Abstract
During development of pancreatic cancer, alternatively-activated macrophages contribute to fibrogenesis, pancreatic intraepithelial neoplasia (PanIN) lesion growth, and generation of an immunosuppressive environment. Here we show that the immunomodulatory agent pomalidomide depletes pancreatic lesion areas of alternatively-activated macrophage populations. Pomalidomide treatment resulted in downregulation of interferon regulatory factor 4 (IRF4), a transcription factor for M2 macrophage polarization. Pomalidomide-induced absence of alternatively-activated macrophages led to a decrease in fibrosis at PanIN lesions and in syngeneic tumors; this was due to generation of an inflammatory, immune-responsive environment with increased expression of IL-1α and presence of activated (IFNγ positive) CD4+ and CD8+ T cell populations. Our results indicate that pomalidomide could be used to decrease fibrogenesis in pancreatic cancer and may be ideal as a combination treatment with chemotherapeutic drugs or other immunotherapies.
Keywords: pancreatic cancer, PanIN lesions, pomalidomide, macrophages, polarization, fibrosis
INTRODUCTION
The microenvironment surrounding pancreatic lesions and cancer cells is mainly composed of fibroblasts and immune cells, and is an important factor affecting tumor development and growth, tumor cell dissemination, and treatment efficacy. Recently, it was shown for different stages of pancreatic ductal adenocarcinoma (PDA) development or progression that targeting alternatively-activated or tumor-associated macrophages (TAMs) and/or their redirection to macrophages with antitumor activities could be an efficient immunomodulation strategy for this cancer (1–3).
For example, targeting TAMs and intratumoral immunosuppressive monocytes with an antagonist of CCR2 improves chemotherapeutic and radiotherapy responses in PDA (4–6); and a combination of CCR2 inhibition with FOLFIRINOX has already been tested in a phase 1b trial for patients with locally advanced cancer (7). Another promising approach that has been tested in phase I clinical trials is the use of CD40 agonists (8). CD40 agonists transform the tumor microenvironment by enhancing macrophage-mediated destruction of tumor stroma and induction of clonal expansion of T cells (9). Eventually, CSF1R blockage and combination of this strategy with PD1 antagonists has been proven successful in preclinical pancreatic cancer models (10).
Recent work from our laboratory demonstrated that inhibition of IL-13 signaling prevents the occurrence of alternatively-activated macrophages and fibrosis in developing pancreatic cancer (2). While this strategy focuses on the prevention of occurrence of M2-polarized macrophages and TAMs, a more intriguing approach would be to reprogram these populations into tumor suppressive macrophages in order to shift the pro-tumorigenic environment into an inflammatory, immune-responsive environment.
Thalidomide analogs such as lenalidomide and pomalidomide are FDA-approved immunomodulatory drugs that have been developed and tested for hematologic cancers including multiple myeloma (MM), myelodysplastic syndrome, non-Hodgkin lymphoma and chronic lymphocytic leukemia (11,12). However, besides a role in mediating inhibition of NF-κB (13), proteasomal downregulation of the transcription factor IKFZ1/Ikaros (14) and effects on cytoskeletal organization (15), there is only little understanding of how they affect intracellular signaling (16). In MM, pomalidomide downregulates IL-1β and IL-6 in monocytes and increases the presence of NK cells (11), but also enhances tumor antigen uptake by dendritic cells and the cross-priming of naïve CD8+ T cells (17). Most recently, it was suggested that in central nervous system (CNS) lymphoma, treatment with pomalidomide can lead to a conversion of M2-polarized macrophages to M1-polarized macrophages, by a yet unidentified mechanism (18).
Based on these findings, we here tested in a precancerous p48cre;LSL-KrasG12D (KC) animal model for pancreatic cancer as well as in a syngeneic tumor model, if treatment with pomalidomide can decrease alternative-activated tumor promoting macrophages or reprogram them towards an anti-tumorigenic phenotype. We found that pomalidomide, by downregulating IRF4, a transcription factor for M2 macrophage polarization, induces a shift from alternatively-activated macrophage populations towards anti-tumorigenic populations. The outcome of such signaling was a decrease in fibrosis at PanIN lesions and generation of an immune-responsive environment with increased presence of activated (IFNγ positive) CD4+ and CD8+ T cells. The fact that it is already an FDA-approved drug, and our results showing its efficacy on fibrosis and immune-responsiveness at pancreatic lesions, suggest that pomalidomide may be an ideal option for combination treatment with chemotherapeutic drugs or other immunotherapy.
MATERIALS AND METHODS
Cell lines
U937 cells were obtained from ATTC (Manassas, VA) in 2011; and are routinely authenticated via their short tandem repeat (STR) profile (latest verification: June, 2018). U937 were cultured in RPMI 1640 supplemented with 10% FBS. Lipofectamine 2000 (Life Technologies, Carlsbad, CA) in serum-free OptiMEM (Life Technologies) was used for transient transfection. After thawing cells were used for up to 25 passages.
Antibodies, expression constructs and reagents
Antibodies used for Western blotting, immunohistochemistry and immunofluorescence are described in detail in Supplemental Table 1. IRF4-shRNA plasmid (TL315501) and IRF4 (NM_002460) human cDNA ORF clone (RC204876) were purchased from Origene (Rockville, MD). Recombinant human IL-4 was purchased from ThermoFisher Scientific (Waltham, MA). DAPI was from Sigma-Aldrich (St. Louis, MO). Pomalidomide was from Abcam (Cambridge, MA). All other chemicals were from ThermoFisher Scientific.
Isolation of KPC tumor cells and PSC for syngeneic tumor implantation
Primary tumor cells (KPC1) for the syngeneic tumor model were isolated from pancreatic carcinoma of a 4 month old LSL-KrasG12D/+;Trp53R172H/+;PdxCretg/+ mouse (BL6/129). In short: The pancreatic tissue was removed under sterile conditions, washed with PBS, cut into 1–5 mm pieces and digested for 30 min (37 °C, 220 rpm) in 5 ml of 2 mg/ml collagenase (Sigma-Aldrich) in Hank’s Balanced Salt Solution (HBSS) (GE Healthcare, Marlborough, MA). The digestion was stopped with cold HBSS and centrifuged at 2000 rpm, 4 °C for 2 min. The pellet was resuspended in DMEM-F12 (Corning, Corning, NY) + 5% Bovine Calf Serum-iron supplemented (Hyclone, GE Healthcare) + 0.5 μg/ml hydrocortisone (Sigma) + 5 μg/ml ITS (Corning), transferred to a 10 cm dish, and adherent cells were cultured weekly. Cells continued to be viable after 100 passages. Genotype of the cell line was confirmed using real-time PCR (Transnetyx, Cordova, TN).
Primary pancreatic stellate cells (PSC) were isolated from pancreata of 3 month old LSL-KrasG12D/+;PdxCretg/+ mice (BL6/129) using a previously described method (19) with slight modifications. Briefly, the pancreatic tissue was washed with PBS, minced into 1–5 mm pieces and digested for 1.5 hrs (37 °C, 220 rpm) in 5 ml of HBSS + 2 mg/ml collagenase + 200 units DNase I (Sigma-Aldrich). The digestion was stopped with cold HBSS + 5% Fetal Bovine Serum (FBS) and centrifuged at 2000 rpm, 4 °C for 2 min. After two additional washes, cold 10 ml 5% FBS in HBSS was added to the pellet and the cell suspension was filtered through 500 μm and 105 μm nylon meshes, respectively, into a 50 ml tube. The cells were washed and resuspended in cold 9.5 ml HBSS + 0.3 % BSA then mixed with 8 ml 30 % (w/v) Nycodenz + HBSS (Accurate, Westbury, NY) solution. The 17.5 ml cell suspension was carefully layered under 6 ml of HBSS + 0.3 % BSA in a 50 ml tube using a 10 ml serological pipet. The tube was centrifuged (1400 g, 4 °C for 20 min), and the cells were carefully collected from the thin white band above the Nycodenz interface using a P1000 micropipette. The cells were then washed with HBSS + 0.3 % BSA, centrifuged (480 g, 4 °C for 5 min), resuspended in DMEM HG + 10% FBS + 1% Pen/Strep and transferred to a 60 mm dish. After 48 hours, the culture medium was changed daily and cells were cultured after 7 days. The cells were viable for 6 passages.
Isolation of primary macrophages
Primary peritoneal macrophages for cell culture studies were isolated as previously described (20). In brief, mice were intraperitoneally injected with 2 ml of 5% aged thioglycollate solution. Five days after injection, peritoneal macrophages were collected through a single injection of 10 ml RPMI-1640 plus 10% FBS into the peritoneal cavity and subsequent withdrawal. The peritoneal exudate was centrifuged and washed with RPMI-1640 plus 10% FBS before plating onto tissue culture dishes. After one hour in an incubator (37 °C, 5% CO2), plates were washed with PBS for three times to remove non-adherent cells.
