Abstract
Background:
Alcohol dependence and long-term excessive alcohol use may cause liver damage, but only some patients develop cirrhosis. Similarly, high alcohol intake without evident liver disease often but not always produces abnormal enzymatic liver function tests, particularly gamma-glutamyl transferase (GGT). We postulate that the factors predisposing to cirrhosis in alcoholics and to liver enzyme abnormality in drinkers are similar, and that biochemical liver function tests could therefore be useful as markers of risk of alcoholic liver disease in excessive drinkers.
Methods:
Data from participants in twin and twin-family studies on alcohol use and dependence were used to identify 1003 people who had reported excessive alcohol intake (28 drinks or more per week). 962 of these provided blood for biochemical tests at the same time. Body mass index (BMI) and biomarkers of metabolic syndrome, inflammation, and iron stores, were used in logistic regression with abnormality in serum GGT, alanine aminotransferase (ALT), or aspartate aminotransferase (AST) as outcomes. We conducted genome-wide association analyses for GGT, ALT and AST separately in the group reporting excessive alcohol intake (N=951) and a low-intake group reporting 14 drinks or fewer per week (N=8716), and compared results.
Results:
Abnormal GGT and ALT among excessive drinkers were associated with higher BMI, triglycerides, insulin, uric acid, C-reactive protein, ferritin and transferrin saturation; and with lower HDL cholesterol. Abnormal AST was associated with triglycerides, ferritin and transferrin saturation. ALT was significantly associated with variants at reported genetic loci for alcoholic liver disease (PNPLA3, rs738409 p = 0.0076; TM6SF2, rs10401969 p = 0.0076; HSD17B13, rs10433879 p = 0.0024).
Conclusions:
Known risk factors for alcoholic cirrhosis including obesity and markers of metabolic syndrome, iron overload, and inflammation are associated with liver enzyme abnormality in excessive drinkers.
Keywords: Alcohol, ALT, AST, cirrhosis, GGT
INTRODUCTION
Excessive alcohol use is associated with risks including intoxication, development of addiction, organ damage and increased mortality. The liver is particularly susceptible to alcohol-related damage, possibly because it is the main site of alcohol metabolism.
Elevation of the serum enzymes used as liver function tests (LFTs) is a common consequence of high alcohol intake. This is most evident for gamma glutamyl transferase (GGT) but also occurs with alanine aminotransferase (ALT) and aspartate aminotransferase (AST) (Rosalki et al., 1970, Whitfield et al., 1978, Conigrave et al., 2003, McDonald et al., 2013, Agarwal et al., 2016, Niemela et al., 2017). The dose-response relationships in the general population show increases in mean values and in the prevalence of abnormal values with increasing alcohol intake (Whitehead et al., 1996, Steffensen et al., 1997, Sillanaukee et al., 2000, Alatalo et al., 2009, Liangpunsakul et al., Whitfield et al., 2013). Mean differences between drinker and abstainer groups become apparent when alcohol intake is above 20–40 grams per day or 14–28 drinks per week (Whitfield et al., 2013).
There are similar dose-related effects of drinking on risk of alcoholic liver disease (Pequignot et al., 1978, Tuyns and Pequignot, 1984, Becker et al., 1996). Meta-analysis of data from nine studies on cirrhosis (Corrao et al., 2004) showed that people consuming more than 25 grams of alcohol per day are at significantly increased risk compared to non-drinkers. However not all alcoholics, or drinkers exceeding the thresholds for hazardous intake, show clinical or laboratory evidence of liver damage (Lelbach, 1976, Grant et al., 1988, Mann et al., 2003). Despite the statistical association between intake and cirrhosis, alcoholics with cirrhosis may have consumed similar amounts of alcohol to those who have only steatosis (Grove et al., 1998) or no detectable liver disease (Whitfield et al., 2015). It has been estimated that over half of excessive drinkers develop fatty liver but less than 20% develop cirrhosis (Gramenzi et al., 2006). About half show abnormality of LFTs, particularly GGT (Conigrave et al., 2003). This suggests that there is variation in susceptibility to liver damage from alcohol, both in the occurrence of clinical problems and in the occurrence of test abnormality, and it would be useful to know whether abnormality of GGT, AST or ALT in at-risk drinkers is a precursor and potential predictor of alcoholic liver disease.
Known risk factors for alcoholic liver disease include obesity and metabolic syndrome (Naveau et al., 1997, Iturriaga et al., 1988) and iron overload (Ganne-Carrie et al., 2000). A variant in the PNPLA3 gene, known to be associated with fatty liver and with serum ALT activity, is associated with risk of alcoholic liver disease and cirrhosis (Tian et al.)2010). This was confirmed and additional loci near TM6SF2 and MBOAT7 were identified in a genome-wide study (Buch et al., 2015). A subsequent genome-wide study investigating loci which affect serum AST or ALT (Abul-Husn et al., 2018) found that a locus on chromosome 4 near the HSD17B13 gene contains a protective variant affecting risk of both alcoholic and non-alcoholic liver diseases, including cirrhosis.
A combination of biomarker and genomic data from our twin/family studies on alcohol use in the general population allows us to address the causes of the observed variation in GGT, AST, and ALT in excessive drinkers. We focus on these because the dose-response relationships in our data between reported alcohol intake and other LFTs are weaker or biphasic (for bilirubin, alkaline phosphatase) or, for albumin, in the opposite direction to that expected (Whitfield et al., 2013). Our primary aim was to investigate whether liver enzyme abnormalities in at-risk drinkers who are not known to have significant liver disease at the time of blood collection are associated with risk of developing alcoholic cirrhosis later (assuming they continue drinking). We approach this by assessing whether and how far (a) the existence of liver enzyme abnormalities in an at-risk drinker and (b) alcoholic cirrhosis, share the same predisposing factors. The rationale is that if they do, then the group who show liver enzyme abnormality will overlap with those who develop cirrhosis, and the liver enzyme abnormalities may be useful as predictors, or as surrogates for clinical outcomes in studies of differences in vulnerability. To address this question we first focus on obesity or metabolic syndrome and on iron overload, and then test whether genetic loci associated with alcoholic cirrhosis are important in determining the LFT response to alcohol in hazardous drinkers. Secondly, we aim to identify any novel risk factors or loci whose effects only become apparent under conditions of high alcohol exposure. We approach this by testing whether the biomarker associations and genetic causes of GGT, AST and ALT variation in excessive drinkers are similar to those in low-risk drinkers.
MATERIALS AND METHODS
Participants and data were from our studies on the genetics of alcohol and nicotine dependence and on the biological consequences of excessive alcohol use. Initial contact was through a voluntary twin registry (the Australian Twin Registry), with invitations to participate later extended to twins’ relatives and to their spouses and spouses’ families. Patterns of alcohol use, psychiatric morbidity and parental education (before selection on criteria outlined below) were consistent with those expected for the Australian population. All individuals gave informed consent, and the studies were approved by the appropriate ethics review committees.
