Supplemental digital content is available in the text.
Abstract
Background
For the growing numbers of older transplant patients, increased incidence of infection and death compared with younger patients may limit the many benefits provided by transplantation. However, little is known about age-associated immune dysfunction in the older transplant recipient.
Methods
A cohort of 60 kidney transplant recipients, 23 older (≥ 60y) and 37 younger (30-59y), matched on antithymocyte induction and donor type (living vs deceased) was evaluated. Gene expression in peripheral blood mononuclear cells 3 months after kidney transplantation was analyzed to compare differences between older and younger patients.
Results
Proinflammatory genes were upregulated in older kidney transplant patients, including cytokines IL1-β and IL-6. Downregulated genes were associated with B-cell and T-cell function, including CCR7 and CD27. Analysis of predicted transcription factor binding suggested an increase in proinflammatory transcription factor CCAAT/enhancer binding protein β-binding sites in older patients, whereas interferon regulatory factor 2 transcription factor binding sites were less prevalent.
Conclusions
Older kidney transplant recipients exhibited multiple differences in gene expression compared with younger patients, with upregulation of proinflammatory genes and downregulation of adaptive immune response genes. These findings may explain the mechanism of increased vulnerability to infection and malignancy observed in older transplant patients.
Although the number of older transplant recipients continues to grow, little is known about immune dysfunction associated with aging in patients receiving immunosuppression. Under the current standard of care management, many older patients may be overimmunosuppressed.1,2 Reaching a better understanding of the mechanisms of immune dysfunction, including changes in transcriptional regulation between older and younger transplant recipients, can shed light on the increased vulnerability of older patients to infection. Ultimately, developing the ability to noninvasively measure immune function can lead to the ability to individualize immunosuppression regimens and risk stratify patients at increased risk for adverse clinical outcomes. This approach has been applied successfully for the prediction of allograft rejection in heart transplant recipients.3,4
Older transplant recipients experience increased rates of infection and malignancy, but less rejection after transplantation.1,2,5 Increased incidence of infection includes bacterial and fungal as well as viral infections.6,7 This observation suggests that the immune dysfunction described in older adults also plays an important role in posttransplant outcomes and that improving our understanding of the interaction between age-associated immune dysfunction and administration of immunosuppression could improve our ability to individualize immunosuppression regimens for older transplant recipients.8 The types of immune dysfunction described in older adults include terminal differentiation, immune senescence, and exhaustion.9-12 At the same time, this immune dysfunction is associated with increased inflammation and “inflammaging,” which has been implicated as the cause behind many age-associated illnesses.13-16 This question is important to study, given the increasing numbers of older patients with chronic kidney disease and other end-stage organ diseases, increasing the numbers of candidates for solid organ transplantation. The number of older kidney transplant recipients older than 65 years increased more than 4-fold between 1988 and 2012, with 3315 patients older than 65 years undergoing kidney transplantation in 2014.1,17,18
Our previous work has identified immune phenotypes associated with older kidney transplant recipients, including decrease in the frequency of naive T cells, increased terminal differentiation and immunosenescence, and changes in the frequency of monocyte subtypes. Other researchers have demonstrated a decreased incidence of rejection in kidney transplant recipients with an increased frequency of senescent T cells.19,20 These observations, however, do not address the etiology of changes in immune phenotype in older transplant recipients. Measuring changes in gene expression offers the ability to determine the mechanism behind the immune dysfunction in older transplant recipients. Evaluation of changes in gene expression has revealed transcriptional changes involved in aging.21,22
Although other researchers have evaluated transcriptional changes between older and younger community-dwelling adults, it remains unknown as to what impact the administration of immunosuppression would have on age-associated patterns of transcriptional regulation. Administration of triple immunosuppression, including calcineurin inhibitor, mycophenolate mofetil, and prednisone, might be predicted to exert a “leveling” effect on patterns of gene transcription, leading to similar patterns of immune gene regulation regardless of patient age. Therefore, it is important to evaluate whether transcriptional changes of aging persist during maintenance of immunosuppression. We present here an evaluation of changes in gene transcription between a cohort of older and younger transplant recipients receiving immunosuppression to examine whether age-associated differences in gene expression can be detected in older patients compared with younger patients on identical immunosuppression regimens.