Mouse lines, syngeneic animal model and treatment
Ptf1a/p48cre/+, PdxCretg/+, Trp53R172H/+ and LSL-KrasG12D/+ mouse strains and genotyping of mice have been described previously (2,21,22). For treatment of Ptf1a/p48cre/+,LSL-KrasG12D/+ (KC; labeling in figures: p48cre;LSL-KrasG12D) mice (Scheme, see Supplemental Fig. S1A), 8 week old mice were administered pomalidomide (or vehicle control) every day for 4 weeks at a dose of 5 mg/kg (based on (18)) mixed with food (15 g DietGel, ClearH2O, Portland, ME). For syngeneic orthotopic tumor implantation, immunocompetent B6/129 mice were anesthetized and the pancreas was exposed through a left flank incision. Cells (16,000 KPC1+ 84,000 PSC) were suspended in 30 μl Matrigel (BD Biosciences, Sand Diego, CA) diluted 50% in PBS and injected into the pancreas using a U-100 insulin syringe with a 28G needle. The pancreas was placed back into the peritoneal cavity and the incision was closed using 4–0 Vicryl suture and 7 mm wound clips. After tumor establishment, mice were treated every day for two weeks with pomalidomide (or vehicle control), at a dose of 5 mg/kg mixed with food (15g DietGel, ClearH2O). After two weeks of treatment, blood was collected via intracardiac puncture while mice were under terminal anesthesia, and processed for various analyses including FACS, complete blood count and coagulation profile (IDEXX, North Grafton, MA). Pancreatic tissues were collected and used directly (flow cytometry) or were fixed in formalin or flash frozen and processed for IHC, Western blotting and other assays. For all animal experiments, the sex of the animal subjects was random, since we do not observe gender-specific differences using the KC or syngeneic animal models. All animal experiments were approved by the Mayo Clinic IACUC committee and were performed in accordance with relevant institutional and national guidelines and regulations.
Tissue homogenates, cell lysates and Western blotting
Pancreatic tissue was homogenized in 4 ml ice-cold lysis buffer (Biovision, Milpitas, CA), containing protease inhibitor cocktail (Sigma-Aldrich), using an electric homogenizer, followed by incubation for 2 hours at 4 °C with gentle agitation. Subsequently, tissue lysates were centrifuged at 13,000 rpm, 4 °C for 20 min, and the supernatant was collected and assayed or stored at −80°C. The total protein concentration of the samples was measured using Bio-Rad Protein Assay (Bio-Rad, Hercules, CA) and normalized to 2 mg/ml. For generation of cell lysates, cells were washed twice with ice-cold PBS (140 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.2), lysed with lysis buffer (25 mM Tris HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS), and incubated on ice for 5 min. Samples were subjected to SDS-PAGE and then transferred to nitrocellulose membrane. Proteins of interest were detected using indicated specific primary antibodies and HRP-conjugated secondary antibodies.
Quantitative PCR (qPCR)
For live cells, RNA was extracted with the RNeasy PLUS Mini Kit (Qiagen, Germantown, MD). For fixed cells, RNA was extracted with Arcturus Paradise Plus RNA Extraction and Isolation Kit (Applied Biosystems, Foster City, CA) and then amplified with Arcturus Paradise Plus RNA Amplification Kit (Applied Biosystems). The High Capacity cDNA RT Kit (Applied Biosystems) was used to convert RNA to cDNA and quantitative PCR was performed using a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems). qPCR reactions utilized TaqMan Fast Mix 2x (Applied Biosystems) in conjunction with the following probe/primer sets from Applied Biosystems: Gapdh (Mm99999915_g1), Chil3/Ym1 (Mm00657889_mH), Il10 (Mm01288386_m1), Retnla (Mm0045109_m1), Mrc1 (Mm01329362_m1), Clec10a/Cd301 (Mm00546125_g1), Il1rn (Mm00446186_m1), Il1a (Mm00439620_m1), and Arg1 (Mm00475988_m1). Calculation of mRNA abundance was via the ΔΔCT method and normalization was to Gapdh.
Flow cytometric analyses and fluorescence-activated cell sorting (FACS)
Resected pancreas (tumor) tissue was dissociated using mouse tumor dissociation kit (Miltenyi, Bergisch Gladbach, Germany) via autoMACS® Pro Separator (Miltenyi) per manufacturer protocol. Dissociated tissue was passed through 35 μm filters (Corning) to obtain single cell suspensions. For cytometric analyses, the total leukocyte population was isolated from dissociated tissues with mouse CD45+ MicroBeads (Miltenyi) per manufacturer protocol, using LS Columns (Miltenyi) on a QuadroMACS magnet (Miltenyi). Peripheral blood and tissue leukocyte samples were fixed and stained using PerFix-nc kit (Beckman Coulter) per manufacturer protocol. Multicolor fluorescence detection and analysis was performed on a 4-laser CytoFLEX flow cytometer using CytExpert software (Beckman Coulter, Pasadena, CA). Fluor-conjugated antibodies were from Biolegend (San Diego, CA) and include CD45, CD3, CD4, CD8, F4/80, and IFNγ (details in Supplemental Table 1). Single-stained samples were used to generate compensation matrixes and determine pos/neg fluorescence as compared to unstained control. Minimum 100,000 events were recorded per sample.
For fluorescence-activated cell sorting the digestion of the pancreas was performed as described above for the isolation of KPC cells. Then pancreatic cell suspensions were labelled with LIVE/DEAD Fixable Violet Dead Cell Stain Kit (ThermoFisher Scientific), blocked with α-CD32 (1:50, BD Biosciences), and labelled with α-F4/80 conjugated to Alexa Fluor 488 (1:50, Bio-Rad). In sorts to distinguish Ym1- and Ym1+ populations, fixation and permeabilization (Fix & Perm kit, Life Technologies) followed α-F4/80 labelling. Then, cells were labelled with α-Ym1 conjugated to Alexa Fluor 647 (1:50, Abcam).
Immunohistochemistry (DAB and IF)
Slides were deparaffinized and rehydrated as previously described (2,21) and antigen retrieval was performed in sodium citrate buffer (10 mM, pH 6.0). Tissue samples were then treated with 3% H2O2 (5 min), washed with 0.5% Tween 20/PBS, and blocked with Protein Block Serum-Free Solution (Agilent, Santa Clara, CA; 5 min, RT).
For DAB immunohistochemistry (IHC), primary antibodies were diluted in Antibody Diluent Background Reducing Solution (Agilent). Specific antibodies used and dilutions are listed in Supplemental Table 1. Staining was visualized using EnVision Plus Anti-Rabbit Labelled Polymer Kit (Agilent), or biotin-streptavidin (Biocare Medical, Concord, CA) 2-step conjugation when primary goat antibodies were used. H&E staining was performed as described (2,21). Trichrome staining was performed with Masson Trichrome Stain Kit (Sigma-Aldrich). ScanScope XT scanner and ImageScope software (Aperio, Vista, CA) were used to capture images.
For fluorescent IHC (IF-IHC), slides were incubated with primary antibodies (dilutions listed in Supplemental Table 1) in Antibody Diluent Background Reducing solution (Agilent) at 4 °C, overnight. After 3 washes with 0.05% Tween-20/PBS, Alexa Fluor 488 or Alexa Fluor 594 labeled secondary antibodies (Invitrogen) at a 1:500 dilution were added, and samples incubated at RT for 1 hr. For nuclear staining, 125 μg/ml DAPI was added for 5 min at RT, after incubation with secondary antibodies. After five washes with 0.05% Tween-20/PBS, LabVision PermaFluor (ThermoFisher Scientific) was used as mounting medium. Images were captured using a ScanScope FL scanner and ImageScope software (Aperio).
RNAscope in situ hybridization (ISH) and IHC
In-situ hybridization (ISH) was performed using RNAscope® Assay 2.5 HD Reagent Kit-Brown (Advanced Cell Diagnostics [ACD], Hayward, CA) according to the manufacturer’s protocol (23), with some modifications. Briefly, formalin fixed paraffin embedded (FFPE) 5 μm sections were baked at 60 °C for 1 hour, deparaffinized in xylene for 15 min, dehydrated in 100% ethanol and dried at room temperature (RT) overnight in a desiccator. The slides were then treated with hydrogen peroxide (provided by the kit) for 10 min, rinsed in deionized water, boiled in target retrieval reagents for 8 min (time optimized for pancreas), followed by protease treatment for 15 min at 40 °C in a hybridization oven. Slides were then incubated for 2 hours at 40 °C with one of the following ACD RNAscope® mouse target probes: Mm-Il1a (NM_010554.4, region 2–1284) for IL-1α, Mm-Il1b (NM_008361.3, region 2–950) for IL-1β, or Mm-Il13 (NM_008355.3, region 20–632) for IL-13. After hybridization, six amplification steps were performed using amplification buffers (provided by the kit: Amp 1–6, with 2 min washes in between amplification steps; Amp 5 was modified to 1 hour incubation), and the mRNA signal was detected with DAB staining. After five quick washes with water, the slides were counterstained with hematoxylin, dehydrated in alcohol and mounted. Images were captured using ScanScope XT scanner and ImageScope software (Aperio).
