Abstract
Wollmuth highlights recent work identifying two cysteine substitutions in kainate receptors that result in direct activation by cadmium.
The transition of ion channels from the closed, nonconducting conformation to the open, conducting conformation—a process referred to as gating—is fundamental to physiology. It is in the open state, when the water-filled ion conduction pathway is formed, that ions cross the membrane to impact membrane, cellular, and ultimately, organ physiology. In this issue of the Journal of General Physiology, Wilding and Huettner probe this gating process in glutamate-gated ion channels, a ubiquitous and functionally critical class of ion channels. They discover something quite perplexing yet extremely useful: a means to pry open a gate.
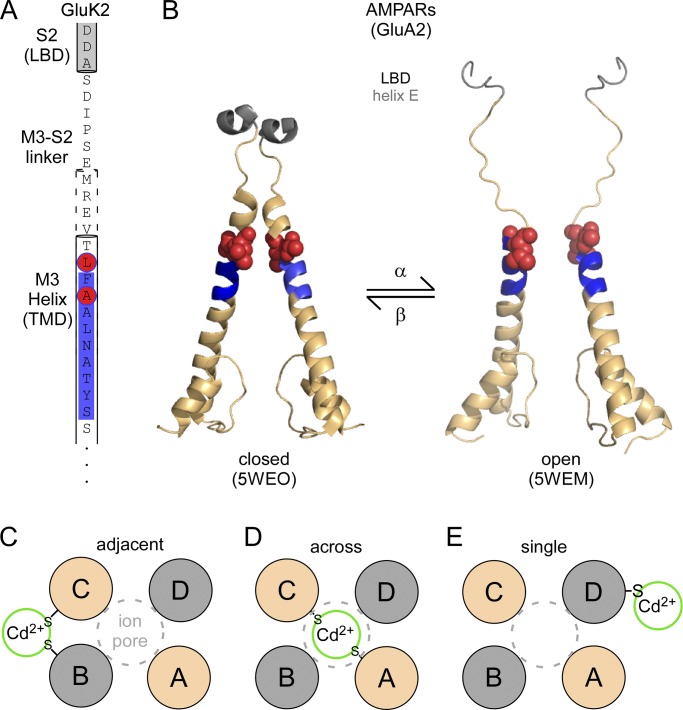
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that are gated by the neurotransmitter glutamate, the most prominent neurotransmitter in the central nervous system. The core of the ion channel in iGluRs shares homology with K+ channels. The major pore-lining transmembrane segment, the M3 segment, is homologous to TM2 or S6 in K+ channels (Wo and Oswald, 1995). However, the orientation of iGluRs is inverted with respect to K+ channels, so the M3 segments form a bundle helical crossing, or a gate, at their extracellular ends, which occludes the flux of ions in the closed state (Chang and Kuo, 2008; Sobolevsky et al., 2009; Ladislav et al., 2018). Agonist binding to the extracellular ligand-binding domain (LBD) pulls the M3 segments away from the central axis of the pore and therefore leads to ion channel opening (Twomey and Sobolevsky, 2018). Structures exist of the closed and open states of iGluRs, at least for one subtype, and they have been transformative in understanding the mechanism of ion channel gating in iGluRs (Fig. 1, A and B; Chen et al., 2017; Twomey et al., 2017). Still, these structures define endpoints and do not reveal the dynamics and energetics of how the M3 segments, as well as other transmembrane segments, are rearranged to transition from the closed to the open state.
Figure 1.
Opening of the M3 gate. (A) Primary sequence of the upper M3 segment (M3 helix), the linker connecting M3 to the ligand-binding domain (LBD; M3-S2 linker), and the most proximal elements of the LBD (helix E, located in S2). The SYTANLAAF, the most highly conserved motif in iGluRs, is highlighted in blue. Positions A8 and L10 that, when substituted with cysteine, show current potentiation when exposed to Cd2+, are highlighted in red. (B) Structures of the corresponding elements either in the closed state (left, PDB accession no. 5WEO) or in the open state (right, PDB accession no. 5WEM; Twomey et al., 2017). (C–E) Cartoon representations of the possible configurations for coordination of Cd2+ by A8C or L10C. The same subunits (A/C or B/D) are colored the same.
