Abstract
Gut microbiota and their metabolites are instrumental in regulating homeostasis at intestinal and extra-intestinal sites. However, the complex effects of prenatal and early postnatal microbial exposure on adult health and disease outcomes remain incompletely understood. Here, we showed that mice raised under germ-free conditions until weaning and then transferred to specific-pathogen free conditions, harbored altered microbiota composition, augmented inflammatory cytokine and chemokine expression, and were hyper-susceptible to colitis-associated tumorigenesis later in adulthood. Increased number and size of colon tumors and intestinal epithelial cell proliferation in recolonized germ-free mice were associated with augmented intratumoral CXCL1, CXCL2, CXCL5 expression and granulocytic myeloid-derived suppressor cell accumulation. Consistent with these findings, CXCR2 neutralization in recolonized germ-free mice completely reversed the exacerbated susceptibility to colitis-associated tumorigenesis. Collectively, our findings highlight a crucial role for early-life microbial exposure in establishing intestinal homeostasis that restrains colon cancer in adulthood.
Keywords: microbiota, colitis-associated cancer, chemokines, myeloid-derived suppressor cell
Introduction
The gut microbiota consists of trillions of bacteria that co-develop with the host and foster physiological and immunological homeostasis (1–3). Microbiota-induced immune responses are particularly important early in life (4,5) when the maternal microbiota can influence prenatal and early post-natal host development (6). Exposure to microbiota and associated metabolites during this time may have long-lasting effects on immune and non-immune homeostasis of the gut and other organs, and ultimately disease outcomes. Because colitis-associated cancer (CAC) is influenced by the microbiota and inflammatory signaling pathways (7), we explored the contribution of early-life microbiota exposure to the development of CAC later in life.
Here, we show that mice raised under germ-free conditions until weaning and then conventionalized with complex gut microbiota into adulthood (exGF mice) were hyper-susceptible to CAC when compared to specific pathogen-free (SPF) mice. The increased number and size of colon tumors and intestinal epithelial cell (IEC) proliferation in exGF mice were associated with augmented intratumoral chemokine expression and granulocytic myeloid-derived suppressor cell (G-MDSC) accumulation. Accordingly, CXCR2 neutralization in exGF mice reversed the hyper-susceptibility to CAC. Collectively, these findings highlight a crucial role for early-life microbiota in establishing gut homeostasis that restrains colon cancer in adulthood.
Methods
Mice
C57BL/6 mice were obtained from the Jackson Laboratories and housed in SPF conditions (SPF mice). GF C57BL/6 mice were kept under GF conditions in Park Bioservices isolators. Conventionalization of GF mice was performed after weaning at the age of 3 weeks (21 days) by transferring the GF mice out of GF isolators and co-housing them with SPF mice for 3 weeks (exGF mice). Animal protocols were approved by the IACUC of Georgia State University.
Colitis-associated cancer model
CAC was induced as previously described (8). Briefly, as shown in Fig. 1A, SPF and exGF mice were intraperitoneally injected with AOM (10 mg/kg body weight) (Sigma) diluted in PBS. Mice were then subjected to two cycles of dextran sodium sulfate (DSS) treatment (MP Biomedicals), in which each cycle consisted of 2.5% DSS for 7 days followed by 14 days with regular water. After the colitis-associated cancer protocol was complete (day 122 of life; day 80 following initiation of the AOM/DSS regimen), mice were euthanized, and colon length, colon weight, and spleen weights were measured. At day 98 and 122 of life, mice were fasted for 15 hours, and the colonoscopy procedure was performed under general anesthesia with isoflurane (Karl Storz Endoskope, El Segundo, CA).