MATERIALS AND METHODS
Clinical Care
We enrolled kidney transplant recipients after transplantation at Ronald Reagan Medical Center. The University of California, Los Angeles (UCLA) Institutional Review Board approved this observational study. All patients signed an informed consent. As described previously, inclusion criteria were any adult kidney transplant recipient willing and able to provide informed consent, and exclusion criteria were the presence of active infection or rejection at the time of the 3-month blood collection.23 Blood was collected for peripheral blood mononuclear cell (PBMC) isolation at 3 months after transplantation during outpatient clinic visits, as previously described. We identified 23 older patients (older than 60 y) who had PBMC available for analysis. We additionally identified an additional 37 kidney transplanted recipients between the ages of 30 and 51 years for whom PBMC were available, transplanted during the same era as the older patients; these younger control patients were selected to ensure a similar proportion of deceased versus living donor and antithymocyte globulin (ATG) versus basiliximab induction therapy to create a total cohort of 60 older and younger patients. Patients were clinically stable at the time of PBMC collection, without evidence of infection or rejection. See Liang et al23 for full details regarding patient care, including immunosuppression and antibiotic prophylaxis. Patients received similar maintenance immunosuppression regimens with protocolized target drug levels and monitoring for infection as previously described. Donor information was obtained by review of the UCLA electronic medical record to obtain donor age, sex, cold ischemia time, and Kidney Donor Profile Index (KDPI). Donor age was available for all patients, whereas donor sex was missing for 13 patients and cold ischemia time was missing for 6 patients. KDPI was available for 26 of 27 deceased donors.
Transcriptome Analysis
PBMCs were isolated and frozen for storage using standard techniques,24 followed by isolation of total ribonucleic acid (RNA) (RNeasy, Qiagen) in the UCLA Biological Samples Processing Core. Quality of the purified RNA was verified on an Agilent 2100 Bioanalyzer; RNA Integrity Number ranged from 7.8 to 9.7, with a median of 8.9. RNA concentrations were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA was converted to fluorescent cRNA and hybridized to Illumina Human HT-12 v4 BeadArrays following the manufacturer's standard protocol. Genomewide transcriptional profiling was performed on isolated PBMC from all 60 subjects in 1 batch. Standard quality assurance metrics were applied to ensure validity of results. Assays were performed by microarray for transcriptome analysis.25 Assays were performed as previously described at the UCLA Neuroscience Genomics Core.26
Statistical Analysis
Demographic differences between older patients and younger patients were analyzed by Fisher's exact test using JMP Pro 11 (SAS Software).
Gene expression values were quantile-normalized and log2-transformed. Thirty-four thousand six hundred and seventy-four gene transcripts were analyzed. Log2-transformed values were subjected to general linear model analyses relating the expression of each assayed gene transcripts to patient age while also controlling for the following variables: sex, transplant type (deceased vs living donor), and induction type (basiliximab vs ATG). Because the sample size was limited, we did not attempt to perform any genomewide discovery analysis to identify statistically significant associations between specific individual gene transcripts and age (this sample is underpowered for that type of analysis). Instead, we used point estimates of association magnitude for each gene as input into higher-order bioinformatics analyses testing for age-related variations in the activity of predefined sets of genes involved in a narrow range of specifically hypothesized biological processes (eg, inflammation, interferon-related antiviral responses, activity of specific transcription factors, etc.), as described previously.27 These gene set-based analyses have substantially greater statistical power than analyses testing for individual transcript associations and were therefore judged to be more feasible and appropriate for the limited sample size available in this study. We list specific genes with point estimates of association greater than a predefined cutoff, but these lists should be treated as descriptive only and not interpreted as statistically significant at an individual gene level.
Gene transcripts showing >1.25-fold difference in average expression in older versus younger transplant patient groups served as input into higher-order bioinformatics analyses (fold-change threshold was established a priori to provide a suitable number of input genes for well-powered gene set analyses). Primary analyses examined specific transcription control pathways that may have contributed to the observed differences in gene expression. In these analyses, the list of genes, which was upregulated (or downregulated) in older compared with younger patients, served as an input into higher-order bioinformatics analyses involving the Transcription Element Listening System promoter-based bioinformatic analysis.28 This analysis assessed the presence of NF-κB, AP-1, interferon regulatory factor (IRF), and CREB and GR family transcription factors linked to transcriptional regulation of conserved transcriptional response to adversity (CTRA) genes, as previously described.26
Transcript origin analysis was also applied to these gene lists to identify the leukocyte subtypes mediating the observed differences in gene expression, as previously described.29 As described, every single gene in the genome has a set of numerical scores indicating how predominately it is expressed in each cell type relative to the other cell types examined. These scores are derived from a separate reference study in which the different cell types were physically separated and subject to individual transcriptome profiling (Gene Expression Omnibus GSE1133), with score computation as described (essentially expressing the average expression of each gene in each cell type in terms of its standard deviation difference from the average expression level in all other PBMC cell types).29 These scores are averaged for all genes identified as differentially expressed to provide a cell type specificity score for each cell type analyzed, and that average score is tested for statistically significant difference from the mean cell type specificity score computed across all genes assayed by the microarray (the population null hypothesis value) using standard errors derived from bootstrap resampling.