Nicotine-Alcohol Study
The largest and most comprehensive source of data and samples was a study on the genetics of alcohol and nicotine use and dependence, which recruited twins who had participated in our previous studies on alcohol consumption and their spouses, parents, siblings, and adult children (Heath et al., 2011, Whitfield et al., 2013). This group of studies, each having the same questionnaire and sample handling protocols but differing in targeting of potential participants, took place between 2002 and 2005. The sub-studies focused on families of probands known (from previous information) to be (a) heavy smokers, or (b) alcohol-dependent or scoring highly for a heaviness-of-drinking factor score, or (c) from large sibships (with 4–14 full siblings). This group will be referred to as the Nicotine-Alcohol Study.
Information was collected in a telephone interview which included questions on lifetime and past-year alcohol or nicotine dependence, and on the usual quantity and frequency of alcohol use over the previous twelve months. Blood was collected between 2002 and 2005 from 9031 people (3998 men and 5033 women) aged 18–92 (mean 51.6, SD 13.8) years. At the time of blood collection, participants reported on their alcohol intake and smoking over the past seven days, their height and weight, and the time since their last meal. Alcohol and smoking data were compiled from the self-report diaries by summing the number of alcoholic drinks or the number of times tobacco products were used over the 7-day period. Anyone who used any tobacco products, including snuff and chewing tobacco, was characterized as a “smoker” but practically all tobacco use (98%) was as cigarettes. Body mass index (BMI) was calculated as kg/m2 from self-reports of weight and height.
The participants lived throughout Australia, and blood samples were sent for processing to the Queensland Institute of Medical Research in Brisbane via courier for next-day delivery. After centrifugation, serum was obtained from tubes without anticoagulant and plasma from fluoride-oxalate tubes (for glucose measurement). Aliquots were stored at −70 °C until analysis. Serum ALT, AST, cholesterol, C-reactive protein (CRP), ferritin, high-density-lipoprotein cholesterol (HDL-C), GGT, glucose, iron, transferrin, triglycerides and urate were measured using Roche reagents on a Roche 917 or Modular P analyser. Serum insulin, hepatitis B surface antigen and hepatitis C antibodies were measured using Abbott reagents on an Abbott AxSym analyser. We calculated low-density-lipoprotein cholesterol (LDL-C) using the Friedewald equation, and transferrin saturation from iron and transferrin.
Additional Studies
Additional data and samples, including information on alcohol use, biochemical test results and DNA, were available from two earlier studies. The range of tests performed and the information on smoking status was less complete than for the Nicotine-Alcohol Study described above. Data from these two additional studies were mainly used to maximize the number of people available for genome-wide association analysis in sub-groups defined by alcohol intake (as described below). They also allowed us to assess repeatability across occasions for GGT, AST and ALT results in excessive drinkers.
In the first of these additional studies, data on alcohol use and potentially co-morbid conditions were collected using the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) interview. In the SSAGA Blood Study (Whitfield et al., 2002), which took place between 1993 and 1996, blood was collected from a subset of the interview participants comprising 1048 male and 2012 female twins between 28 and 89 years old (mean 44.4, SD 11.7). At the time of blood collection participants reported on their alcohol intake over the past seven days, their height and weight, and the time since their last meal. BMI was calculated from weight and height. Alcohol data were compiled from the self-report diaries by summing the number of alcoholic drinks over the 7-day period. Blood was processed and stored as described above, except that approximately a third of the samples were processed within two hours of collection rather than being sent to a central laboratory. Serum ALT, AST, and GGT were measured using Boehringer reagents on a Hitachi 747 analyser.
The Anxiety Study took place between 1993 and 1996 and included 943 male and 1533 female subjects, twins and their siblings, between 24 and 96 years old (mean 40.8, SD 11.2) at the time of blood collection. Serum ALT, AST, and GGT were measured using Roche reagents on a Roche 917 analyser. Information on alcohol intake and smoking was taken from earlier questionnaire-based studies.
Genotyping
Genotype data were derived from several genotyping projects with Illumina 317K, 370K, 610K, Omni Express or Infinium CoreExome chips. After quality control of sample and SNP data, imputation of SNP genotypes was performed using SNPs common to these platforms and haplotype data on ~65,000 chromosomes from the Haplotype Reference Consortium (http://www.haplotype-reference-consortium.org/, accessed 2018–02-13). Imputed SNPs with a minor allele frequency below 1%, or with the imputation quality measure R2 less than 0.3, were excluded, leaving approximately 7.8m autosomal and X-chromosome markers.
Data analysis
Frequency distributions of the variables were checked and where necessary log-transformation was carried out (for ALT, AST, CRP, ferritin, GGT, insulin, triglycerides). Reported alcohol intake was transformed as log(N of drinks + 1) to allow inclusion of people who reported no drinks. Glucose and insulin results were adjusted for time between the last meal and blood collection, by creating intervals of fasting time and normalising the results for each person by subtracting the mean and dividing by the standard deviation for that time period. Because of the possible effect of past or current hepatitis on liver function tests, data from 13 people in the excessive drinking group with positive results for either Hepatitis B or C tests were excluded.
Data analysis initially used IBM SPSS, release 22 (IBM Corp., Armonk, NY, USA) for data management and estimation of means and correlations. However, because our studies emphasized twins and their families, there is a genetic overlap between many of the subjects. This means that, to the extent that family members are similar to each other for genetic reasons (which will vary according to the heritability of the phenotypes), the number of independent observations is less than the number of participants and the standard errors for calculated statistics will be underestimated. However the estimates themselves, including means, correlation coefficients and hazard ratios (HR), are not changed; only the standard errors and therefore the confidence intervals and P-values are affected. To overcome this problem, we repeated the logistic regression analysis using Stata (StataCorp LLC, College Station, TX, USA) with the ‘clustered robust standard error’ option, grouping subjects by family to generate robust standard errors for the regression coefficients and for estimation of confidence intervals for hazard ratios.
Allelic association analysis for log-transformed GGT, AST and ALT was done using an additive model in GEMMA (Genome-wide Efficient Mixed Model Association algorithm) (Zhou and Stephens, 2012) with sex, age, BMI, number of drinks in previous week, smoking status and 10 ancestry-informative principal components as covariates.
RESULTS
There were 11,985 people with data on alcohol intake and biochemical results from one or more of the three studies, of whom 1003 reported alcohol intake ≥ 28 drinks/week in any study. Out of the 1003, 962 had GGT, ALT and AST results, and 951 had information on alcohol consumption, liver enzyme results from the time of reporting high alcohol intake, and genotyping. Because the Nicotine-Alcohol study had the most comprehensive set of test results and self-report data, our main focus is on this.
