Abstract
The hypocretin/orexin neuropeptide system coordinates the regulation of various physiological processes. Our previous study reported that a reduction in the expression of pleomorphic adenoma gene-like 1 (Plagl1), which encodes a C2H2 zinc-finger transcription factor, occurs in hypocretin neuron-ablated transgenic mice, suggesting that PLAGL1 is co-expressed in hypocretin neurons and regulates hypocretin transcription. The present study examined whether canonical prepro-hypocretin transcription is functionally modulated by PLAGL1. Double immunostaining indicated that the majority of hypocretin neurons were positive for PLAGL1 immunore-activity in the nucleus. Notably, PLAGL1 immunoreactivity in hypocretin neurons was altered in response to several conditions affecting hypocretin function. An uneven localization of PLAGL1 was detected in the nuclei of hypocretin neurons following sleep deprivation. Chromatin immunoprecipitation revealed that endogenous PLAGL1 may bind to a putative PLAGL1-binding site in the proximal region of the hypocretin gene, in the murine hypothalamus. In addition, electroporation of the PLAGL1 expression vector into the fetal hypothalamus promoted hypothalamic hypocretin transcription. These results suggested that PLAGL1 may regulate hypothalamic hypocretin transcription.
Keywords: orexin, hypocretin, pleomorphic adenoma gene-like 1, sleep, hypothalamus
Introduction
Hypocretin/orexin is a hypothalamic neuropeptide, which is involved in various physiological processes, including emotion, energy homeostasis, feeding behavior, reward, and sleep/vigilance maintenance and regulation (1). Hypocretin expression is almost exclusively localized in the lateral hypothalamus (2,3). Furthermore, two conserved promoter regions, orexin regulatory element (OE)1 and OE2, have been identified within a 3.2-kb fragment located upstream of the prepro-hypocretin gene. These regions have been reported to target specific expression within the lateral hypothalamus (3).
To detect and characterize the proliferation of hypocretin neurons during embryonic development, 5-bromo-2-deoxyuri-dine was administered into the developing ventral diencephalon of embryos of pregnant rats; this region is where the embryonic hypothalamus is located (4). Hypocretin neuron proliferation peaked sharply on day 12 of embryonic development. In mice, our previous study detected prepro-hypocretin transcription from embryonic day (E)10, using reverse transcription-polymerase chain reaction (PCR) (5). A recent study using mice expressing enhanced green fluorescent protein (EGFP) under the control of the prepro-hypocretin promoter demonstrated that EGFP and the hypocretin peptide are detectable from E12 and E14, respectively (6). These findings suggest that translational repression persists until the hypocretin neurons migrate to a predetermined position, similar to other neurons (7). In utero electroporation of an EGFP expression vector into the third ventricle at E12 revealed that some hypocretin neurons co-localize with EGFP in the adult hypothalamus (8). These signals suggest that some hypocretin neurons originate from the progenitor cells of the third ventricular zone, at least at E10-E12. Following migration to the designated area, these cells probably differentiate into hypocretin-producing cells with the help of various factors, including the hedgehog protein and its downstream transcription factors, which are essential for anterior hypothalamic patterning (9).
The co-expressed transcription factors in hypocretin neurons regulate hypocretin transcriptional activities under several conditions. Forkhead box A2 (FOXA2) regulates prepro-hypocretin expression in the lateral hypothalamus during fasting (10). Furthermore, co-expression with LIM homeobox 9 (LHX9) has been observed in a subset of developmental hypocretin-positive cells in the hypothalamus (9) and adult murine hypocretin neurons (11). LHX9 is important for the normal development of a subset of hypocretin neurons and canonical sleep behavior; however, to the best of our knowledge, no direct effect on adult hypocretin expression has been found (11). In our previous study, several transcription factors were identified that are downregulated in the hypothalamus of hypocretin neuron-ablated transgenic mice (12); specifically, early B-cell factor 2 (Ebf2); fifth ewing sarcoma variant (Fev); insulin-like growth factor-binding protein 3 (Igfbp3); pleomorphic adenoma gene-like 1 (Plagl1); POU domain class 2, transcription factor 1 (Pou2f1); paired-related homeobox gene 1 (Prrx1); nuclear receptor subfamily 6 group A member 1 (Nr6a1); and visual system homeobox gene 2 (Vsx2). Ebf2-null mice exhibit a significant reduction in hypocretin neurons in the lateral hypothalamus (13), and EBF2 modulates hypocretin promoter activities via the olf-1 motif (14). Igfbp3, Plagl1 and Pou2f1 have also been identified as co-localized genes using comprehensive translational profiling of all ribosome-bound transcripts (11). Our previous study revealed that IGFBP3 reduces promoter activity of prepro-hypocretin (12), and NR6A1 upregulates hypocretin transcription in embryos (8).
The present study aimed to examine the involvement of these downregulated transcription factors (12) in hypo-cretin transcription. This study further identified PLAGL1 as a co-expressed transcription factor in hypothalamic hypocretin neurons. PLAGL1 has seven zinc fingers of the C2H2 class (15), recognizes DNA and RNA, and possesses transcriptional activity (16). In humans, PLAGL1 undergoes parental genomic imprinting and is paternally expressed (17). Plagl1-null mice exhibit intrauterine growth restriction, altered bone formation and neonatal lethality (18); therefore, null mouse lines are difficult to obtain. In the present study, the functional relevance of PLAGL1 in hypocretin transcription was investigated and the results revealed that PLAGL1 may be associated with the canonical development and modulation of various physiological processes in hypocretin neurons via newly identified PLAGL1 binding sites (G4N2G4) (19) within the hypocretin upstream promoter region.
Materials and methods
Ethics statement
All animal experiments were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals of the National Institute of Health (20), and were approved by the Ethics committee on Animal Experiments of the Kansai Medical University (Hirakata, Japan; approval ID: 17-050) and the Tokyo Metropolitan Institute of Medical Science (Tokyo, Japan).
Animals
A total of 41 male C57 black/6J mice were obtained from Shimizu Laboratory Supplies Co., Ltd. (Kyoto, Japan) housed under a 12-h light/dark cycle, with the lights on between 8:00 a.m. and 8:00 p.m., corresponding to Zeitgeber time (ZT) 0-12. The mice were maintained at 22-24°C, and were provided food and water ad libitum. A total of 20 mice (weight, 28-30 g; age, 12 weeks) were sacrificed at ZT6, ZT18, or ZT6 with 6 h of total sleep deprivation (SD6h) for immunohistochemical analysis. SD6h experiments were conducted using the Seesaw shaker (shaking angle, 10°; Wave-SI; Taitec Corporation, Koshigaya, Japan) for examination of sleep/wakefulness organization. To examine the effects of fasting, nine mice (weight, 28-30 g; age, 12 weeks) were deprived of food at ZT6 and were sacrificed within the following 2 days at ZT6. Another group of 12 mice were fed a high-fat diet (HFD; D12451; Research Diets Inc., New Brunswick, NJ, USA) beginning at 8 weeks of age. A normal chow diet (MF; Oriental Yeast Co., Ltd., Tokyo, Japan) provided 3.6 kcal/g of energy (61% carbohydrate, 26% protein and 13% fat), whereas the HFD provided 4.7 kcal/g of energy (35% carbohydrate, 20% protein and 45% fat). After 4 weeks of the HFD, the mice were sacrificed at ZT6. There was no difference in food intake between the control and HFD groups (28.3±1.4 vs. 26.2±3.8 g at 8 weeks of age); however, a significant difference was observed in body weight between the two groups following each diet intake (28.0±2.4 vs. 35.3±2.1 g at 12 weeks of age; P<0.01).
Double immunohistochemical staining combined with RNAscope (in situ hybridization) to determine specificity of the PLAGL1 antibody
To confirm the specificity of the PLAGL1 antibody [ZAC1 antibody (C-7): cat. no. sc-166944, Santa Cruz Biotechnology, Inc., Dallas, TX, USA], double immunohistochemical staining was performed combined with in situ hybridization.
Mice were deeply anesthetized with pentobarbital (50 mg/kg, i.p.), and transcardially perfused with PBS, followed by 4% formaldehyde in 0.1 M phosphate buffer (pH 7.4). The whole brains were removed, immersed in 4% formaldehyde in 0.1 M phosphate buffer (pH 7.4) for 12 h at 4°C, cryoprotected in 20% sucrose in 0.1 M phosphate buffer (pH 7.4) for 6 h at 4°C, and cryosectioned into 25-µm coronal sections. Sections were stored in PBS/0.3% Triton X-100 (PBST) with 60% glycerol at −20°C until use. The free-floating sections were blocked with PBST/5% bovine serum albumin (BSA; cat. no. A-4503; Sigma-Aldrich; Merck KGaA) at room temperature for 2 h and incubated overnight at 4°C with a rabbit anti-hypocretin-1 antibody (1:5,000; cat. no. H-003-30; Phoenix Pharmaceuticals, Inc., Burlingame, CA, USA) and mouse anti-PLAGL1 antibody [1:1,000; ZAC1 antibody (C-7)] in PBST/5% BSA. The next day, sections were incubated with a donkey Cy5-labeled anti-mouse immunoglobulin (Ig)G (1:500; cat. no. 715-175-150, Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) and a goat Alexa Fluor® 488-labeled anti-rabbit IgG (1:3,000; cat. no. A-11008, Molecular Probes; Thermo Fisher Scientific, Inc., Waltham, MA, USA) for 30 min at room temperature. After washing with PBST, the stained sections were mounted on Superfrost Plus slides (Thermo Fisher Scientific, Inc.) and air-dried. Once washed with distilled water, slides were baked at 60°C for 30 min. Subsequently, in situ hybridization was conducted with the RNAscope® 2.5 HD Reagent kit-RED (cat. no. 322350; Advanced Cell Diagnostics, Inc., Newark, CA, USA), and probe Mm-Plagl1 (cat. no. 462941; Advanced Cell Diagnostics, Inc.), positive control probe Mm-RNA polymerase II subunit A (cat. no. 312471; Advanced Cell Diagnostics, Inc.) or negative control probe DapB (cat. no. 310043; Advanced Cell Diagnostics, Inc.), according to the manufacturer's protocols. After final amplification, Fast Red chromogenic detection was performed. The slides were coverslipped with DAPI Fluoromount-G (cat. no. 0100-20; Southern Biotech, Birmingham, AL, USA), and the resulting hypocretin, PLAGL1 protein and Plagl1 mRNA signals were visualized using confocal laser microscopy (LMS700; Zeiss GmbH, Jena, Germany). Bright-field images of Fast Red staining were captured using the Eclipse E-1000M (Nikon Corporation, Tokyo, Japan) with a digital camera.