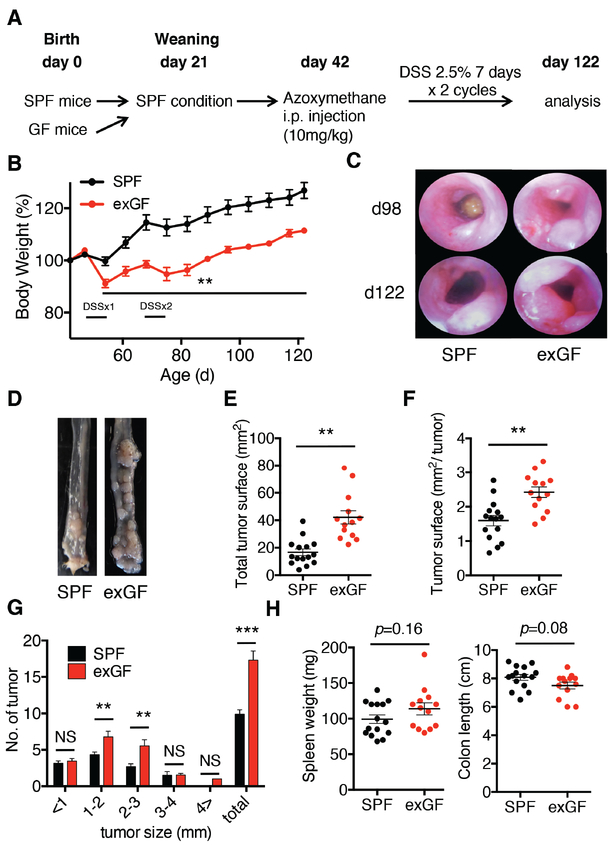
Figure 1. Absence of microbiota prior to weaning exacerbates CAC in adult mice.
A, Experimental flow chart. SPF mice (n=15; 8 male and 7 female) and exGF mice (n=13; 7 male and 6 female) were injected intraperitoneally with AOM (10mg/kg body weight) at day 42 of life followed by two 7-day cycles of 2.5% DSS in the drinking water. Mice were euthanized at day 122 of life. B, Body weight of SPF and exGF mice treated with AOM/DSS. C. (DSSx1; 1st cycle of DSS; DSSx2 2nd cycle of DSS) Representative colonoscopy from each group at day 98 and 122. D, Representative images of the distal colon at day 122. E-F, Total number of tumors and tumor surface areas in the colon at day 122. G, Tumor number and size were measured under a dissecting microscope. H, Spleen weight and colon length from each group. Student’s t test was used to determine significance. Data are the means ± SEM. **P < 0.01, ***P < 0.001; NS, not significant.
CXCR2 neutralization in vivo
Mice were injected with anti-CXCR2 (polyclonal goat anti-mouse; a gift from Dr. Lukacs) or goat IgG (isotype control; Bio X Cell) via the intraperitoneal route (500 μg twice a week) beginning 4 days after the AOM injection. Antibodies against CXCR2 were generated by immunization of a goat with murine CXCR2 peptide in multiple intradermal sites and purified over a protein A column as previously described (9).
Isolation of colonic lamina propria (LP) cells and flow cytometry
Isolation of colonic LP cells was performed as previously described (10). Briefly, colonic tissue was cut into pieces 0.5 cm in length and placed in an orbital shaker for 20 minutes at 37°C in Hank’s Balanced Salt Solution (HBSS) with 10% fetal bovine serum (FBS) and 2 mM EDTA in order to remove epithelial cells. This shaking step was repeated and then the remaining tissue was minced and placed in RPMI with 10% FBS, collagenase IV (1 mg/mL; Sigma), and DNase I (40 U/mL; Roche) and shaken for 15 minutes at 37°C. The contents are then filtered through a 100 μm cell strainer directly into a 50-mL conical tube. Each 50-mL conical tube was topped off with HBSS with FBS and centrifuged at 4 °C twice. The cell pellet was resuspended into a cell suspension, which was stained with specific antibodies and analyzed by flow cytometry. Fluorescence-labeled antibodies were from Thermo Fisher Scientific: CD11b (M1/70), CD11c (N418), CD127 (A7R43), F4/80 (BM8), CD90.2 (53–2.1), Ly6C (HK1.4), Ly6G (1A8), Ki67 (16A8); from Becton Dickinson (BD): MHC-II (M5/114.15.2), CD3 (145–2C11), CD4 (L3T4), CD8α (5H10), CD45 (30F11), E-cadherin (DECMA-1); Biolegend: NK1.1 (PK136), TCRβ (H57–597). Fc block (2.4G2) was purchased from BD. LIVE/DEAD fixable aqua dead cell stain (Thermo Scientific) was used as a viability dye.
