OBJECTIVES:
Biliary tract cancer (BTC) is an aggressive malignant tumor, and biomarker-based clinical trials for this cancer are currently ongoing. Endoscopic ultrasound–guided fine needle aspiration (EUS-FNA) is a safe procedure and enables pathological diagnoses; however, it is uncertain whether a tiny tumor sample of BTC obtained through EUS-FNA can be analyzed for diverse genetic alterations in the development and tolerance of BTC. Thus, we aimed to verify the feasibility of genetic analyses with EUS-FNA samples of BTC.
METHODS:
Targeted amplicon sequencing using a cancer gene panel with 50 genes was performed with tissue samples of 21 BTC patients obtained through EUS-FNA with a novel rapid on-site process compared with paired peripheral blood samples.
RESULTS:
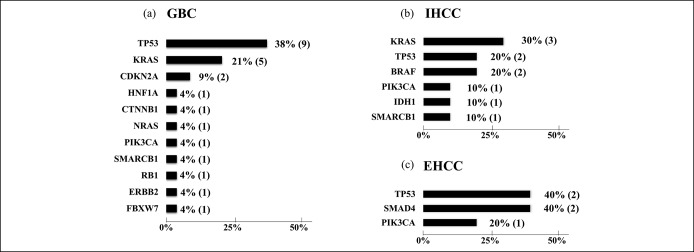
Pathogenic gene alterations were successfully identified in 20 out of 21 patients (95.2%) with EUS-FNA specimens of BTC, which included 19 adenocarcinomas and 2 adenosquamous carcinomas. Eighty single nucleotide variants and 8 indels in 39 genes were identified in total, and 28 pathogenic alterations in 14 genes were identified (average, 1.4 alterations per patient). The most common alterations were TP53, KRAS, and CDKN2A in gallbladder carcinoma; TP53, KRAS, PIK3CA, and BRAF in intrahepatic cholangiocarcinoma; and TP53 and SMAD4 in extrahepatic cholangiocarcinoma. Actionable gene alterations (BRAF, NRAS, PIK3CA, and IDH1) were identified in 7 out of 21 patients.
CONCLUSIONS:
A novel approach in genetic analysis using targeted amplicon sequencing with BTC specimens obtained through EUS-FNA was feasible and enabled us to identify genomic alterations.
INTRODUCTION
Biliary tract cancers (BTCs), including cholangiocarcinoma and gallbladder carcinoma, have been recognized as troublesome and aggressive tumors, and the global incidence is increasing with greater frequency in Asian countries than in Western countries (1–3). Although surgical resection is the only treatment for complete cure, many patients are diagnosed with unresectable tumors due to the difficulty of early diagnosis and thus cannot survive for long by conventional chemotherapies with gemcitabine, cisplatin, and 5-fluorouracil alone (4,5). No effective targeted molecular therapies have been established; however, genomic spectra of BTC have recently been reported. Nakamura et al. (6) characterized a large BTC cohort composed of Japanese patients through whole-exome and transcriptome sequencing. The cohort revealed that molecular alteration of BTC has variety and organ-specific spectra, which include therapeutic targets in nearly 40% of BTC cases. A phase II study in patients with fibroblast growth factor receptor 2 (FGFR2) fusions revealed that a selective pan-FGFR kinase inhibitor showed significant clinical effect against cholangiocarcinoma (7). Pan-FGFR inhibitors are also being investigated in other clinical trials for advanced BTCs harboring the FGFR2 gene (8). In addition, other biomarker-driven clinical trials for BTC, which target BRAF, MEK, and IDH1/2, are currently ongoing (NCT02034110, NCT01242605, NCT02989857, NCT02273739, NCT02428855, and NCT02073994) (8).
Endoscopic ultrasound–guided fine needle aspiration (EUS-FNA) is a safe procedure in the diagnosis and staging of BTC with minimal complications (9). The genetic analyses of EUS-FNA specimens from some organs using targeted amplicon sequencing (TAS) has already been reported (10–14). The specimens of pancreatic tumor, metastatic lymph node in rectal cancer, gastric gastrointestinal stromal tumors, and non–small cell lung cancer, all of which were obtained through EUS-FNA, have been analyzed using TAS with a cancer gene panel. Meanwhile, genetic analysis of BTC specimens obtained through EUS-FNA has never been reported. Therefore, the evaluation of gene mutations in BTC specimens obtained through EUS-FNA is significant and can contribute to the development of personalized targeted molecular therapy for patients with BTC.
In this study, we performed targeted deep-sequencing analyses of the BTC specimens obtained through EUS-FNA from 21 BTC patients with a high coverage depth of sequencing using a next-generation sequencer for 50 cancer-related genes that are likely related to the molecular alterations in BTC (6).