Additional analyses were tested whether age was associated with differential expression of an a priori–defined set of 53 genes involved in the CTRA, including hypothesized upregulated expression of 19 proinflammatory genes (IL1A, IL1B, IL6, IL8, TNF, PTGS1, PTGS2, FOS, FOSB, FOSL1, FOSL2, JUN, JUNB, JUND, NFKB1, NFKB2, REL, RELA, and RELB, each weighted +1 to reflect their positive contribution to the CTRA profile); downregulated expression of 31 genes involved in type I IFN responses (GBP1, IFI16, IFI27, IFI27L1-2, IFI30, IFI35, IFI44, IFI44L, IFI6, IFIH1, IFIT1-3, IFIT5, IFIT1L, IFITM1-3, IFITM4P, IFITM5, IFNB1, IRF2, IRF7-8, MX1-2, OAS1-3, and OASL, weighted −1 to reflect their inverse contribution to the CTRA profile); and 3 genes involved in antibody synthesis (IGJ, IGLL1, and IGLL3, weighted −1).26 Contrast coefficient-weighted association statistics were averaged to summarize the magnitude of association over the entire CTRA gene set (as well as the inflammatory, antiviral, and antibody-related gene subsets), and standard errors were derived from 200 cycles of bootstrap resampled residual vectors (to account for potential correlation among residuals across genes).
To confirm that results were not confounded by differences between leukocyte subtypes within the total PBMC pool, ancillary analyses also adjusted for leukocyte subset frequency by controlling for the prevalence of transcripts marking T lymphocytes (CD3D, CD3E, CD4, CD8A), B lymphocytes (CD19), natural killer cells (CD16/FCGR3A, CD56/NCAM1), and monocytes (CD14).30 Repeating the reported analyses with adjustment for leukocyte frequency did not affect the results.
RESULTS
Demographic Characteristics of Patients
Twenty-two patients older than 60 years who had undergone kidney transplantation (age, 60-80 y; median age, 67 y) and 38 younger patients (age, 34-51 y; median age, 43 y) were evaluated, as previously reported (Table 1).23 Older patients had similar frequencies of deceased donors compared with the cohort of younger patients (44% compared with 46%, P = 1.000), and there were similar frequencies of induction with ATG in the older compared with younger patients (both 30%). There were, however, differences in underlying cause of renal disease, with increased incidence of diabetes and decreased incidence of polycystic kidney disease in older patients (Table 1). Frequencies of demographic characteristics were not significantly different in older compared with younger patients, with 74% of older recipients of male sex compared with 60% of younger patients (P = 0.282); 65% of older patients were white compared with 68% of younger patients (P = 1.000). Donor characteristics differed slightly by recipient age, with older patients generally receiving transplants from older donors, although the difference was not very large (median donor age, 42 y for older compared with 36 y for younger recipients; P = 0.034). Donor sex and median cold ischemia time were not significantly different by recipient age group, although there was a trend toward higher KDPI in older patients (Table 1). The CMV antibody positivity of recipients was also similar between older patients and younger patients (78% compared with 70%; P = 0.561). No patient was experiencing an acute episode of infection or rejection at the time of blood sample collection. Although there was a trend toward increased rates of infection and decreased rates of rejection in the older patient group compared with the younger patient group, these differences did not reach statistical significance (Table 2).
TABLE 1.
Demographic and clinical characteristics and of older compared with younger kidney transplant recipients analyzed
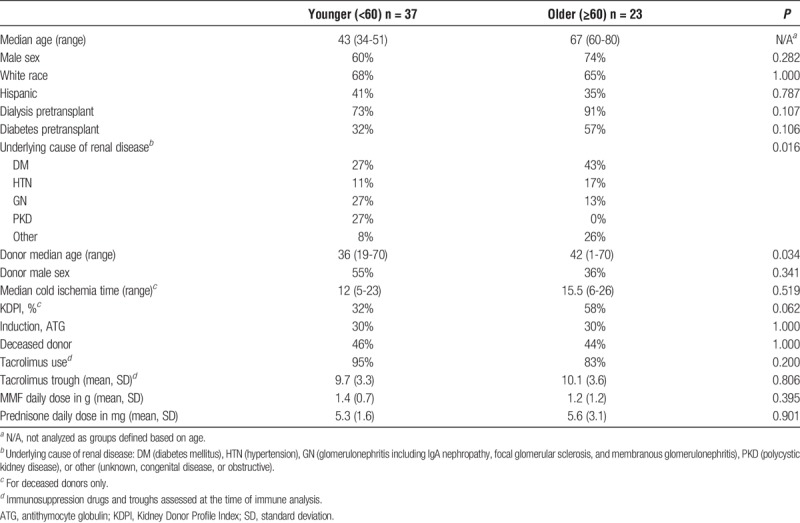
TABLE 2.