16S rRNA and mRNA sequencing
Ilumina 16S ribosomal (r)RNA gene sequencing was performed on feces isolated from SPF and exGF mice at days 42, 98, and 122, as previously described (11). Briefly, quality filtering was performed using the Quantitative Insights Into Microbial Ecology (QIIME; http://qiime.org) software package (12), and sequences were aligned to operational taxonomic units (OTUs) using the UCLUST algorithm (13) and classified taxonomically using the Greengenes reference database 13_8 (14). A single representative sequence for each OTU was aligned, and a phylogenetic tree was built using FastTree (15). The phylogenetic tree was used for computing the unweighted UniFrac distances between samples, rarefaction was performed, and used to compare abundances of OTUs across samples. Rarefaction was performed at 12962 sequences per sample, for which a total of 5650 OTUs were observed (818 +/− 203 per sample). Principal coordinates analysis (PCoA) plots were generated through QIIME and used to assess the variation between experimental groups (beta diversity). The PCoA plot representations are 3D and the dot sizes are dependent on the relative localization. Alpha diversity was analyzed using the Shannon index. LEfSe (LDA Effect Size) was used to investigate bacterial members that drive differences between groups, with a significance level of 2 used as a threshold (16). The mRNA-seq experiments were performed by Novogene (Beijing, China). Volcano plots were generated using R, and heatmaps were generated using Morpheus (https://software.broadinstitute.org/morpheus/). Pie chart representation of pathways was performed using Panther Classification System (http://pantherdb.org) to gain insight into specific signaling pathways. The accession numbers for the unprocessed transcriptomic sequencing data and the unprocessed 16S sequencing data reported in this paper are in the European Nucleotide Archive: PRJEB30607 and PRJEB30606, respectively.
Histology
Mouse distal colons with tumors were fixed in 10%-buffered formalin for 24 hours at room temperature and subsequently embedded in paraffin. Sectioning, hematoxylin/eosin staining, slide scanning, and assessment of histology were subsequently performed at the Institute for Biomedical Sciences histology core at Georgia State University by using a BX61 microscope (OLYMPUS).
TUNEL assay
To quantitate the number of apoptotic cells in colonic epithelial cells, mouse proximal colon devoid of tumor were fixed in 10%-buffered formalin for 24 hours at room temperature, embedded in paraffin, sectioned at 5-μm thickness, deparaffinized, and stained for apoptotic nuclei according to the manufacturer’s instructions using the In-Situ Cell Death Detection Kit (Sigma Aldrich, St. Louis, MO). TUNEL-positive cells overlapping with DAPI nuclear staining were counted per crypt by using a BZ-X710 microscope (Keyence).
RNA isolation and qPCR
Total RNA was isolated from murine colonic tissues using the Qiagen RNeasy Mini Kit and QIAcube with on-column DNase digestion. The RNA concentration was measured using NanoDrop 8000 (Thermo Scientific). cDNA was generated from 1μg of RNA using the Superscript First-Strand Synthesis System for RT-PCR and random hexamer primers (Invitrogen). qPCR was performed with SYBR Green on a StepOnePlus real-time PCR system (Applied Biosystems). Gene expression was normalized to Gapdh and analyzed using 2−ΔΔCT method with duplicates.
Primers used were:
cxcl1 (F: GCTTGAAGGTGTTGCCCTCAG; R: AAGCCTCGCGACCATTCTTG)
cxcl2 (F: CGCTGTCAATGCCTGAAG; R: GGCGTCACACTCAAGCTCT)
cxcl5 (F: GGTCCACAGTGCCCTACG; R: GCGAGTGCATTCCGCTTA)
gapdh (F: TGGCAAAGTGGAGATTGTTGCC; R: AAGATGGTGATGGGCTTCCCG)
Human microarray analysis
Microarray datasets (GSE37283; (17)) were downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). Detailed patient demographic information is available in Table 1 of (17). Microarray analysis was performed as previously described (18). Microarray assay were performed on 20 RNA samples isolated from colon mucosa of 20 patients, including 5 normal controls, 4 quiescent UC, and 11 UC with neoplasia. Inclusion criteria is as follows: a previous clinical diagnosis of UC confirmed by an expert GI pathologist, a disease duration > 7 years, and an extent of disease >20 cm proximal to the anal verge. For microarray analysis, expression and raw expression data were summarized and normalized using the Robust Multi-array Average algorithm from the Bioconductor library for the R statistical programming system. All samples were clustered using Ward’s method in Spotfire (TIBCO Software, Palo Alto, CA).