To address the dynamics of the M3 transmembrane segments during pore opening, Wilding and Huettner (2018) took advantage of the chemistry of cysteine. Substituting individual residues with cysteine has been an invaluable approach to study the dynamics of ion channels because its thiol side chain is highly and specifically reactive. One set of tools that interact with substituted cysteines is the methanethiolsulfonate (MTS) reagents, which covalently react with water-accessible thiol side chains (Karlin and Akabas, 1998). Another inherent tool, in terms of defining protein dynamics, is that cysteines can form disulfide bonds under oxidizing conditions if proximal enough (Kazi et al., 2013). Wilding and Huettner (2018) took advantage of an additional aspect of the diverse reactivity of thiol side chains: coordination of various monovalent and divalent cations. Wilding and Huettner (2018) used Cd2+, which is highly advantageous over other thiol-reactive approaches: First, it requires multiple, presumably at least two, proximal cysteines to coordinate Cd2+ and hence alter receptor function (there is always the possibility of a silent effect; coordination occurs, but there is no functional effect). Second, the coordination of Cd2+ occurs rapidly, minimizing the trapping of rarely visited conformations, and is readily reversible. This is in contrast to the slow and irreversible actions of MTS reagents and the slow actions and need for multiple reagents for disulfide bond formation/breakage. Finally, unlike some MTS reagents, Cd2+ has a permanent positive charge so it will not cross the membrane or enter hydrophobic crevices. Of course, the question that must always be considered when using substituted cysteines is whether any effect is due solely to the introduced cysteines.
There are three major iGluR subtypes: AMPA (AMPARs), kainate, and NMDA (NMDAR) receptors. All three subtypes share a common overall membrane topology (Plested, 2016). Nevertheless, the various subtypes display differences in biophysical properties that contribute to their unique roles in synaptic physiology (Traynelis et al., 2010). In the present study, the authors focused on kainate receptors and mainly on the GluK2 subunit. In the unedited form, GluK2(Q), a glutamine (Q) is present at the Q/R site. In the edited form, GluK2(R), an arginine (R) is present at the Q/R site. These subunits form functional homotetramers, hence any introduced cysteines would be present in all four subunits. The authors substituted cysteines throughout the M3 transmembrane segment and probed these cysteine-substituted receptors with bath-applied Cd2+. For the majority of these cysteine-substituted receptors, Cd2+ had an effect comparable to that for wild type, indicating that the cysteines introduced at these positions were unable to coordinate Cd2+ (or that coordination was functionally silent). In contrast, dramatic effects on receptor function occurred when cysteines were substituted at two positions located at the apex of the M3 segment: at an alanine (A; referred to as A8) located in the most highly conserved motif in iGluRs, the SYTANLAAF motif, and at a leucine (L; referred to as L10) just external to the SYTANLAAF motif (Fig. 1 A). Both of these positions are located around the bundle helical crossing that forms a gate in iGluRs.
Identifying an effect of Cd2+ on substituted cysteines, especially two independent positions, represents a fantastic new tool to probe the dynamics of the M3 gate. Still, what was most unexpected was the specific effect of Cd2+ on receptors containing A8C or L10C. In the presence of agonist (kainate), Cd2+ strongly potentiated current amplitude above and beyond that generated by agonist alone. More surprisingly, Cd2+ by itself could activate receptors containing either A8C or L10C independent of any added agonist. Thus, Cd2+ coordination at the bundle helical crossing facilitates transition to the open state and can do so independent of agonist. That Cd2+ can open the channel by itself highlights the critical role the bundle helical crossing plays in the energetics of pore opening.
