilled symbols represent apparently unaffected individuals. Proband is marked with an arrow. Clinical diagnosis and segregation of the NRXN3 deletion are shown.
Case presentation
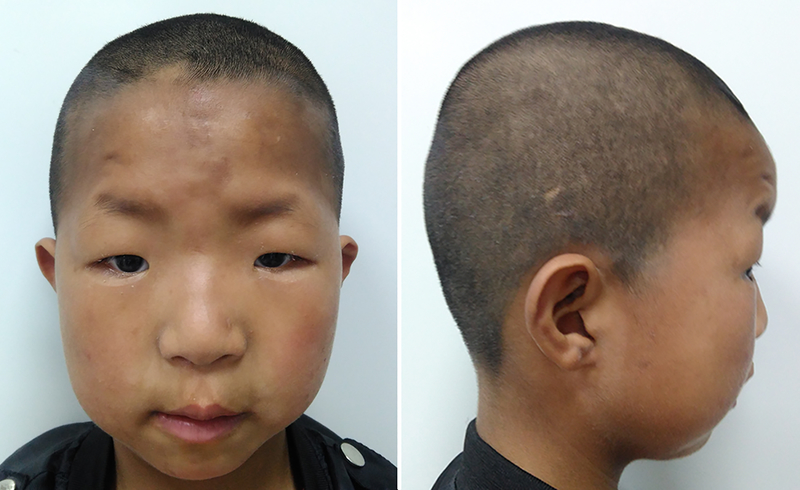
The proband (F-001) was one of two children of nonconsanguineous parents, the product of an uneventful pregnancy. His younger brother (F-002) was healthy. The proband was delivered at 38 weeks of gestation with normal birth measurements: weight 3.2 kg (40th centile), length 51 cm (65th centile) and head circumference 34.2 cm (40th centile). Apgar scores were all 9. No feeding problem was noted after birth. His growth development was within normal range: he weighed 24 kg (60th centile), his height was 121.8 cm (45th centile), and his head circumference was 51.4 cm (50th centile) at age of 7 years, but his motor development was delayed: he raised his head at 4 months, sat alone at 1 year and could walk independently at 2 years. His language development was significantly delayed, he only developed two-word sentences and displayed moderate intellectual disability with Intelligence Quotient (IQ) 50 by the Wechsler Intelligence Scale. The proband’s abnormal behaviors included autism spectrum disorder (ASD) and Attention Deficit Hyperactivity Disorder (ADHD). He demonstrated poor eye contact, social difficulties and stereotyped behaviors as well as restricted interests, such as playing with building blocks and sand. He showed aggressive, depressive and anxious open field behaviors and was diagnosed with ASD after a clinical assessment at the age of 2-year-5-month. Reassessment at the age of 3-year-8-month confirmed that he met the clinical diagnostic criteria for autism based on the Autism Behavior Checklist (ABC), Childhood Autism Rating Scale (CARS) and Modified Checklist for Autism in Toddlers Revised (M-CHAT-R). He had distinctive facial features including high forehead, flat facial profile, auricle dysplasia, widely spaced eyes, blepharoptosis, flat nose bridge, long broad philtrum, dental dysplasia, micrognathia, sparse eyebrows and hair (Fig. 2). Brain magnetic resonance imaging (MRI) was normal.
Fig. 2:
Clinical pictures of the proband at 7 years old age. Note cranial dysmorphism, high forehead, flat facial profile, auricle dysplasia, widely spaced eyes, blepharoptosis, flat nose bridge, long broad philtrum, dental dysplasia, micrognathia, sparse eyebrows and hair.
The proband’s father (F-003) was apparently healthy. The mother (F-004) presented with language delay, learning difficulties, anxiety, slightly impaired socialization skills as well as difficulty in communication. She had good eye contact, did not show stereotyped behaviors, and did not meet the criteria for ASD upon a clinical assessment. The maternal grandmother (F-009) was obviously unaffected. The maternal grandfather (F-008) was assessed by clinical review and was found to have language delay, mild impaired communication and social interaction issues, schizophrenia, temper tantrums, aggressive behavior and anxiety in unfamiliar situations, but had good eye contact and no evidence of stereotyped behaviors or restricted interests. An evaluation for autism was not performed. The maternal aunt (F-005) and her 10-year-old son (F-007) were both normal.