Quantification and Statistical Analysis
All cell biological and biochemical experiments have been performed at least 3 times. For animal experiments, if not stated otherwise in the figure legends, pancreatic samples form n = 3 mice have been used for quantification analyses. 4–6 fields per sample were subject to quantification. IHC data was quantified by manual counting of positive cells or by using the Aperio Positive Pixel Count Algorithm; ISH was quantified using the Aperio Positive Pixel Count Algorithm (Aperio). Co-expression of proteins in cells was judged by analyses of IF for each protein on the same slide. Data are presented as mean ± SD. P values (if not stated otherwise in the figure legends) and were acquired with the unpaired student’s t-test with Welch’s correction using Graph Pad software (GraphPad Inc., La Jolla, CA). p < 0.05 was considered statistically significant.
RESULTS
Pomalidomide decreases the expansion of KRasG12D-induced abnormal areas in the pancreas
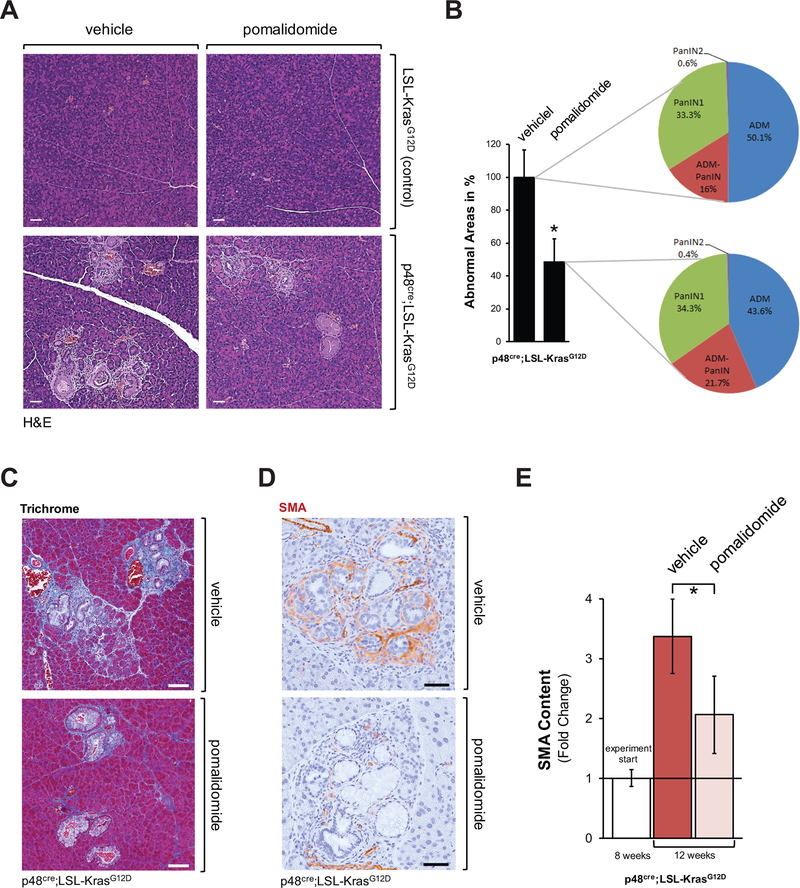
Inflammatory monocytes (M1) can deplete fibrosis at pancreatic lesions (24), while alternatively-activated (M2, Ym1 positive) macrophages can drive fibrosis (2). In CNS lymphoma, pomalidomide has been shown to convert the polarization status of IL4-stimulated macrophages from M2 to M1 in vitro (18). Therefore, we tested the effect of pomalidomide on fibrosis and PanIN formation in the precancerous p48cre;LSL-KrasG12D (KC) animal model for pancreatic cancer. Pomalidomide (5 mg/kg) or vehicle was orally administered to 8 week old mice every day for 4 weeks (treatment scheme in Supplemental Fig. S1A). Treatment with pomalidomide over this time period led to a significant (approximately 50%) reduction in pancreatic abnormal structures in KC mice (Figs. 1A and 1B), indicating that it affected the expansion of these regions. However, the relative presence of acinar-to-ductal metaplasia (ADM), ADM-PanIN or PanIN1A/B regions was not significantly shifted (Fig. 1B). Most significant effects observed after treatment with pomalidomide were on the stroma in the lesion areas. Staining of collagen using Masson’s Trichrome (Fig. 1C), or immunohistochemical staining for smooth muscle actin (SMA) as a marker for pancreatic stellate cells (PSCs) (Fig. 1D), indicated a significant prevention of fibrosis (Fig. 1E). This was also confirmed by Western blot analyses of total pancreas homogenates and staining for SMA, as well as desmin another marker for PSCs (Supplemental Fig. S1B). In some areas of the pancreata of pomalidomide treated mice, PanINs were not surrounded by stroma at all (Supplemental Fig. S1C).
Figure 1: Pomalidomide decreases fibrosis in the lesion areas of the pancreas of KC mice.
A-D: Control mice or p48cre;LSL-KrasG12D (KC) mice at an age of 8 weeks were treated with pomalidomide, every day for 4 weeks (treatment scheme depicted in Supplemental Fig. S1). At an age of 12 weeks pancreata were harvested and abnormal areas analyzed. A: H&E staining of representative areas. The scale bar indicates 50 μm. B: The bar graph shows a quantification of the abnormal areas in control or pomalidomide treated KC mice (n=4 per treatment group). The asterisk indicates statistical significance (p<0.05) as compared to control. The pie graphs show grading of pancreatic lesions and relative presence of ADM, ADM-PanIN transition areas, and PanIN1. C, D: Trichrome staining or IHC staining for smooth muscle actin (SMA) of lesion areas in KC mice after control or pomalidomide treatment. Representative areas are shown. The scale bar indicates 100 μm in C and 50 μm in D. E: Quantification of SMA content of abnormal pancreatic areas. IHC stained samples (n=3) from each treatment group were analyzed. The white bars show the SMA content at treatment start as a reference (n=3 samples). The asterisk indicates statistical significance (p<0.05).
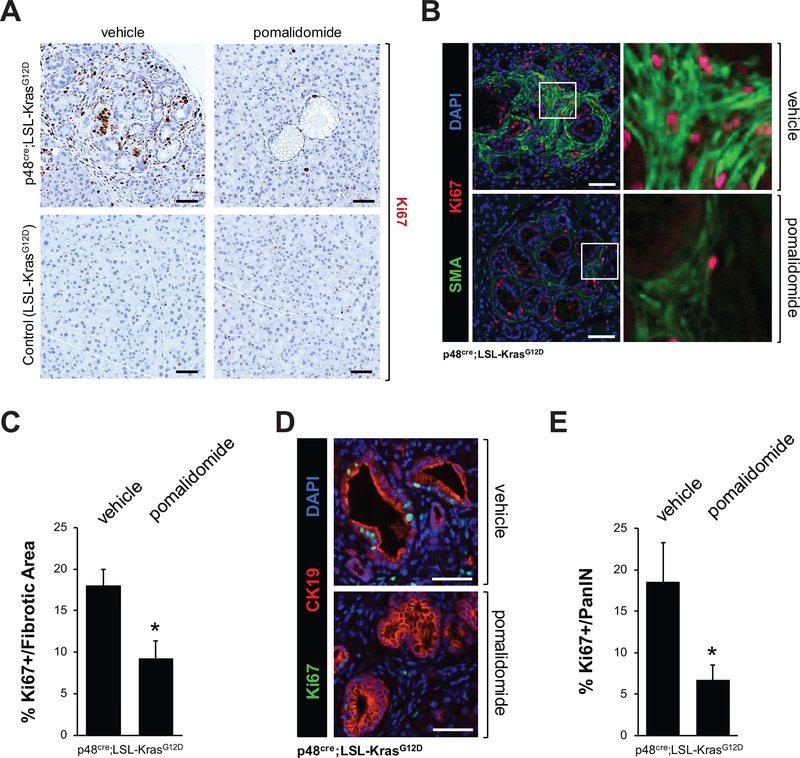
The reduction in fibrotic areas was not due to an increase in apoptotic events, as judged by the absence of cleaved caspase 3 (Supplemental Fig. S2A). We therefore determined the presence of proliferating cells in lesion areas by immunohistochemistry for presence of the proliferation markers Ki67 or Cyclin D1 (Figs. 2A and Supplemental Figs. 2B–2D), and found a significant decrease in cells positive for these markers (Ki67 positive cells decrease to 29% ± 6% and Cyclin D1 positive cells decrease 50% ± 5%). A more in depth analysis of Ki67 positive cells in fibrotic areas (IF-IHC for SMA combined with Ki67) as well as PanIN (IF-IHC for CK19 combined with Ki67) indicated a significant decrease of proliferative cells in both the stromal areas and ductal cells of the lesions (Figs. 2B–2E). Overall, our data indicated that treatment of KC mice with pomalidomide decreases stromal/fibrotic areas around the preneoplastic lesions and prevents their expansion.