Clinical outcomes of older and younger kidney transplant recipients during the first year after transplant

Differences in Gene Transcript Expression in Older Compared With Younger Patients
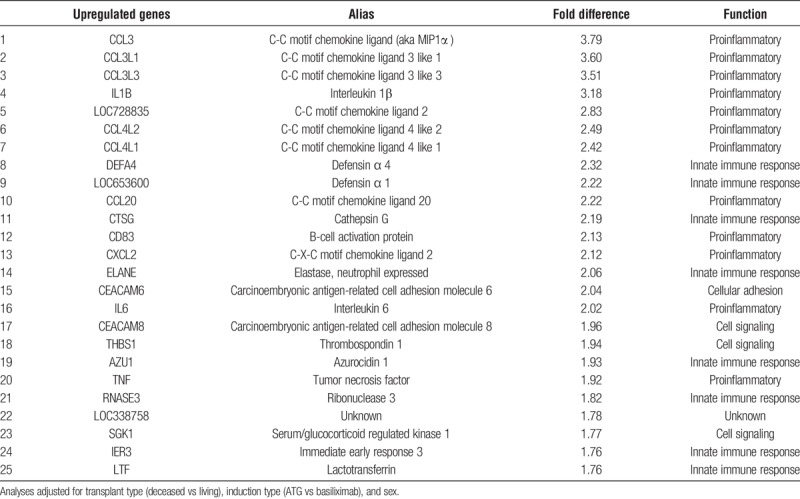
Genomewide transcriptional profiling revealed multiple differences in PBMC gene expression in older compared with younger kidney transplant recipients. Review of the top 25 upregulated genes revealed important proinflammatory genes or important in the innate immune response (Table 3). These included chemokine ligands, such as CCL3, CCL4, and CCL20. In addition, defensins, cathepsin G, elastase, and azurocidin, important in the innate immune response, and CD83, important for B cell activation, were upregulated. Proinflammatory cytokines, such as IL1-β, IL-6, and TNF, were also identified. Additional genes up to 1.5× upregulated are shown in Supplementary Material (Table S1, http://links.lww.com/TXD/A188).
TABLE 3.
Top 25 upregulated genes in older compared with younger kidney transplant recipients
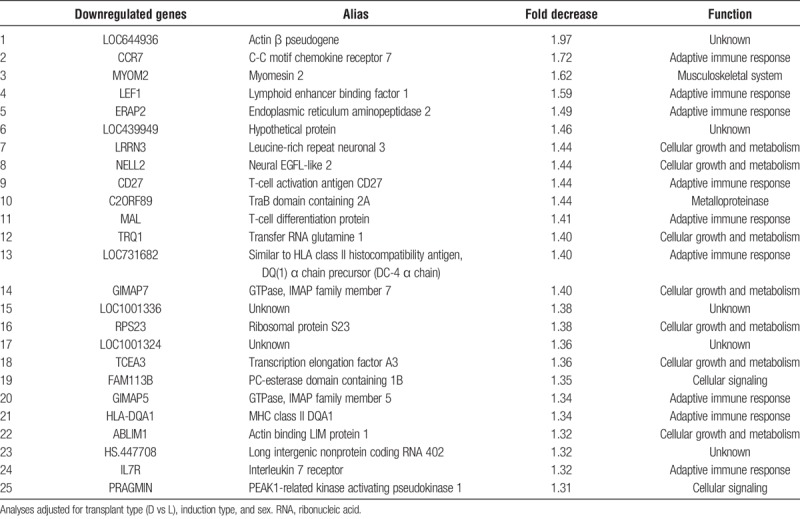
Review of the top 25 downregulated genes revealed many transcripts important in B-cell and T-cell function (Table 4). These included chemokine receptor CCR7, the surface cell-signaling molecule CD27 important in T-cell activation, lymphoid enhancer binding factor 1, HLA class I peptide generation (ERAP2), and T-cell differentiation protein (MAL) important in the adaptive immune response. Other downregulated genes were involved in cellular growth and metabolism, such as the Leucine Rich Repeat Neuronal 3 (LRRN3), important for brain development, protein C kinase binding protein Neural EGFL Like 2 (NELL2), and transfer RNA glutamine 1 (TRQ1). Additional genes up to 1.5× downregulated are shown in Supplementary Material (Table S2, http://links.lww.com/TXD/A189).