The antibody used to identify hypocretin-immunoreactive neurons exhibits 100% cross-reactivity with human, bovine, mouse and rat hypocretin-1, but no cross-reactivity with human hypocretin-2 or other related peptides. Control experiments included the omission of the primary antibody, as well as extensive tests for cross-reactivity of the secondary antibodies. All control experiments resulted in the absence of staining.
Double immunohistochemical staining
The procedure of fixing, brain sectioning, blocking and probing with primary antibodies of male C57 black/6J mice (age, 12 and 18 weeks), was the same as the aforementioned procedure. After blocking, free-floating sections were incubated overnight at 4°C with rabbit anti-hypocretin-1 antibody (1:5,000) and mouse anti-PLAGL1 antibody (1:1,000), as well as goat anti-PLAGL2 antibody (1:500; cat. no. sc-19907; Santa Cruz Biotechnology, Inc.), goat anti-PLAG1 zinc finger antibody (1:500; cat. no. sc-20320; Santa Cruz Biotechnology, Inc.), mouse anti-FEV antibody (1:500; cat. no. H00054738-A01; Abnova, Taipei, Taiwan), goat anti-PRRX1 antibody (1:500; cat. no. LS-B2380; LifeSpan BioSciences, Inc., Seattle, WA, USA) and sheep anti-VSX2 antibody (1:500; cat. no. PAB9843; Abnova) in PBST/5% BSA. The next day, free-floating sections were incubated with goat, chicken or donkey Alexa Fluor® 594-labeled anti-mouse (cat. no. A11005), anti-goat (cat. no. A21468), or anti-sheep (cat. no. A11016) IgG, and goat (cat. no. A11008) or donkey (cat. no. R37118) Alexa Fluor® 488-labeled anti-rabbit IgG (1:3,000; Molecular Probes) for 30 min at room temperature followed by mounting with DAPI Fluoromount-G. The resulting immunopositive signals were visualized under a fluorescence microscope mounted with a digital camera (Olympus BX51 and DP70; Olympus Corporation, Tokyo, Japan).
Expression vectors and reporter plasmids
The CAG promoter derived from the pCA-pA expression vector was cloned into the pcDNA3 vector (Invitrogen; Thermo Fisher Scientific, Inc.), resulting in the development of the pCAGGS vector as a control for in utero electroporation (mock). High-fidelity PCR amplification of murine Plagl1 from the embryonic brain at E10-E12 was performed using KOD plus DNA polymerase (Toyobo Life Science, Osaka, Japan), betaine and either an mPlagl1_546F primer with a KpnI site (5′-CGGGGTACCAAAGGCCATGGCTCCATTCCGCTGTCA-3′) or an mPlagl1_2671R primer with a BamHI site (5′-TAGATCCGGTGGATCCTTATCTAAATGCGTGATGGA-3′). The KpnI and BamHI sites are underlined in the sequences provided. Following PCR amplification, as follows: One cycle at 98°C for 2 min, followed by 20 cycles of denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec and extension at 68°C for 2 min, and a final extension step at 68°C for 5 min, Plagl1 was sub-cloned into the pCAGGS vector (pCAGGS-mPlagl1). The expression vector pCAGGS-EGFP, which contains EGFP cDNA under the control of the CAG promoter, was prepared to confirm the introduced locations and check the introduction efficiencies for in utero electroporation in the murine hypothalamus (8).
The reporter plasmids pGL3-basic and pGL4.74 [hRluc/TK] were purchased from Promega Corporation (Madison, WI). The pGL4.74 [hRluc/TK], encoding Renilla luciferase, was used as an internal control of transfection efficiency for the reporter assay. The human prepro-hypocretin promoter sequence from the position −3,278/+87 was subcloned into the upstream region of the firefly luciferase gene in the pGL3-basic plasmid (3.2 kb-basic) (8). The murine prepro-hypocretin promoter sequence from the position −382/+91 amplified with KOD plus DNA polymerase, mHcrt_+92_BglII (5′-GGAAGATCTGGAACCTTTGTAGAAGGAAAG-3′; the BglII site is underlined), and mHcrt_-385_XhoI (5′-CCGCTCGAGAGGTACCCTCCCTACCTTCAA-3′; the XhoI site is underlined) was subcloned into the pGL3-basic plasmid (473 bp-basic). In the 473 bp-basic plasmid, the PLAGL1 binding site was deleted from the prepro-hypocretin promoter sequence.
All the constructs were confirmed to have no mutation, insertion or deletion by sequencing analysis using the BigDye Terminator Cycle Sequencing Reaction kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) and the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.). Both strands were read with sequence primers (Table SI).
Chromatin immunoprecipitation (ChIP)-PCR
The brains of 12-week-old C57 black/6J mice were rapidly removed at ZT6, and the hypothalamus was dissected coronally from the optic chiasma to the mammillary bodies (−3.5 mm from the bregma) using a brain slicer (Zivic Instruments, Pittsburgh, PA, USA). The dorsal limit of the hypothalamus was the roof of the third ventricle, while the lateral limit was the amygdala (21). The hypothalamus was finely minced with a razor blade and crosslinked with 1% formaldehyde for 10 min at room temperature; a final concentration of 0.125 M glycine was added to block further crosslinking. Tissues were washed twice with ice-cold PBS, and a crude nuclear extract was prepared according to a previously reported method (22). To obtain digested genomic DNA ranging between 150 and 900 bp, the nuclear extract was incubated with micrococcal nuclease (Cell Signaling Technology, Inc., Tokyo, Japan) for 20 min at 37°C. The size of the DNA fragments was confirmed by gel electrophoresis following treatment with RNase I for 30 min at 37°C and Proteinase K for 2 h at 65°C. The lysate was sonicated at 43 kHz three times (20 sec/sonication) at 4°C, centrifuged and the obtained supernatant was diluted in ChIP buffer (Cell Signaling Technology, Inc.) overnight at 4°C on a wheel rotator with either 2 µg mouse anti-ZAC1 antibody (C-7)X (cat. no. sc-166944 X; Santa Cruz Biotechnology, Inc.), which was used to capture the PLAGL1 protein, or 2 µg normal rabbit IgG (cat. no. 2729; Cell Signaling Technology, Inc.), which was used as a negative control. Samples were then incubated with Protein G agarose beads for 2 h at 4°C. Beads were washed with low- and high-salt wash buffers, and DNA templates for PCR were purified from the resulting DNA-protein complexes. ChIP-PCR analysis was performed using KOD plus DNA polymerase (Toyobo Life Sciences), primers (Table SI) and a thermal cycler, with the following settings: One cycle at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 58°C for 30 sec and extension at 68°C for 30 sec.
Luciferase reporter assay
NIH3T3 (murine embryonic fibroblast; American Type Culture Collection, Manassas, VA, USA) cells were grown in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere containing 5% CO2.
Cells were seeded at a density of 1x104 cells/well in 96-well cell culture plates. Two types of luciferase plasmids and one expression vector were co-transfected into the cells with Lipofectamine® 3000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The amounts of co-transfected plasmids and vectors per well were as follows: 100 ng firefly luciferase-encoding reporter plasmid (pGL3), 10 ng Renilla luciferase-encoding internal control plasmid (pGL4.74), and 100 ng expression vector pCAGGS (control plasmid as mock) or pCAGGS-mPlagl1. Approximately 48 h post-transfection, luciferase activities were measured sequentially in duplicate using Dual-Glo Luciferase Reporter assay reagents (Promega Corporation) and an EnSpire plate reader (PerkinElmer, Inc., Waltham, MA, USA), according to the manufacturer's protocol. The activity of firefly luciferase luminescence (FLU) from the pGL3 plasmid and the activity of Renilla luciferase luminescence (RLU) from the pGL4.74 plasmid were measured independently. The relative luciferase activity per well was determined as FLU divided by RLU values. Data for the relative activities were expressed as the mean of at least eight independent experiments. Mouse Plagl1 mRNA expression was confirmed after transfection by PCR with gene specific primers (Table SI) (data not shown).