How can the Cd2+-induced current potentiation be explained? One possibility is that coordination occurred between adjacent subunits in the tetrameric complex (Fig. 1 C). But one of the limitations of working with non-NMDARs is that there is no way to distinguish the four subunits in these homotetramers (in contrast, NMDARs are obligate heterotetramers in which the GluN1 and GluN2 subunits sit across from each other in the tetrameric complex; Salussolia et al., 2011b). To circumvent the homotetrameric problem, Wilding and Huettner (2018) took advantage of chimeras in which the transmembrane domain (TMD) of GluK2(Q) is attached to the LBD of either GluN1 (N1/K2(Q)) or GluN2B (N2B/K2(Q)) subunits. These chimeras, identified by the authors previously (Wilding et al., 2014), are an underappreciated and brilliant insight. Previous iGluRs chimeras expressed poorly (Villmann et al., 1999), making them of limited use experimentally. Huettner and colleagues solved this problem by including all transmembrane segments from the same subtype in the chimeric construct, taking advantage of the finding that the eukaryotic-specific M4 transmembrane segment specifically interacts with the inner pore domain (Salussolia et al., 2011a).
These N1/K2(Q) and N2B/K2(Q) chimeric subunits when coexpressed form, like NMDARs, obligate heterotetramers (Wilding et al., 2014). Hence, the authors could now introduce A8 or L10 cysteines into either chimeric subunit and test which specific subunits were involved in coordination. Alas, the original explanation for potentiation—that it occurred with adjacent subunits—was rejected. Cysteines introduced in either N1/K2(Q) (A/C conformation) or N2B/K2(Q) (B/D conformation), which sit across from each other in the tetrameric complex, still showed Cd2+-induced potentiation (though it was less than that observed when all four subunits had a cysteine). This is a rather surprising outcome, as one might surmise that coordination of subunits that sit across from each other would inhibit current (Fig. 1 D). There are several possible explanations for this surprising result. First, perhaps it is possible that the N1/K2(Q) and N2B/K2(Q) do not position across from each other in the tetrameric complex, though this alternative seems highly unlikely (Wilding et al., 2014). Second, perhaps only a single introduced cysteine is mediating the Cd2+-induced potentiation (Fig. 1 E). In this scenario, either an endogenous cysteine or another side chain could be facilitating coordination, or perhaps just a single cysteine mediates the coordination (Raghuraman et al., 2012) and the strong potentiation reflects the critical structural roles of A8 and L10 in the energetics of the bundle helical crossing. One approach to test the idea that a single introduced cysteine mediates the Cd2+-induced potentiation would be to use a triheteromeric system (e.g., Hansen et al., 2014) to introduce a single cysteine in the tetrameric complex. That only a single introduced cysteine participates in coordination, leading to a more unstable complex, might account for the rather rapid reversibility of the Cd2+-induced potentiation effect.
The most likely arrangement, given the available data, is that Cd2+ is coordinated across the pore and potentiates channel activity (Fig. 1 D; the authors did show, using noise analysis, that the potentiation most likely reflects an increase in open probability). Simplistically, it is hard to image how coordinating across subunits would potentiate currents. But maybe our view of the channel’s gate and its dynamics are too simplistic (Fig. 1 C–E); perhaps the bundle helical crossing has more conformational freedom than we typically envision. Indeed, unexpectedly, in the transition from the closed to the open state, helices in the M3 segment and associated LBD (helix E) bend and unravel (Fig. 1 B).
Regardless of whether Cd2+-induced potentiation is arising from coordination across subunits or caused by a single cysteine, Wilding and Huettner (2018) have introduced a new tool to explore the M3 gate and hence the gating process in iGluRs in general. Beyond the present work, the most obvious question is whether Cd2+ would have a similar effect in AMPARs and NMDARs containing a substituted cysteine at A8 or L10. Although iGluRs share a common overall topology, including having the M3 segment as the central pore-lining segment, there are differences in gating between the subtypes. One notable difference is concerted versus subunit-specific gating. NMDARs require all four agonists to bind (two glycines and two glutamates) before the channel enters into an open state and typically, with single-channel recordings, shows a single conductance level. In contrast, non-NMDARs display subunit-specific gating with single-channel recordings showing three to four subconductance levels (Smith and Howe, 2000), which are presumably related to the four subunits. One possibility is that these subconductance states arise from variations in opening of the M3 gate. Using Cd2+ to trap and control the M3 gates may be one approach to start addressing the contribution of different M3 segments to subunit-specific gating.