Material and methods
Chromosome karyotype analysis
Cytogenetic investigation (GTG banding) of 20 metaphases obtained from PHA-stimulated peripheral lymphocytes of the proband was performed following standard protocols.
FMR1 gene testing
FMR1 testing was carried out for the proband following the manufacturer’s instructions as described elsewhere [Biancalana and Macpherson, 2004].
Chromosomal microarray analysis (CMA)
Genomic DNAs were isolated from peripheral blood samples using a commercial kit (Qiagen Valencia, CA, USA). Chromosomal microarray analysis was carried out for all members of the large Chinese family using Affymetrix Cytoscan 750K platform (Affymetrix, Santa Clara, CA, USA). Labeling and hybridization procedures were performed according to the manufacturer’s instructions. The raw chromosomal microarray data were analyzed by Affymetrix Chromosome Analysis Suite Software. The pathogenicity of the copy number variants was interpreted according to the American College of Medical Genetics (ACMG) guidelines [Kearney et al., 2011].
Whole Exome Sequencing (WES)
Whole exome sequencing (Illumina, San Diego, CA, USA) was performed for F-001, −004, and −008. Library preparation, cluster generation and sequencing were performed according to manufacturer’s protocols. Bcl2fastq tool (v2.15.0.4) was used for extracting Fastq files from Illumina bcl sequencing file. BWA (0.7.10-r789), Picard (v1.128) and Genome Analysis Toolkit (GATK v3.5) were employed for genome alignments and variant detection. The Annovar tool was applied for variant annotation and the copy number variants (CNVs) were analyzed using ExomeDepth (v1.1.4). Common variants were filtered based on their frequencies in the databases of the Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org), the Exome Sequencing Project (https://esp.gs.washington.edu), or 1000G (http://www.1000genomes.org), and our internal database. The pathogenicity of the sequence variants was interpreted according to the American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) guidelines [Richards et al., 2015].
All data were collected with the informed consent of the patients. Ethical approval for the present study was obtained from Dongguan Maternal and Child Health Care Hospital
Results
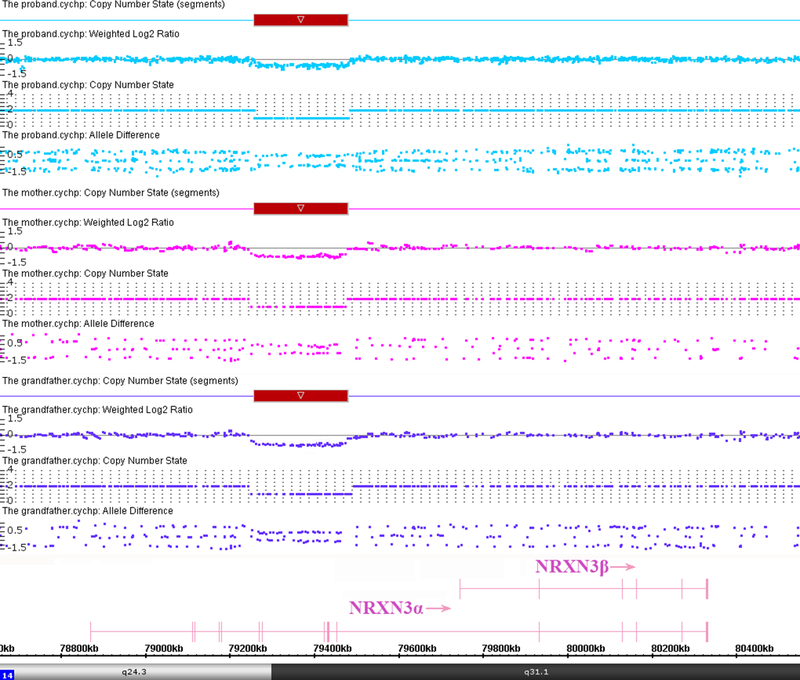
The proband had a normal karyotype and a normal FMR1 repeat number but chromosomal microarray analysis detected a 222 Kb heterozygous microdeletion at 14q24.3q31.1 with genomic coordinates 79,258,267–79,480,738 (GRCh37/hg19), encompassing exons 6–12 of the alpha isoform of NRXN3 (Fig. 3). The NRXN3 deletion was also detected in the mother and the maternal grandfather. The other members of the large Chinese family showed normal copy number in this region. We also performed WES tests for F-001, −004, and −008 to search for variants that might contribute to the phenotype. No clinical significant sequence variants were identified.