Statistics
Statistical analyses were performed with GraphPad Prism software, version 6.0b (Graphpad Software). Student’s t test or one-way ANOVA and Tukey’s Multiple Comparison Test was used to determine significance. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant.
Results
Absence of microbiota prior to weaning exacerbates colitis-associated cancer in adult mice
To investigate the contribution of early-life microbiota exposure to CAC, we compared SPF mice to exGF mice (born and raised in GF conditions until weaning at day 21 when they were co-housed with age- and sex-matched SPF mice for an additional 21 days). Both groups of mice were subsequently administered AOM at day 42 of life followed by two 7-day cycles of DSS to induce CAC (Fig. 1A). We observed that exGF mice lost significantly more body weight compared to SPF mice, beginning during the first cycle of DSS (Fig. 1B). ExGF mice also developed significantly larger colonic tumors, as assessed by endoscopy at day 98 and 122 (Fig. 1C) and macroscopic examination (Fig. 1D). Similar differences between exGF and SPF mice were observed for total tumor surface area (Fig. 1E), tumor surface area per individual tumor at day 122 (Fig. 1F), and in the total number of colonic tumors, specifically in the 1–2mm and 2–3mm size range (Fig. 1G). We did not observe significant differences in spleen weights and colon lengths between the groups (Fig. 1H). These data demonstrated that the lack of microbiota before weaning favors CAC development in adulthood.
Enhanced colonic tumor size and epithelial turnover in AOM/DSS-treated exGF mice
Histological examination confirmed the presence of larger adenomas particularly in the middle to distal colonic region and rectum in AOM/DSS-treated exGF mice compared to SPF controls (Supplementary Fig. S1A-B). Given this increased tumor burden observed in the exGF group, we investigated whether colonic epithelial cell death and proliferation were altered in these mice. We observed a significantly increased number of TUNEL+ cells in tissue sections (Supplementary Fig. S1C-D), and increased Ki67 staining of isolated intestinal epithelial cells (IECs) (Supplementary Fig. S1E-F) among exGF mice compared to SPF controls. These results suggest that increased IEC turnover in exGF mice may enhance CAC.
ExGF mice exhibit altered microbiota composition and colonic gene expression
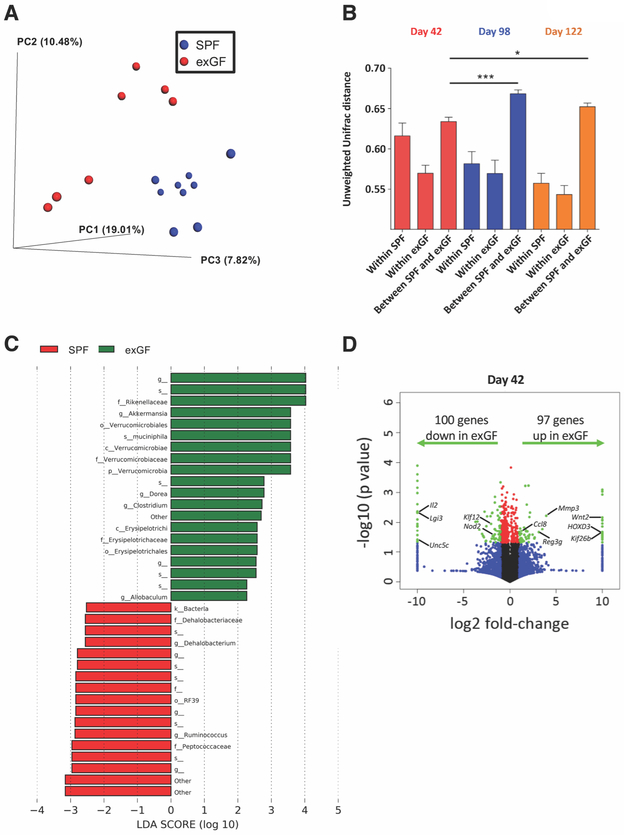
We next analyzed the microbiota and host gene expression that may be associated with and/or contribute to the enhanced colonic tumor burden in exGF mice. To analyze microbiota composition, 16S rRNA gene sequencing was performed on feces isolated from SPF and exGF mice at days 42, 98, and 122. Principal coordinate analysis (PCoA) of the unweighted Unifrac distance revealed separate clustering of SPF (blue dots) and exGF samples (red dots) at day 42 preceding initiation of AOM/DSS (Fig. 2A). We observed distinct clustering between SPF and exGF mice (blue and red dots, respectively; Supplementary Fig. S2A) at day 42 (red dots; Supplementary Fig. S2B), with this microbiota clustering becoming more distinct at day 98 and day 122 (blue and orange dots, respectively; Supplementary Fig. S2B). We observed two subgroups for both SPF and exGF animals at day 42 (Fig. 2A), which could be explained by the samples originating from two independent experiments. Shannon index