Table 1 shows the characteristics of 8382 participants from the Nicotine-Alcohol Study who had results for GGT, AST and ALT and who had provided data for the 7-day alcohol recall, and also for sub-groups of 860 excessive drinkers who reported 28 drinks or more in the seven days before blood collection and 4908 low-risk drinkers who reported 14 drinks or fewer. Similar information is given for 3060 participants in the SSAGA study and 2351 participants in the Anxiety study in Supplementary Table 1; because some people participated in more than one study, the total number of results from Table 1 and Supplementary Table 1 can be greater than the total number of participants.
Table 1.
Characteristics of the Nicotine-Alcohol Study participants. Excessive drinkers are defined as those who reported 28 drinks or more in the previous week at the time of blood collection, and Low-risk drinkers as those who reported 14 drinks or fewer. The 90% ranges are based on the 5th and 95th centiles for each variable.
| All participants | Excessive Drinkers only | Low-risk Drinkers only | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MALE | FEMALE | MALE | FEMALE | MALE | FEMALE | |||||||||||||
| N | Median | 90% range | N | Median | 90% range | N | Median | 90% range | N | Median | 90% range | N | Median | 90% range | N | Median | 90% range | |
| Age (years) | 3741 | 49 | 34–76 | 4641 | 48 | 33–76 | 729 | 47 | 33–71 | 131 | 43 | 33–69 | 1583 | 51 | 34–78 | 3325 | 49 | 34–77 |
| Total Drinks (Past week) | 3741 | 10 | 0–53 | 4641 | 3 | 0–22 | 729 | 41 | 28–93 | 131 | 35 | 28–73 | 1583 | 1 | 0–7 | 3325 | 1 | 0–7 |
| Current smokers (%) | 3741 | 20% | 4641 | 18% | 729 | 37% | 131 | 54% | 1583 | 15% | 3325 | 15% | ||||||
| BMI (kg/m2) | 3741 | 26.6 | 21.4–34.6 | 4641 | 25.4 | 19.8–37.1 | 729 | 26.6 | 21.2–34.1 | 131 | 25.8 | 19.7–36.1 | 1583 | 26.6 | 21.2–35.2 | 3325 | 25.7 | 19.8–37.8 |
| GGT (units/L) | 3741 | 27 | 13–100 | 4639 | 17 | 9–60 | 729 | 39 | 16–174 | 131 | 29 | 12–132 | 1583 | 24 | 12–81 | 3325 | 17 | 8–58 |
| ALT (units/L) | 3741 | 26 | 13–61 | 4641 | 18 | 10–40 | 729 | 29 | 15–75 | 131 | 20 | 11–72 | 1583 | 25 | 13–59 | 3325 | 18 | 10–40 |
| AST (units/L) | 3741 | 25 | 17–43 | 4641 | 21 | 14–34 | 729 | 27 | 18–58 | 131 | 24 | 16–55 | 1583 | 25 | 16–40 | 3325 | 21 | 14–34 |
| Triglyceride (mmol/L) | 3740 | 1.89 | 0.86–4.85 | 4640 | 1.44 | 0.69–3.46 | 729 | 1.92 | 0.84–5.77 | 131 | 1.43 | 0.76–5.31 | 1583 | 1.86 | 0.88–4.61 | 3325 | 1.5 | 0.69–3.56 |
| HDL-C (mmol/L) | 3741 | 1.31 | 0.85–2.03 | 4640 | 1.63 | 1.04–2.44 | 729 | 1.53 | 0.98–2.32 | 131 | 1.88 | 1.10–2.83 | 1583 | 1.2 | 0.8–1.85 | 3325 | 1.57 | 1.01–2.28 |
| LDL-C (mmol/L) | 3535 | 3.33 | 1.94–4.90 | 4565 | 3.15 | 1.88–4.83 | 671 | 3.44 | 2.05–4.92 | 123 | 2.98 | 1.57–4.76 | 1508 | 3.26 | 1.86–4.86 | 3270 | 3.19 | 1.92–4.89 |
| Glucose (mmol/L) | 3034 | 4.9 | 3.7–8.6 | 3765 | 4.6 | 3.6–7.0 | 535 | 5.0 | 4.0–8.9 | 93 | 4.8 | 3.9–6.2 | 1286 | 4.91 | 3.64–9.30 | 2668 | 4.62 | 3.50–7.22 |
| Insulin (pmol/L) | 1046 | 60 | 14–371 | 1099 | 57 | 18–265 | 722 | 52 | 13–324 | 129 | 50 | 16–250 | 158 | 112 | 26–495 | 354 | 70 | 23–297 |
| Urate (μmol/L) | 3741 | 0.340 | 0.230–0.480 | 4639 | 0.250 | 0.160–0.390 | 729 | 0.360 | 0.230–0.500 | 131 | 0.270 | 0.176–0.400 | 1583 | 0.33 | 0.22–0.47 | 3325 | 0.25 | 0.16–0.39 |
| C-reactive protein (mg/L) | 3731 | 1.8 | 0.3–10.4 | 4631 | 2.2 | 0.3–13.7 | 728 | 1.9 | 0.3–10.6 | 131 | 2.6 | 0.4–15.1 | 1578 | 1.80 | 0.3–12.6 | 3320 | 2.3 | 0.3–14.7 |
| Ferritin (mg/L) | 3740 | 218 | 44–634 | 4640 | 79 | 15–320 | 729 | 268 | 83–752 | 131 | 121 | 23–574 | 1582 | 200 | 36–617 | 3325 | 77 | 14–320 |
| Transferrin Saturation (%) | 3738 | 31 | 18–51 | 4637 | 27 | 12–45 | 728 | 33 | 20–56 | 131 | 30 | 14–51 | 1582 | 30.2 | 16.9–49.2 | 3323 | 25.8 | 11.5–43.9 |
Alcohol intake was assessed by the seven-day recall method, which has good repeatability across occasions, correlates with quantity-frequency measures, and is associated with all-cause mortality (Whitfield et al., 2004, Whitfield et al., 2018). The choice of the cut-off values of ≥ 28 for ‘excessive’ drinking and ≤ 14 drinks per week for ‘low-risk’ drinking for both men and women was based on the dose-response curves presented below.
Effect of reported alcohol intake on liver enzymes
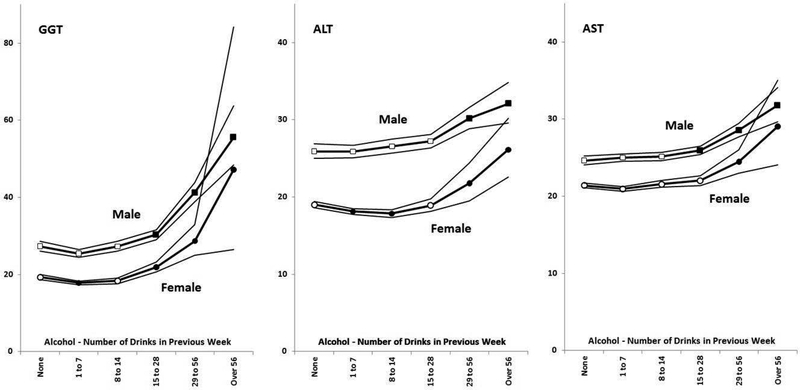
Participants in the Nicotine-Alcohol Study were divided into six alcohol intake groups based on the reported number of alcoholic drinks taken in the seven days before blood collection (none, 1 to 7, 8 to 14, 15 to 28, 29 to 56 and over 56). The means and 95% confidence intervals for log-transformed GGT, ALT and AST were calculated for each group and then back-transformed to the original units. The dose-response relationships for men and women (Figure 1) were essentially parallel and there was no significant sex by alcohol group interaction for GGT, ALT or AST. Based on data shown in Figure 1, we defined excessive drinkers (for the purpose of identifying those at risk of abnormal liver enzymes) as those reporting ≥ 28 drinks in the previous week (40 g alcohol per day or more), for both men and women.