In situ hybridization of embryos
In situ hybridization using digoxigenin-labeled cRNA probes was performed on cryosections. The procedure of fixing and sectioning brain samples at E13, E14, and E18 were the same as aforementioned. Briefly, at E10-E12, embryos were immersed in 4% formaldehyde in 0.1 M phosphate buffer (pH 7.4) for 16 h at 4°C, cryoprotected in 20% sucrose in 0.1 M phosphate buffer (pH 7.4) for 6 h at 4°C and cryosectioned into 25-µm coronal sections. Riboprobes for murine Plagl1 and negative control riboprobes for murine Plagl2 were generated by PCR with either the mPlagl1_606F (5′-GAAGTTCACCATTCACAATTATTCC-3′) and mPlagl1_1992R (5′-AATGGCTATATTCACAGCATCTACC-3′) primer pair, the mPlagl1_1968F (5′-GGTAGATGCTGTGAATATAGCCATT-3′) and mPlagl1_3513R (5′-TGAGTATCACAGTCCACAAAACACT-3′) primer pair, the mPlagl2_587F (5′-GAGTGTGGTAAGAATTATAATACAAAGCTG-3′) and mPlagl2_1822R (5′-TCCATATAGACACTAGAATTTTCTTTTTGA-3′) primer pair, or the mPlagl2_3352F (5′-ACTAGGGTTTCAGGACAGACTACCT-3′) and mPlagl2_4857R (5′-AACACCTACCTAAGGCAAAGTTTCT-3′) primer pair, with subsequent sub-cloning of the PCR products into the pGEMT-easy vector (Promega Corporation) and in vitro transcription with DIG-11-UTP (cat. no. 3359247910; Sigma-Aldrich; Merck KGaA). Brain sections were post-fixed in 4% formaldehyde for 10 min at 20-22°C, acetylated with 0.5% acetic anhydride in 0.1 M triethanolamine (pH 8.0) for 10 min and rinsed in PBS. The slides were pre-hybridized in a hybridization buffer without cRNA probe at room temperature for 2 h and hybridized using a hybridization buffer (50% formamide, 5X SSPE, 1 mg/ml yeast tRNA, 0.2% sodium dodecyl sulfate) containing 1 µg/ml cRNA probe at 60°C overnight. The next day, slides were washed in 2X SSC containing 50% formamide at 60°C for 1 h. Hybridization was detected using an anti-digoxigenin Fab (1:2,000; cat. no. 11093274910; Sigma-Aldrich; Merck KGaA) for 1 h at 20-22°C coupled to alkaline phosphatase using NBT/BCIP. The resulting digoxigenin signals with Plagl probes were captured using a microscope mounted with a digital camera (Olympus BX51 and DP70; Olympus Corporation).
In utero electroporation
In utero electroporation was performed according to a previously reported method (8), with some modifications. Briefly, ~1 µl pCAGGS-mPlagl1 and pCAGGS-EGFP expression vectors (2 µg/µl) for the PLAGL1 group, or 1 µl pCAGGS and pCAGGS-EGFP expression vectors (2 µg/µl) for the mock group were mixed with 0.05% Fast Green as a tracer and injected into the third ventricle of each C57 black/6J mouse embryo (E12) using free-hand injections with a glass capillary pipette through the uterine wall. Electrodes (3 mm round shape; CUY650P3; Nepa Gene Co., Ltd., Ichikawa, Japan) were placed on both sides of the embryo, near the head of the embryo, and electro-poration was performed using a square-pulse electroporator CUY21 EDIT in vivo electroporator (Nepa Gene Co., Ltd.) (four pulses; amplitude, 39V; duration 50 msec; interval between pulses, 950 msec), resulting in gene transfer into one side of the hypothalamus. Control mice were electropor-ated with the same vectors from the lateral ventricle into the cortex. Intact embryos or gene electroporated-embryos were harvested at E14, E15, E18 or postnatal day (P)0, and the embryos were transcardially perfused with 4% formaldehyde in a 0.1 M phosphate buffer (pH 7.4). The whole brains were removed, post-fixed, and cryosectioned into 30-µm coronal sections. Hypocretin and PLAGL1 expression levels were detected by immunofluorescence methods using either a rabbit anti-hypocretin-1 or a mouse ZAC1 antibody, and a goat Alexa Fluor® 594-labeled anti-rabbit or anti-mouse IgG. The resulting hypocretin and PLAGL1 signals were visual-ized under a fluorescence microscope mounted with a digital camera (Olympus BX51 and DP70; Olympus Corporation). To reduce the background signals in embryo staining, the Mouse on Mouse Immunodetection kit (Vector Laboratories, Inc., Burlingame, CA, USA) was used, according to the manufacturer's protocols.
Quantitative PCR (qPCR)
Total RNA was isolated from the hypothalamus of each of the control animals (n=8, 12 weeks old), HFD animals (n=8) at ZT6, P0 embryos in the mock group (n=4) or P0 embryos in the PLAGL1 group (n=6) using Sepasol-RNA I Super G reagent (Nacalai Tesque, Inc., Kyoto, Japan). Single-stranded cDNA was synthesized using the PrimeScript RT reagent kit with gDNA Eraser (Takara Bio, Inc., Otsu, Japan), according to manufacturer's protocol. The expression levels of each mRNA were determined by qPCR with the Rotor-Gene Q system (Qiagen GmbH, Hilden, Germany) using the THUNDERBIRD qPCR mix (Toyobo Life Science) and gene-specific primers (Table SI). PCR products were amplified using the following thermocycling conditions: One cycle at 95°C for 1 min, followed by 40 cycles at 95°C for 10 sec and 60°C for 60 sec. The housekeeping gene with minimum diurnal variation in the hypothalamus was identified in a previous study by checking gene expression every 4 h over a 24-h period (23). Subsequently, the relative level of target gene expression was evaluated using the 2-ΔΔCq method (24) with hypoxanthine phosphoribosyltransferase 1 as an internal control.
Statistical analyses
Statistical analyses were conducted using IBM SPSS version 25 (IBM Corp., Armonk, NY, USA) or Microsoft Excel 2013 software (Microsoft Corporation, Redmond, WA, USA). Data are presented as the means ± standard deviation. Normality was analyzed by the Shapiro-Wilk normality test for all groups. F-test was used to determine homoscedasticity for all comparisons. All groups were normally distributed in the hypocretin neuron counts, PLAGL1 counts and PLAGL1 immunopositivity experiments (P>0.05). One-way analysis of variance (ANOVA), followed by Steel's post hoc test against the control group (ZT6), was used to analyze the differences; P<0.05 was considered to indicate a statistically significant different (n=4-5). The differences in hypocretin and Plagl1 expression due to HFD compared to the control group were compared using two-tailed Student's t-tests. To evaluate alterations in β-2-microglobulin (B2m) expression due to HFD, the two-tailed Welch's t-test was used due to a lack of homoscedasticity; P<0.05 was considered to indicate a statistically significant difference (n=8). Alterations in promoter activity for each reporter plasmid with/without the PLAGL1 expression vector, or from pGL3-basic, were compared using one-way ANOVA followed by Tukey's post hoc test; P<0.05 was considered to indicate a statistically significant difference (n=8). The qPCR data of the in utero electroporation experiments were not normally distributed (P<0.05); therefore, the Mann-Whitney U-test was used to compare differences; P<0.05 compared to the contralateral side (mock group, n=4; PLAGL1 group, n=6) was considered to indicate a statistically significant difference.
Results
Localization of PLAGL1 in adult hypothalamic hypocretin neurons
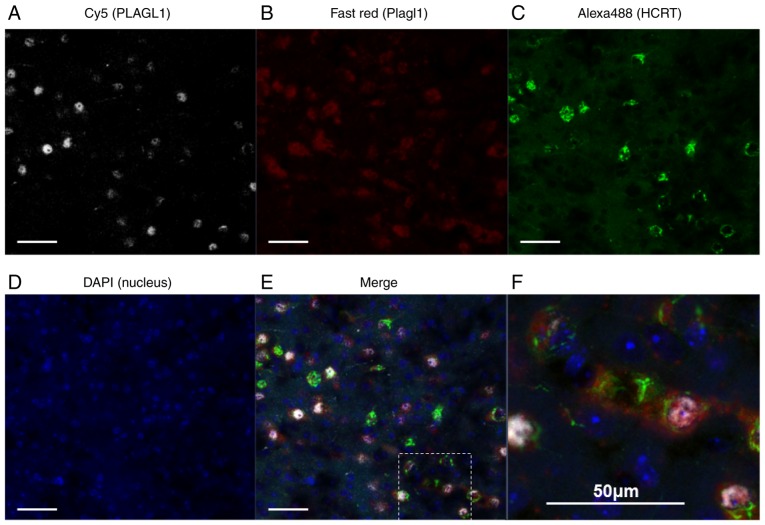
Plagl1 mRNA expression was clearly detected in the lateral hypothalamic area (Fig. S1), which has also been reported in the Allen Brain Atlas database (25). FEV PRRX1, VSX2 and other PLAG family proteins, including PLAG1 and PLAGL2, were not detected in the lateral hypothalamic area in this study (data not shown). To confirm the specificity of the PLAGL1 antibody [ZAC1 antibody (C-7)], double immunohis-tochemical staining was performed in combination with in situ hybridization. Neurons with PLAGL1 signals (i.e., neurons immunopositive for the anti-PLAGL1 antibody) exhibited 100% merge with RNAscope Plagl1 signals in three different fields. A total of 85 cells positive for PLAGL1 were detected in 85 cells with RNAscope Plagl1 signals. However, neurons with RNAscope Plagl1 signals were not always merged with the PLAGL1 signals stained by anti-PLAGL1 antibody. The results demonstrated that 15% of RNAscope Plagl1-positive neurons were not merged with the PLAGL1 signals stained by anti-PLAGL1 antibody (Fig. S2). In the RNAscope experiments, the hypothalamus sections were treated with Proteinase K; therefore, it is possible that PLAGL1 proteins were partly degraded, resulting in weak or no immunopositive signals. Notably, hypocretin signals were weaker in Proteinase K-treated sections (Figs. 1 and S3) than in non-treated sections (Fig. 2).