TABLE 4.
Top 25 downregulated genes in older compared with younger kidney transplant recipients
Expression-based Monitoring of Transcription Factor Activity
Application of a “Transcription Element Listening System” analysis to upregulated and downregulated transcripts revealed enrichment of several transcription factors known to be important in promotion or suppression of inflammation. In results consistent with the gene expression findings, we found an increased prevalence of transcription factor binding sites for CCAAT/enhancer binding protein β, an important transcription factor regulating proinflammatory responses, with a 3.38 ± 0.27 fold elevation of transcription factor binding sites in genes upregulated in older patients (P < 0.001). In contrast, analyses indicated age-related decreases in the activity of IRF2 transcription factor binding (0.33 ± 0.43 fold, P < 0.001). Similarly, there was a predicted decrease in AP1 activator protein, which is important for cellular proliferation and differentiation (0.37 ± 0.39 fold, P = 0.003) and early growth response (EGR)2 and EGR3, which are important in T-cell anergy (0.33 ± 0.58 fold, P = 0.009, and 0.25 ± 0.59 fold, P = 0.003, respectively). In addition, 2 transcription factors important for B-cell function were also found to be downregulated, namely IK2, IKAROS B signaling (0.74 ± 0.16 fold, P = 0.39), and interferon-stimulatory element (0.31 ± 0.80 fold, P = 0.045), which may account for some of the downregulation of genes important in B-cell function as described above.
Evaluation of Cellular Origin of Transcripts
Analysis of transcript abundance differences associated with specific cell types can reveal which cell type is the primary source of age-related differences in the overall PBMC transcriptome profile. Transcript origin result analysis identified monocytes and B cells the likely source of the upregulated genes observed in older compared with younger patients (Figure 1). Analysis of downregulated genes identified CD8+ and CD4+ T cells, as well as B cells as primary cellular origins (Figure 1). The fact that B cells are identified as being the source for both upregulation and downregulation suggests that different B-cell subsets that are responsible for the changes are observed.
FIGURE 1.
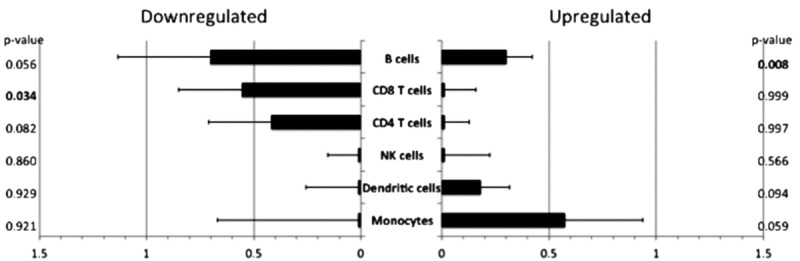
Transcript origin analysis for genes differentially expressed in older compared to younger patients. See Materials and Methods section for details regarding the cell-type identification. Bar graph indicates mean diagnosticity score for each cell type plus standard error. P values shown on right hand of figure. P values <0.05 are bolded for emphasis. A, Transcript origin analysis for genes upregulated in older compared to younger patients. B, Transcript origin analysis for genes downregulated in older patients compared with younger patients.
We further analyzed cell type abundance to determine whether these differences might be responsible for the changes observed in terms of changes in gene transcript abundance (Table S3, http://links.lww.com/TXD/A189). This analysis revealed no statistically significant differences in cell type abundance for CD8+ T cells, CD4+ T cells, NK cells, dendritic cells, or monocytes, indicating that differences in the numbers of these cell types were not responsible for the differences in gene transcript upregulation or downregulation reported above. The 1 exception to this observation was for B cells, which were estimated to be reduced by 3.7% in older compared with younger patients (P = 0.025). However, this relatively small percentage change is unlikely to fully explain the many changes in gene transcript abundance observed (Tables 3 and 4).