Figure 1.
Dose-response relationships for serum GGT, ALT and AST activity by alcohol intake in men and women. Points and lines show geometric means and 95% CIs for those means, estimated from the means and standard errors for log-transformed enzyme activity and converted back to enzyme units for clarity. Note the different scale on the y-axis for GGT. Open squares (for men) and circles (for women) show groups where means do not differ significantly (p > 0.01) from the group reporting no drinks, and filled squares and circles show groups whose means do differ (at p < 0.01) from the no-drinks means.
Reference Ranges
A reference group was defined, using conservative criteria with respect to both alcohol and obesity, as people with low or no alcohol intake (reporting ≤ 7 drinks/week) and also with normal BMI (≤ 25.0 kg/m2). The upper reference intervals (95th centile) were estimated for each enzyme and separately for men and women. For GGT, ALT and AST these were 61, 44 and 36 units/L in men and 43, 32 and 32 units/L in women, respectively. These limits were used in the definition of abnormal GGT, ALT and AST for logistic regression analysis.
Phenotypic predictors of abnormal enzyme results in hazardous drinkers
Using logistic regression to define the variables associated with abnormal enzyme results in the excessive drinkers (Table 2), we found that a cluster of variables related to obesity and metabolic syndrome, the two measures of iron status, and the inflammation marker CRP, each had significant effects. This pattern was consistent across the three enzymes, although the Odds Ratio estimates and p-values suggested stronger associations for GGT and ALT than for AST. The significant predictors at p < 0.01 were (in increasing order of p-values, i.e. starting with the most robust association) triglycerides, CRP, ferritin, BMI, insulin, transferrin saturation, urate and glucose for GGT; triglycerides, ferritin, insulin, BMI, CRP, transferrin saturation, HDL-C and LDL-C for ALT; and transferrin saturation, ferritin, triglycerides and urate for AST. When analysed separately, results were similar in men and women but the preponderance of men among the excessive drinkers led to less statistically significant associations in the women. The reported number of drinks, even in this group selected for high alcohol intake, showed significant associations with GGT and AST but smoking status (smoker or non-smoker at the time of blood collection) did not. Multivariate analysis, in which all the postulated predictors were entered in order to identify independent effects, gave results shown in Supplementary Table 2.
Table 2.
Results of logistic regression with abnormality in GGT, ALT or AST as the outcome and predictor variables as listed. Excessive drinkers (28 or more drinks per week) only. Glucose and insulin are adjusted for time between blood collection and the last meal (fasting time).
| Univariate | N | GGT | AST | ALT | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | p | OR | 95% CI | p | OR | 95% CI | p | ||
| Age (years) | 860 | 1.019 | 1.009–1.030 | 4.16 × 10−4 | 0.996 | 0.983–1.010 | 0.617 | 0.989 | 0.975–1.003 | 0.114 |
| Total drinks (log) | 860 | 27.45 | 10.91–69.04 | 1.92 × 10−12 | 8.043 | 2.937–22.02 | 4.91 × 10−5 | 2.452 | 0.864–6.959 | 0.091 |
| Non-smoker/Smoker | 860 | 1.314 | 0.977–1.768 | 0.070 | 0.925 | 0.664–1.290 | 0.646 | 1.000 | 0.715–1.399 | 1.000 |
| BMI | 860 | 1.107 | 1.062–1.154 | 1.67 × 10−6 | 1.054 | 1.009–1.100 | 0.018 | 1.128 | 1.079–1.180 | 9.82 × 10−8 |
| Triglyceride (log) | 860 | 16.61 | 8.856–31.16 | < 1.0 × 10−15 | 3.279 | 1.649–6.521 | 6.99 × 10−4 | 11.18 | 5.222–23.93 | 4.97 × 10−10 |
| HDL-XSC | 860 | 0.676 | 0.482–0.947 | 0.023 | 0.988 | 0.655–1.489 | 0.952 | 0.483 | 0.290–0.804 | 0.0051 |
| LDL-C | 794 | 1.145 | 0.957–1.371 | 0.139 | 1.167 | 0.932–1.462 | 0.180 | 1.374 | 1.103–1.712 | 0.0047 |
| Glucose | 628 | 1.256 | 1.075–1.467 | 0.0041 | 1.126 | 0.964–1.315 | 0.136 | 1.217 | 1.026–1.443 | 0.024 |
| Insulin (log) | 851 | 1.445 | 1.238–1.686 | 3.01 × 10−6 | 0.972 | 0.804–1.176 | 0.772 | 1.604 | 1.368–1.881 | 6.25 × 10−9 |
| Urate (mmol/l) | 860 | 44.84 | 6.144–327.3 | 1.77 × 10−4 | 24.64 | 2.677–226.8 | 0.0047 | 38.26 | 4.193–349.1 | 0.0012 |
| C-reactive protein (log) | 857 | 2.974 | 2.158–4.098 | 2.74 × 10−11 | 1.501 | 1.043–2.161 | 0.029 | 2.138 | 1.513–3.021 | 1.63 × 10−5 |
| Ferritin (log) | 860 | 5.545 | 3.322–9.258 | 5.75 × 10−11 | 7.156 | 3.492–14.67 | 7.45 × 10−8 | 9.263 | 4.643–18.48 | 2.62 × 10−10 |
| Transferrin saturation (%) | 859 | 1.026 | 1.013–1.040 | 1.23 × 10−4 | 1.043 | 1.029–1.058 | 2.68 × 10−9 | 1.026 | 1.013–1.040 | 1.04 × 10−4 |
In excessive drinkers who participated in one of the other studies (Anxiety or SSAGA Blood) as well as in the Nicotine-Alcohol Study, enzyme levels showed significant repeatability across occasions (Supplementary Figure 1); results on one occasion predicted results on another.
Comparison of predictors of abnormal enzyme results in excessive and low-risk drinkers
Information on factors associated with abnormal results in low-risk drinkers (those reporting 14 drinks or fewer in the preceding seven days) is shown in Supplementary Table 3. Comparing these results with those for the excessive drinkers, we find that once again the obesity, metabolic and lipid results are associated with GGT, AST and ALT. Higher CRP, and ferritin but not transferrin saturation, are also associated with increased probability of abnormal enzyme results.