Figure 1.
Double immunohistochemical staining and RNAscope in the murine lateral hypothalamic area at ZT6. (A) PLAGL1 was visualized by Cy5 (white), (B) Plagl1 was visualized by Fast Red (red), (C) HCRT was visualized by Alexa Fluor® 488 (green), (D) and DAPI labeled the nuclei (blue). (E) Merged image of A-D; (F) expanded views of the square field in E. Scale bar, 50 µm. ZT, Zeitgeber time; HCRT, hypocretin; PLAGL1, pleomorphic adenoma gene-like 1.
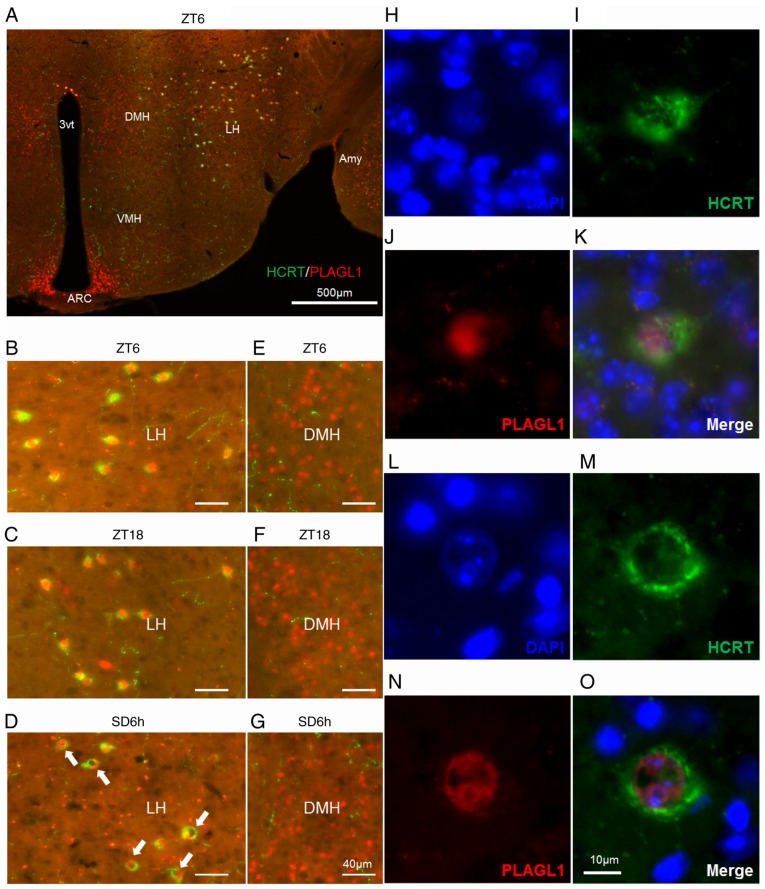
Figure 2.
Localization of PLAGL1 in murine hypocretin neurons under several conditions. (A-G) Murine HCRT was visualized by staining with Alexa Fluor® 488 (green) and murine PLAGL1 was visualized by staining with Alexa Fluor® 594 (red). (A) Murine hypothalamic sections at ZT6. The lateral hypothalamic area under (B) ZT6, (C) ZT18 and (D) SD6h conditions, with a relatively specific and uneven distribution of PLAGL1 (arrows). The dorsomedial hypothalamic nucleus under (E) ZT6, (F) ZT18 and (G) SD6h conditions. (H) DAPI-labeled nuclei at ZT6. (I) Alexa Fluor®488-labeled murine HCRT at ZT6. (J) Alexa Fluor® 594-labeled murine PLAGL1 at ZT6. (K) Merged image of (H, blue), (I, green) and (J, red) immunofluorescence at ZT6. (L) DAPI-labeled nuclei at SD6h. (M) Alexa Fluor®488-labeled murine HCRT at SD6h. (N) Alexa Fluor®594-labeled murine PLAGL1 at SD6h. (O) Merged image of (L, blue), (M, green), and (N, red) immunofluorescence at SD6h. Scale bar, (A) 500 µm, (B-G) 40 µm, (O) 10 µm. (H-O) These images are displayed at the same magnification. 3vt, third ventricle; Amy, amygdala; ARC, arcuate nucleus; DMH, dorsomedial hypothalamic nucleus; HCRT, hypocretin; LH, lateral hypothalamus; PLAGL1, pleomorphic adenoma gene-like 1; SD6h, 6 h of sleep deprivation; VMH, ventromedial nucleus of the hypothalamus; ZT, Zeitgeber time.
PLAGL1 is expressed in the lateral hypothalamic area, dorsomedial hypothalamic nucleus and arcuate nucleus of the adult male mouse hypothalamus at ZT6 (Fig. 2A) and is localized within hypocretin neurons (Fig. 2A and B). A relatively specific and uneven nuclear localization of PLAGL1 was detected in the hypocretin neurons of the lateral hypothalamic area in mice with SD6h (Fig. 2D, arrows) compared with ZT6 (Fig. 2B) and ZT18 (Fig. 2C) in the dorsomedial hypothalamic nucleus at ZT6, ZT18 and SD6h (Fig. 2E-G), or the arcuate nucleus (data not shown). Subsequently, DAPI staining revealed diffuse nuclear staining of PLAGL1 at ZT6 (Fig. 2B and H-K) and a heterochromatin-like structure in the hypocretin neurons with SD6h. The uneven nuclear localization of PLAGL1 was not merged with the heterochromatin-like structure (Fig. 2L-O), suggesting that PLAGL1 was specifically related to the euchromatin structure and transcription in hypocretin neurons following SD6h. Furthermore, a significant increase was noted in the number and frequency of PLAGL1-positive hypocretin neurons at ZT18 compared with the control group (ZT6) (Fig. 3).
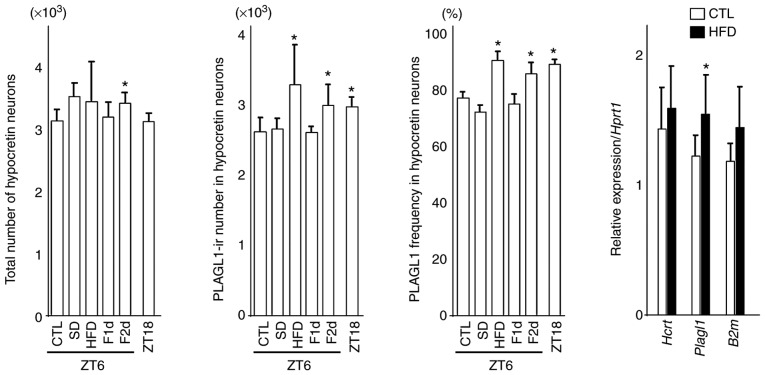
Figure 3.
Number of hypocretin neurons, PLAGL1-ir number in hypocretin neurons, frequency of PLAGL1 and relative gene expression in hypocretin neurons. Differences in the count of hypocretin neurons, PLAGL1 counts and PLAGL1 immunopositivity were compared using one-way analysis of variance followed by Steel's post hoc test. *P<0.05 vs. the control (CTL) group (ZT6). Four or five animals from each group were used for each immunohistochemical analysis. The differences in relative gene expression were compared using two-tailed Student's t-tests for hypocretin and Plagl1 expression, and two-tailed Welch's t-test for B2m. Eight animals from each group were used for gene expression analysis. B2m, β-2-microglobulin; CTL, control (mice sacrificed at ZT6); F1d, 1 day of fasting; F2d, 2 days of fasting; Hcrt, hypocretin; HFD, high-fat diet; ir, immunoreactive; Plagl1, pleomorphic adenoma gene-like 1; SD, 6 h of sleep deprivation; ZT, Zeitgeber time; ZT18, mice sacrificed at ZT18. Bars indicate means, and error bars represent standard deviations.
Notably, 1 month of a HFD slightly increased the number of hypothalamic hypocretin neurons and induced a significant increase in the number and frequency of PLAGL1-positive hypocretin neurons compared with the control group (ZT6). In addition, 2 days of fasting affected the number of hypothalamic hypocretin neurons, and induced a significant increase in the number and frequency of PLAGL1-positive hypocretin neurons compared with the control group (ZT6) (Fig. 3).
There was a significant increase in Plagl1 expression in the HFD group (n=8) (mean ± standard deviation, 1.55±0.30; P<0.05) compared with the control group (n=8) (1.22±0.15), although there was no significant difference in hypocretin (HFD vs. control, 1.58±0.32 vs. 1.44±0.31) and B2m (1.44±0.32 vs. 1.18±0.13) expression (Fig. 3). B2m was examined as a reference gene.