METHODS
Patients and samples
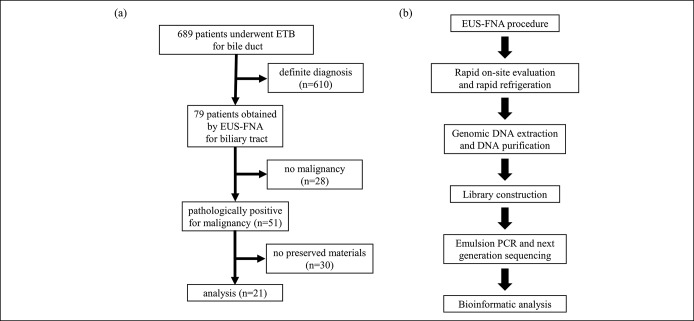
From September 2013 to April 2018, 689 patients with a biliary disease who underwent endoscopic transpapillary biopsy (ETB) for their biliary tract lesion were identified. Among these patients, 79 suspected of having BTC underwent EUS-FNA for the bile duct, gallbladder, or lymph node because of an indefinite diagnosis by ETB. Of the 79 patients, 51 were pathologically diagnosed to have adenocarcinoma or adenosquamous carcinoma with the remaining 28 having benign disease, by experienced pathologists at Hokkaido University Hospital. Thirty of the 51 malignant samples were available in quantities that were insufficient for preservation. Finally, 21 patients (21 samples) were enrolled and analyzed (Figure 1a). The BTC specimens were obtained through EUS-FNA before treatment including chemotherapy and surgery. At the same time, paired peripheral blood samples from the same patients were collected and peripheral blood mononuclear cells were isolated by centrifugation for TAS. Because the specimen obtained through EUS-FNA usually includes both tumor cells and blood cells, analysis requires high depth to identify somatic alterations. The TAS analysis of normal paired samples is useful to minimize sequencing noise and to identify pathogenic alterations more accurately. Furthermore, TAS analysis helps identify whether there are somatic or germline alterations in the genes (15). All BTC specimens and paired peripheral blood mononuclear cells were stored in 1.5-mL tubes at −30 °C until use. Participants provided written informed consent or consent for the disclosure of study information as an opt-out.
Figure 1.
(a) Case–cohort design. (b) The workflow from EUS-FNA to bioinformatic analysis. ETB, endoscopic transpapillary biopsy; EUS-FNA, endoscopic ultrasound-guided fine needle aspiration; PCR, polymerase chain reaction.
The ethics committee at Hokkaido University Hospital approved the study. All samples and medical data used in this study were irreversibly anonymized.
EUS-FNA procedure and preservation of the samples
EUS-FNA was performed by experienced endoscopists using a linear echoendoscope (GF-UCT260; Olympus Medical Systems, Tokyo, Japan) and 22 or 25 gauge needles (EXPECT or Acquire; Boston Scientific) with the patient under conscious sedation. After the target lesion was visualized by EUS, the needle was advanced into the lesion through the gastric or duodenal wall. The central stylet was removed, and a syringe was attached to the needle hub to apply negative suction pressure. The needle was then moved back and forth within the lesion at least 10 times and then removed through the scope. The specimen obtained through aspiration was placed on a slide, air-dried, alcohol-fixed, and used to prepare smears that were stained using the rapid Romanowsky technique for quick interpretation and assessment of sample adequacy (Diff-Quik stain; Kokusai Shiyaku, Kobe, Japan). Diff-Quik staining was performed on all specimens by an experienced cytotechnologist. We also stored 0.5–1.0-mm3 portions of the EUS-FNA white samples, which were considered to include malignant cells near the area of Diff-Quik staining, in RNAlater (Life Technologies, Carlsbad, CA) at 4 °C immediately after confirmation of malignancy (Figure 1b), and at −30 °C after a few days. Thereafter, all specimens obtained through EUS-FNA were evaluated for malignancy by cytological and pathological examinations by an expert pathologist.
Genomic DNA extraction of the EUS-FNA tissue and blood cell
Genomic DNA was extracted from the samples using an AllPrep DNA/RNA/Protein Mini Kit (Qiagen, Valencia, CA) and then purified using AMPure XP (BECKMAN COULTER) and 70% ethanol. The genomic DNA samples were quantified using NanoDrop (Life Technologies), Qubit dsDNA HS Assay Kit (Life Technologies) designed to be accurate for sample concentrations from 10 to 100 ng/μL, and StepOnePlus qPCR system with TaqMan assays (Life Technologies) according to the manufacturer's instructions. Samples with low DNA concentration (<2.0 ng/μL) were concentrated using the Nucleospin Genomic DNA Clean-up XP protocol (MACHEREY-NAGEL GmbH & Co. KG, Neumann, Germany).
