Abstract
As the complexity of biomedical data increases, so do the opportunities to leverage them to advance science and clinical care. Electronic health records form a rich but complex source of large amounts of data gathered during routine clinical care. Through the use of codified and free-text concepts identified using clinical informatics tools such as natural language processing, disease phenotyping can be performed with a high degree of accuracy. Technologies such as genome sequencing, gene expression profiling, proteomic and metabolomic analyses, and electronic devices and wearables are generating large amounts of data from various populations, cell types, and disorders (big data). However, to make these data useable for the next step of biomarker discovery, precision medicine, and clinical practice, it is imperative to harmonize and integrate these diverse data sources. In this article, we introduce important building blocks for precision medicine, including common data models, text mining and natural language processing, privacy-preserved record linkage, machine learning for predictive modeling, and health information exchange.
INTRODUCTION
Adoption and meaningful use of electronic health records (EHRs) has continued to increase, spurred by federal mandates in the United States. These electronic systems collect vast amounts of clinical data either as structured elements (vital parameters, laboratory data, etc.) or unstructured clinical notes. In addition, these data are intended to facilitate effective clinical decision support (CDS), as defined by HealthIT.gov (1) as systems or processes that “(provide) clinicians, staff, patients or other individuals with knowledge and person-specific information, intelligently filtered or presented at appropriate times, to enhance health and health care.” These data, currently used primarily for clinical care and administrative purposes, hold tremendous potential for advancing biomarker discovery and personalized medical decision-making at point of care in patients with chronic diseases like inflammatory bowel diseases (IBD).
In parallel with widespread EHR adoption, there have been tremendous advancements in computational biology techniques with proliferation of standardized genetic platforms and sequencing technologies, explosion of multiomics approaches, along with streamlined analytic pipelines, facilitating pooling of research data across populations. However, such efforts have relied on carefully curated cohorts with research teams manually identifying patients from clinical care by review of individual charts to identify eligible individuals, which requires significant personnel support and is resource intensive. Moving forward, utilizing the EHR to curate large disease-based cohorts in a short amount of time with modest resources, carefully performing automated detailed disease phenotyping utilizing text mining and natural language processing (NLP), and then integrating these diverse “big data” sources through privacy-preserved linkage can promote effective and efficient discovery research, rapid translation and integration, and adoption at point of care. In this chapter, we discuss important concepts of clinical informatics, a rapidly evolving field at the cross-section of information technology and healthcare, required to facilitate such advancement. Figure 1 summarizes the approach to precision medicine using EHRs.
Figure 1.
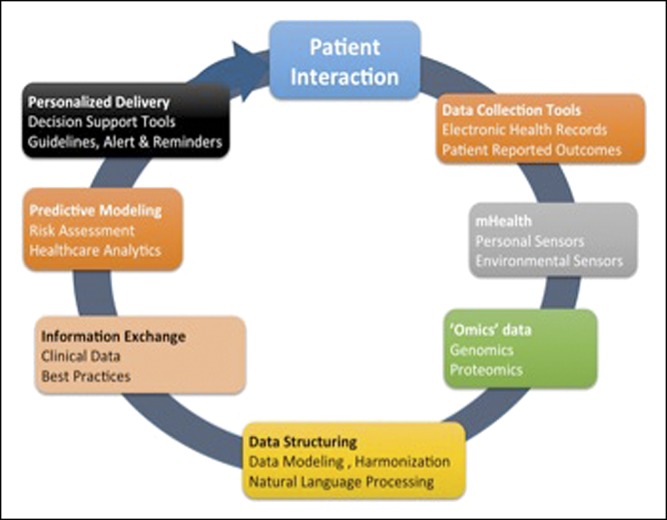
Key tools for precision medicine using electronic health records.