Analysis of Gene Expression in Relation to Physiological Stress
Analysis of gene expression was performed in relation to a priori–defined transcripts identified in the CTRA, which includes increased expression of genes involved in inflammation (such as proinflammatory cytokines) and decreased expression of genes involved in type I IFN antiviral responses, and IgG antibody synthesis was performed. This analysis revealed a statistically significant 1.13 ± 0.06 fold elevation of CTRA gene expression in older patients (P = 0.036). Breakdown of the 3 components of the CTRA signal revealed a statistically significant elevation of proinflammatory transcripts in older patients (1.28 ± 0.11 fold, P = 0.010) (Figure 2). In contrast, there was a trend toward downregulation of antiviral and antibody-related genes in older patients (Figure 2). This analysis confirms the analysis of transcript abundance shown in Tables 1 and 2.
FIGURE 2.
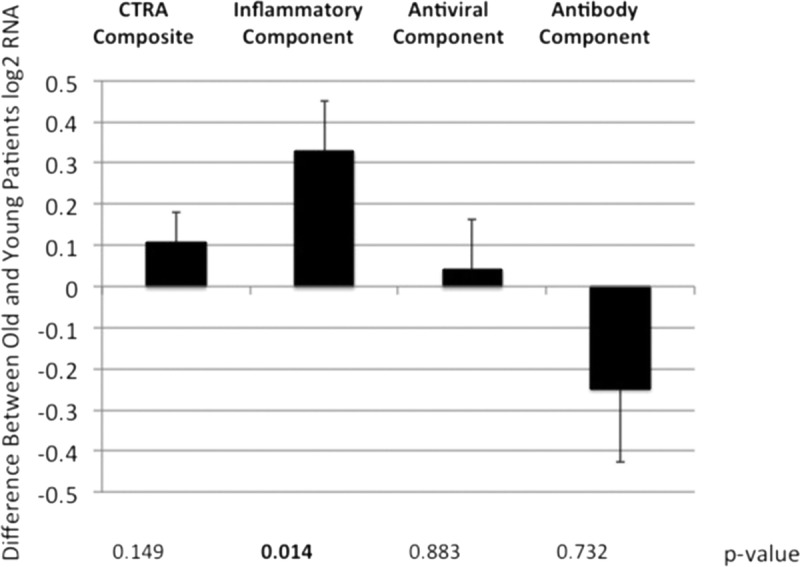
Impact of patient age on CTRA gene expression. See Materials and Methods section for the list of genes involved in the calculation of each component. Bar graph indicates mean fold difference for either composite CTRA gene expression or its subsets: Inflammatory-, antiviral-, or antibody-related gene expression, as indicated, plus standard error. P values shown below figure. P values <0.05 are bolded for emphasis. CTRA, conserved transcriptional response to adversity; RNA, ribonucleic acid.
DISCUSSION
We analyzed differences in gene expression in kidney transplant recipients to compare the impact of age in older patients compared with younger patients. This represents, to our knowledge, the first analysis of the impact of patient age on gene expression in the setting of chronic immunosuppression. Surprisingly, despite receipt of comparable immunosuppression regimens, a significant impact of patient age was detected: older transplant recipients demonstrated increased abundance of proinflammatory transcripts, including cytokines and chemokines. Interestingly, components important in the innate immune response were also upregulated (Table 3). This suggests the possibility that increased levels of proinflammatory cytokines may be stimulating monocytes and other innate immune cells known to be associated with the aging immune system.31 Conversely, genes important in the antiviral immune response were observed to be downregulated (Table 4). This important observation may explain some of the increased vulnerability noted in older transplant recipients to infection. In addition, although older transplant patients experience overall lower rates of rejection, when rejection occurs they are less likely to respond, which may be related to the increased proinflammatory gene expression observed.
To try to determine the mechanism behind differential gene regulation in older compared with younger patients, we evaluated abundance of transcription factor binding sites upstream for the differentially regulated genes. This revealed increased enrichment of the CCAAT/enhancer binding protein β transcription factor binding site, an important proinflammatory factor. Decreased enrichment of binding sites for transcription factors important for interferon response and T-cell function was also observed. These analyses are consistent with and underscore the findings of differential transcriptional regulation seen in older patients, with an increased proinflammatory footprint but a decreased antiviral immune response. This analysis points to the biological pathways most affected by patient age.
Interestingly, the cellular source of the upregulated gene transcripts in older patients was found to be monocytes and B cells. The cellular source of downregulated gene transcripts was found to be CD4+ and CD8+ T cells. B cells were also the source of downregulated gene transcripts, suggesting that different B-cell subtypes are responsible for either differential upregulation or downregulation of gene expression in older transplant patients. That said, although we were able to identify cell types that served as a source for changes in gene transcription, further analysis suggested that changes in cell type abundance were not the cause of the differences observed.