Library preparation
Ten nanograms of DNA was used for library construction with the Ion Ampliseq Cancer Hotspot Panel v2 (Life Technologies), which targets 2,790 COSMIC alteration hotspots in 50 cancer-related genes: KRAS, NRAS, TP53, BRAF, FGFR1, FGFR2, FGFR3, IDH1, IDH2, SMAD4, EGFR, PIK3CA, CDKN2A, HRAS, ATM, RET, PTEN, PTPN11, HNF1A, FLT3, RB1, AKT1, CDH1, ERBB2, ERBB4, STK11, JAK2, JAK3, ALK, SRC, GNAS, SMARCB1, VHL, MLH1, CTNNB1, PDGFRA, KIT, KDR, FBXW7, APC, CSF1R, NPM1, MPL, MET, SMO, ABL1, NOTCH1, EZH2, GNA11, and GNAQ.
The DNA library was prepared by amplifying target regions using multiple polymerase chain reactions (PCRs) followed by adapter DNA ligation. The library concentration was evaluated using an Agilent 2,100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Emulsion PCR and ion torrent personal genome machine sequencing
Pooled 100 pmol/L libraries were clonally amplified using the Ion OneTouch 2 instrument with the Ion personal genome machine Template OT2 200kit (Life Technologies), and then, the samples were enriched using Ion OneTouch ES (Life Technologies). The enriched templates were loaded on an Ion 318 chip and sequenced using the Ion personal genome machine system (Life Technologies) according to the manufacturer's protocol. Human genome built 19 (hg19) was used as a reference, and variant calls and annotations were made on the Ion Torrent Suite v5.6 software (Life Technologies). When matched normal controls were not available, other control sequence data (Thermo Fisher Scientific) was used as a control. Somatic alterations were detected using statistical approaches in tumor and normal samples from the AmpliSeq tumor–normal pair workflow with customized filters. Filters included removing common single nucleotide polymorphisms, nonimpactful events (synonymous, intron, or reference allele), and mutation frequencies lower than 2% (recommended by manufacturer protocol). The filter threshold covered 99% of the captured region in each case.
Ethics statement
This study on human subjects was approved by the institutional review boards of Hokkaido University Hospital (Clinical Research approval number 017–0072). Written informed consent was obtained from each participant. All experiments were done in accordance with the ethical guideline of the 2013 Declaration of Helsinki.
RESULTS
Patient and tumor demographics
The age of the included patients ranged from 39 to 85 years (median 66.5 years), and fifteen (71.4%) of the patients were men. At the time of EUS-FNA, CA19-9 level ranged from 1.0 to 8,512.5 U/mL (median, 221.0), and CEA level ranged from 1.4 to 168.0 ng/mL (median, 7.9). The details of the BTCs were as follows: gallbladder cancers (GBCs; N = 12, 57.1%), intrahepatic cholangiocarcinomas (IHCCs; N = 6, 28.6%), and extrahepatic cholangiocarcinomas (EHCCs; N = 3, 14.3%) (Table 1).
Table 1.
Clinicopathological characteristics in the patients with BTC
In 12 GBCs, 10 primary lesion tissues and 2 metastatic lymph node tissues were obtained through EUS-FNA. In 6 IHCCs, one primary lesion tissue and 5 metastatic lymph node tissues were obtained, and in 3 cases of EHCCs, one primary lesion tissue and 2 metastatic lymph node tissues were obtained through EUS-FNA. Two samples of GBCs were diagnosed as adenosquamous carcinoma, and the others were diagnosed as adenocarcinoma. According to the eighth edition of the Union for International Cancer Control clinical staging system, 17 BTC cases were stage IV, 2 were stage III, and 2 were stage II. Both patients with Union for International Cancer Control stage II had EHCC; in one case, the lymph node was targeted, and in the other case, the primary lesion was targeted due to failure of harvesting BTC tissue by ETB. No adverse events such as bleeding, infection, perforation, and bile leakage associated with EUS-FNA occurred in all patients, including the 2 patients who underwent curative surgery.