While M3 controls an extracellular gate to open the ion channel, a remaining question is what controls M3? Clearly, the direct action is the clam-shell closure induced by agonist binding to the LBD (Zhang et al., 2008), which mechanically pulls on the M3 segments (Kazi et al., 2014). However, the M3 segments do not reside in isolation in the membrane, and it is now becoming clear that the other transmembrane segments —M1 and M4 and the linkers that connect them to the LBD—are not passive and can influence the dynamics of pore opening. This idea was initially suggested by experiments using MTS reagents targeted to linkers connecting the LBD to the TMD (Talukder et al., 2010), but more recently has been highlighted by disease-associated missense mutations in M1, M4, and associated linkers (Yuan et al., 2014; Ogden et al., 2017; Amin et al., 2018). Again, the experiments of Wilding and Huettner (2018) provide invaluable additional tools to study how these more peripherally located helices impact the dynamics of the inner M3 helices. Indeed, if Cd2+ is being coordinated by a nonthiol side chain in addition to that of the introduced cysteine, the most likely location would be from the S2-M4 (Twomey et al., 2017). If correct, and if this interacting side chain could be identified, trapping of the open state by Cd2+ could provide a real time index of the dynamics of M3 and the outer structures.
In terms of understanding the dynamics of pore opening in iGluRs, progress has been tremendous. High-resolution open and closed states have been invaluable and give guidance to the endpoints (Twomey and Sobolevsky, 2018). However, as illustrated by Wilding and Huettner (2018), there remain many nuances of this gating process that still need to be resolved before we have a full picture of how iGluRs transition from agonist binding to the open state and hence how they impact membrane physiology.
Acknowledgments
Kenton J. Swartz served as editor.
References
- Amin J.B., Leng X., Gochman A., Zhou H.-X., and Wollmuth L.P.. 2018. A conserved glycine harboring disease-associated mutations permits NMDA receptor slow deactivation and high Ca2+ permeability. Nat. Commun. 9:3748 10.1038/s41467-018-06145-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.R., and Kuo C.C.. 2008. The activation gate and gating mechanism of the NMDA receptor. J. Neurosci. 28:1546–1556. 10.1523/JNEUROSCI.3485-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhao Y., Wang Y., Shekhar M., Tajkhorshid E., and Gouaux E.. 2017. Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM. Cell. 170:1234–1246. 10.1016/j.cell.2017.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K.B., Ogden K.K., Yuan H., and Traynelis S.F.. 2014. Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron. 81:1084–1096. 10.1016/j.neuron.2014.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin A., and Akabas M.H.. 1998. Substituted-cysteine accessibility method. Methods Enzymol. 293:123–145. 10.1016/S0076-6879(98)93011-7 [DOI] [PubMed] [Google Scholar]
- Kazi R., Gan Q., Talukder I., Markowitz M., Salussolia C.L., and Wollmuth L.P.. 2013. Asynchronous movements prior to pore opening in NMDA receptors. J. Neurosci. 33:12052–12066. 10.1523/JNEUROSCI.5780-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi R., Dai J., Sweeney C., Zhou H.X., and Wollmuth L.P.. 2014. Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nat. Neurosci. 17:914–922. 10.1038/nn.3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladislav M., Cerny J., Krusek J., Horak M., Balik A., and Vyklicky L.. 2018. The LILI Motif of M3-S2 Linkers Is a Component of the NMDA Receptor Channel Gate. Front. Mol. Neurosci. 11:113 10.3389/fnmol.2018.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden K.K., Chen W., Swanger S.A., McDaniel M.J., Fan L.Z., Hu C., Tankovic A., Kusumoto H., Kosobucki G.J., Schulien A.J., et al. . 2017. Molecular Mechanism of Disease-Associated Mutations in the Pre-M1 Helix of NMDA Receptors and Potential Rescue Pharmacology. PLoS Genet. 13:e1006536 10.1371/journal.pgen.1006536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plested A.J. 2016. Structural mechanisms of activation and desensitization in neurotransmitter-gated ion channels. Nat. Struct. Mol. Biol. 23:494–502. 10.1038/nsmb.3214 [DOI] [PubMed] [Google Scholar]