Fig. 3:
Affymetrix Cytoscan 750K analysis including weighted log2 ratio (upper), copy number state (middle) and allele difference (lower) are shown for chromosome 14. The result shows hemizygous microdeletion at 14q24.3q31.1, extending from chr14:79,258,267–79,480,738 (GRCh37/hg19), a region that overlaps exons 6–12 of the alpha isoform of NRXN3. The microdeletion region is denoted by a red bar. The NRXN3 deletions were detected in the proband, the mother and the maternal grandfather.
Discussion
CNVs involving NRXN1 have been associated with intellectual disability, language delay, autism, and several psychiatric disorders, in particular, schizophrenia [Feng et al., 2006; Kim et al., 2008; Glessner et al., 2009; Rujescu et al., 2009; Ching et al., 2010; Gauthier et al., 2011; Liu et al., 2012; Gjørlund et al., 2012; Stessman et al., 2017], indicating a broad phenotypic spectrum for NRXN mutations. In the four previously reported pedigrees with NRXN3 deletion, the deletion carrying probands were all diagnosed with autism. In addition, three of the probands also presented with aggression, anger and anxiety. One deletion-carrying father presented with subclinical autism and one deletion-carrying mother presented with social difficulties, language delay and anxiety but no autism [Vaags et al., 2012]. These data indicated variable clinical presentations of individuals with NRXN3 deletion, as well as reduced penetrance of NRXN3 deletion for autism. The exonic NRXN3 deletion detected in the Chinese family was absent from 5,163 healthy individuals of Chinese ethnicity (http://database.gdg-fudan.org/) and 3,397 normal controls in our internal database. We also excluded to the degree possible other known genetic causes of autism in the proband. Based on current evidence, it is reasonable to conclude that this deletion is likely to be causally linked with proband’s phenotype. Interestingly, the same CNV deletion was present in the mother, who did not meet the criteria of ASD diagnosis but had language delay, learning difficulties, anxiety, slightly impaired socialization skills as well as difficulty in communication. The proband’s deletion-negative younger brother had a normal development. The deletion was also detected in the maternal grandfather who had deficits in language, mild impaired communication and social interaction issues, schizophrenia, temper tantrums, aggressive behavior and anxiety in unfamiliar situations, but did not have a formal ASD diagnosis. The unaffected maternal aunt did not inherit the deletion and had a 10-year-old healthy son. In view of the phenotypes displayed by the proband, mother and maternal grandfather, this pedigree demonstrates co-segregation of this NRXN3 deletion with neurodevelopmental/neuropsychiatric phenotypes, but also provides evidence for variable expressivity of phenotype associated with NRXN3 haploinsufficiency.
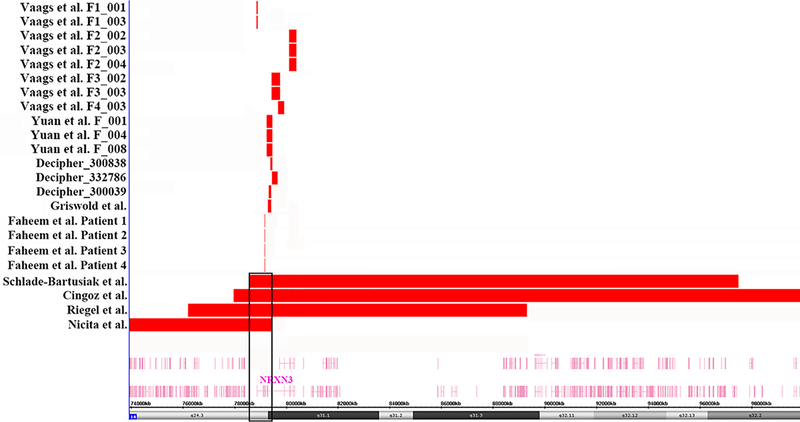
Deletions involving NRXN3 have rarely but sporadically been reported in databases and publications, so we compiled and compared these cases in Table 1 and Fig. 4. We compared the phenotypes of all 23 cases carrying NRXN3 deletions and found that 9 patients (39%) had ASD, as the most consistent feature of individuals with NRXN3 deletion. Other features included language delay, social difficulties, intellectual disability, ADHD, temper tantrums, anxious, aggressive and obsessive behaviors [Schlade-Bartusiak et al., 2008; Cingoz et al., 2011; Vaags et al., 2012; Griswold et al., 2012; Riegel et al., 2014; Faheem et al., 2015; Nicita et al., 2015; https://decipher.sanger.ac.uk/]. Our proband’s maternal grandfather was diagnosed with schizophrenia, which was a rare phenotype in NRXN3 deletion patients. It was interesting to note that our proband had distinctive facial features which had not been described in those patients who carried exonic deletions within NRXN3. However, we found that four patients who carried large deletions at 14q24.3-q32 that overlap NRXN3 and other genes also presented with facial dysmorphism. We observed that our proband and these four patients shared similar facial features, such as hypertelorism, flat nose bridge, long broad philtrum, auricle dysplasia and mild micrognathia [Schlade-Bartusiak et al., 2008; Cingoz et al., 2011; Riegel et al., 2014; Nicita et al., 2015]. Whether or not the NRXN3 deletion is responsible for facial dysmorphism requires further investigation.