analysis demonstrated that the alpha diversity of exGF mice was not significantly altered compared to SPF mice (Supplementary Fig. S2C). We next analyzed the unweighted Unifrac distance separating these groups in order to quantitate the differences, which confirmed that alterations in the microbiota composition between SPF and exGF mice was present at day 42 and significantly exacerbated at day 98 and 122 (Fig. 2B). Microbiota alteration in exGF mice at day 42 was characterized by increases in members of the families Verrucomicrobiaceae, Rikenellaceae, and Erysipelotrichaceae and reductions in Dehalobacteriaceae, Peptococcaceae, and Ruminococcaceae (Fig. 2C, Supplementary Fig. S2D-E).
Figure 2. ExGF mice exhibit altered microbiota composition and intestinal gene expression.
A, Microbiota composition was analyzed by 16S rRNA sequencing at day 42. PCoA of the unweighted Unifrac distance, with samples colored by group (SPF: blue, n=9 and exGF: red, n=7). ANOSIM P < 0.01. B, Unweighted Unifrac distance within and between SPF and exGF groups at day 42 (red), day 98 (blue), and day 122 (orange). Data are means ± SEM. *P < 0.05, ***P < 0.01. C, LEfSe analysis investigating the bacterial taxa significantly increased (green) or decreased (red) in exGF mice compared to SPF mice. D, RNA-seq data were visualized on a volcano plot comparing gene expression in SPF and exGF mice. For each gene, the difference in abundance between the two groups is indicated in log2 fold-change on the x-axis, with positive values corresponding to increased expression in exGF mice compared to SPF mice and negative values corresponding to decreased expression in exGF mice compared to SPF mice. Significance between the two groups is indicated by –log10 P-value on the y-axis. Red dots correspond to genes with a P-value <0.05 between exGF and SPF mice. Blue dots correspond to genes with at least a 2-fold decreased or increased expression in exGF and SPF mice. Green dots correspond to genes with at least a 2-fold decreased or increased abundance in exGF and SPF mice and a P-value <0.05. One-way ANOVA and Bonferroni post-hoc test was used to determine significance. Data are from (C,D) one or (A,B) two independent experiments.
We next performed RNA-seq analysis of total colon tissue at days 7, 14, 21, and 42 of life, all prior to the initiation of AOM-DSS. Volcano plots indicated that the number of genes significantly upregulated or downregulated in exGF mice by at least 2-fold at day 14 (96 genes up in exGF; 253 genes down in exGF) and day 21 (375 genes up in exGF; 216 genes down in exGF) were higher than at day 7 (96 genes up in exGF; 82 genes down in exGF) and day 42 (97 genes up in exGF; 100 genes down in exGF) (Fig. 2D, Supplementary Fig. S3A). These data are consistent with the expansion of microbiota in SPF mice, but not GF mice, around the time of transitioning away from breast milk to eating solid food (~day 14) and the time of weaning (day 21). At day 42, just prior to the initiation of AOM/DSS, analysis of specific genes that were significantly upregulated in exGF mice revealed higher expression of the monocyte chemokine Ccl8, the antimicrobial peptide Reg3g, and several pro-proliferative genes (Mmp3, Wnt2, HOXD3, Kif26b). Alternatively, lower expression of anti-inflammatory genes (Il2, Unc5c) and tumor suppressor genes (Nod2, Lgi3, Klf12) were observed in exGF mice (Fig. 2D).