COMMON DATA MODELS
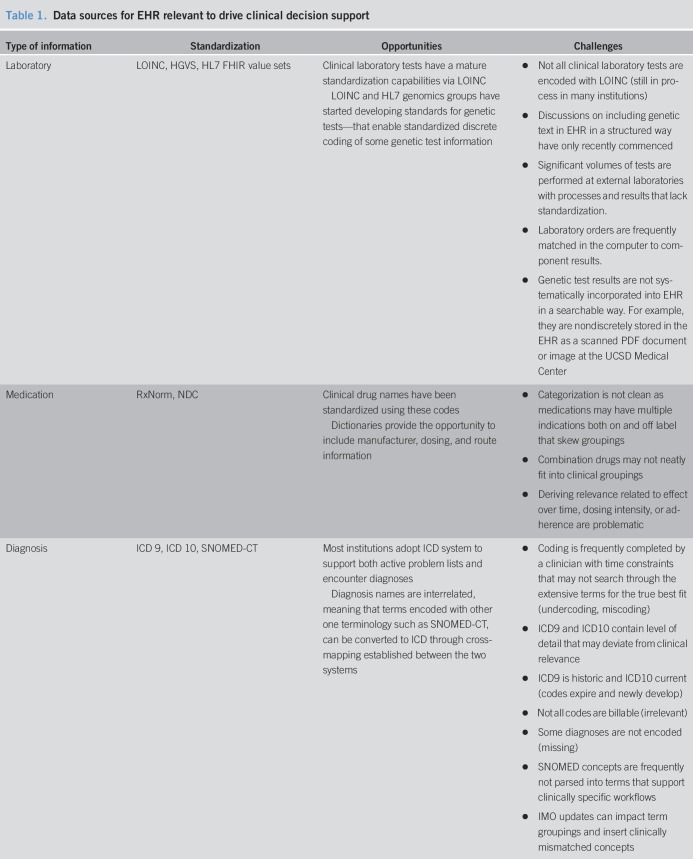
An intrinsic limitation to any big data approach is the issue of data quality in terms of volume, variety, velocity, and veracity (2–4). Hence, to make EHR data usable across formats and institutions, it is critical to develop a common data model with the use of standard terminology. Each type of data has an associated terminology that enables the vocabulary to be operationalized within the context of the EHR. These terminology systems have unique data formatting, coding, domain coverages, and hierarchical relationships between a specific instantiation, such as amoxicillin capsule 250 mg, and a concept, such as penicillin. Table 1 shows the common EHR data sources relevant to CDS. Precision medicine is developing a new vocabulary related to genetic conditions, which has yet to be standardized in the EHR. Genetic test results should follow relevant data standards, such as Logical Observation Identifiers Names and Codes, Health Level 7 Genomics, Human Genome Variation Society, that contain information about test findings and potential risk; yet, this is a challenge since these standards are not adopted by all laboratories. The rapid evolution of tests makes this challenging for the field of genetics, posing challenges for discrete data retrieval of this information in the EHR. Precision medicine also relies on other types of data that were not traditionally recorded in EHRs, such as patient-generated data such as wearables, electronic devices, social media, which are still early in standardization and the reporting is highly variant according to socioeconomic status, race, literacy, etc.
Table 1.