PLAGL1 binds to the hypocretin upstream region in vivo
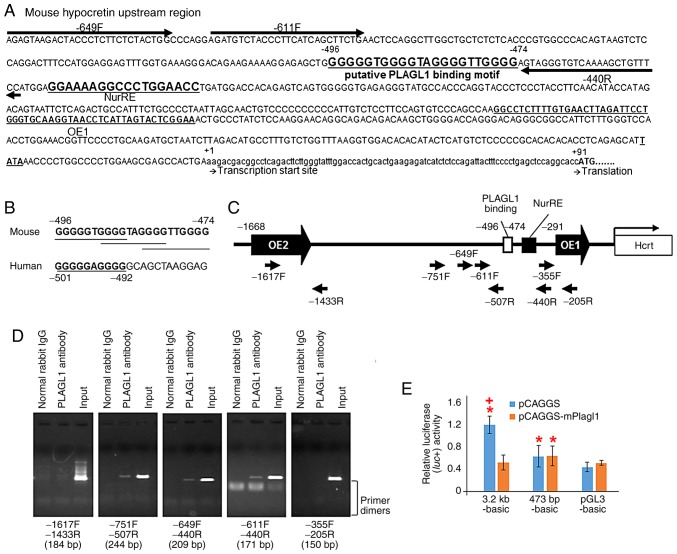
The 'find' tool in Microsoft Word 2013 software (Microsoft Corporation) was used to detect the known consensus binding site for PLAGL1 (19,26) in the hypocretin gene upstream region, which is highly conserved between humans and mice and includes OE1 and OE2 (2,3). Newly identified PLAGL1-binding sites (G4N2G4) (19) were found and are denoted as GGGGGTGGGGTAGGGGTTGGGG at position −496/−474, relative to the transcription start site, in the upstream region of the murine hypocretin gene (Fig. 4A), and as GGGGGAGGGG at position −501/−492 in the human sequence (Fig. 4B). Subsequently, a ChIP-PCR analysis was performed using primers specific to the murine hypocretin upstream region around the G4N2G4 motifs (Fig. 4A and C) and negative control primers (−1617F and −1433R pair, and −355F and −205R pair) (Fig. 4C), with purified DNA immu-noprecipitated with an anti-PLAGL1 antibody. ChIP-PCR analysis with the mHcrt_-611F and mHcrt_-440R primer pair revealed a positive reaction for the murine hypocretin upstream region around the G4N2G4 motifs (Fig. 4C and D). An additional positive reaction was found in the experiment using the mHcrt_-649F and mHcrt_-440R primer pair and the mHcrt_-751F and mHcrt_-507R primer pair (Fig. 4C and D). No amplification was detected from the purified DNA incubated with normal rabbit IgG (Fig. 4C and D) or water, which were used as negative control templates, or negative control experiments with the mHcrt_-1617F and mHcrt_-1433R primer pair and the mHcrt_-355F and mHcrt_-205R primer pair (Fig. 4D).
Figure 4.
Schematic representation of the murine prepro-hypocretin gene regulatory region, putative PLAGL1 binding sites in mice and humans, ChIP-PCR and luciferase reporter assay. (A) Sequence of the murine prepro-hypocretin gene regulatory region. Highly conserved region orexin regulatory element 1, which has been previously reported (2,3). The first residues of the transcription start site and the translation start site are marked as +1 and +91, respectively. NurRE (8), the putative PLAGL1 binding site and primers for ChIP-PCR analysis are shown. (B) Putative PLAGL1 binding sites in mouse and human promoter sequences are shown in bold and are underlined. (C) Schematic representation of the murine prepro-hypocretin gene regulatory region, and the position of ChIP-PCR primers. (D) ChIP-PCR analysis of endogenous PLAGL1 binding using ChIP-PCR primers. The length of each PCR product is represented at the bottom of each image. (E) Transcriptional activities of the human prepro-hypocretin promoter and murine deletion mutants in NIH3T3 cells. *P<0.05 vs. pGL3-basic with pCAGGS vector. +P<0.05 vs. each reporter with pCAGGS-mPlagl1. ChIP, chromatin immunoprecipitation; IgG, immunoglobulin G; NurRE, nuclear receptor response element; PLAGL1, pleomorphic adenoma gene-like 1; PCR, polymerase chain reaction.
PLAGL1 overexpression suppresses hypocretin promoter activity in NIH3T3 cells
Co-transfection of the pGL3-basic plasmid with the pCAGGS (mean ± standard deviation, 0.444±0.090) or pCAGGS-mPlagl1 (0.517±0.044) vectors exhibited similar luciferase expression in NIH3T3 cells, indicating an absence of confounding endogenous transcriptional regulatory elements in the pGL3-basic plasmid (Fig. 4E). The luciferase activity from 3.2 kb-basic with the pCAGGS vector (mock) exhibited a significant three-fold increase compared with the pGL3-basic with pCAGGS vector (mock) (1.205±0.161; P<0.001) in NIH3T3 cells (Fig. 4E). The observed activities in the 3.2 kb-basic group were suppressed by co-transfection with the pCAGGS-mPlagl1 vector compared to mock in NIH3T3 cells (0.521±0.135; P<0.05; Fig. 4E).
Subsequently a deletion mutant of the PLAGL1 binding site was generated (473 bp-basic). This deletion mutant reporter plasmid with mock led to significant increases in relative lucif-erase activity compared to pGL3-basic with mock in NIH3T3 cells (0.637±0.198; P<0.05; Fig. 4E). No response to PLAGL1 was detected in the deletion mutant 473 bp-basic reporter plasmid with the pCAGGS-mPlagl1 vector compared to the deletion mutant with mock (0.644±0.183; Fig. 4E), suggesting that the PLAGL1 binding ability to the preprohypocretin promoter region disappeared in the 473 bp-basic reporter plasmid; however, the promoter activities remained. Notably, the transcriptional activities of the prepro-hypocretin promoter sequence may be suppressed by PLAGL1 via the putative PLAGL1 binding site in NIH3T3 cells.
Localization of PLAGL1 within embryonic hypocretin neurons
Murine Plagl1 was detected in the periventricular zone of the mesencephalic, forebrain and telencephalic vesicle at E10, using either the mouse Plagl1 cRNA 606F-1992R probe (Fig. 5) or the 1968F-3513R probe (data not shown). Plagl1 signals were subsequently detected in periventricular areas and the hypothalamic sulcus at E11-E13. The co-expressed signals of hypocretin and PLAGL1 were also detected after E14 using immunofluorescence detection. Hypocretin neurons were positive for PLAGL1 in their nuclei at E18 (Fig. 5). There was no signal with Plagl2 probes (data not shown).
Figure 5.
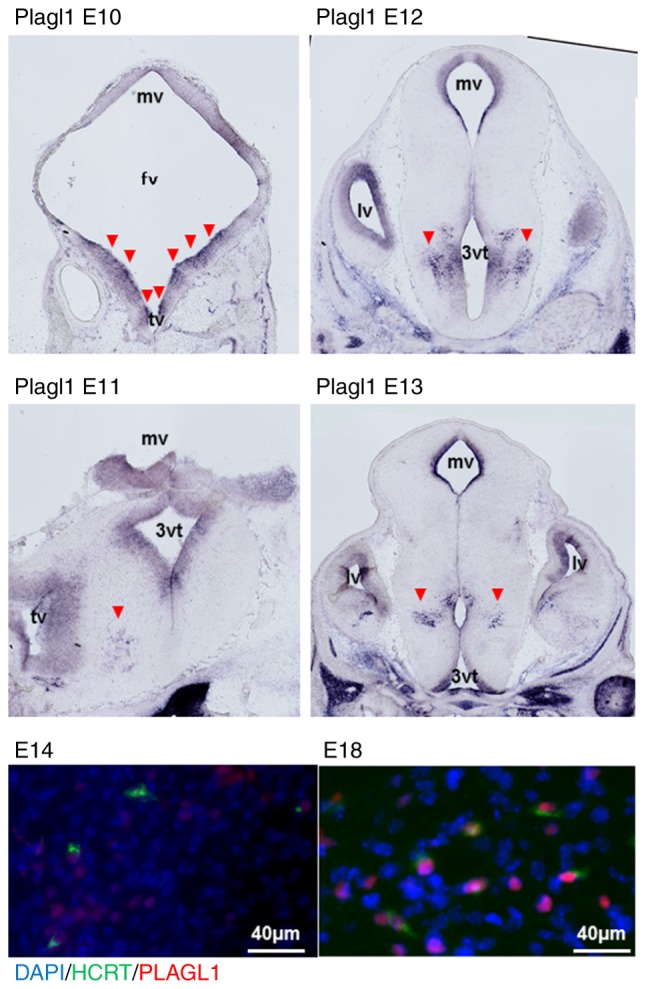
Plagl1 expression in embryonic murine brains and localization of PLAGL1 in hypocretin neurons of the embryonic murine hypothalamus. The Plagl1 RNA probe detected Plagl1 expression in periventricular zones and the hypothalamus at E10-E13 by in situ hybridization (red arrowheads) (magnification, x20). Murine HCRT is visualized by staining with Alexa Fluor®488 (green) and murine PLAGL1 is visualized by staining with Alexa Fluor® 594 (red) at E14 and E18. 3vt, third ventricle; E, embryonic day; fv, forebrain vesicle; HCRT, hypocretin; mv, mesencephalic vesicle; PLAGL1, pleomorphic adenoma gene-like 1; tv, telencephalic vesicle.