Figure 2: Pomalidomide decreases proliferation in the lesion areas of the pancreas of KC mice.
KC or control mice at an age of 8 weeks were treated with pomalidomide for 4 weeks. At an age of 12 weeks pancreas tissues were harvested and analyzed. A: Representative IHC staining of pancreatic abnormal areas for presence of Ki67 as marker for proliferating cells. Representative areas are shown. The scale bar indicates 50 μm. B, D: IHC-IF analyses of fibrotic areas (B, Ki67 in red and SMA (fibrosis marker) in green) and PanIN lesions (D, Ki67 in green and CK19 (lesion cell marker) in red). The scale bar indicates 50 μm. C, E: Show quantification of Ki67 positive cells in fibrotic areas (C) or PanIN lesions (E). IHC stained samples of n=4 mice from each treatment group were analyzed. The asterisk indicates statistical significance (p<0.05).
Pomalidomide decreases the presence of Ym1 positive macrophages at PanIN lesions
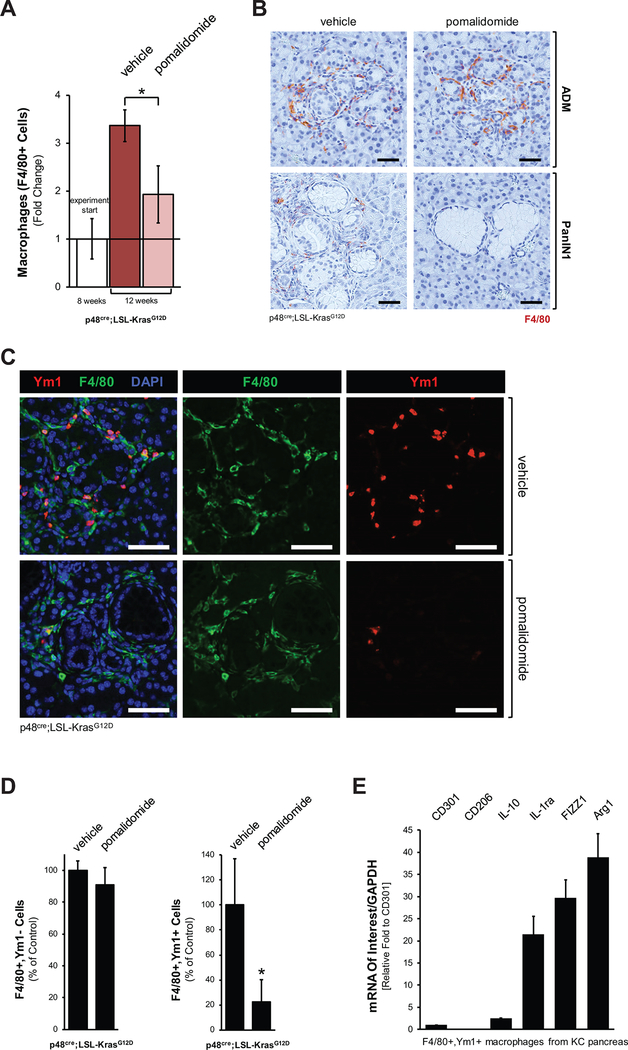
Since alternatively-activated macrophages have been shown to mediate fibrosis and lesion growth in the KC animal model (2), we next tested if pomalidomide affects macrophage populations within the pancreas. Using the pan macrophage marker F4/80 we found that in the time period between treatment start (week 8) and endpoint (week 12) in vehicle-treated mice total macrophage populations increased approximately 3.5-fold, which is most likely due to lesion growth. However, treatment with pomalidomide showed a significant decrease in macrophages at the endpoint (Fig. 3A). This decrease in macrophages mainly occurred in the stromal regions at the PanIN lesions (Fig. 3B), where mainly Ym1+ M2 macrophages reside (2), while we did not observe significant decrease in macrophages at classical ADM regions (Fig. 3B), where mainly M1 macrophages reside (20,25). Immunohistochemical staining for Ym1, which is a marker for alternatively-activated (M2) macrophages in the pancreas, confirmed that pomalidomide decreases this population at PanIN lesions (Supplemental Fig. S3A). Moreover, to demonstrate that pomalidomide specifically targets the Ym1 positive population we performed a co-immunofluorescence staining for Ym1 and F4/80 (Fig. 3C, control shown in Supplemental Fig. S3B). Quantification of this data suggests that while F4/80+,Ym1- macrophage populations in typical ADM/PanIN areas remain at comparable levels, the F4/80+,Ym1+ macrophage population decreases with pomalidomide (Fig. 3D). Quantitative PCR analyses for markers of alternatively activated macrophages after isolation of the F4/80+,Ym1+ population per FACS indicated that they express Arg1 and FIZZ1 as typical M2 and TAM markers (Fig. 3E). In addition, F4/80+,Ym1+ also express the IL-1 antagonist IL-1ra, which we previously had identified as factor secreted by pancreatic macrophages that promote lesion growth (2).
Figure 3: Pomalidomide decreases the presence of M2 macrophages in pancreata of KC mice.
A: Quantification of presence of F4/80 positive cells (macrophages) in the pancreata of n=3 mice per experimental group. The white bars show the SMA content at treatment start as a reference (n=3 samples). The asterisk indicates statistical significance (p<0.05) as compared to vehicle control. B: Analysis of ADM and PanIN areas of vehicle treated or pomalidomide treated mice for presence of macrophages (IHC for F4/80, brown staining). Shown are representative areas. The scale bar indicates 50 μm. C: Pomalidomide decreases the presence of Ym1 positive (M2) macrophages in ADM/PanIN regions. ADM/PanIN areas were analyzed for presence of alternatively-activated macrophages (IF-IHC: YM1 and F4/80, as indicated). Nuclei were stained with DAPI. Representative staining of PanIN areas is shown. The scale bar indicates 50 μm. D: Quantification of F4/80+,Ym1- and F4/80+,Ym1+ macrophages under both treatment conditions. IF-IHC stained samples from n=3 mice of each treatment group were analyzed. The asterisk indicates statistical significance (p<0.05). E: Characterization of F4/80+,Ym1+ macrophages. Pancreatic macrophages from three KC mice were labeled with F4/80- and Ym1-specific antibodies and sorted for the F4/80+,Ym1+ population. Cells were analyzed by quantitative PCR for expression of indicated mRNAs and GAPDH (normalization). Expression of CD301 was set 1 fold (CD301 is expressed in both F4/80,Ym1+ and F4/80,Ym1- macrophages populations at similar levels), and fold expression of analyzed markers was plotted relative to CD301 expression.
Since it recently was shown that PanIN cells can express interleukin-13 (IL-13) to induce the presence of Ym1+ macrophages at these lesions (2), using in situ hybridization we next tested if pomalidomide affects IL-13 mRNA levels. However, we found no evidence that IL-13 expression is regulated by pomalidomide in our experimental system (Supplemental Fig. S3C). Therefore, we determined if pomalidomide engages other mechanisms that can affect macrophage polarity.