Data sources for EHR relevant to drive clinical decision support
Text mining and natural language processing
While several elements handled through common data models are based on structured or codified elements, free text or narratives still dominate in terms of clinically relevant information contained in EHRs. While free narrative is effective and convenient for medical record keeping, its unprocessed form is difficult to search, summarize, or analyze for secondary purposes such as research or quality improvement. NLP is any computer-based algorithm that handles, augments, and transforms natural language so that it can be represented for computation. Because a computer cannot comprehend meaning from a block of text, a series of operations must be defined to transform the data into usable information, which is the essence of NLP. In elegant use of this combination of codified data and NLP to develop an EHR-based cohort, Ananthakrishnan et al. created a cohort of 11,000 patients with IBD within 2 hospitals in Boston (4–6). From among all patients with at least 1 billing code for Crohn's disease or ulcerative colitis, a chart review revealed a positive predictive value of only 60% with frequent misclassification. Extraction of codified data ascertaining disease complications as well as narrative free text data comprising number of mentions of individual disease names (“Crohn's disease”) or disease-related terms in clinical notes (“abdominal pain”, “diarrhea”), radiology reports (“ileal wall thickening”), endoscopy (“ileitis” “aphthous ulcer”), and pathology (“crypt abscess”) allowed for development of a classification algorithm using machine learning that was able to achieve a positive predictive value of 97%. The addition of free-text data to the codified information not only improved the accuracy of identifying cases but also increased the number of patients who could be classified as having disease. In addition, this approach also allowed identification of phenotypes of disease, such as primarily sclerosing cholangitis, which is limited by lack of specific diagnostic codes or high frequency of use of codes for competing diagnosis (e.g., cholelithiasis), determining status of disease activity in relapsing and remitting disorders, or identifying response to treatment. NLP software is increasingly sophisticated to be able to distinguish positive findings (“has diarrhea”) from negative ones (“does not have diarrhea”), assign specific contexts for occurrence of phrases (“abdominal pain” from “joint pain”), separate personal from family history (“family history of colon cancer”), and search within specific components of the note (such as indication for procedures). Despite the inherent variability in structure and content of EHR data and differences in quality of provider documentation across institutions, disease-defining algorithms created at one institution are portable to other institutions using distinct EHRs and retain their accuracy, which is key for multi-institutional consortia, such as the Electronic Medical Records and Genomics Network. With advances in the field of NLP, detailed phenotyping is feasible, allowing performance of large scale, integrated genome- and phenome-wide studies to promote biomarker discovery and precision medicine.
Privacy-preserved record linkage
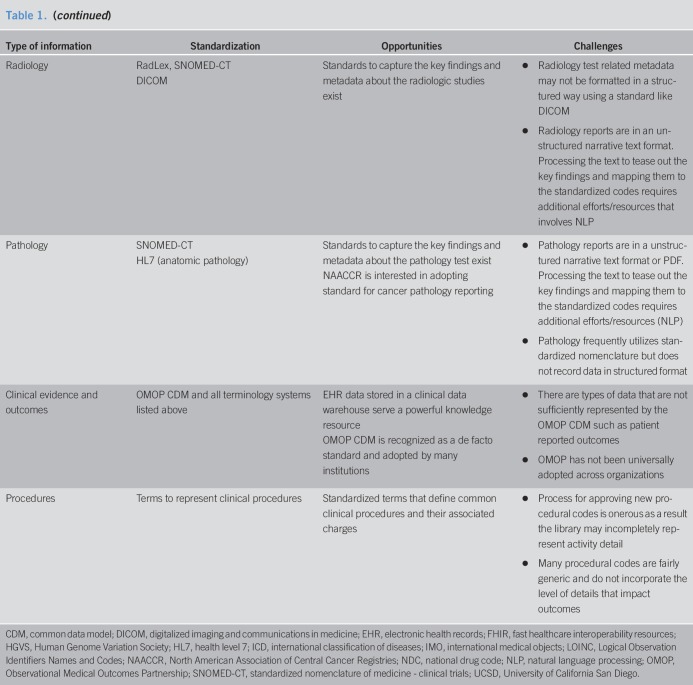
One of the greatest challenges to utilizing big data for research has been data partitioning from diverse sources. In a research network, information from the same individual may be partitioned among several sites such as healthcare providers, sequencing facilities, insurance companies, research institutions. There are mainly 2 types of patient data partitioning across institutions: (i) horizontal partitioning, where different institutions hold information on the same and (ii) vertical partitioning, where different institutions hold information on different attributes. The former one consists of records with the same features, for an overlapping or nonoverlapping set of individuals. Feature values are the same in the case of true overlap, or they can differ when patients switch healthcare systems or receive complementary care in different health systems (e.g., patients cared for primarily at the Veterans Health Administration system but receiving specialty care in another system). In vertical partitioning, there is information about different features for the same individual at different sites. In both situations, patient record linkage is an essential step to combine data in cross-institutional studies. For example, if the truly duplicated records across different institutions cannot be sufficiently removed, the estimation could become biased in the study with horizontally partitioned data. For the case of vertically partitioned data, the genome data of a particular group hosted in a sequencing facility can be significantly enriched by linking the data to EHRs. In addition to linking patient records across research networks, the existing clinical data research networks can link their data to publicly available databases of vital statistics (such as the National Death Index), pharmaceutical databases, etc., allowing comprehensive and simultaneous capture of multiple exposures, health status, interventions, and outcomes. The existing record linkage methods can be categorized into 2 approaches: deterministic and probabilistic (7). If there exist explicit identifiers (e.g., name and social security number) among different datasets, deterministic record linkage methods are used. Probabilistic linkage methods are more complex, as they assign different weights for different discriminative linkage variables to compute an overall score that indicates how likely it is that a record pair comes from the same patient. Furthermore, due to concerns of invasion of privacy, institutions and patients alike may be hesitant to share personal health information outside the health system. Hence, robust privacy-preserving record linkage tools are clearly needed before this rich environment is ripe for research use. Figure 2 depicts an example of a record linkage system for vertically partitioned data between a hospital and a biobank where DNA data are available.