Genomic DNA and library status
For all samples, appropriate DNA concentrations for TAS (>2.0 ng/μL, recommended by Life Technologies) were obtained in 17 EUS-FNA samples and 21 samples of peripheral blood mononuclear cells using the AllPrep DNA/RNA/Protein mini kit (Qiagen, Valencia, CA). The remaining 4 samples with low DNA concentration (<2.0 ng/μL) were concentrated using the Nucleospin gDNA Clean-up XP protocol (MACHEREY-NAGEL GmbH & Co. KG) to achieve the appropriate DNA concentrations (Table 2). Most of the needle calibers used in EUS-FNA were of 22 gauge (19/21), and there was no relationship between needle caliber used in EUS-FNA and yield of genomic DNA. All DNA samples were purified using AMPure XP and 70% ethanol immediately after DNA extraction and significantly improved the quality of the DNA samples (Figure 2). These samples were evaluated for fragmentation using the StepOnePlus qPCR system with TaqMan assays (Life Technologies), and the values measured by both the Qubit dsDNA HS assay kit and the NanoDrop system for used for library construction. Non-specific product contamination and measurable concentrations (>100 pmol/L, recommended by Life Technologies) (Table 2) were verified using the Agilent 2,100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) in all library samples. In all cases, DNA extraction and library preparation were completed without problems, such as deterioration of quality due to DNA fragmentation, despite the old age of some specimens that were used.
Table 2.
The genomic DNA (gDNA) and library dose in all patients with biliary tract cancer
Figure 2.
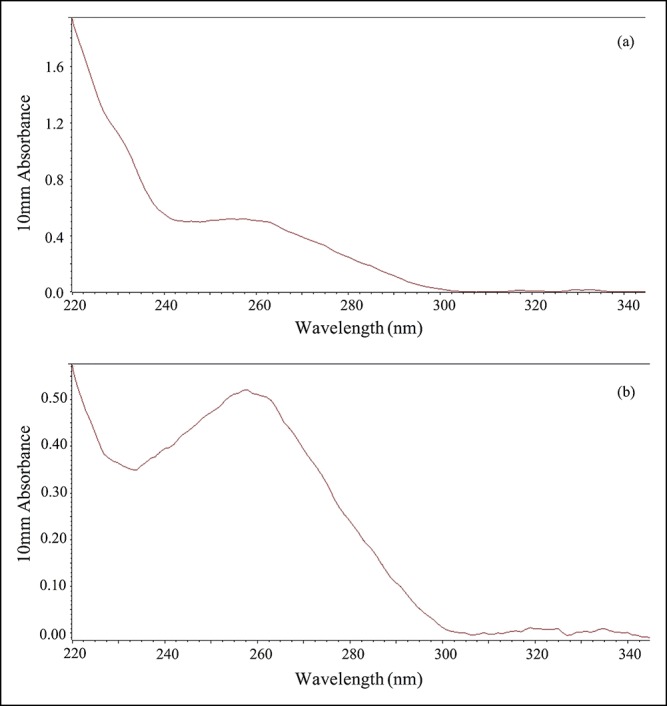
The measurement of the purity of extracted the DNA using NanoDrop system. (a) Absorbance graph before the purification of extracted DNA. High absorbance around 230 nm indicated the contamination of guanidine hydrochloride. (b) Absorbance graph after the purification of extracted DNA. The graph shows the absorbance peak around 260 nm and indicated the removal of contamination other than the DNA.
Gene alteration profiles
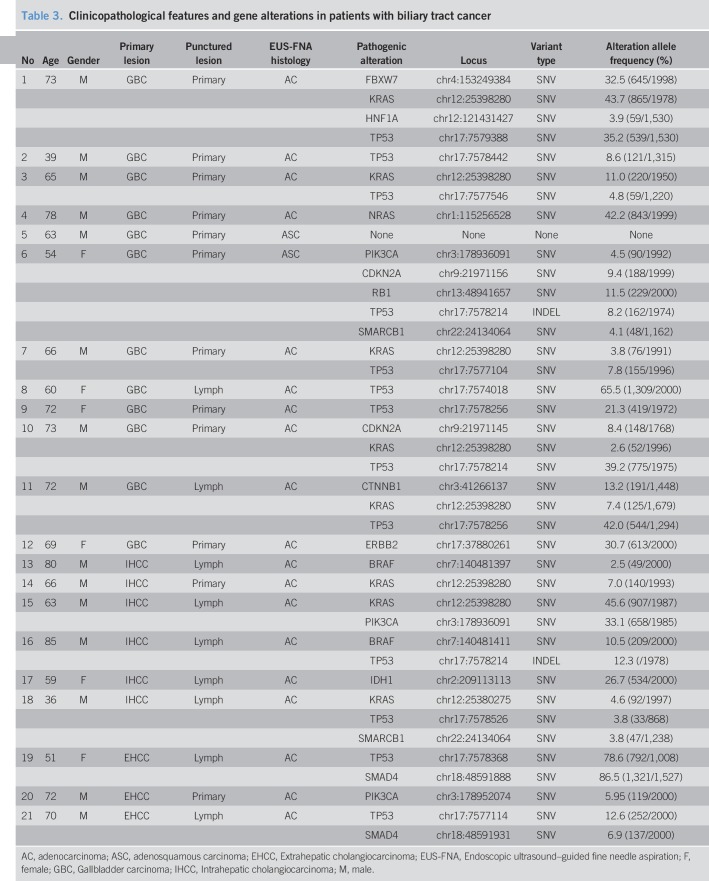
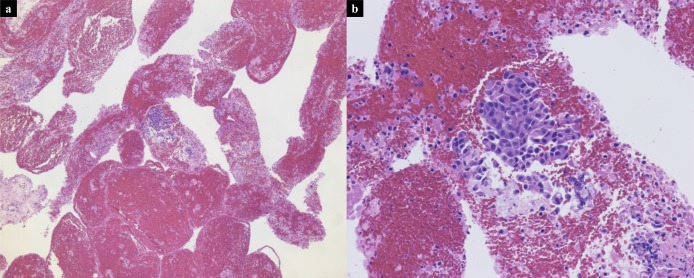
Analysis with Cancer Hotspot Panel v2 indicated that the average sample loading was 72% (range, 67%–77%) and the average number of total reads was 5,595,584 with an average read length of 119 bp. The average sequencing depth of coverage for the 21 BTC specimens was ×2855 (×1617–×7467). We identified 80 SNVs and 8 INDELs in 39 genes in total, and the filtered normal–tumor analyses revealed 28 pathogenic alterations in 14 genes. The pathogenic alterations were identified in 20 (95.2%) of 21 patients, with a median of 