We further performed heatmap analyses of SPF and exGF colon tissue samples from day 42 and found that 551 genes were differentially expressed (Supplementary Fig. S3B). Pie chart representation of pathways was used to gain insight into specific signaling pathways that may have been altered by early-life microbiota by analyzing genes with a more than 2-fold increase expression in exGF mice compared to SPF mice. This approach identified the ‘inflammation-mediated by chemokine and cytokine signaling pathway’ as one of the most enriched pathways (Supplementary Fig. S3C). Collectively, these data indicated a steady-state shift towards a pro-proliferative, pro-inflammatory milieu in the colons of exGF mice, which is associated with distinct microbiota composition and enhanced CAC.
Enhanced CXCR2 ligands and granulocytic MDSCs in AOM/DSS-treated exGF mice
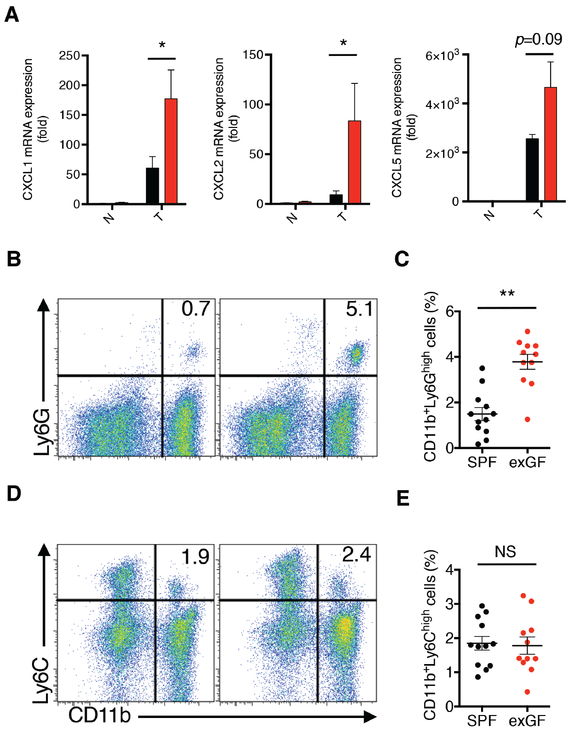
In light of enhanced pro-inflammatory gene expression and the alteration in the expression of cytokine/chemokine signaling pathways in exGF mice at day 42, we next explored how specific chemokine signaling may contribute to increased CAC. We focused on the CXCL2/CXCR2 axis because CXCR2-dependent recruitment of myeloid-derived suppressor cells (MDSCs) has been implicated in CAC (19). Using qRT-PCR we observed higher CXCL1 and CXCL2 mRNA expression in tumor tissues (T), but not non-tumor tissue (N), obtained from exGF mice compared to SPF mice, and CXCL5 mRNA followed a similar trend (Fig. 3A). To examine whether increased CXCR2 ligand expression was associated with enhanced MDSCs, we next analyzed granulocytic MDSCs (G-MDSCs; CD11b+Ly6Ghigh) and monocytic MDSCs (M-MDSCs; CD11b+Ly6C+) (20) in colons from AOM/DSS-treated exGF and SPF mice. Following treatment with AOM/DSS, FACS analyses of colonic tissues revealed that exGF mice displayed significantly increased accumulation of G-MDSCs, but not M-MDSCs, compared to SPF mice (Fig. 3B-E).
Figure 3. CXCR2 ligands and G-MDSCs are enhanced in colons of AOM/DSS-treated exGF mice.
A, The mRNA expression of CXCL1, CXCL2, and CXCL5 in colonic tumor (T) and matched non-tumor (N) mucosa were determined by qPCR at day 122. Black and red bars represent the samples from SPF mice and exGF mice, respectively. B-E, Infiltration of MDSC subsets into the colons in exGF mice in response to AOM/DSS. B,D, Lamina propria cells were isolated from colonic tissue and the frequencies of MDSCs in SPF (n=12; 6 males and 6 females) and exGF mice (n=11; 6 males and 5 females) were analyzed using flow cytometry by staining for Ly6G, Ly6C, CD11b. C,E, Each dot represents the frequency of the indicated MDSC subpopulation in the colonic mucosa for an individual mouse. One-way ANOVA and Tukey’s Multiple Comparison Test was used to determine significance. Data are from two independent experiments showing the means ± SEM. *P < 0.05, **P < 0.01; NS, not significant.