- Raghuraman H., Cordero-Morales J.F., Jogini V., Pan A.C., Kollewe A., Roux B., and Perozo E.. 2012. Mechanism of Cd2+ coordination during slow inactivation in potassium channels. Structure. 20:1332–1342. 10.1016/j.str.2012.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salussolia C.L., Corrales A., Talukder I., Kazi R., Akgul G., Bowen M., and Wollmuth L.P.. 2011a Interaction of the M4 segment with other transmembrane segments is required for surface expression of mammalian α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors. J. Biol. Chem. 286:40205–40218. 10.1074/jbc.M111.268839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salussolia C.L., Prodromou M.L., Borker P., and Wollmuth L.P.. 2011b Arrangement of subunits in functional NMDA receptors. J. Neurosci. 31:11295–11304. 10.1523/JNEUROSCI.5612-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.C., and Howe J.R.. 2000. Concentration-dependent substate behavior of native AMPA receptors. Nat. Neurosci. 3:992–997. 10.1038/79931 [DOI] [PubMed] [Google Scholar]
- Sobolevsky A.I., Rosconi M.P., and Gouaux E.. 2009. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 462:745–756. 10.1038/nature08624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder I., Borker P., and Wollmuth L.P.. 2010. Specific sites within the ligand-binding domain and ion channel linkers modulate NMDA receptor gating. J. Neurosci. 30:11792–11804. 10.1523/JNEUROSCI.5382-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis S.F., Wollmuth L.P., McBain C.J., Menniti F.S., Vance K.M., Ogden K.K., Hansen K.B., Yuan H., Myers S.J., and Dingledine R.. 2010. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62:405–496. 10.1124/pr.109.002451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey E.C., and Sobolevsky A.I.. 2018. Structural Mechanisms of Gating in Ionotropic Glutamate Receptors. Biochemistry. 57:267–276. 10.1021/acs.biochem.7b00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey E.C., Yelshanskaya M.V., Grassucci R.A., Frank J., and Sobolevsky A.I.. 2017. Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature. 549:60–65. 10.1038/nature23479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villmann C., Strutz N., Morth T., and Hollmann M.. 1999. Investigation by ion channel domain transplantation of rat glutamate receptor subunits, orphan receptors and a putative NMDA receptor subunit. Eur. J. Neurosci. 11:1765–1778. 10.1046/j.1460-9568.1999.00594.x [DOI] [PubMed] [Google Scholar]
- Wilding T.J., and Huettner J.E.. 2018. Cadmium opens GluK2 kainate receptors with cysteine substitutions at the M3 helix bundle crossing. J. Gen. Physiol. 10.1085/jgp.201812234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding T.J., Lopez M.N., and Huettner J.E.. 2014. Radial symmetry in a chimeric glutamate receptor pore. Nat. Commun. 5:3349 10.1038/ncomms4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wo Z.G., and Oswald R.E.. 1995. Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci. 18:161–168. 10.1016/0166-2236(95)93895-5 [DOI] [PubMed] [Google Scholar]
- Yuan H., Hansen K.B., Zhang J., Pierson T.M., Markello T.C., Fajardo K.V., Holloman C.M., Golas G., Adams D.R., Boerkoel C.F., et al. . 2014. Functional analysis of a de novo GRIN2A missense mutation associated with early-onset epileptic encephalopathy. Nat. Commun. 5:3251 10.1038/ncomms4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Cho Y., Lolis E., and Howe J.R.. 2008. Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating. J. Neurosci. 28:932–943. 10.1523/JNEUROSCI.3309-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]