Pomalidomide shifts macrophage populations by affecting interferon regulatory factor 4 levels
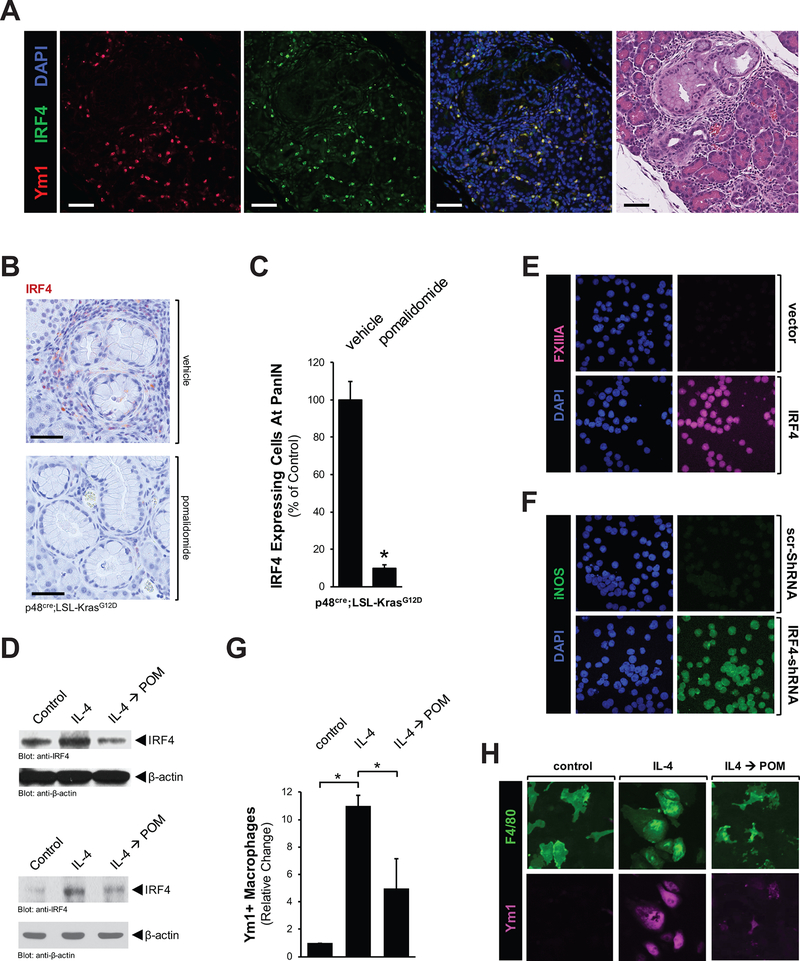
Recent work indicates that interferon regulatory factor 4 (IRF4) is a key transcription factor regulating M2 macrophage polarization (26); and lenalidomide, a compound related to pomalidomide, has been shown to inhibit the expression of IRF4 (27). Therefore, we tested if pomalidomide affects macrophage populations by regulating IRF4. An immunofluorescence analysis on pancreata from KC mice indicated that Ym1-positive cells in PanIN areas express IRF4 (Fig. 4A). Treatment with pomalidomide decreased the IRF4 expression level at PanIN areas approximately 90% (Figs. 4B and 4C), while levels of its upstream regulator Ikaros (IKZF1) remained unchanged (Supplemental Fig. S4A).
Figure 4: Pomalidomide decreases the expression of IRF4 in M2 macrophages to drive their phenotype conversion to an inflammatory phenotype.
A: Ym1 positive macrophages express IRF4. PanIN/ADM areas were analyzed by IF-IHC for co-localization of IRF4 (green) and Ym1 (red). Nuclei were stained with DAPI. The scale bar indicates 50 μm. B: Immunohistochemical analysis of IRF4 expression in PanIN regions of vehicle or pomalidomide treated mice. Shown are representative areas. The scale bar indicates 50 μm. C: The bar graph shows the quantification of presence of IRF4 positive cells in pancreata of n=3 mice per experimental group. The asterisk indicates statistical significance (p<0.05) as compared to vehicle control. D: U937 cells (top panel) or primary mouse macrophages (bottom panel) were treated with IL-4 (20 ng/ml, 48 hours), and then for another 48 hours with pomalidomide (10 μg/ml). Shown are Western blots for IRF4 and β-actin (loading control). E: U937 cells were transfected with IRF4 and after 2 days subjected to IF analysis for expression of FXIIIA (M2 marker) combined with DAPI staining. F: U937 cells were transfected with IRF4-shRNA and after 2 days subjected to IF analysis for expression of iNOS (M1 marker) combined with DAPI staining. G, H: Primary macrophages were treated as described in F and then visualized with F4/80 and Ym1 staining. The bar graphs (G) show a quantitative analysis of Ym1+ macrophages under different treatment conditions. The asterisk indicates statistical significance (p<0.05).
Next, we investigated if pomalidomide directly affects IRF4 levels in alternatively-activated macrophages. We first differentiated U937 monocytes into M2 macrophages using IL-4, and then treated them with pomalidomide. While the differentiation to M2 correlated with an increase in IRF4 expression, the sequential treatment with pomalidomide depleted the expression of this transcription factor below basal levels (Fig. 4D, top panel). Similarly, isolated mouse primary macrophages, after M2 polarization with IL-4, show increased levels of IRF4, which can be depleted with pomalidomide (Fig. 4D, bottom panel).
In order to investigate the importance of IRF4 for M2 macrophage identity, we again utilized the human monocyte cell line U937 which, in response to different stimuli, can differentiate to mature macrophages. First we tested if altered levels of IRF4 can regulate macrophage polarization. Therefore, we either overexpressed IRF4, or decreased basal expression by a reverse genetics approach. As judged with FXIIIA (Fig. 4E) and pY641-STAT6 (Supplemental Fig. S4B) as markers for alternatively-activated macrophages (18,28), ectopic (increased) expression of IRF4 (control shown in Supplemental Fig. S4C) in U937 monocytes induced their M2 polarization. On the other hand, depleting IRF4 below basal levels using IRF4-specific shRNA (control shown in Supplemental Fig. S4D) led to differentiation of U937 monocytes to inflammatory macrophages as judged by increased expression of iNOS (Fig. 4F), and phosphorylation of STAT1 at pY701 (Supplemental Fig. S4E) as markers (2,29). Taken together, this suggested that levels of IRF4 determine if monocytes differentiate to inflammatory or alternatively-activated macrophages. Eventually, we found that M2 polarization of isolated mouse primary macrophages and as induced by IL-4 was reverted by pomalidomide (Fig. 4G and 4H).
In summary, our data indicate a mechanism of how M2 dedifferentiation is initiated by pomalidomide, namely by down-regulating their IRF4 expression levels.
The downregulation of M2 macrophages by pomalidomide generates an anti-tumorigenic environment
Next, we determined if pomalidomide-induced alterations in macrophage populations affect the pro-tumorigenic environment at the pancreatic lesion areas that can be responsible for immune escape. Therefore, we focused on the presence of modulators of inflammatory signaling and T cell exclusion.
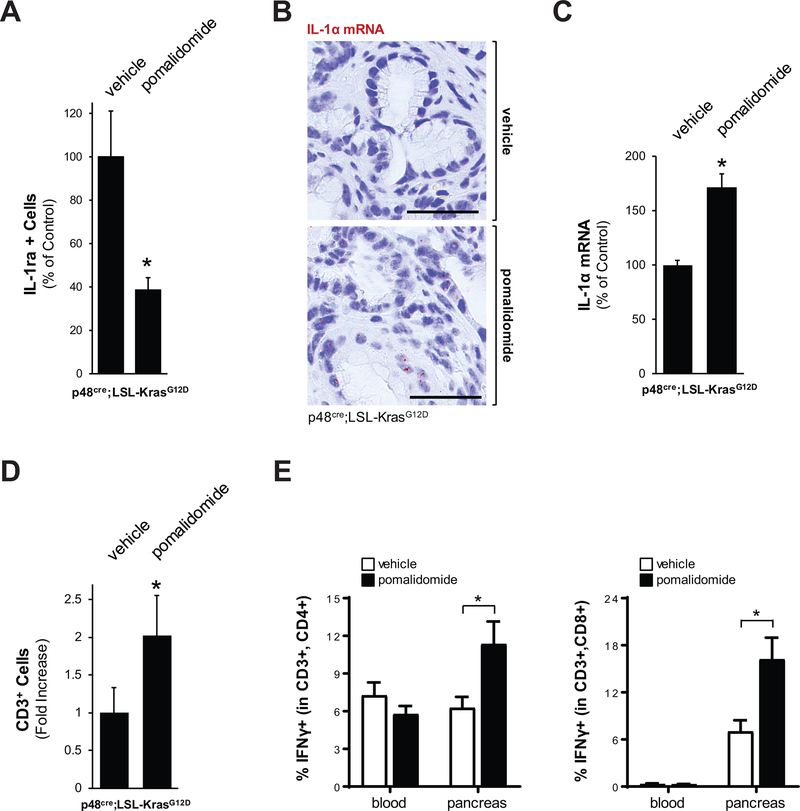
Interleukin 1 signaling is responsible for the production of inflammation. PanIN lesion cells and stroma cells in pancreatic abnormal areas express the interleukin-1 receptor (IL-1R) (Supplemental Fig. S5A), and we previously have shown that Ym1+ M2 macrophages can block IL-1R signaling by producing interleukin-1 receptor antagonist (IL-1ra), and that such signaling promotes lesion growth (2). As expected by the observed decrease in Ym1+ cells, treatment of mice with pomalidomide led to downregulation of IL-1ra expression (Fig. 5A and Supplemental Fig. S5B). Moreover, ISH analyses for IL-1R ligands IL-1α and IL-1β indicated that treatment with pomalidomide increased IL-1α expression in cells residing in PanIN lesion areas (Figs. 5B and 5C), while IL-1β expression was unaffected (Supplemental Fig. S5C). Since activated inflammatory macrophages produce IL-1α, which upregulates TNF signaling via activation of IL-1R, this suggests a shift to an inflammatory, anti-tumorigenic environment. Indeed, isolated pancreatic F4/80+ cells showed a dramatic increase in IL-1α expression in response to pomalidomide indicating a shift to pro-inflammtory populations (Supplemental Fig. S5D).