Table 1.
Clinical features observed in patients with deletion involving NRXN3. The genomic coordinates are based on GRCH37/hg19.
| Reference | Genomic location | Size | Methods | Origin | NRXN3 exons | Austism | Other phenotype |
|---|---|---|---|---|---|---|---|
| Our proband | chr14:79258267–79480738 | 222Kb | microarray | Inherited from mother | exons 6–12 (α) | Yes | ID, ADHD, facial dysmorphism, motor delay, aggression, depression, anxiety |
| The mother | chr14:79258267–79480738 | 222Kb | microarray | Inherited from father | exons 6–12 (α) | No | Language delay, social impairments, learning difficulties and anxiety |
| The maternal grandfather | chr14:79258267–79480738 | 222Kb | microarray | Unknown | exons 6–12 (α) | No | Schizophrenia, social impairments, temper tantrums, aggression, anxiety |
| Vaags et al., F1-proband | chr14:78864063–78927002 | 63Kb | microarray | Inherited from mother | exon 1 (α) | Yes | Aggression, anger, anxiety, depression |
| Vaags et al., F1-mother | chr14:78864063–78927002 | 63Kb | microarray | Unknown | exon 1 (α) | No | High energy, social difficulties, anxiety, language-processing difficulties |
| Vaags et al., F2-proband | chr14:80125165–80416882 | 292Kb | microarray | Inherited from father | exons 14–17 (α) and 3–7 (β) | Yes | None |
| Vaags et al., F2-sister | chr14:80125165–80416882 | 292Kb | microarray | Inherited from father | exons 14–17 (α) and 3–7 (β) | Yes | None |
| Vaags et al., F2-father | chr14:80125165–80416882 | 292Kb | microarray | Inherited from father | exons 14–17 (α) and 3–7 (β) | No | Color blind |
| Vaags et al., F3-proband | chr14:79433698–79769716 | 336Kb | microarray | Inherited from father | exons 10–12 (α) and 1 (β) | Yes | Aggression, self harm, obsession, delusional and persecutory ideas, sleep issues and obesity |
| Vaags et al., F3-father | chr14:79433698–79769716 | 336Kb | microarray | Unknown | exons 10–12 (α) and 1 (β) | No | Learning difficulties, social avoidance, depression, alcohol-related issues |
| Vaags et al., F4-proband | chr14:79696374–79943510 | 247Kb | microarray | De novo | exon 13 (α) and 1–2 (β) | Yes | Aggression, anger, anxiety, temper tantrums |
| Faheem et al., Patient 1 | chr14:79176037–79183213 | 7.2Kb | microarray | De novo | exon 5 (α) | No | Developmental delay, seizure |
| Faheem et al., Patient 2 | chr14:79176037–79183213 | 7.2Kb | microarray | De novo | exon 5 (α) | No | ID, ADHD, difficulty in communication |
| Faheem et al., Patient 3 | chr14:79176037–79183213 | 7.2Kb | microarray | Unknown | exon 5 (α) | No | Seizure, motor delay, obsession |
| Faheem et al., Patient 4 | chr14:79176037–79183213 | 7.2Kb | microarray | Unknown | exon 5 (α) | Yes | ID, seizure |
| Decipher 300039 | chr14:79328112–79433755 | 106Kb | microarray | Inherited from mother | exons 8–10 (α) | Yes | GDD |
| Decipher 300838 | chr14:79388255–79473346 | 85Kb | microarray | Unknown | exons 8–12 (α) | No | ID, behavioral abnormality |
| Decipher 332786 | chr14:79448118–79683009 | 235Kb | microarray | De novo | exons 12 (α) | No | GDD, hypotonia, seizures |
| Griswold et al., patient | chr14:79303752–79436470 | 133kb | microarray | Inherited from mother | exons 8–11 (α) | Yes | NR |
| Nicita et al., patient | chr14:73939850–79446043 | 5.5Mb | microarray | De novo | exon 1–11 (α) | No | Developmental delay, language delay, dysmorphic face, seizures |
| Riegel et al., patient | chr14:76211806–89327656 | 13Mb | microarray | De novo | the whole NRXN3 | No | ID, ADHD, motor delay, language delay, dysmorphic features, hypotonia |
| Schlade-Bartusiak et al., patient | 14q24.3q32.2 | 19Mb | FISH | De novo | the whole NRXN3 | No | GDD, short stature, absent speech and auditory neuropathy, minor dysmorphic features, feeding difficulty, hypotonia |