These results based on logistic regression, with the emphasis on classification of enzyme results as normal or abnormal, were supplemented by analyses based on correlation analyses (Supplementary Table 4).
Candidate Genes & GWAS
We assessed the effects of SNPs in PNPLA3, TM6SF2, MBOAT7 and HSD17B13, which have been shown to affect risk for alcoholic cirrhosis, on GGT, ALT and AST results in excessive drinkers. These results are summarised in Table 3, and results for the same SNPs in low-risk drinkers are shown for comparison. In the excessive drinker group, variation at rs738409 at PLPNA3 showed at least nominally significant (p < 0.05) association with ALT (p = 0.0076) and AST (p = 0.018); in each case the less frequent G allele was associated with higher enzyme results. SNPs at TM6SF2 and HSD17B13 were associated with ALT (p = 0.0076 and 0.024, respectively) but not AST; for rs10401969 at TM6SF2 the less frequent C allele was associated with higher ALT but for rs10433879 at HSD17B13 the less frequent C allele was associated with lower ALT results. None of these alcoholic cirrhosis loci significantly affected GGT in the excessive drinkers. Conversely, we used publicly available data to assess whether SNPs shown to affect ALT or GGT at p < 5 × 10−8 in the general population (Chambers et al., 2011) showed associations in the summary statistics of a GWAS of alcoholic cirrhosis (Buch et al., 2015). No data were available for AST. Results are shown in Supplementary Table 5. The only overlap after allowing for multiple testing was for the PNPLA3 SNP rs738409 which affected ALT at p = 1.17 × 10−28 and alcoholic cirrhosis at p = 1.54 × 10−48; none of the significant GGT SNPs showed even nominal association with alcoholic cirrhosis.
Table 3.
Allelic associations for serum GGT, AST and ALT in excessive drinkers and in low-risk drinkers, at loci which have been shown to affect risk of alcoholic cirrhosis. Covariates were sex, age, number of drinks in previous week, BMI and smoking status (current versus never or ex-smoker).
| Locus | SNP | Drinking | GGT β ± SE | AST β ± SE | ALT β ± SE |
|---|---|---|---|---|---|
| PNPLA3 | rs738409 | Excessive | 0.02139 ± 0.01688, p = 0.205 | 0.02099 ± 0.00887, p = 0.018 | 0.03256 ± 0.01216, p = 0.0076 |
| Low-risk | 0.00033 ± 0.00502, p = 0.947 | 0.01403 ± 0.00262, p = 9.11 × 10−8 | 0.02335 ± 0.00399, p = 4.85 × 10−9 | ||
| TM6SF2 | rs10401969 | Excessive | 0.04399 ± 0.02755, p = 0.111 | 0.02130 ± 0.01460, p = 0.145 | 0.05341 ± 0.01996, p = 0.0076 |
| Low-risk | −0.00655 ± 0.00797, p = 0.412 | 0.00358 ± 0.00417, p = 0.391 | 0.00336 ± 0.00634, p = 0.596 | ||
| MBOAT7 | rs626283 | Excessive | 0.00309 ± 0.01411, p = 0.827 | −0.00445 ± 0.00744, p = 0.549 | −0.00445 ± 0.01020, p = 0.663 |
| Low-risk | 0.00171 ± 0.00413, p = 0.679 | −0.00087 ± 0.00217, p = 0.689 | −0.00072 ± 0.00329, p = 0.827 | ||
| HSD17B13 | rs10433879 | Excessive | −0.02701 ± 0.01572, p = 0.086 | −0.00921 ± 0.00830, p = 0.268 | −0.03446 ± 0.01134, p = 0.0024 |
| Low-risk | −0.00814 ± 0.00468, p = 0.082 | −0.00144 ± 0.00245, p = 0.557 | −0.01155 ± 0.00372, p = 0.0019 |
After performing genome-wide association analyses for GGT, AST and ALT separately in the excessive and low-risk drinking groups, no SNPs reached genome-wide significance in the excessive-drinkers group. Four loci for GGT and one for ALT were significant in the low-risk group. The four GGT loci were in or near the genes HNF1A (rs7979478, chr. 12, p = 3.02 × 10−15), EXOC3L4 (previously C14orf73) (rs7151779, chr. 14, p = 2.06 × 10−12), RORA (rs340005, chr. 15, p = 3.66 × 10−9) and GGT1 (rs28509371, chr. 22, p = 1.84 × 10−28), which had been recognised in previous studies. The significant locus for ALT was again at PNPLA3 (p = 2.68 × 10−9 for rs3747207, which has r2 = 0.983 with rs738409) and this SNP was also marginally significant (p = 5.71 × 10−8) for AST.
To contrast the allelic association results in the two groups, we selected ‘suggestive’ SNPs (with p < 5 × 10−6) for each enzyme either in excessive drinkers or in low-risk drinkers (listed in Supplementary Table 6), and compared their effect sizes between the two groups. Results of these comparisons are shown in Supplementary Figures 2, 3 and 4, restricted to SNPs with p < 5 × 10−7 for clarity. There are loci which appear to show differences in allelic effects between excessive and low-risk drinkers but none reach genome-wide statistical significance (p < 5 × 10−8) for heterogeneity, and the apparent differences in allelic associations between the two drinking groups may be due to the large number of variants tested.
DISCUSSION
Relationships between alcohol intake and GGT, ALT, AST
All three enzymes showed increasing mean values as self-reported alcohol intake increased above 14 drinks per week or an average of two drinks per day. As we reported previously (Whitfield et al., 2013) and as expected from previous studies, the alcohol-induced change was greater for GGT than for ALT or AST. Our selection of 28 drinks per week (four drinks or 40 grams per day) as a definition of excessive drinkers was based on several considerations. This was the lowest level of alcohol intake giving a significant difference in mean results from the no-alcohol group for all three enzymes and in both sexes. The epidemiological data on alcoholic liver disease risk (Pequignot et al., 1978, Becker et al., 1996, Corrao et al., 2004), and most previous data on liver enzymes in drinkers, are consistent with our alcohol/enzyme dose-response curves (Figure 1). We chose to combine data from men and women into one analysis for each enzyme with separate cut-off values defining abnormality, rather than sex-specific analyses. The parallel curves for men and women and the absence of significant sex x alcohol group interaction support this approach for the enzyme results, even though sex differences in susceptibility are expected for alcoholic liver disease.