Comparing our findings with previously published analyses of CTRA gene profiles similarly revealed upregulation of proinflammatory and a trend toward downregulation of antiviral- and antibody-related genes (Figure 2). Chemokine expression has been associated with inflammation and “inflammaging” thought to be contributory to disease associated with aging.16 Comparing our analyses in transplant recipients on immunosuppression with previously published results in older patients demonstrates a similar finding of decreased antiviral immune response genes and increased frequency of proinflammatory genes.
These findings have important implications for clinical transplantation regarding vulnerability to infection and specifically viral infections, as well as development of alloreactivity. In addition, the persistence of significant age-associated differences despite receipt of identical immunosuppression regimens suggests that adjustment of immunosuppression intensity based on patient “biological age” might reduce overimmunosuppression in many older patients, reducing adverse clinical outcomes, including viral infection.8,32 Future studies will evaluate changes in pretransplant compared with posttransplant cohorts of older patients compared with younger patients to determine if there is a differential impact of transplantation and initiation of immunosuppression in patients of different ages and with different levels of gene expression at baseline.
Limitations to this study include its cross-sectional approach, with evaluation of a single posttransplant time point. We hope in future studies to perform longitudinal analysis, including pretransplant evaluation, to determine the impact of transplantation and start of immunosuppression on older compared with younger patients, as well as whether the differences observed become stronger over time with ongoing immunosuppression receipt. The variation in transplant type of induction also raises difficulties for analysis. However, we have corrected for these differences in our statistical approach, and the protocolized patient care management in terms of immunosuppression, patient monitoring, and antibiotic prophylaxis meliorates this issue somewhat. Differences in donor characteristics were similar between older and younger recipients and therefore unlikely to have had a significant impact in the differences observed. In addition, although the sample size analyzed is similar to other translational studies in transplant recipients, the inclusion of 60 patients raises questions as to whether the findings are representative of all transplant populations. This limitation should be addressed in larger, multicenter cohort studies to validate the findings presented. Finally, this study was not powered for genomewide discovery analyses of statistically significant associations between age and specific individual gene transcripts. Specific differentially expressed genes are noted solely for descriptive purposes and the list of genes showing point estimates of differential expression served only as inputs into higher-order bioinformatics that maintain their own statistical control over false-positive errors at the level of aggregate gene sets. Individual genes should not be interpreted as statistically significant correlates of age until replicated in larger studies specifically powered for discovery of individual transcript associations.
In addition to repeating analysis in an independent cohort of younger and older patients to validate the findings presented in an independent patient cohort, future studies will evaluate transcriptional changes for correlation with the clinical outcomes of rejection, infection, and death. Future studies will also be performed to analyze gene expression on the CD8+ T-cell subset identified as most strongly associated with patient age and adverse clinical outcomes in our previous studies.20 We also propose to assess how changes in gene expression impact immunosenescent and proinflammatory phenotypes. Our goal is to identify genes associated with increased patient age and age-associated immune dysfunction in transplant patients and use this approach to risk stratify and predict infection, malignancy, and death after transplantation. By improving our understanding of gene expression differences in immune cells, we will reach a better understanding of the mechanism behind vulnerability to infection and death in the growing numbers of older transplant recipients.
Supplementary Material
Footnotes
Published online 4 March, 2019.
The authors declare no conflicts of interest.
This work was supported by the National Institutes of Health [R03AG050946] (J.S.), [U01AI124319] (E.F.R.), [U19AI128913] (E.F.R.), the IDSA ERF and NFID Young Investigator Award (J.S.), the Older Americans Independence Center Career Development Award (J.S.), and the NIH National Center for Advancing Translational Science (NCATS) [UL1TR001881] (J.S.).
J.S. designed the study, participated in data analysis and interpretation, and wrote the article. M.R. and E.R. participated in research design and data interpretation. B.A., E.L., S.B., T.P.P., and G.D. contributed to performance of the research and research design. S.C. participated in research design and interpretation, performed data analysis, and wrote the article.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantationdirect.com).