| Cingoz et al., patient | 14q24.3q32.2 | 21Mb | FISH | De novo | the whole NRXN3 | No | ID, no speech, motor delay, facial dysmorphism, short stature, hypotonia |
ID intellectual disability, ADHD attention deficit hyperactivity disorder, GDD global developmental delay
Fig. 4:
The panel shows a genome view of all deletion cases (red colored custom tracks) relative to the genomic coordinates at 14q24.3q31.1 region, extracted from Human Genome Build 37 (hg19). The minimal overlapping region of deletion in the four patients who carry large deletions involves only NRXN3, which is surrounded by black boxes.
In conclusion, we identified a rare exonic deletion of NRXN3 in a three-generation Chinese family. The co-segregation evidence further supported a role for NRXN3 in autism and other neurodevelopmental/neuropsychiatric disorders. By comparing the clinical phenotypes of all 23 patients with NRXN3 deletions, we confirmed that patients carrying exonic NRXN3 deletions display variable clinical features and reduced penetrance for autism. Facial dysmorphism and schizophrenia might be novel phenotypes which need to be further studied.
Acknowledgments
We would like to thank the family of the proband for their cooperation with this study. This study was supported by the Dongguan Social development project (2013108101021; 20161081101023).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Autism Genome Project Consortium, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39(3):319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancalana V, Macpherson J. 2004. Fragile X disease. Methods Mol Med 92:157–182. [DOI] [PubMed] [Google Scholar]
- Born G, Grayton HM, Langhorst H, Dudanova I, Rohlmann A, Woodward BW, et al. 2015. Genetic targeting of NRXN2 in mice unveils role in excitatory cortical synapse function and social behaviors. Front Synaptic Neurosci 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle MI, Jespersgaard C, Nazaryan L, Ravn K, Brøndum-Nielsen K, Bisgaard AM, et al. 2015. Deletion of 11q12.3–11q13.1 in a patient with intellectual disability and childhood facial features resembling Cornelia de Lange syndrome. Gene 572(1):130–134. [DOI] [PubMed] [Google Scholar]
- Ching MS, Shen Y, Tan WH, Jeste SS, Morrow EM, Chen X, et al. 2010. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet B Neuropsychiatr Genet 153B(4):937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingöz S, Bache I, Bjerglund L, Ropers HH, Tommerup N, Jensen H, et al. 2011. Interstitial deletion of 14q24.3-q32.2 in a male patient with plagiocephaly, BPES features, developmental delay, and congenital heart defects. Am J Med Genet A 155A(1):203–206. [DOI] [PubMed] [Google Scholar]
- Dachtler J, Glasper J, Cohen RN, Ivorra JL, Swiffen DJ, Jackson AJ, et al. 2014. Deletion of α-neurexin II results in autism-related behaviors in mice. Transl Psychiatry 25;4:e484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faheem M, Naseer MI, Chaudhary AG, Kumosani TA, Rasool M, Algahtani HA, et al. 2015. Array-comparative genomic hybridization analysis of a cohort of Saudi patients with epilespsy. CNS Neurol Disord Drug Targets 2015;14(4):468–475. [DOI] [PubMed] [Google Scholar]
- Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A, et al. 2006. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett 409(1):10–13. [DOI] [PubMed] [Google Scholar]
- Gauthier J, Siddiqui TJ, Huashan P, Yokomaku D, Hamdan FF, Champagne N, et al. 2011. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum Genet 130(4):563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. 2009. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459(7246):569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjørlund MD, Nielsen J, Pankratova S, Li S, Korshunova I, Bock E, et al. 2012. Neuroligin-1 induces neurite outgrowth through interaction with neurexin-1β and activation offibroblast growth factor receptor-1. FASEB J 26(10):4174–4186 [DOI] [PubMed] [Google Scholar]