1 alteration (range, 1–5 alterations) per patient (Table 3). Genomic profiles revealed pathogenic alterations in TP53 (12 patients; 57.1%), KRAS (8 patients; 38.1%), PIK3CA (3 patients; 14.3%), SMAD4 (2 patients; 9.5%), CDKN2A (2 patients; 9.5%), BRAF (2 patients; 9.5%), SMARCB1 (1 patient; 4.8%), NRAS (1 patient; 4.8%), CTNNB1 (1 patient; 4.8%), IDH1 (1 patient; 4.8%), FBXW7 (1 patient; 4.8%), HNF1A (1 patient; 4.8%), and RB1 (1 patient; 4.8%), ERBB2 (1 patient; 4.8%), respectively (Figure 3). Thus, in this study, TAS identified genomic alterations sensitively even though the malignant cell aggregates obtained through EUS-FNA were small (Figure 4a,b).
Table 3.
Clinicopathological features and gene alterations in patients with biliary tract cancer
Figure 3.

Pathogenic gene alteration profile of the patients with BTC. All displayed detected alterations of variant impacts were filtered for common single nucleotide polymorphisms, nonimpactful events (synonymous, intron, or reference allele), and a <2% mutation frequency. BTC, biliary tract cancer.
Figure 4.
Scanned low magnification (a, ×5) and high magnification (b, ×20) images of hematoxylin-and-eosin–stained slide from the primary GBC specimen obtained through EUS-FNA demonstrating small amount of adenocarcinoma. EUS-FNA, endoscopic ultrasound-guided fine needle aspiration; GBC, gallbladder carcinoma.
Pathogenic gene alterations in each organ
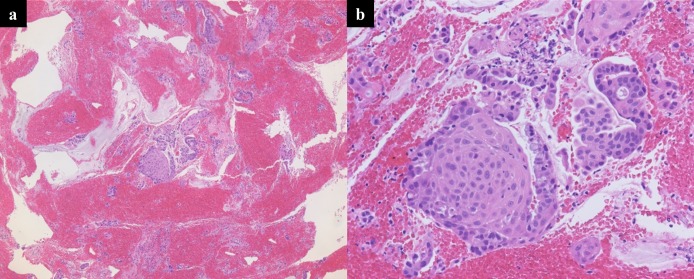
In GBC specimens, 11 pathogenic gene alterations were identified (TP53, KRAS, CDKN2A, NRAS, PIK3CA, FBXW7, HNF1A, RB1, ERBB2, CTNNB1, and SMARCB1). Alterations in TP53 (75.0%) and KRAS (41.7%) were frequently identified as pathogenic alterations (Figure 5a). In intrahepatic cholangiocarcinoma specimens, 6 pathogenic gene alterations were identified (KRAS, TP53, BRAF, PIK3CA, IDH1, and SMARCB1) (Figure 5b). In extrahepatic cholangiocarcinoma specimens, 3 pathogenic gene alterations were identified (TP53, PIK3CA, and SMAD4) (Figure 5c). Alterations in BRAF and SMARCB1 were identified only in intrahepatic cholangiocarcinoma, and alterations in SMAD4 were identified only in extrahepatic cholangiocarcinoma. In the 2 GBC samples diagnosed as adenosquamous carcinoma, pathogenic gene alterations in TP53, CDKN2A, RB1, PIK3CA, and SMARCB1 were identified in one sample, while no gene alteration was identified in the other despite the large amount of malignant cells observed upon hematoxylin and eosin staining (Figure 6). The difference in the gene alteration status and the number of gene alterations between the primary lesions (12 cases) and the metastatic lymph nodes (9 cases) targeted by EUS-FNA was not significant.