Figure 5: Pomalidomide induces anti-tumorigenic signaling in pancreata of KC mice.
A: Quantification of presence of IL-1ra positive cells in pancreata of n=3 mice per experimental group. The asterisk indicates statistical significance (p<0.05) as compared to vehicle control. B: Pomalidomide increases IL-1α mRNA expression in ADM lesion areas. Shown is a representative in situ hybridization (ISH) for IL-1α mRNA (brown dots) in KC mice either vehicle treated (control) or treated with pomalidomide. The bar represents 50 μm. C: Quantification of cells positive for IL-1α mRNA in pancreata of n=3 mice per experimental group. The asterisk indicates statistical significance (p<0.05) as compared to vehicle control. D: Quantification of presence of CD3 positive cells in pancreata of n=3 mice per experimental group. The asterisk indicates statistical significance (p<0.05) as compared to vehicle control. E: Flow cytometry of blood or pancreas/tumor tissue from control or pomalidomide treated mice (n=3 per treatment group) to determine the percentage of IFNγ-positive cells within the CD3+,CD4+ and CD3+,CD8+ T cell populations. The asterisk indicates statistical significance (p<0.05) as compared to vehicle control.
One function of IL-1α is that it regulates the presence of cytotoxic T cells (CTL) in tumors to generate an immuno-responsive environment (30,31). Therefore, we determined if the pomalidomide-induced increase in IL-1α leads to an accumulation of this T cell population. As judged by IHC, in the pancreatic lesion areas of mice treated with pomalidomide we observed a significant increase of CD3 positive cells (Fig. 5D and Supplemental Fig. S5E). Further analyses did not indicate significant changes in total numbers of CD4+ cells (Supplemental Fig. S5F), but an approximately 2-fold increase in CD8+ cells (Supplemental Fig. S5G). A more detailed flow cytometric analysis in which we determined the relative percentage of IFNγ positive populations within CD3+,CD4+ or CD3+,CD4+ T cells indicated that in response to pomalidomide CD4+ effector and CD8+ effector T cell populations significantly increased (approximately 2-fold and 3-fold) in pancreas tissue samples (Fig. 5E). Taken together, our data indicates that pomalidomide shifts the immuno-suppressive, pro-tumorigenic environment at pancreatic lesions to an inflammatory, immune-responsive environment.
Pomalidomide generates an immune-responsive, anti-tumorigenic environment in orthotopic tumors.
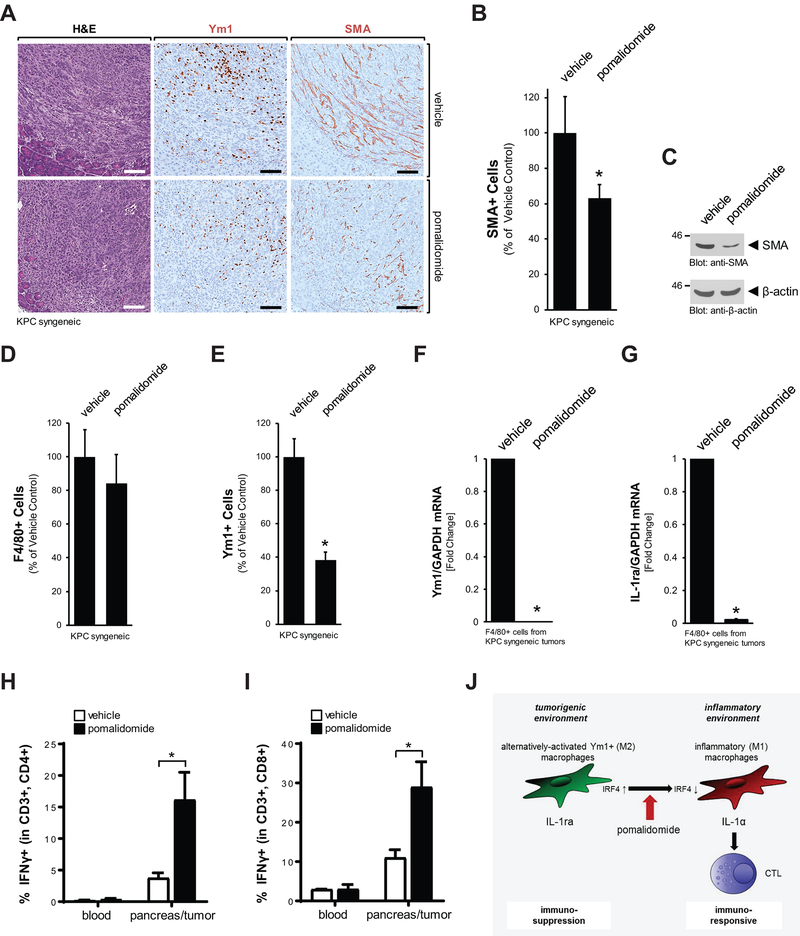
A key question is if the effects of pomalidomide on the lesion microenvironment that were observed in the KC model can be obtained in established pancreatic tumors. In order to test this we utilized a syngeneic model, in which we established pancreatic tumors by orthotopic implantation of a KPC/PSC cell mix. After tumor establishment, mice were treated with vehicle or pomalidomide every day for a period of 2 weeks (Supplemental Figs. S6A). While we did not observe significant effects on the tumor size at the endpoint, we saw a significant decrease in the presence of fibrotic cells as judged by IHC or Western blotting for SMA (Fig. 6A–6C). IHC analyses for F4/80 and Ym1 indicated that unlike in the precancerous KC model, overall numbers of macrophages remained unchanged with pomalidomide (Fig. 6D), but Ym1+ TAM population decreased significantly about 60% (Fig. 6E). To confirm this decrease with another method, we sorted F4/80 positive cells from tumors and probed them via quantitative PCR for expression of Ym1. We found a dramatic decrease in both, Ym1+ expression and the Ym1+ cell marker IL-1ra in the tumor macrophage populations (Figs. 6F and 6G). Additional flow cytometric analyses did not detect significant changes in tissue-derived total lymphocyte (CD45+) numbers after treatment with pomalidomide (Supplemental Figs. S6B and S6C). Moreover, no significant changes in total macrophages or CD4+ and CD8+ T cell composition were detected in blood or pancreas/tumor samples after treatment (Supplemental Figures S6D–S6F). However, similar as seen with the KC model, in response to pomalidomide significant increases were observed in the CD4+ effector and CD8+ effector T cell populations in pancreas/tumor tissue-derived samples, but not in blood-derived samples (Figs. 6H and 6I), indicating activated T cell responses in the tumors. Taken together the data with the syngeneic model show that pomalidomide, similar as observed and analyzed in detail with the KC model, can decrease Ym1+ tumor-associated macrophages and fibrosis in tumors. This goes along with an increase of presence of activated T cells, thus switching the immune-suppressive into an immune-responsive environment.
Figure 6: Pomalidomide generates an immune-responsive, anti-tumorigenic environment in orthotopic tumors.
A: H&E or IHC staining of representative orthotopic tumors of vehicle or pomalidomide-treated mice for Ym1 or SMA. The scale bar indicates 100 μm. B: Quantification of presence of SMA positive cells in the pancreata of n=4 mice per experimental group. The asterisk indicates statistical significance (p<0.05) as compared to vehicle control. C: Tissue homogenates of pancreata/tumors of indicated treatment groups were analyzed by Western blot for expression of SMA or for β-actin (loading control). D: Quantification of presence of F4/80 positive cells in orthotopic pancreatic tumors of n=4 mice per experimental group. E: Quantification of presence of Ym1 positive cells in the pancreata of n=4 mice per experimental group. The asterisk indicates statistical significance (p<0.05) as compared to vehicle control. F, G: F4/80+ cells were isolated from orthotopic tumors of vehicle or pomalidomide-treated mice via FACS and then analyzed by quantitative PCR for expression of Ym1 and IL-1ra. H, I: Flow cytometry of blood or pancreas/tumor tissue from control or pomalidomide treated mice (n=3 per treatment group) to determine the percentage of IFNγ-positive cells within the CD3+,CD4+ and CD3+,CD8+ T cell populations. The asterisk indicates statistical significance (p<0.05) as compared to vehicle control. J: Schematic of how pomalidomide shifts macrophage populations and modulates the lesion environment from immuno-suppressive and pro-tumorigenic to immuno-responsive and anti-tumorigenic.