In utero electroporation
To confirm the physiological relevance of PLAGL1 in hypothalamic hypocretin transcription, Plagl1 overexpression in the murine hypothalamus was generated using in utero electroporation. Electroporated EGFP expression was detected on one side of the hypothalamus at E15 (Fig. S4, arrows). The present study confirmed the co-expression of PLAGL1 and EGFP following electroporation into the cortex using immunofluorescence detection (Fig. S5); however, it was noted that some background signals did not overlap with the EGFP signals even when using the Mouse on Mouse Immunodetection kit, which could be considered either endogenous neural expression, or the presence of blood cells and vesicles in the cortex. Although most EGFP-positive cells did not express hypocretin in the vicinity of the third ventricle, some EGFP-positive cells that moved to the cerebral parenchyma from the vicinity of the third ventricle were hypocretin-positive (Fig. S6, arrows). Hypocretin expression in the cortex could not be detected after in utero electroporation of pCAGGS-mPlagl1 and pCAGGS-EGFP into the lateral ventricle (Fig. S5), suggesting that PLAGL1 specifically affects hypothalamic hypocretin transcription. In some cases, EGFP/Plagl1 might have been electroporated from the bottom of the lateral ventricle or the outer portion of the dura matter, inducing hypocretin expression in the subthalamic nucleus and lateral hypothalamic area (Fig. S7). These findings suggested that PLAGL1 may suppress hypocretin expression in the vicinity of the third ventricle as no hypocretin expression was detected in EGFP-positive cells in this zone, whereas the effects of PLAGL1 on transcription may be sufficient in some cells of the cerebral parenchyma, the lateral hypothalamic area and the subthalamic nucleus.
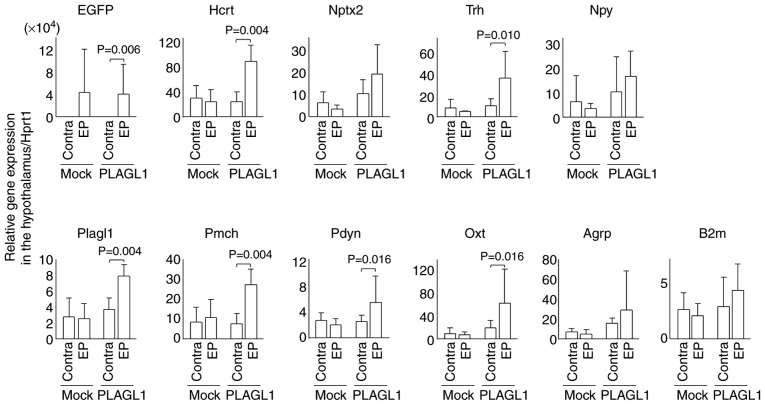
In order to avoid false positives associated with background signals in immunohistochemical staining, qPCR was performed to confirm the transcriptional effects of PLAGL1 on hypocretin. The EGFP expression levels were elevated on the electroporated side as compared with the contralateral side (contralateral side: mean ± standard deviation, 1,383.9±1,978.6 in the mock group, 723.4±1,057.6 in the PLAGL1 group; electroporated side: 42,006.1±75,966.2 in the mock group, 38,586.3±53,484.7 in the PLAGL1 group) in both the mock and PLAGL1 groups (Fig. 6). There was no significant difference in Plagl1 expression between the contralateral side (2.8±2.4) and the electroporated side (2.6±1.8) in the mock group, but there was a significant difference in Plagl1 expression (P<0.005) between the contralateral side (3.7±1.4) and electroporated side (7.9±1.5) in the PLAGL1 group. There was no significant difference in hypocretin expression between the contralateral side (27.6±20.1) and electroporated side (22.7±18.6) in the mock group, but there was a significant difference (P<0.01) in hypocretin expression between the contralateral side (21.9±15.3) and electroporated side (83.5±25.9) in the PLAGL1 group. Therefore, these data suggested that PLAGL1 may upregulate hypocretin transcription. PLAGL1 slightly affected neuronal pentaxin 2 transcription and significantly affected prodynorphin transcription. These two genes are concurrently expressed in hypocretin neurons (27,28). There was no effect of PLAGL1 on the housekeeping gene, B2m.
Figure 6.
Relative expression levels of hypothalamic genes following in utero electroporation. In utero electroporation was performed with pCAGGS-EGFP and pCAGGS-mPlagl1 (PLAGL1 group, n=6) to evaluate the effect of PLAGL1 on Hcrt transcription. Control mice were electroporated with pCAGGS-EGFP and pCAGGS (mock group, n=4). Animals were harvested at P0 stage. Following total RNA extraction from the hypothalamus and cDNA synthesis, quantitative polymerase chain reaction was performed. The relative expression levels of each gene were normalized to the relative expression levels of Hprt1. Alterations in transcription levels between the electroporated and contra sides in each group were compared using the Mann-Whitney U-test. Data are presented as the means ± standard deviation. Agrp, agouti related neuropeptide; B2m, β-2-microglobulin; Contra, contralateral side (no electroporation); EGFP, enhanced green fluorescent protein; EP, electroporated side; Hcrt, hypocretin; Hprt1, hypoxanthine phosphoribosyltransferase 1; Nptx2, neuronal pentaxin 2; Npy, neuropeptide Y; Oxt, oxytocin; Pdyn, prodynorphin; Pmch, pro-melanin-concentrating hormone; Trh, thyrotropin-releasing hormone.
Since PLAGL1 is expressed in the lateral hypothalamic area, the dorsomedial hypothalamic nucleus and the arcuate nucleus of the hypothalamus, this study examined whether PLAGL1 affects transcription of other neuropeptides in hypothalamic neurons that project to/from hypocretin neurons. PLAGL1 overexpression affected the expression of pro-melanin-concentrating hormone, thyrotropin-releasing hormone (Trh) and oxytocin (Oxt), but not that of neuro-peptide Y (Npy) and agouti related neuropeptide (Agrp) (Fig. 6 and Table SII).
Discussion
Hypocretin contributes to energy expenditure and feeding behavior by regulating the activation of wakefulness and the sympathetic system (29,30), as well as the promotion of food-seeking behaviors (31-33). During fasting, hypocretin expression is regulated by FOXA2, a downstream target of insulin signaling (10). The present study demonstrated that hypocretin-immunoreactive neurons increased after fasting. It was hypothesized that PLAGL1, whose expression is also altered in response to fasting, may contribute to food-seeking behaviors after fasting. An increase in PLAGL1 frequency in the hypocretin nucleus was detected in the HFD group. Since an alteration in c-fos expression has been observed in 2-3-month-old mice fed a HFD (34), PLAGL1 may contribute to a change in hypocretin neuronal activity in response to a HFD. In addition, hypocretin-immunoreactive neurons and hypocretin mRNA were increased after a 1-month HFD. Therefore, PLAGL1 may be associated with hypocretin transcription even under a HFD condition. The role of hypo-cretin in the maintenance and stability of normal sleep and wakefulness is well known (35). The present study revealed an increase in PLAGL1 in the active phase (ZT18) of hypo-cretin neurons, but no increase in hypocretin-immunoreactive cells in the active phase (ZT18); these findings suggested that PLAGL1 may have no effect on hypocretin transcription at night. Furthermore, the changes in hypocretin neuronal activity and the fluctuation of cerebrospinal fluid hypocretin-1 levels across the light-dark cycle and sleep-wake activities have been well documented (36-38); therefore, PLAGL1 may be involved in the modulation of hypocretin neuronal activity or the regulation of the hypocretin secretion-related protein at night. Increases in cerebrospinal fluid hypocretin levels following sleep deprivation (39,40) are accompanied by an increase in hypocretin-immunoreactive neurons (41), c-fos expression (42) and hypocretin mRNA expression (43), thus suggesting that hypocretin neurons may act to sustain vigilance during sleep deprivation. However, the present study did not detect a significant increase in hypocretin-immunoreactive neurons following sleep deprivation. Instead, sleep deprivation induced an uneven nuclear localization of PLAGL1. Because uneven nuclear localization of PLAGL1 also existed in control conditions (ZT6) and in ZT18, this change in distribution to an uneven position in the nucleus may be associated with an unknown or undefined physiological condition in hypocretin neurons. In the majority of interphase cells, DAPI-bright heterochromatin is present in the peripheral compartment, chromocenters and around the nucleoli. Heterochromatin regions hold a packed conformation and the included genes are silent. Euchromatin is a dim area in the inner compartment of the nucleus (44-46). In this area, tissue-specific genes are transcribed. PLAGL1 was localized in the euchromatin region of hypocretin neuronal nuclei upon sleep deprivation. Since there was no direct effect of PLAGL1 on hypocretin transcription in mice with sleep deprivation, it may be suggested that PLAGL1 binds the euchromatin region upon sleep deprivation in order to regulate other mediators of hypocretin neuron activities.
The consensus binding site for PLAGL1 was identified as the sequence G6C4 by the Systematic Evolution of Ligands by EXponential (SELEX) enrichment method (26); however, there were no G6C4 sequences within the 0 to 2.0 kb upstream of the transcription start site in the murine hypocretin gene, as determined by the present manual search. Because SELEX aptamers consist of randomly generated sequences, there is a possibility that some genomic sequences were not examined using the SELEX method. Novel PLAGL1 binding sites (G4N2G4) have been identified by ChIP and genome-wide transcriptomics in PLAGL1 vector-transfected Neuro2a cells (19). However, no hypocretin locus, including the ±20-kb region, was immunoprecipitated in these experiments (19). The hypocretin gene locus might not be in the active state, thus explaining transient expression of PLAGL1 in Neuro2a cells. PLAGL1 cannot reach the hypocretin upstream region and, therefore, the hypocretin locus would not appear in the list of genes of Neuro2a cells (19). It has been established that conversion from the inactive state to the active state by epigenetic switching is critical to the transcriptional regulation of hypocretin, but that requires time (47,48). Conversely, the present in vivo ChIP assay using hypothalamic tissue revealed PLAGL1 binding to the hypocretin locus. PLAGL1 binding sites could be present in a constant active state and activated by several conditions. This G4N2G4 consensus sequence is conserved even in the upstream region of the human hypo-cretin gene; therefore, PLAGL1 might also be important in human hypocretin transcription.