- Griswold AJ, Ma D, Cukier HN, Nations LD, Schmidt MA, Chung RH, et al. 2012. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet 21(15):3513–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Zhang J, Jin C, Mi W, Wang F, Ma W, et al. 2013. Association study of NRXN3 polymorphisms with schizophrenia and risperidone-induced bodyweight gain in Chinese Han population. Prog Neuropsychopharmacol Biol Psychiatry 43:197–202. [DOI] [PubMed] [Google Scholar]
- Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST; Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. 2011. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med 13(7):680–685. [DOI] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, et al. 2008. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet 82(1),199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hu Z, Xun G, Peng Y, Lu L, Xu X, et al. 2012. Mutation analysis of the NRXN1 gene in a Chinese autism cohort. J Psychiatr Res 46(5):630–634. [DOI] [PubMed] [Google Scholar]
- Lowther C, Speevak M, Armour CM, Goh ES, Graham GE, Li C, et al. Molecular characterization of NRXN1 deletions from 19,263 clinical microarray cases identifies exons important for neurodevelopmental disease expression. Genet Med. 2017;19(1):53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann I, Gillessen-Kaesbach G, Siebert R, Caliebe A, Hellenbroich Y. 2011. A de novo 0.57Mb microdeletion in chromosome 11q13.1 in a patient with speech problems, autistic traits, dysmorphic features and multiple endocrine neoplasia type 1. Eur J Med Genet 54(4):e461–464. [DOI] [PubMed] [Google Scholar]
- Nicita F, Di Giacomo M, Palumbo O, Ferri E, Maiorani D, Vigevano F, et al. 2015. Neurological features of 14q24-q32 interstitial deletion: report of a new case. Mol Cytogenet 8:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissner C, Klose M, Fairless R, Missler M. 2008. Mutational analysis of the neurexin/neuroligin complex reveals essential and regulatory components. Proc Natl Acad Sci U S A 105(39):15124–15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegel M, Moreira LM, Espirito Santo LD, Toralles MB, Schinzel A. 2014. Interstitial 14q24.3 to q31.3 deletion in a 6-year-old boy with a non-specific dysmorphic phenotype. Mol Cytogenet 7(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, Toulopoulou T, et al. 2009. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet 18(5):988–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlade-Bartusiak K, Macintyre G, Zunich J, Cox DW. 2008. A child with deletion (14)(q24.3q32.13) and auditory neuropathy. Am J Med Genet A 146A(1):117–123. [DOI] [PubMed] [Google Scholar]
- Stessman HA, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, et al. 2017. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet 49(4):515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. 2008. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455(7215):903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Sudhof TC. 2002. Structure and evolution of neurexin genes: Insight into the mechanism of alternative splicing. Genomics 79(6):849–859. [DOI] [PubMed] [Google Scholar]
- Vaags AK, Lionel AC, Sato D, Goodenberger M, Stein QP, Curran S, et al. 2012. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am J Hum Genet 90(1):133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury-Smith M, Nicolson R, Zarrei M, Yuen RKC, Walker S, Howe J, et al. 2017. Variable phenotype expression in a family segregating microdeletions of the NRXN1 and MBD5 autism spectrum disorder susceptibility genes. NPJ Genom Med 2 pii: 17. doi: 10.1038/s41525-017-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]