Quantitative predictors of liver enzyme abnormality among excessive drinkers
Three groups of quantitative variables are postulated to affect risk of abnormal enzyme results: alcohol intake, variables associated with obesity and metabolic syndrome, and markers of iron status (ferritin, transferrin saturation). Higher reported amount of alcohol (even when above the 28 drinks limit) increased risk of abnormal results for all three enzymes. Obesity- or lipid-related phenotypes affected the risk of abnormal results; triglyceride for all three enzymes and HDL-C and BMI for one or more. Iron status (ferritin, transferrin saturation) affected risk of abnormality for all three. In addition CRP, a marker of inflammation, was strongly associated with abnormal results for GGT. Two of these three domains affecting enzyme abnormality, obesity and its associated metabolic changes and iron status, were predicted from published work on risk factors for liver disease in drinkers (Iturriaga et al., 1988, Naveau et al., 1997, Ganne-Carrie et al., 2000, Majumdar et al., 1991). Inflammation is also recognised as part of the pathological process leading to alcoholic liver disease (Wang et al., 2012, Gao and Tsukamoto, 2016), and both high CRP and positive correlations between CRP and GGT, AST and ALT in excessive drinkers have been reported previously (Qu et al., 2016). From this pattern of associations, we conclude that the quantitative risk factors for alcoholic liver disease and for enzyme abnormality in excessive drinkers are similar.
We also tested whether the known risk factors for alcoholic liver disease (obesity- and iron-related measures) affected the probability of abnormal liver enzymes in a group of participants who reported low-risk alcohol intake (defined for our purposes as people who reported 14 or fewer drinks in the week preceding blood collection). Similar to results in the excessive drinkers, risk of abnormal GGT, AST and ALT results in the low-risk drinkers was associated with BMI, triglycerides, glucose, insulin, urate, CRP and ferritin (Supplementary Table 3).
Genetic variation in relation to liver enzyme abnormality among excessive drinkers
We next tested whether the small number of genetic variants previously reported to affect risk of cirrhosis in alcoholics had significant effects on GGT, AST or ALT results in the excessive drinkers. Compared with a genome-wide approach, this diminishes the multiple-testing problem and allows a less stringent p-value. The only notable association was for SNPs in PNPLA3 (Table 3), which are already known to affect ALT (Chambers et al., 2011) and also to affect risk for both alcoholic cirrhosis (Buch et al., 2015) and non-alcoholic fatty liver disease (Xu et al., 2015). Our results confirm that this locus affects ALT and AST in both excessive and low-risk drinkers.
A recent report (Abul-Husn et al., 2018) implicated rs72613567, an insertion-deletion variant in HSD17B13, in liver disease (including alcoholic liver disease and alcoholic cirrhosis) and also showed it is associated with variation in AST and ALT (but not GGT) levels. This variant was not imputed in our data but a close proxy (rs10433879, r2=0.958) was available and it was nominally associated (at p < 0.05) with ALT in both excessive (p = 0.0025) and low-risk (p = 0.0019) drinkers but not with AST or GGT.
Turning to genome-wide association results, no SNPs reached the accepted significance level of p < 5 × 10−8 among the excessive-drinking participants. However the number of people in this group with genotype and enzyme data was 951 and although this is a substantial cohort given the precondition of excessive drinking, the power to detect effects at genome-wide significance is limited. Only loci accounting for more than about 4% of trait variance would be detectable with 80% power. The larger low-risk drinking group showed significant associations for GGT at four loci which are already known from population-based data without differentiation by alcohol intake (Chambers et al., 2011), and for ALT and probably AST at PNPLA3. Based on the number of subjects available, we did not find evidence for significantly different genetic effects on enzyme results in excessive and low-risk drinkers.
Implications
It would be useful to be able to conclude that factors affecting the liver enzymes also affect development of liver disease in people drinking too much, because the enzyme results might then serve as markers of future risk. Several older papers compared patients’ GGT with their liver histology (Wu et al., 1976, Banciu et al., 1983, Frezza et al., 1989), finding a consistent association between higher GGT and structural abnormality. Despite a large literature on GGT and mortality, information related to liver disease mortality is sparse and even papers which report positive associations between mortality from liver disease and GGT (Breitling et al., 2011, Ruhl and Everhart, 2009) do not distinguish between alcoholic and other causes.
We found that phenotypic risk factors for alcoholic liver disease and for liver enzyme abnormality are similar. We also found that both the phenotypic and genetic influences on the enzyme results are similar between excessive (28 or more drinks per week) and low-risk (14 or fewer drinks per week) drinking groups. However our results (and a literature-based comparison of GWAS findings for enzymes and for alcoholic cirrhosis) provide only limited support for shared genetic risk factors for abnormal liver enzyme results in excessive drinkers and for alcoholic cirrhosis, and mainly for ALT. Ideally, a prospective study which enrolled excessive drinkers without evidence of liver disease at baseline and used clinical liver disease as an endpoint is needed to assess the predictive value of GGT, AST or ALT in relation to development of alcoholic liver disease.
In relation to the use of enzyme results, particularly GGT, as markers of alcohol intake or of abstinence in alcoholics, the quantitative risk factors related to obesity and iron status can indicate which patients are more likely, or less likely, to have abnormal results while drinking. The fact that many of the influences on GGT are common to the excessive and low-risk drinking groups suggests that those who have low results while drinking will also have low results while abstinent, and reinforces the clinical value of within-person comparisons across time rather than a universal reference range.
This study is subject to a number of limitations. The number of people reporting excessive drinking is limited, which means that only substantial effects can be detected at the stringent p-values required for genome-wide genetic studies. The study is not prospective, so although we can say that certain characteristics are associated with abnormal enzyme results we cannot claim that these characteristics are predictive or causative. We do not have data on the enzyme results in the same subjects during both low-risk and excessive drinking, for within-person comparisons. The results refer to liver enzyme abnormality and not directly to effects on alcoholic liver disease. Abnormal results may be signalling the presence of fatty liver, an early phenomenon reversible with abstinence, rather than irreversible changes, although at least for GGT there is previous evidence of association with fibrosis and cirrhosis in alcoholics. Some of these limitations, particularly for power, will be overcome in the foreseeable future: information on the genetics of these liver enzymes (in the general population and in the subset of excessive drinkers) can be expected from UK Biobank data (http://biobank.ctsu.ox.ac.uk/~bbdatan/biomarkers.pdf, accessed 2018–08-22). Prospective or nested case-control data on predictive performance will become more accessible with the spread of electronic medical records, and linkage of laboratory results to outcomes.
Supplementary Material
ACKNOWLEDGEMENTS
We appreciate the contributions of all those who participated in these studies, often on several occasions. We also acknowledge the contributions of project staff at QIMR Berghofer Medical Research Institute, for administration, sample processing and data management; and at Royal Prince Alfred Hospital, Sydney, for biomarker measurements.
Sources of Support
Subject recruitment and interviews, and blood collection and processing, were supported by grants AA013321, AA013326, DA012854 and AA013320 from the US National Institutes of Health to ACH, NGM, PAFM, and the late Richard Todd, MD, PhD. Biomarker measurement was supported by AA014041 to JBW. GWM is supported by the National Health and Medical Research Council of Australia Fellowship Scheme.
Footnotes
Conflict of Interest
The Authors report no conflict of interest.