PLAGL1 can function as a transcription activator or repressor (26,49,50). In this study, functional differences in PLAGL1 were detected; specifically, PLAGL1 acted as a repressor in NIH3T3 cells and as an activator in the embryonic murine hypothalamus for hypocretin transcription. Under 2-day fasting and HFD conditions, PLAGL1 frequency and hypocretin-immunoreactive neurons were increased. One possible explanation for this discrepancy is that PLAGL1 monomer binding to G4N6G4 results in transcriptional repression, while dimer binding to G4N6G4 induces transactivation (49,50). The same discrepancy might occur at G4N2G4 sites. Since there are three G4N2G4 binding sites in the proximal region of the prepro-hypocretin gene, dimer binding of PLAGL1 might occur after 2 days of fasting, a HFD and during embryonic development. There might be different proteins or small-RNAs that bind G4N2G4 sites in NIH3T3 cells. As a result, PLAGL1 can only bind to one site of G4N2G4 as a monomer in NIH3T3 cells.
In the present Plagl1 overexpression model using in utero electroporation, it was revealed that PLAGL1 may induce hypocretin expression, specifically in the hypothalamus. The majority of EGFP signals co-localized with hypocretin were detected in the cerebral parenchyma at a distance from the vicinity of the third ventricle, suggesting that some progenitor cells of the third ventricular zone need to migrate to the designated area from the ependymal layer of the third ventricle. The differentiation of these cells might also be affected by the gradient of some factors, including the hypothalamic patterning factor sonic hedgehog (9), the adult hypocretin neurogenesis inducer fibroblast growth factor 2 (51), generators of hypocretin neurons (47,48), or other unidentified factors at the time of migration or arrival at the designated area.
Neurons expressing Trh and Oxt interact directly with hypocretin neurons (52,53) and were shown to be modulated by PLAGL1 overexpression in this study; however, no effect was detected on Npy or Agrp expression. Neurons expressing Npy and Agrp project to hypocretin neurons from the arcuate nucleus (52,53). It may be possible that there could have been some indirect effects from other neurons affected by PLAGL1 that project to hypocretin neurons.
In conclusion, the present study revealed the involvement of PLAGL1 in hypocretin transcriptional regulation; however, PLAGL1 is also expressed in other areas of the hypothalamus. Therefore, further in vivo studies using cross-hybridization with Plagl1 conditional knockout mice and hypocretin promoter-Cre mice (54) are required to clarify whether PLAGL1 directly or indirectly affects hypocretin expression.
Supplementary Materials
Acknowledgments
The authors would like to thank Ms. Hitomi Okabe (Kansai Medical University) for her technical assistance.
Funding
This study was supported by JSPS KAKENHI [grant nos. 15K08224 (ST) and 16K08533 (ST)] and the Takeda Science Foundation (ST). The funding agency had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SuT, ToK and HY were involved in study concept and design. SuT, YoH, ShT, TaK, TY and NT acquired the data. SuT, YoH and ShT processed the data. SuT, SO, YuH, YT, KK, ToK and HY analyzed and interpreted the data. SuT drafted the manuscript and ShT, TaK, SO, YuH, YT, KK, ToK and HY critically revised it for important intellectual content. SuT conducted the statistical analysis. SuT, YT, ToK and HY provided materials, and YuH, YT, KK, ToK and HY supervised the study. All authors have approved the final draft of the manuscript that was submitted.
Ethics approval and consent to participate
All animal experiments were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals of the National Institute of Health, and were approved by the Ethics committee on Animal Experiments of the Kansai Medical University (approval ID: 17-050) and the Tokyo Metropolitan Institute of Medical Science.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sakurai T. The role of orexin in motivated behaviours. Nat Rev Neurosci. 2014;15:719–731. doi: 10.1038/nrn3837. [DOI] [PubMed] [Google Scholar]
- 2.Sakurai T, Moriguchi T, Furuya K, Kajiwara N, Nakamura T, Yanagisawa M, Goto K. Structure and function of human prepro-orexin gene. J Biol Chem. 1999;274:17771–17776. doi: 10.1074/jbc.274.25.17771. [DOI] [PubMed] [Google Scholar]
- 3.Moriguchi T, Sakurai T, Takahashi S, Goto K, Yamamoto M. The human prepro-orexin gene regulatory region that activates gene expression in the lateral region and represses it in the medial regions of the hypothalamus. J Biol Chem. 2002;277:16985–16992. doi: 10.1074/jbc.M107962200. [DOI] [PubMed] [Google Scholar]
- 4.Amiot C, Brischoux F, Colard C, La Roche A, Fellmann D, Risold PY. Hypocretin/orexin-containing neurons are produced in one sharp peak in the developing ventral diencephalon. Eur J Neurosci. 2005;22:531–534. doi: 10.1111/j.1460-9568.2005.04224.x. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka S. Transcriptional regulation of the hypocretin/orexin gene. Vitam Horm. 2012;89:75–90. doi: 10.1016/B978-0-12-394623-2.00005-6. [DOI] [PubMed] [Google Scholar]
- 6.Ogawa Y, Kanda T, Vogt K, Yanagisawa M. Anatomical and electrophysiological development of the hypothalamic orexin neurons from embryos to neonates. J Comp Neurol. 2017;525:3809–3820. doi: 10.1002/cne.24261. [DOI] [PubMed] [Google Scholar]
- 7.Zahr SK, Yang G, Kazan H, Borrett MJ, Yuzwa SA, Voronova A, Kaplan DR, Miller FD. A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron. 2018;97:520–537 e6. doi: 10.1016/j.neuron.2017.12.045. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka S, Kodama T, Nonaka T, Toyoda H, Arai M, Fukazawa M, Honda Y, Honda M, Mignot E. Transcriptional regulation of the hypocretin/orexin gene by NR6A1. Biochem Biophys Res Commun. 2010;403:178–183. doi: 10.1016/j.bbrc.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, et al. A genomic atlas of mouse hypothalamic development. Nature Neurosci. 2010;13:767–775. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva JP, von Meyenn F, Howell J, Thorens B, Wolfrum C, Stoffel M. Regulation of adaptive behaviour during fasting by hypothalamic Foxa2. Nature. 2009;462:646–650. doi: 10.1038/nature08589. [DOI] [PubMed] [Google Scholar]
- 11.Dalal J, Roh JH, Maloney SE, Akuffo A, Shah S, Yuan H, Wamsley B, Jones WB, de Guzman Strong C, Gray PA, et al. Translational profiling of hypocretin neurons identifies candidate molecules for sleep regulation. Genes Dev. 2013;27:565–578. doi: 10.1101/gad.207654.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honda M, Eriksson KS, Zhang S, Tanaka S, Lin L, Salehi A, Hesla PE, Maehlen J, Gaus SE, Yanagisawa M, et al. IGFBP3 colocalizes with and regulates hypocretin (orexin) PLoS One. 2009;4:e4254. doi: 10.1371/journal.pone.0004254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De La Herrán-Arita AK, Zomosa-Signoret VC, Millán-Aldaco DA, Palomero-Rivero M, Guerra-Crespo M, Drucker-Colín R, Vidaltamayo R. Aspects of the narcolepsy-cataplexy syndrome in O/E3-null mutant mice. Neuroscience. 2011;183:134–143. doi: 10.1016/j.neuroscience.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez-García A, Cabral-Pacheco GA, Zomosa-Signoret VC, Ortiz-López R, Camacho A, Tabera-Tarello PM, Garnica-López JA, Vidaltamayo R. Modular organization of a hypocretin gene minimal promoter. Mol Med Rep. 2018;17:2263–2270. doi: 10.3892/mmr.2017.8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spengler D, Villalba M, Hoffmann A, Pantaloni C, Houssami S, Bockaert J, Journot L. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J. 1997;16:2814–2825. doi: 10.1093/emboj/16.10.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kas K, Voz ML, Hensen K, Meyen E, Van de Ven WJ. Transcriptional activation capacity of the novel PLAG family of zinc finger proteins. J Biol Chem. 1998;273:23026–23032. doi: 10.1074/jbc.273.36.23026. [DOI] [PubMed] [Google Scholar]
- 17.Kamiya M, Judson H, Okazaki Y, Kusakabe M, Muramatsu M, Takada S, Takagi N, Arima T, Wake N, Kamimura K, et al. The cell cycle control gene ZAC/PLAGL1 is imprinted-a strong candidate gene for transient neonatal diabetes. Hum Mol Genet. 2000;9:453–460. doi: 10.1093/hmg/9.3.453. [DOI] [PubMed] [Google Scholar]
- 18.Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Varrault A, Dantec C, Le Digarcher A, Chotard L, Bilanges B, Parrinello H, Dubois E, Rialle S, Severac D, Bouschet T, Journot L. Identification of Plagl1/Zac1 binding sites and target genes establishes its role in the regulation of extracellular matrix genes and the imprinted gene network. Nucleic Acids Res. 2017;45:10466–10480. doi: 10.1093/nar/gkx672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals . Guide for the Care and Use of Laboratory Animals. 8th edition. National Academies Press (US); Washington, DC: 2011. [Google Scholar]