Figure 5.
(a) Alteration ratio in GBC. (b) Alteration ratio in IHCC. (c) Alteration ratio in EHCC. EHCC, extrahepatic cholangiocarcinoma; GBC, gall bladder carcinoma; GBC, gallbladder carcinoma; IHCC, intrahepatic cholangiocarcinoma.
Figure 6.
Scanned low magnification (a, ×5) and high magnification (b, ×20) images of hematoxylin-and-eosin–stained slide from the primary GBC specimen, which included no pathogenic alterations and showed large amount of malignant cells in the tissues. Interstitial, necrotic, or connective tissues were not found in this specimen. GBC, gallbladder carcinoma.
DISCUSSION
This is the first report of genetic analysis of BTC using specimens obtained through EUS-FNA. We achieved deep sequencing coverage and identified pathogenic alterations in 95.2% (20/21) of the patients with BTC using EUS-FNA samples. Previous reports on BTC showed equal or inferior success rates (47.5%, 92.1%, and 90.1%) (16–18) in identifying somatic alterations in BTC specimens obtained through surgical resection or ETB. With regard to using Hotspot Cancer Panel with pulmonary and pancreatic tissue specimens obtained through EUS-FNA, previous reports have shown 61.7%, 50%, and 100% success rates for the analysis of somatic alterations using TAS (10,19,20). There were several factors that contributed to the high success rate of TAS in this study. First, we performed rapid on-site evaluation of all specimens obtained through EUS-FNA before storing white tissues. The white tissues obtained through EUS-FNA sometimes include interstitial, necrotic, or connective tissues, and rapid on-site evaluation can prevent contamination with these unnecessary tissues before storing white tissue samples. Second, rapid tissue preservation in RNAlater and refrigeration immediately after EUS-FNA can preserve DNA quality by preventing fragmentation and chemical modification, which are common in formalin-fixed, paraffin-embedded (FFPE) tissue samples (21). Third, DNA purification with AMPure XP and 70% ethanol improved the DNA quality. The DNA extracted using AllPrep DNA/RNA/Protein mini kit was contaminated with guanidine hydrochloride, which impairs the quality of libraries. Thus, additional purification steps prior to library preparation can help produce superior sequence data.
Regarding the case that yielded no pathogenic alterations upon analysis, there are a few possibilities–the sample truly had no pathogenic alterations, it may have had pathogenic alterations outside the hotspots, or the tissue obtained through EUS-FNA did not have tumor cells. However, we stored a portion of the EUS-FNA samples immediately after the FNA procedure and rapid on-site evaluation; hematoxylin and eosin staining of the tissue sampled showed a large amount of malignant cells. Therefore, the most likely possibility is that the sample had pathogenic alterations outside the hotspots.
In the current study, KRAS alterations were identified in GBC specimens, whereas a previous report using a larger cohort reported the absence of the KRAS alteration in this type of BTC (6). This cohort included 28 cases of GBC. In 2 prior reports involving 26 and 57 cases of GBC, the rate of KRAS alterations was 19% and 8%, respectively (22,23). Presently, CT imaging or MRI clearly excluded metastasis involving other organs, such as the pancreas, and colorectal cancer. Therefore, the identified KRAS alterations in GBC in the present study are considered to be consistent with prior data.
EUS-FNA is also useful as a sensitive and safe diagnostic modality for patients with suspected BTC in addition to pancreatic and mediastinal tumors (24,25). Although endoscopic retrograde cholangiopancreatography with transpapillary biopsy is a common procedure for pathological diagnosis of biliary diseases, transpapillary biopsy is frequently unable to give pathological diagnoses for GBC or IHCC because of their locations. Therefore, EUS-FNA can more effectively contribute to the genetic analysis of patients with BTC. Some case reports of tumor dissemination through the needle tract caused by EUS-FNA procedure have been reported, and include pancreatic cancer, lymph node metastasis from gastric cancer, and malignant melanoma (26–28). Tumor dissemination following EUS-FNA in patients with BTC has never been reported. Nevertheless, the possibility of needle tract seeding should be considered.