DISCUSSION
The fibrotic stroma around pancreatic lesions and tumor areas is an immuno-suppressive environment that generates a barrier for immune cells and chemotherapeutic drugs (32,33). Recent evidence from preclinical animal models suggest that alternatively-activated (M2) macrophages, which represent approximately 85% of TAMs in PDA (34), regulate fibrosis and exclusion of cytotoxic T lymphocytes, two hallmarks of immune escape in PDA (1,2,24,32,35). Multiple strategies have been developed to target these TAMs and have either been tested in preclinical mouse models or have already advanced to clinical trials (3,4,6,7,9). The common idea behind all these strategies is to transform immunosuppressive macrophage populations within the tumor microenvironment into tumor regressing macrophages, which drive destruction of tumor stroma and the presence of cytotoxic T cells. We here show that the immune modulator pomalidomide can fulfill such a function.
Treatment of KC mice with pomalidomide led to an approximately 50% decrease of abnormal pancreatic areas (Fig. 1A). Although there were less lesions in pomalidomide-treated mice, the distribution between ADM areas, ADM-PanIN1 areas and PanIN1A/B lesions was similar between treatment conditions (Fig. 1B), indicating that pomalidomide may not affect the progression of lesion stages. However, we observed a significant decrease in proliferation of stromal and PanIN cells in abnormal regions (Fig. 2), which went along with decreased fibrosis (Fig. 1C–E). Both can be explained by pomalidomide’s effects on the Ym1+ macrophage population (Fig. 3), since similar results are obtained in the same animal model when the occurrence of this population is blocked with neutralizing antibodies for IL-13 (2). Moreover, the effect on the proliferation of PanIN cells in our model for PDA development is in line with the proliferation inhibitory effects seen in orthotopic tumors (13).
Interferon regulatory factors play key roles in hematopoietic development of monocytes and their differentiation into macrophages, but also in regulating polarization phenotype and phenotype switching (36). Of these, IRF4 is highly expressed in alternatively-activated TAMs and mediates M2 polarization. Studies in multiple myeloma (MM) suggest that pomalidomide and lenalidomide inhibit IRF4 gene expression (27,37). In MM, lenalidomide and pomalidomide affect IRF4 levels indirectly, by activating the CRBN-CRL4 E3 ubiquitin ligase to facilitate proteasomal degradation of Ikaros (14) and Aiolos (IKZF3) (38). However, for pancreas-associated M2 macrophages we found that pomalidomide downregulates IRF4 expression levels, while Ikaros expression levels remain unchanged (Fig. 4 and Supplemental Fig. S4). This indicates a possible regulation of IRF4 expression at the transcriptional level, which has to be investigated in future studies.
Using a monocyte cell line we demonstrated that, dependent on the expression level of IRF4, monocytes can differentiate into either M2 or M1 macrophages (Figs. 4D, 4E). Moreover, we showed for U937 cells and primary macrophages that M2 polarization after treatment with IL-4 leads to increased IRF4 levels, and that this is reverted by pomalidomide (Fig. 4D). Overall, such pomalidomide signaling results in a decrease in Ym1-positive macrophages (Figs. 4G, 4H). Thus our data make a compelling case that pomalidomide-induced downregulation of IRF4 may reprogram macrophage populations in the pancreas to an inflammatory phenotype. However, futures studies should investigate if pomalidomide can also regulate other factors that control TAM-induced adaptive immune suppression in PDA, such as NLRP3/IL-10 signaling (39), or factors of the necrosome such as RIP3 and Mincle (40).
Through its effects on macrophage populations and the fibrotic environment, pomalidomide contributes to generation of an anti-tumorigenic environment by activating IL-1R signaling through downregulation of the receptor antagonist IL-1ra, and upregulation of IL-1α expression. We previously have shown that IL-1ra can be produced by a multitude of cells within pancreatic abnormal areas, including Ym1+ M2 macrophages, and that it promotes lesion growth (2). The downregulation of IL-1ra in combination with increased expression of IL-1α (Fig. 5A–C) while IL-1β expression is unchanged (Supplemental Fig. S5C) suggests that the IL-1R mainly is activated via IL-1α in pomalidomide-treated pancreata. This is interesting because IL-1α is a marker of the senescence-associate secretory phenotype (41), and IL-1α deficient tumors have an immunosuppressive environment due to exclusion of cytotoxic T cells (CTL) (30,31). In accordance with this, in our experimental setting, pomalidomide induced an approximately 2-fold increase in CD3+ cells in pancreata of KC mice (Fig. 5D). Flow cytometric analyses of the CD3+ cells for activated T-cells indicated both increased occurrence of IFNγ positive helper and cytotoxic T cells. This is in line with studies that showed that in the pancreas a shift from M2 to M1 populations can orchestrate effective T cell immunotherapy (42).
Eventually, using a syngeneic PDA tumor model, we showed that similar effects of pomalidomide on the microenvironment can be obtained in established tumors (Fig. 6 and Supplemental Fig. S6). This suggests that pomalidomide holds promise for immunotherapy of pancreatic cancer by generating a shift from an immuno-suppressive, pro-tumorigenic to an inflammatory, immune-responsive environment (Fig. 6J). As potential side effects for human use, 4% of patients observed pomalidomide treatment-induced adverse events, including clotting complications and deep vein thrombosis, pulmonary embolism and thrombocytopenia in 2% of patients (43). However, in our animal experiments, after performing complete blood counts (CBC) (Supplemental Figs. S7A, S7B), we did not observe any hematologic side effects of pomalidomide. Additional analyses of coagulation factors such as prothrombin time, partial thromboplastin time or levels of fibrinogen remained in normal range (for mice) between treatment conditions (Supplemental Fig. S7C). Nevertheless, in future clinical trials these potential side effects may require all patients to take thromboprophylaxis with aspirin or an anticoagulant as long as they remain on pomalidomide.
The benefit of pomalidomide is that it is already FDA-approved, and that a recent phase I clinical study (NCT00540579) demonstrated that its use for untreated advanced carcinoma of the pancreas is feasible and safe (44). Thus it may be combined with standard of care chemotherapy, where it had shown to promote chemosensitization in mice (13), as well as other approaches to sensitize pancreatic tumors to T cell immunotherapy such as inhibition of focal adhesion kinase with VS-4718, anti-PD1 therapy, or depletion of CD44 or CCL2 (4–6,8,10,45).
Supplementary Material
SIGNIFICANCE.
Findings reveal new insights into how macrophage populations within the pancreatic cancer microenvironment can be modulated, providing the means to turn the microenvironment from immunosuppressive to immune-responsive.
ACKNOWLEDGEMENTS
We thank Dr. Lizhi Zhang (Mayo Clinic Rochester) for pathological advice and members of the Storz laboratory and the Discovery and Translation Labs Pancreatic Cancer Theme Group at Mayo Clinic Florida for helpful discussions. This work was supported by the Chartrand Foundation, a Mayo Clinic Center for Biomedical Discovery pilot grant, a pilot grant from the Mayo Clinic SPORE in Pancreatic Cancer (CA102701–12DRP3) and the NIH grant CA200572 to P. Storz, as well as Mayo Clinic internal funding to H. W. Tun. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Conflict of interest: The authors declare that there is no conflict of interest.