REFERENCES
- 1.Knoll GA. Kidney transplantation in the older adult. Am J Kidney Dis. 2013;61:790–797. [DOI] [PubMed] [Google Scholar]
- 2.Krenzien F, Elkhal A, Quante M, et al. A rationale for age-adapted immunosuppression in organ transplantation. Transplantation. 2015;99:2258–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng MC, Eisen HJ, Mehra MR, et al. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6:150–160. [DOI] [PubMed] [Google Scholar]
- 4.Deng MC. The AlloMap™ genomic biomarker story: 10 years after. Clin Transpl. 2017;31:e12900. [DOI] [PubMed] [Google Scholar]
- 5.Lehner LJ, Staeck O, Halleck F, et al. Need for optimized immunosuppression in elderly kidney transplant recipients. Transplant Rev (Orlando). 2015;29:237–239. [DOI] [PubMed] [Google Scholar]
- 6.Meier-Kriesche HU, Ojo AO, Hanson JA, et al. Exponentially increased risk of infectious death in older renal transplant recipients. Kidney Int. 2001;59:1539–1543. [DOI] [PubMed] [Google Scholar]
- 7.Trouillhet I, Benito N, Cervera C, et al. Influence of age in renal transplant infections: cases and controls study. Transplantation. 2005;80:989–992. [DOI] [PubMed] [Google Scholar]
- 8.Klinger M, Banasik M. Immunological characteristics of the elderly allograft recipient. Transplant Rev (Orlando). 2015;29:219–223. [DOI] [PubMed] [Google Scholar]
- 9.Weng NP, Akbar AN, Goronzy J. CD28(−) T cells: their role in the age-associated decline of immune function. Trends Immunol. 2009;30:306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McElhaney JE, Effros RB. Immunosenescence: what does it mean to health outcomes in older adults? Curr Opin Immunol. 2009;21:418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pritz T, Weinberger B, Grubeck-Loebenstein B. The aging bone marrow and its impact on immune responses in old age. Immunol Lett. 2014;162(1, pt B):310–315. [DOI] [PubMed] [Google Scholar]
- 12.Martins PN, Tullius SG, Markmann JF. Immunosenescence and immune response in organ transplantation. Int Rev Immunol. 2014;33:162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Martinis M, Franceschi C, Monti D, et al. Inflammation markers predicting frailty and mortality in the elderly. Exp Mol Pathol. 2006;80:219–227. [DOI] [PubMed] [Google Scholar]
- 14.Singh T, Newman AB. Inflammatory markers in population studies of aging. Ageing Res Rev. 2011;10:319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S4–S9. [DOI] [PubMed] [Google Scholar]
- 16.Frasca D, Blomberg BB. Inflammaging decreases adaptive and innate immune responses in mice and humans. Biogerontology. 2016;17:7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abecassis M, Bridges ND, Clancy CJ, et al. Solid-organ transplantation in older adults: current status and future research. Am J Transplant. 2012;12:2608–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart A, Smith JM, Skeans MA, et al. OPTN/SRTR 2014 annual data report: kidney. Am J Transplant. 2015;1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trzonkowski P, Debska-Slizien A, Jankowska M, et al. Immunosenescence increases the rate of acceptance of kidney allotransplants in elderly recipients through exhaustion of CD4+ T-cells. Mech Ageing Dev. 2010;131:96–104. [DOI] [PubMed] [Google Scholar]
- 20.Schaenman JM, Rossetti M, Sidwell T, et al. Increased T cell immunosenescence and accelerated maturation phenotypes in older kidney transplant recipients. Hum Immunol. 2018;79:659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Passtoors WM, Boer JM, Goeman JJ, et al. Transcriptional profiling of human familial longevity indicates a role for ASF1A and IL7R. PLoS One. 2012;7:e27759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters MJ, Joehanes R, Pilling LC, et al. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang E, Rossetti M, Sidwell T, et al. Differences in pro-inflammatory cytokines and monocyte subtypes in older as compared with younger kidney transplant recipients. Transplant Direct. 2017;1–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freer G, Rindi L. Intracellular cytokine detection by fluorescence-activated flow cytometry: basic principles and recent advances. Methods. 2013;61:30–38. [DOI] [PubMed] [Google Scholar]
- 25.Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cole SW, Capitanio JP, Chun K, et al. Myeloid differentiation architecture of leukocyte transcriptome dynamics in perceived social isolation. Proc Natl Acad Sci U S A. 2015;112:15142–15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine ME, Crimmins EM, Weir DR, et al. Contemporaneous social environment and the architecture of late-life gene expression profiles. Am J Epidemiol. 2017;186:503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole SW, Yan W, Galic Z, et al. Expression-based monitoring of transcription factor activity: the TELiS database. Bioinformatics. 2005;21:803–810. [DOI] [PubMed] [Google Scholar]
- 29.Cole SW, Hawkley LC, Aravalo JM, et al. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci U S A. 2011;108:3080–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su AI, Wiltshire T, Batalove S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linehan E, Fitzgerald DC. Ageing and the immune system: focus on macrophages. Eur J Microbiol Immunol (Bp). 2015;5:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seyda M, Quante M, Uehara H, et al. Immunosenescence in renal transplantation. Curr Opin Organ Transplant. 2015;20:417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