Figure 2.
Privacy-preserved record linkage approaches.
Health information exchange
One of the limitations of EHR-based research is that data are contained in silos in health systems, which do not interact adequately with each other. While patients move in and out of health systems, their data do not move and get lost in translation. Not only does this disrupt clinical care, but it also impedes phenome-wide association studies and biomarker discovery due to misclassification of clinical data. However, through approaches of health information exchange (HIE), a special case of privacy-preserved record linkage, this barrier may be overcome (2). The Health Information Technology for Economics and Clinical Health Act of 2009 was proposed to promote interoperable health information. HIE initiatives aim at realizing timely and appropriate level of access to the patient level of health information stored in the EHR by healthcare providers through a secure means to exchanging health data among healthcare organizations. Having complete information about disease progression and treatment data at the point of care helps healthcare providers make better treatment decisions and achieve better patient outcomes. Utilizing information collected from different healthcare systems is an important step toward this goal.
HIE covers 3 types of data exchange: (i) Directed exchange that occurs between healthcare providers to complete the planned healthcare services such as sending and receiving laboratory test orders and results, and exchanging patient referral documents, (ii) Query-based exchange that occurs when a healthcare provider delivers unplanned services and requires accessing necessary health information about the patient. For example, when an emergency room physician needs to access patient's disease history, current medications, allergies, etc. (iii) Consumer-mediated exchange that lets patients control their health information. In this model, patients grant access to their health information to healthcare providers. However, establishing a sustainable HIE is not a trivial task; there are a number of technical and nontechnical barriers that need to be addressed first. For example, lack of business incentives, specifically concerns on losing patients to other hospitals by making their health data available anywhere, has long been recognized as a factor that makes some healthcare systems hesitant to embrace HIEs. Patients and providers sometimes opt out from HIEs due to privacy concerns. Other recognized challenges are poor data standardization, inefficient processes of sorting through overloaded unselective information of a patient, and difficulties in understanding the shared data in the absence of context when detailed clinical notes are withheld due to privacy concerns.