REFERENCES
- Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, Liu Y, Kozlitina J, Stender S, Wood GC, Stepanchick AN, Still MD, McCarthy S, O’Dushlaine C, Packer JS, Balasubramanian S, Gosalia N, Esopi D, Kim SY, Mukherjee S, Lopez AE, Fuller ED, Penn J, Chu X, Luo JZ, Mirshahi UL, Carey DJ, Still CD, Feldman MD, Small A, Damrauer SM, Rader DJ, Zambrowicz B, Olson W, Murphy AJ, Borecki IB, Shuldiner AR, Reid JG, Overton JD, Yancopoulos GD, Hobbs HH, Cohen JC, Gottesman O, Teslovich TM, Baras A, Mirshahi T, Gromada J, Dewey FE (2018) A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N Engl J Med 378:1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, Fulgoni VL 3rd, Lieberman HR (2016) Assessing alcohol intake & its dose-dependent effects on liver enzymes by 24-h recall and questionnaire using NHANES 2001–2010 data. Nutr J 15:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alatalo P, Koivisto H, Puukka K, Hietala J, Anttila P, Bloigu R, Niemela O (2009) Biomarkers of liver status in heavy drinkers, moderate drinkers and abstainers. Alcohol Alcohol 44:199–203. [DOI] [PubMed] [Google Scholar]
- Banciu T, Weidenfeld H, Marcoane E, Berinde L (1983) Serum gamma-glutamyltranspeptidase assay in the detection of alcohol consumers and in the early and stadial diagnosis of alcoholic liver disease. Med Interne 21:23–29. [PubMed] [Google Scholar]
- Becker U, Deis A, Sorensen TI, Gronbaek M, Borch-Johnsen K, Muller CF, Schnohr P, Jensen G (1996) Prediction of risk of liver disease by alcohol intake, sex, and age: a prospective population study. Hepatology 23:1025–1029. [DOI] [PubMed] [Google Scholar]
- Breitling LP, Claessen H, Drath C, Arndt V, Brenner H (2011) Gamma-glutamyltransferase, general and cause-specific mortality in 19,000 construction workers followed over 20 years. J Hepatol 55:594–601. [DOI] [PubMed] [Google Scholar]
- Buch S, Stickel F, Trepo E, Way M, Herrmann A, Nischalke HD, Brosch M, Rosendahl J, Berg T, Ridinger M, Rietschel M, McQuillin A, Frank J, Kiefer F, Schreiber S, Lieb W, Soyka M, Semmo N, Aigner E, Datz C, Schmelz R, Bruckner S, Zeissig S, Stephan AM, Wodarz N, Deviere J, Clumeck N, Sarrazin C, Lammert F, Gustot T, Deltenre P, Volzke H, Lerch MM, Mayerle J, Eyer F, Schafmayer C, Cichon S, Nothen MM, Nothnagel M, Ellinghaus D, Huse K, Franke A, Zopf S, Hellerbrand C, Moreno C, Franchimont D, Morgan MY, Hampe J (2015) A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet 47:1443–1448. [DOI] [PubMed] [Google Scholar]
- Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Holm H, Sanna S, Kavousi M, Baumeister SE, Coin LJ, Deng G, Gieger C, Heard-Costa NL, Hottenga JJ, Kuhnel B, Kumar V, Lagou V, Liang L, Luan J, Vidal PM, Mateo Leach I, O’Reilly PF, Peden JF, Rahmioglu N, Soininen P, Speliotes EK, Yuan X, Thorleifsson G, Alizadeh BZ, Atwood LD, Borecki IB, Brown MJ, Charoen P, Cucca F, Das D, de Geus EJ, Dixon AL, Doring A, Ehret G, Eyjolfsson GI, Farrall M, Forouhi NG, Friedrich N, Goessling W, Gudbjartsson DF, Harris TB, Hartikainen AL, Heath S, Hirschfield GM, Hofman A, Homuth G, Hypponen E, Janssen HL, Johnson T, Kangas AJ, Kema IP, Kuhn JP, Lai S, Lathrop M, Lerch MM, Li Y, Liang TJ, Lin JP, Loos RJ, Martin NG, Moffatt MF, Montgomery GW, Munroe PB, Musunuru K, Nakamura Y, O’Donnell CJ, Olafsson I, Penninx BW, Pouta A, Prins BP, Prokopenko I, Puls R, Ruokonen A, Savolainen MJ, Schlessinger D, Schouten JN, Seedorf U, Sen-Chowdhry S, Siminovitch KA, Smit JH, Spector TD, Tan W, Teslovich TM, Tukiainen T, Uitterlinden AG, Van der Klauw MM, Vasan RS, Wallace C, Wallaschofski H, Wichmann HE, Willemsen G, Wurtz P, Xu C, Yerges-Armstrong LM, Abecasis GR, Ahmadi KR, Boomsma DI, Caulfield M, Cookson WO, van Duijn CM, Froguel P, Matsuda K, McCarthy MI, Meisinger C, Mooser V, Pietilainen KH, Schumann G, Snieder H, Sternberg MJ, Stolk RP, Thomas HC, Thorsteinsdottir U, Uda M, Waeber G, Wareham NJ, Waterworth DM, Watkins H, Whitfield JB, Witteman JC, Wolffenbuttel BH, Fox CS, Ala-Korpela M, Stefansson K, Vollenweider P, Volzke H, Schadt EE, Scott J, Jarvelin MR, Elliott P, Kooner JS (2011) Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 43:1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conigrave KM, Davies P, Haber P, Whitfield JB (2003) Traditional markers of excessive alcohol use. Addiction 98 Suppl 2:31–43. [DOI] [PubMed] [Google Scholar]
- Corrao G, Bagnardi V, Zambon A, La Vecchia C (2004) A meta-analysis of alcohol consumption and the risk of 15 diseases. Prev Med 38:613–619. [DOI] [PubMed] [Google Scholar]
- Frezza M, Pozzato G, Chiesa L, Terpin M, Barbone F, Di Padova C (1989) Abnormal serum gamma-glutamyltranspeptidase in alcoholics. Clues to its explanation. Neth.J.Med 34:22–28. [PubMed] [Google Scholar]
- Ganne-Carrie N, Christidis C, Chastang C, Ziol M, Chapel F, Imbert-Bismut F, Trinchet JC, Guettier C, Beaugrand M (2000) Liver iron is predictive of death in alcoholic cirrhosis: a multivariate study of 229 consecutive patients with alcoholic and/or hepatitis C virus cirrhosis: a prospective follow up study. Gut 46:277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Tsukamoto H (2016) Inflammation in Alcoholic and Nonalcoholic Fatty Liver Disease: Friend or Foe? Gastroenterology 150:1704–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramenzi A, Caputo F, Biselli M, Kuria F, Loggi E, Andreone P, Bernardi M (2006) Review article: alcoholic liver disease--pathophysiological aspects and risk factors. Aliment Pharmacol Ther 24:1151–1161. [DOI] [PubMed] [Google Scholar]
- Grant BF, Dufour MC, Harford TC (1988) Epidemiology of alcoholic liver disease. Semin Liver Dis 8:12–25. [DOI] [PubMed] [Google Scholar]