LITERATURE
- 1.Long KB, Collier AI, Beatty GL. Macrophages: Key orchestrators of a tumor microenvironment defined by therapeutic resistance. Mol Immunol 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liou GY, Bastea L, Fleming A, Doppler H, Edenfield BH, Dawson DW, et al. The Presence of Interleukin-13 at Pancreatic ADM/PanIN Lesions Alters Macrophage Populations and Mediates Pancreatic Tumorigenesis. Cell Rep 2017;19:1322–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchem JB, Brennan DJ, Knolhoff BL, Belt BA, Zhu Y, Sanford DE, et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res 2013;73:1128–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nywening TM, Belt BA, Cullinan DR, Panni RZ, Han BJ, Sanford DE, et al. Targeting both tumour-associated CXCR2(+) neutrophils and CCR2(+) macrophages disrupts myeloid recruitment and improves chemotherapeutic responses in pancreatic ductal adenocarcinoma. Gut 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connolly KA, Belt BA, Figueroa NM, Murthy A, Patel A, Kim M, et al. Increasing the efficacy of radiotherapy by modulating the CCR2/CCR5 chemokine axes. Oncotarget 2016;7:86522–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalbasi A, Komar C, Tooker GM, Liu M, Lee JW, Gladney WL, et al. Tumor-Derived CCL2 Mediates Resistance to Radiotherapy in Pancreatic Ductal Adenocarcinoma. Clin Cancer Res 2017;23:137–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nywening TM, Wang-Gillam A, Sanford DE, Belt BA, Panni RZ, Cusworth BM, et al. Targeting tumour-associated macrophages with CCR2 inhibition in combination with FOLFIRINOX in patients with borderline resectable and locally advanced pancreatic cancer: a single-centre, open-label, dose-finding, non-randomised, phase 1b trial. Lancet Oncol 2016;17:651–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beatty GL, Torigian DA, Chiorean EG, Saboury B, Brothers A, Alavi A, et al. A phase I study of an agonist CD40 monoclonal antibody (CP-870,893) in combination with gemcitabine in patients with advanced pancreatic ductal adenocarcinoma. Clin Cancer Res 2013;19:6286–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrne KT, Vonderheide RH. CD40 Stimulation Obviates Innate Sensors and Drives T Cell Immunity in Cancer. Cell Rep 2016;15:2719–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, et al. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res 2014;74:5057–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chanan-Khan AA, Swaika A, Paulus A, Kumar SK, Mikhael JR, Rajkumar SV, et al. Pomalidomide: the new immunomodulatory agent for the treatment of multiple myeloma. Blood Cancer J 2013;3:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gopalakrishnan R, Matta H, Tolani B, Triche T Jr., Chaudhary PM. Immunomodulatory drugs target IKZF1-IRF4-MYC axis in primary effusion lymphoma in a cereblon-dependent manner and display synergistic cytotoxicity with BRD4 inhibitors. Oncogene 2016;35:1797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirai Y, Saito N, Uwagawa T, Shiba H, Horiuchi T, Iwase R, et al. Pomalidomide promotes chemosensitization of pancreatic cancer by inhibition of NF-kappaB. Oncotarget 2018;9:15292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 2014;343:301–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Li J, Ferguson GD, Mercurio F, Khambatta G, Morrison L, et al. Immunomodulatory drugs reorganize cytoskeleton by modulating Rho GTPases. Blood 2009;114:338–45 [DOI] [PubMed] [Google Scholar]
- 16.Zhu YX, Kortuem KM, Stewart AK. Molecular mechanism of action of immune-modulatory drugs thalidomide, lenalidomide and pomalidomide in multiple myeloma. Leuk Lymphoma 2013;54:683–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry JY, Labarthe MC, Meyer B, Dasgupta P, Dalgleish AG, Galustian C. Enhanced cross-priming of naive CD8+ T cells by dendritic cells treated by the IMiDs(R) immunomodulatory compounds lenalidomide and pomalidomide. Immunology 2013;139:377–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Qiu Y, Personett D, Huang P, Edenfield B, Katz J, et al. Pomalidomide shows significant therapeutic activity against CNS lymphoma with a major impact on the tumor microenvironment in murine models. PLoS One 2013;8:e71754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apte MV, Haber PS, Applegate TL, Norton ID, McCaughan GW, Korsten MA, et al. Periacinar stellate shaped cells in rat pancreas: identification, isolation, and culture. Gut 1998;43:128–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liou GY, Doppler H, Necela B, Krishna M, Crawford HC, Raimondo M, et al. Macrophage-secreted cytokines drive pancreatic acinar-to-ductal metaplasia through NF-kappaB and MMPs. J Cell Biol 2013;202:563–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liou GY, Doppler H, DelGiorno KE, Zhang L, Leitges M, Crawford HC, et al. Mutant KRas-Induced Mitochondrial Oxidative Stress in Acinar Cells Upregulates EGFR Signaling to Drive Formation of Pancreatic Precancerous Lesions. Cell Rep 2016;14:2325–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doppler H, Panayiotou R, Reid EM, Maimo W, Bastea L, Storz P. The PRKD1 promoter is a target of the KRas-NF-kappaB pathway in pancreatic cancer. Sci Rep 2016;6:33758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 2012;14:22–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long KB, Gladney WL, Tooker GM, Graham K, Fraietta JA, Beatty GL. IFNgamma and CCL2 Cooperate to Redirect Tumor-Infiltrating Monocytes to Degrade Fibrosis and Enhance Chemotherapy Efficacy in Pancreatic Carcinoma. Cancer Discov 2016;6:400–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liou GY, Doppler H, Necela B, Edenfield B, Zhang L, Dawson DW, et al. Mutant KRAS-induced expression of ICAM-1 in pancreatic acinar cells causes attraction of macrophages to expedite the formation of precancerous lesions. Cancer Discov 2015;5:52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 2010;11:936–44 [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Girona A, Heintel D, Zhang LH, Mendy D, Gaidarova S, Brady H, et al. Lenalidomide downregulates the cell survival factor, interferon regulatory factor-4, providing a potential mechanistic link for predicting response. Br J Haematol 2011;154:325–36 [DOI] [PubMed] [Google Scholar]
- 28.Torocsik D, Bardos H, Nagy L, Adany R. Identification of factor XIII-A as a marker of alternative macrophage activation. Cell Mol Life Sci 2005;62:2132–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barros MH, Hauck F, Dreyer JH, Kempkes B, Niedobitek G. Macrophage polarisation: an immunohistochemical approach for identifying M1 and M2 macrophages. PLoS One 2013;8:e80908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marhaba R, Nazarenko I, Knofler D, Reich E, Voronov E, Vitacolonna M, et al. Opposing effects of fibrosarcoma cell-derived IL-1 alpha and IL-1 beta on immune response induction. Int J Cancer 2008;123:134–45 [DOI] [PubMed] [Google Scholar]
- 31.Nazarenko I, Marhaba R, Reich E, Voronov E, Vitacolonna M, Hildebrand D, et al. Tumorigenicity of IL-1alpha- and IL-1beta-deficient fibrosarcoma cells. Neoplasia 2008;10:549–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vonderheide RH, Bajor DL, Winograd R, Evans RA, Bayne LJ, Beatty GL. CD40 immunotherapy for pancreatic cancer. Cancer Immunol Immunother 2013;62:949–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 2009;324:1457–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Partecke LI, Gunther C, Hagemann S, Jacobi C, Merkel M, Sendler M, et al. Induction of M2-macrophages by tumour cells and tumour growth promotion by M2-macrophages: a quid pro quo in pancreatic cancer. Pancreatology 2013;13:508–16 [DOI] [PubMed] [Google Scholar]
- 35.Beatty GL, Winograd R, Evans RA, Long KB, Luque SL, Lee JW, et al. Exclusion of T Cells From Pancreatic Carcinomas in Mice Is Regulated by Ly6C(low) F4/80(+) Extratumoral Macrophages. Gastroenterology 2015;149:201–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chistiakov DA, Myasoedova VA, Revin VV, Orekhov AN, Bobryshev YV. The impact of interferon-regulatory factors to macrophage differentiation and polarization into M1 and M2. Immunobiology 2018;223:101–11 [DOI] [PubMed] [Google Scholar]
- 37.Li S, Pal R, Monaghan SA, Schafer P, Ouyang H, Mapara M, et al. IMiD immunomodulatory compounds block C/EBP{beta} translation through eIF4E down-regulation resulting in inhibition of MM. Blood 2011;117:5157–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bjorklund CC, Lu L, Kang J, Hagner PR, Havens CG, Amatangelo M, et al. Rate of CRL4(CRBN) substrate Ikaros and Aiolos degradation underlies differential activity of lenalidomide and pomalidomide in multiple myeloma cells by regulation of c-Myc and IRF4. Blood Cancer J 2015;5:e354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daley D, Mani VR, Mohan N, Akkad N, Pandian G, Savadkar S, et al. NLRP3 signaling drives macrophage-induced adaptive immune suppression in pancreatic carcinoma. J Exp Med 2017;214:1711–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seifert L, Werba G, Tiwari S, Giao Ly NN, Alothman S, Alqunaibit D, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2016;532:245–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 2008;133:1019–31 [DOI] [PubMed] [Google Scholar]
- 42.Klug F, Prakash H, Huber PE, Seibel T, Bender N, Halama N, et al. Low-dose irradiation programs macrophage differentiation to an iNOS(+)/M1 phenotype that orchestrates effective T cell immunotherapy. Cancer Cell 2013;24:589–602 [DOI] [PubMed] [Google Scholar]
- 43.Rios-Tamayo R, Martin-Garcia A, Alarcon-Payer C, Sanchez-Rodriguez D, de la Guardia A, Garcia Collado CG, et al. Pomalidomide in the treatment of multiple myeloma: design, development and place in therapy. Drug Des Devel Ther 2017;11:2399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Infante JR, Jones SF, Bendell JC, Spigel DR, Yardley DA, Weekes CD, et al. A phase I, dose-escalation study of pomalidomide (CC-4047) in combination with gemcitabine in metastatic pancreas cancer. Eur J Cancer 2011;47:199–205 [DOI] [PubMed] [Google Scholar]
- 45.Jiang H, Hegde S, Knolhoff BL, Zhu Y, Herndon JM, Meyer MA, et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat Med 2016;22:851–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.