Statistical approaches including machine learning
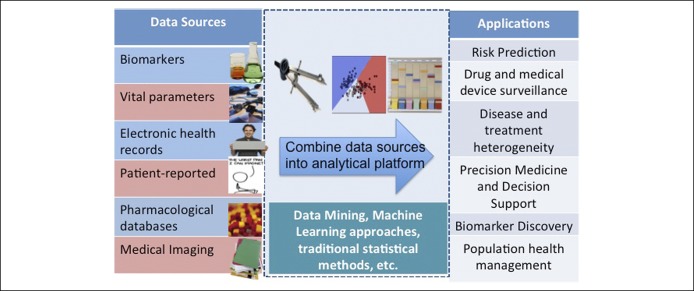
With this vast amount of data being generated from diverse sources, novel and powerful analytic approaches are needed. Figure 3 summarizes different approaches to analysis. Machine-learning methods consist of computational algorithms to relate all or some of a set of predictor variables to an outcome (8). To estimate the model, they search, either stochastically (randomly) or deterministically, for the best fit. This searching process differs across the different algorithms. However, through this search, each algorithm attempts to balance 2 competing interests: bias and variance. In the machine-learning context, bias is the extent to which the fitted predictions correspond to the true values—that is, how accurately does the model predict the “true” risk of death in the population? Variance is the sensitivity of the predictions to perturbations in the input data, that is, how does sampling variability impact the predictions? Even though it is not possible to separately quantify a model's bias and variance, these 2 values are summarized together by loss functions. Many machine-learning methods can be grouped into different families based on their underlying structure. The 2 largest families are those that amend the traditional regression model (such as regularized methods, including common ridge regression and Lasso) and tree-based methods (such as classification and regression trees), and others including artificial neural networks, nearest neighbors, and support vector machines.
Figure 3.
Analytic approaches for big data.
In summary, marrying EHR-based clinical research approaches with advancements in computational biology is immensely promising for biomarker discovery and promoting personalized medical decision-making. One can readily envision this approach being applicable across a wide swath of diseases relevant to gastroenterology, including IBD, chronic liver diseases, and gastrointestinal cancers. All of these diseases have in common varying, and often poor, accuracy of the existing administrative coding-based diagnoses, but can be readily identified in the EHRs using data (e.g., serology, pathology, and endoscopy) that are a routine part of clinical care and that can be mined using clinical informatics tools. Linkage of such disease registries to bio-banked genotyped samples, ensuring appropriate data protection and de-identification, can be enormously valuable to advance scientific discovery. This, however, is contingent on standardization of reporting methods and attributes and the ability to receive structured data from outside sources. Once these data are standardized across formats, it can readily be used to populate specific patients attributes in integrated CDS tools directly and seamlessly promoting personalized medical decision-making at point of care.
CONFLICTS OF INTEREST
Guarantor of the article: Siddharth Singh, MD, MS.
Specific author contributions: S.S. conceived the idea and drafted the article.
Financial support: S.S. is supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under award number K23DK117058, the American College of Gastroenterology Junior Faculty Development Award, and the Crohn's and Colitis Foundation Career Development Award (#404614). He has received research grants from Pfizer and AbbVie, and consulting fees from AbbVie, Takeda, Pfizer, and AMAG Pharmaceuticals.
Potential competing interests: None.
REFERENCES
- 1.The Office of the National Coordinator for Health Information Technology. Clinical Decision Support. HealthIT.gov. https://www.healthit.gov/topic/safety/clinical-decision-support. Accessed on March 8, 2019.
- 2.Sitapati A, Kim H, Berkovich B, et al. Integrated precision medicine: The role of electronic health records in delivering personalized treatment. Wiley Interdiscip Rev Syst Biol Med 2017;9(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rumsfeld JS, Joynt KE, Maddox TM. Big data analytics to improve cardiovascular care: Promise and challenges. Nat Rev Cardiol 2016;13(6):350–9. [DOI] [PubMed] [Google Scholar]
- 4.Ananthakrishnan AN, Lieberman D. Patient electronic health records as a means to approach genetic research in gastroenterology. Gastroenterology 2015;149(5):1134–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ananthakrishnan AN, Cai T, Savova G, et al. Improving case definition of Crohn's disease and ulcerative colitis in electronic medical records using natural language processing: A novel informatics approach. Inflamm Bowel Dis 2013;19(7):1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao KP, Cai T, Savova GK, et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ 2015;350:h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Jiang X, Singh S, et al. Genome privacy: Challenges, technical approaches to mitigate risk, and ethical considerations in the United States. Ann N Y Acad Sci 2017;1387(1):73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein BA, Navar AM, Carter RE. Moving beyond regression techniques in cardiovascular risk prediction: Applying machine learning to address analytic challenges. Eur Heart J 2017;38(23):1805–14. [DOI] [PMC free article] [PubMed] [Google Scholar]