- Grove J, Brown AS, Daly AK, Bassendine MF, James OF, Day CP (1998) The RsaI polymorphism of CYP2E1 and susceptibility to alcoholic liver disease in Caucasians: effect on age of presentation and dependence on alcohol dehydrogenase genotype. Pharmacogenetics 8:335–342. [DOI] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW (2011) A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatry 70:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriaga H, Bunout D, Hirsch S, Ugarte G (1988) Overweight as a risk factor or a predictive sign of histological liver damage in alcoholics. Am.J.Clin Nutr 47:235–238. [DOI] [PubMed] [Google Scholar]
- Lelbach WK (1976) Epidemiology of alcoholic liver disease. Prog.Liver Dis 5:494–515. [PubMed] [Google Scholar]
- Liangpunsakul S, Qi R, Crabb DW, Witzmann F (2010) Relationship Between Alcohol Drinking and Aspartate Aminotransferase:Alanine Aminotransferase (AST:ALT) Ratio, Mean Corpuscular Volume (MCV), Gamma-Glutamyl Transpeptidase (GGT), and Apolipoprotein A1 and B in the U.S. Population. J Stud Alcohol Drugs 71:249–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar SK, Dias N, Aps EJ (1991) Relationship between hepatic histology and conventional biochemical liver function test in chronic alcoholic patients. Drug Alcohol Depend 28:211–214. [DOI] [PubMed] [Google Scholar]
- Mann RE, Smart RG, Govoni R (2003) The epidemiology of alcoholic liver disease. Alcohol Res Health 27:209–219. [PMC free article] [PubMed] [Google Scholar]
- McDonald H, Borinskya S, Kiryanov N, Gil A, Helander A, Leon DA (2013) Comparative performance of biomarkers of alcohol consumption in a population sample of working-aged men in Russia: the Izhevsk Family Study. Addiction 108:1579–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveau S, Giraud V, Borotto E, Aubert A, Capron F, Chaput JC (1997) Excess weight risk factor for alcoholic liver disease. Hepatology 25:108–111. [DOI] [PubMed] [Google Scholar]
- Niemela O, Niemela M, Bloigu R, Aalto M, Laatikainen T (2017) Where should the safe limits of alcohol consumption stand in light of liver enzyme abnormalities in alcohol consumers? PLoS One 12:e0188574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pequignot G, Tuyns AJ, Berta JL (1978) Ascitic cirrhosis in relation to alcohol consumption. Int.J.Epidemiol 7:113–120. [DOI] [PubMed] [Google Scholar]
- Qu BG, Bi W, Jia YG, Liu YX, Wang H, Su JL, Liu LL, Wang ZD, Wang YF, Han XH, Pan JD, Ren GY, Hu WJ (2016) Association between circulating inflammatory molecules and alcoholic liver disease in men. Cell Stress Chaperones 21:865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosalki SB, Rau D, Lehmann D, Prentice M (1970) Gamma-glutamyl transpeptidase in chronic alcoholism. Lancet 2:1139. [DOI] [PubMed] [Google Scholar]
- Ruhl CE, Everhart JE (2009) Elevated serum alanine aminotransferase and gamma-glutamyltransferase and mortality in the United States population. Gastroenterology 136:477–485 e411. [DOI] [PubMed] [Google Scholar]
- Sillanaukee P, Massot N, Jousilahti P, Vartiainen E, Sundvall J, Olsson U, Poikolainen K, Ponnio M, Allen JP, Alho H (2000) Dose response of laboratory markers to alcohol consumption in a general population. Am.J.Epidemiol 152:747–751. [DOI] [PubMed] [Google Scholar]
- Steffensen FH, Sorensen HT, Brock A, Vilstrup H, Lauritzen T (1997) Alcohol consumption and serum liver-derived enzymes in a Danish population aged 30–50 years. Int.J.Epidemiol 26:92–99. [DOI] [PubMed] [Google Scholar]
- Tian C, Stokowski RP, Kershenobich D, Ballinger DG, Hinds DA (2010) Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet 42:21–23. [DOI] [PubMed] [Google Scholar]
- Tuyns AJ, Pequignot G (1984) Greater risk of ascitic cirrhosis in females in relation to alcohol consumption. Int.J.Epidemiol 13:53–57. [DOI] [PubMed] [Google Scholar]
- Wang HJ, Gao B, Zakhari S, Nagy LE (2012) Inflammation in alcoholic liver disease. Annu Rev Nutr 32:343–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead TP, Robinson D, Allaway SL (1996) The effects of cigarette smoking and alcohol consumption on serum liver enzyme activities: a dose-related study in men. Ann.Clin.Biochem 33 ( Pt 6):530–535. [DOI] [PubMed] [Google Scholar]
- Whitfield JB, Heath AC, Madden PA, Pergadia ML, Montgomery GW, Martin NG (2013) Metabolic and Biochemical Effects of Low-to-Moderate Alcohol Consumption. Alcohol Clin Exp Res 37:575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB, Heath AC, Madden PAF, Landers JG, Martin NG (2018) Effects of high alcohol intake, alcohol-related symptoms and smoking on mortality. Addiction 113:158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB, Hensley WJ, Bryden D, Gallagher H (1978) Some laboratory correlates of drinking habits. Ann.Clin.Biochem 15:297–303. [DOI] [PubMed] [Google Scholar]
- Whitfield JB, Rahman K, Haber PS, Day CP, Masson S, Daly AK, Cordell HJ, Mueller S, Seitz HK, Liangpunsakul S, Westerhold C, Liang T, Lumeng L, Foroud T, Nalpas B, Mathurin P, Stickel F, Soyka M, Botwin GJ, Morgan TR, Seth D, Genom ALCC (2015) Brief report: genetics of alcoholic cirrhosis-GenomALC multinational study. Alcohol Clin Exp Res 39:836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB, Zhu G, Madden PA, Neale MC, Heath AC, Martin NG (2004) The genetics of alcohol intake and of alcohol dependence. Alcohol Clin.Exp.Res 28:1153–1160. [DOI] [PubMed] [Google Scholar]
- Whitfield JB, Zhu G, Nestler JE, Heath AC, Martin NG (2002) Genetic covariation between serum gamma-glutamyltransferase activity and cardiovascular risk factors. Clin Chem. 48:1426–1431. [PubMed] [Google Scholar]
- Wu A, Slavin G, Levi AJ (1976) Elevated serum gamma-glutamyl-transferase (transpeptidase) and histological liver damage in alcoholism. Am.J.Gastroenterol 65:318–323. [PubMed] [Google Scholar]
- Xu R, Tao A, Zhang S, Deng Y, Chen G (2015) Association between patatin-like phospholipase domain containing 3 gene (PNPLA3) polymorphisms and nonalcoholic fatty liver disease: a HuGE review and meta-analysis. Scientific reports 5:9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M (2012) Genome-wide efficient mixed-model analysis for association studies. Nat Genet 44:821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.