Effective targeted molecular therapies for BTC, as well as gastric cancer with HER2 mutations, or colorectal cancer with KRAS mutations have never been established due to the wide variety of pathogenic alterations involved (6). Thus, personalized targeted molecular therapy may be more suitable for addressing more variations in several patients with BTC. In the present study, 7 out of 21 cases had actionable alterations (BRAF, NRAS, PIK3CA, and IDH1). In practice, clinical trials for therapies targeting these mutations are currently ongoing (NCT 02304809, NCT01501604, NCT02989857, and NCT03118817). The present study showed nearly 33% of the analyzed BTC cases harbored genetic alterations in potential therapeutic targets, which was nearly equal to that identified in a previous study (40%) (6). In the future, the analysis of pathogenic gene alterations should be required in all BTC cases to provide appropriate targeted molecular therapies. Our results clearly showed that EUS-FNA greatly contributed toward diagnosis and provided appropriate modes of therapy. Furthermore, TAS analysis of EUS-FNA specimens in standard chemotherapy–refractory cases may be useful to detect the changes in gene profiles compared to that prior to treatment. Detection of genetic determinants of response to chemotherapy may also help provide opportunities to receive personalized targeted molecular therapy. In addition, if the specimens that were pathologically diagnosed as normal were genetically analyzed and determined as having genetic evidence of cancer, the approach to therapy or choice of chemotherapy and overall survival could change accordingly.
The present study has several limitations. First, the analyzed samples were not the exact same samples that were evaluated rapidly on-site due to difficulties in sample storage for TAS. Second, the sample size of the present study was small. Although we were able to perform genetic analysis using this novel approach, more cases are required to improve the reproducibility of our results. Third, the current study eliminated the results of germline alterations due to the tumor–normal pair workflow of the analysis software. In consideration of clinical feasibility, we focused on the analysis of somatic alterations in this study. Fourth, we were unable to compare the quality or quantity of DNA between the RNAlater stored specimens and FFPE specimens because of the low number of stored samples.
In conclusion, the present study suggests a novel approach for genetic analysis with TAS using EUS-FNA specimens of BTC, and this may help in developing personalized targeted molecular therapy for patients with BTC.
CONFLICTS OF INTEREST
Guarantor of the article: Masaki Kuwatani, MD, PhD, is accepting full responsibility for the conduct of the study. He had access to the data and has control of the decision to publish.
Specific author contributions: K.H., and M.K., and G.S. contributed to planning and conducting the study, collecting and interpreting data, and drafting the manuscript. M.I., R.S., S.K., K.K., and N.S. contributed to collecting and interpreting data. All authors have approved the final draft submitted.
Financial support: We disclose that there was no funding source and no financial support for the publication.
Potential competing interests: None.
Study Highlights.
WHAT IS KNOWN
✓ Molecular alterations in BTC occur in variety and organ-specific spectra.
✓ Several clinical trials for targeted molecular therapies for BTC have been undergoing.
✓ EUS-FNA can obtain BTC tissues that are difficult to harvest through other procedures.
WHAT IS NEW HERE
✓ Genetic analysis of BTC using specimens obtained through EUS-FNA can successfully be performed.
✓ A novel rapid on-site process before the library preparation can produce superior sequence data using TAS.
TRANSLATIONAL IMPACT
✓ EUS-FNA can contribute to genetic analysis and personalized targeted molecular therapy in patients with BTC.