- 21.Terao A, Wisor JP, Peyron C, Apte-Deshpande A, Wurts SW, Edgar DM, Kilduff TS. Gene expression in the rat brain during sleep deprivation and recovery sleep: An Affymetrix GeneChip study. Neuroscience. 2006;137:593–605. doi: 10.1016/j.neuroscience.2005.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takizawa N, Tanaka S, Oe S, Koike T, Matsuda T, Yamada H. Hypothalamo-hypophysial system in rats with autotransplantation of the adrenal cortex. Mol Med Rep. 2017;15:3215–3221. doi: 10.3892/mmr.2017.6375. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 26.Varrault A, Ciani E, Apiou F, Bilanges B, Hoffmann A, Pantaloni C, Bockaert J, Spengler D, Journot L. hZAC encodes a zinc finger protein with antiproliferative properties and maps to a chromosomal region frequently lost in cancer. Proc Natl Acad Sci USA. 1998;95:8835–8840. doi: 10.1073/pnas.95.15.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou TC, Lee CE, Lu J, Elmquist JK, Hara J, Willie JT, Beuckmann CT, Chemelli RM, Sakurai T, Yanagisawa M, et al. Orexin (hypocretin) neurons contain dynorphin. J Neurosci. 2001;21:RC168. doi: 10.1523/JNEUROSCI.21-19-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reti IM, Reddy R, Worley PF, Baraban JM. Selective expression of Narp, a secreted neuronal pentraxin, in orexin neurons. J Neurochem. 2002;82:1561–1565. doi: 10.1046/j.1471-4159.2002.01141.x. [DOI] [PubMed] [Google Scholar]
- 29.Tsuneki H, Wada T, Sasaoka T. Role of orexin in the central regulation of glucose and energy homeostasis. Endocr J. 2012;59:365–374. doi: 10.1507/endocrj.EJ12-0030. [DOI] [PubMed] [Google Scholar]
- 30.Nattie E, Li A. Respiration and autonomic regulation and orexin. Prog Brain Res. 2012;198:25–46. doi: 10.1016/B978-0-444-59489-1.00004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamanaka A, Beuckmann CT, Willie JT, Hara J, Tsujino N, Mieda M, Tominaga M, Yagami Ki, Sugiyama F, Goto K, et al. Hypothalamic orexin neurons regulate arousal according to energy balance in mice. Neuron. 2003;38:701–713. doi: 10.1016/S0896-6273(03)00331-3. [DOI] [PubMed] [Google Scholar]
- 32.Burdakov D, Jensen LT, Alexopoulos H, Williams RH, Fearon IM, O'Kelly I, Gerasimenko O, Fugger L, Verkhratsky A. Tandem-pore K+ channels mediate inhibition of orexin neurons by glucose. Neuron. 2006;50:711–722. doi: 10.1016/j.neuron.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 33.González JA, Jensen LT, Doyle SE, Miranda-Anaya M, Menaker M, Fugger L, Bayliss DA, Burdakov D. Deletion of TASK1 and TASK3 channels disrupts intrinsic excitability but does not abolish glucose or pH responses of orexin/hypocretin neurons. Eur J Neurosci. 2009;30:57–64. doi: 10.1111/j.1460-9568.2009.06789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valdivia S, Patrone A, Reynaldo M, Perello M. Acute high fat diet consumption activates the mesolimbic circuit and requires orexin signaling in a mouse model. PLoS One. 2014;9:e87478. doi: 10.1371/journal.pone.0087478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakurai T. The neural circuit of orexin (hypocretin): Maintaining sleep and wakefulness. Nat Rev Neurosci. 2007;8:171–181. doi: 10.1038/nrn2092. [DOI] [PubMed] [Google Scholar]
- 36.Fujiki N, Yoshida Y, Ripley B, Honda K, Mignot E, Nishino S. Changes in CSF hypocretin-1 (orexin A) levels in rats across 24 hours and in response to food deprivation. Neuroreport. 2001;12:993–997. doi: 10.1097/00001756-200104170-00026. [DOI] [PubMed] [Google Scholar]
- 37.Lee MG, Hassani OK, Jones BE. Discharge of identified orexin/hypocretin neurons across the sleep-waking cycle. J Neurosci. 2005;25:6716–6720. doi: 10.1523/JNEUROSCI.1887-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida Y, Fujiki N, Nakajima T, Ripley B, Matsumura H, Yoneda H, Mignot E, Nishino S. Fluctuation of extracellular hypocretin-1 (orexin A) levels in the rat in relation to the light-dark cycle and sleep-wake activities. Eur J Neurosci. 2001;14:1075–1081. doi: 10.1046/j.0953-816x.2001.01725.x. [DOI] [PubMed] [Google Scholar]
- 39.Pedrazzoli M, D'Almeida V, Martins PJ, Machado RB, Ling L, Nishino S, Tufik S, Mignot E. Increased hypocretin-1 levels in cerebrospinal fluid after REM sleep deprivation. Brain Res. 2004;995:1–6. doi: 10.1016/j.brainres.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 40.Wu MF, John J, Maidment N, Lam HA, Siegel JM. Hypocretin release in normal and narcoleptic dogs after food and sleep deprivation, eating, and movement. Am J Physiol Regul Integr Comp Physiol. 2002;283:R1079–R1086. doi: 10.1152/ajpregu.00207.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allard JS, Tizabi Y, Shaffery JP, Manaye K. Effects of rapid eye movement sleep deprivation on hypocretin neurons in the hypothalamus of a rat model of depression. Neuropeptides. 2007;41:329–337. doi: 10.1016/j.npep.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Modirrousta M, Mainville L, Jones BE. Orexin and MCH neurons express c-Fos differently after sleep deprivation vs. recovery and bear different adrenergic receptors. Eur J Neurosci. 2005;21:2807–2816. doi: 10.1111/j.1460-9568.2005.04104.x. [DOI] [PubMed] [Google Scholar]
- 43.Martins PJ, Marques MS, Tufik S, D'Almeida V. Orexin activation precedes increased NPY expression, hyperphagia, and metabolic changes in response to sleep deprivation. Am J Physiol Endocrinol Metab. 2010;298:E726–E734. doi: 10.1152/ajpendo.00660.2009. [DOI] [PubMed] [Google Scholar]
- 44.Holmquist GP, Ashley T. Chromosome organization and chromatin modification: Influence on genome function and evolution. Cytogenet Genome Res. 2006;114:96–125. doi: 10.1159/000093326. [DOI] [PubMed] [Google Scholar]
- 45.Sadoni N, Langer S, Fauth C, Bernardi G, Cremer T, Turner BM, Zink D. Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments. J Cell Biol. 1999;146:1211–1226. doi: 10.1083/jcb.146.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Tomaso MV, Liddle P, Lafon-Hughes L, Reyes-Ábalos A, Folle G. Chromatin damage patterns shift according to Eu/Heterochromatin replication. The mechanisms of DNA replication. D S, editor. InTech. 2013 [Google Scholar]
- 47.Hayakawa K, Hirosawa M, Tabei Y, Arai D, Tanaka S, Murakami N, Yagi S, Shiota K. Epigenetic switching by the metabolism-sensing factors in the generation of orexin neurons from mouse embryonic stem cells. J Biol Chem. 2013;288:17099–17110. doi: 10.1074/jbc.M113.455899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayakawa K, Sakamoto Y, Kanie O, Ohtake A, Daikoku S, Ito Y, Shiota K. Reactivation of hyperglycemia-induced hypocretin (HCRT) gene silencing by N-acetyl-d-mannosamine in the orexin neurons derived from human iPS cells. Epigenetics. 2017;12:764–778. doi: 10.1080/15592294.2017.1346775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffmann A, Ciani E, Boeckardt J, Holsboer F, Journot L, Spengler D. Transcriptional activities of the zinc finger protein Zac are differentially controlled by DNA binding. Mol Cell Biol. 2003;23:988–1003. doi: 10.1128/MCB.23.3.988-1003.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuasa S, Onizuka T, Shimoji K, Ohno Y, Kageyama T, Yoon SH, Egashira T, Seki T, Hashimoto H, Nishiyama T, et al. Zac1 is an essential transcription factor for cardiac morphogenesis. Circ Res. 2010;106:1083–1091. doi: 10.1161/CIRCRESAHA.109.214130. [DOI] [PubMed] [Google Scholar]
- 51.Xu Y, Tamamaki N, Noda T, Kimura K, Itokazu Y, Matsumoto N, Dezawa M, Ide C. Neurogenesis in the ependymal layer of the adult rat 3rd ventricle. Exp Neurol. 2005;192:251–264. doi: 10.1016/j.expneurol.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 52.Broberger C, De Lecea L, Sutcliffe JG, Hökfelt T. Hypocretin/orexin- and melanin-concentrating hormone-expressing cells form distinct populations in the rodent lateral hypothalamus: Relationship to the neuropeptide Y and agouti gene-related protein systems. J Comp Neurol. 1998;402:460–474. doi: 10.1002/(SICI)1096-9861(19981228)402:4<460::AID-CNE3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 53.Elias CF, Saper CB, Maratos-Flier E, Tritos NA, Lee C, Kelly J, Tatro JB, Hoffman GE, Ollmann MM, Barsh GS, et al. Chemically defined projections linking the mediobasal hypothalamus and the lateral hypothalamic area. J Comp Neurol. 1998;402:442–459. doi: 10.1002/(SICI)1096-9861(19981228)402:4<442::AID-CNE2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 54.Matsuki T, Nomiyama M, Takahira H, Hirashima N, Kunita S, Takahashi S, Yagami K, Kilduff TS, Bettler B, Yanagisawa M, Sakurai T. Selective loss of GABA(B) receptors in orexin-producing neurons results in disrupted sleep/wakefulness architecture. Proc Natl Acad Sci USA. 2009;106:4459–4464. doi: 10.1073/pnas.0811126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.