REFERENCES
- 1.de Groen PC, Gores GJ, LaRusso NF, et al. Biliary tract cancers. N Engl J Med 1999;341:1368–78. [DOI] [PubMed] [Google Scholar]
- 2.Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis 2004;24:115–25. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 4.Hezel AF, Zhu AX. Systemic therapy for biliary tract cancers. Oncologist 2008;13:415–23. [DOI] [PubMed] [Google Scholar]
- 5.Valle J, Wasan H, Palmer DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 2010;362:1273–81. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura H, Arai Y, Tomoki Y, et al. Genetic spectra of bilially tract cancer. Nat Genet 2015;47:1003–10. [DOI] [PubMed] [Google Scholar]
- 7.Milind J, Maeve L, Rachna T, et al. Phase II study of BGJ398 in patients with FGFR-altered advanced cholangiocarcinoma. J Clin Oncol 2018;36:276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao DY, Lim KH. Current biologics for treatment of biliary tract cancers. J Gastrointest Oncol 2017;8:430–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammoud GM, Almashhrawi A, Ibdah JA. Usefulness of endoscopic ultrasound-guided fine needle aspiration in the diagnosis of hepatic, gallbladder and biliary tract lesions. World J Ganstrointest Oncol 2014;6:420–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kameta E, Sugimori K, Kaneko T, et al. Diagnosis of pancreatic lesions collected by endoscopic ultrasound-guided fine-needle aspiration using next-generation sequencing. Oncol Lett 2016;12:3875–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gleeson FC, Kerr SE, Kipp BR, et al. Targeted next generation sequencing of endoscopic ultrasound acquired cytology from ampullary and pancreatic adenocarcinoma has the potential to aid patient stratification for optimal therapy selection. Oncotarget 2016;7(34):54526–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gleeson FC, Kipp BR, Voss JS, et al. Endoscopic ultrasound fine-needle aspiration cytology mutation profiling using targeted next-generation sequencing: Personalized care for rectal cancer. Am J Clin Pathol 2015;143(6):879–88. [DOI] [PubMed] [Google Scholar]
- 13.Gleeson FC, Kipp BR, Kerr SE, et al. Kinase genotype analysis of gastric gastrointestinal stromal tumor cytology samples using targeted next-generation sequencing. Clin Gastroenterol Hepatol 2015;13(1):202–6. [DOI] [PubMed] [Google Scholar]
- 14.Reynolds JP, Zhou Y, Jakubowski MA, et al. Next-generation sequencing of liquid-based cytology non-small cell lung cancer samples. Cancer Cytopathol 2017;125:178–87. [DOI] [PubMed] [Google Scholar]
- 15.Saunders CT, Wong WS, Swamy S, et al. Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012;28:1811–7. [DOI] [PubMed] [Google Scholar]
- 16.Yoo KH, Kim NK, Kwon WI, et al. Genomic alterations in biliary tract cancer using targeted sequencing. Transl Oncol 2016;9:173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farshidfar F, Zheng S, Gingras MC, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep 2017;18:2780–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yadav S, DE Sarkar N, Kumari N, et al. Targeted gene sequencing of gallbladder carcinoma identifies high-impact somatic and rare germline mutations. Cancer Genomics Proteomics 2017;14:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gleeson FC, Kipp BR, Levy MJ, et al. Lung cancer adrenal gland metastasis: Optimal fine-needle aspirate and touch preparation smear cellularity characteristics for successful theranostic next-generation sequencing. Cancer Cytopathol 2014;122:822–32. [DOI] [PubMed] [Google Scholar]
- 20.Young G, Wang K, He J, et al. Clinical next-generation sequencing successfully applied to fine-needle aspirations of pulmonary and pancreatic neoplasms. Cancer Cytopathol 2013;121:688–94. [DOI] [PubMed] [Google Scholar]
- 21.Chin SF, Santonja A, Grzelak M, et al. Shallow whole genome sequencing for robust copy number profiling of formalin-fixed paraffin-embedded breast cancers. Exp Mol Pathol 2018;104:161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simbolo M, Fassan M, Ruzzenente A, et al. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget 2014;5:2839–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Zhang Z, Li X, et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet 2014;46:872–6. [DOI] [PubMed] [Google Scholar]
- 24.Navaneethan U, Njei B, Venkatesh PG, et al. Endoscopic ultrasound in the diagnosis of cholangiocarcinoma as the etiology of biliary strictures: A systematic review and meta-analysis. Gastroenterol Rep (Oxf) 2015;3:209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohshima Y, Yasuda I, Kawakami H, et al. EUS-FNA for suspected malignant biliary strictures after negative endoscopic transpapillary brush cytology and forceps biopsy. J Gastroenterol 2011;46:921–8. [DOI] [PubMed] [Google Scholar]
- 26.Sakamoto U, Fukuba N, Ishihara S, et al. Correction to: Postoperative recurrence from tract seeding after use of EUS-FNA for preoperative diagnosis of cancer in pancreatic tail. Clin J Gastroenterol 2018;11:260. [DOI] [PubMed] [Google Scholar]
- 27.Shah JN, Fraker D, Guerry D, et al. Melanoma seeding of an EUS-guided fine needle track. Gastrointest Endosc 2004;59(7):923–4. [DOI] [PubMed] [Google Scholar]
- 28.Doi S, Yasuda I, Iwashita T, et al. Needle tract implantation on the esophageal wall after EUS-guided FNA of metastatic mediastinal lymphadenopathy. Gastrointest Endosc 2008;67:988–90. [DOI] [PubMed] [Google Scholar]