We next investigated whether CXCR2 ligands were also enhanced during CAC in humans using datasets for colonic tissues obtained from healthy controls (HC) and ulcerative colitis (UC) patients with or without remote colonic neoplasia. In UC patients, the expression of the CXCR2, as well as the CXCR2 ligands CXCL1, CXCL2, CXCL3, CXCL5, and CXCL8, were all elevated in colonic tissue isolated from patients harboring remote colonic neoplasia compared those without remote neoplasia or HC (Supplementary Fig. S4). Taken together, these data suggest that a lack of microbiota in early-life may enhance expression of CXCR2 ligands accumulation of G-MDSCs in colonic tissue.
CXCR2 blockade inhibits colonic tumorigenesis in AOM/DSS-treated exGF mice
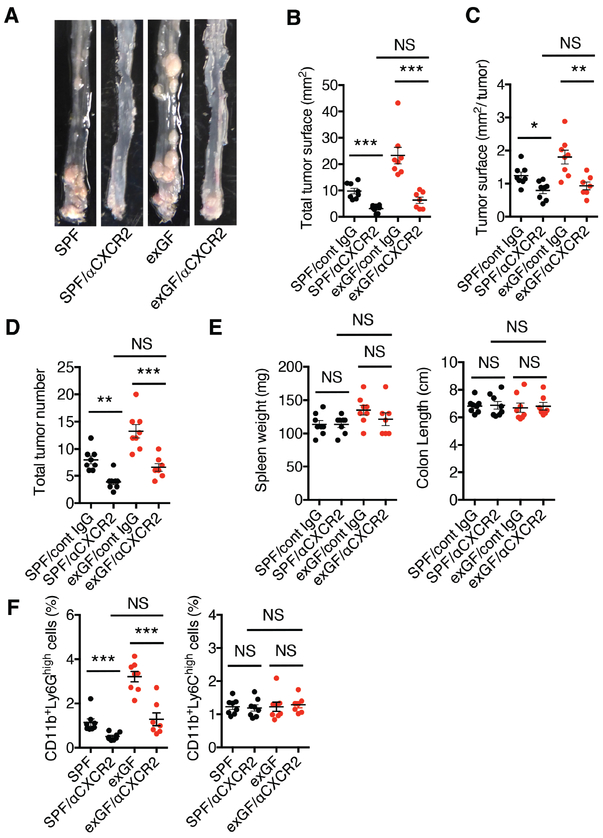
We further explored whether in vivo neutralization of CXCR2 could limit the enhanced recruitment of MDSCs, as well as CAC observed in these mice. Indeed, anti-CXCR2 treatment inhibited the development of AOM/DSS-induced macroscopic colonic tumors in exGF mice to a similar extent as that observed in anti-CXCR2-treated SPF mice, when compared to either group treated with control IgG (Fig. 4A). Similar effects of anti-CXCR2 treatment were observed with regards to inhibiting total tumor surface area (Fig. 4B), tumor surface area per individual tumor at day 122 (Fig. 4C), and the total number of colonic tumors (Fig. 4D), whereas no significant differences in spleen weights and colon lengths between the two groups were seen (Fig. 4E). We confirmed that injection of anti-CXCR2 into AOM/DSS-treated SPF and exGF mice prevented G-MDSC accumulation in colonic tissue (Fig. 4F).
Figure 4. Blockade of CXCR2 ameliorates carcinogenesis in exGF mice.
A, Gross view of colonic tumors in AOM/DSS-treated SPF (n=16; 8 males and 8 females) and exGF mice (n=15; 8 males and 7 females) injected with control antibody (IgG) or a CXCR2 neutralizing antibody (500 μg twice a week; SPF/cont IgG, n=8; SPF/aCXCR2, n=8; exGF/cont IgG, n-8; exGF/aCXCR2, n=7). B, Total tumor surface area and C, tumor surface area per tumor in the colon at day 80 after initiation of the AOM/DSS regimen (day 122 of life). D, total tumor number was quantified using a dissecting microscope. E, Spleen weight and colon length from each experimental group were determined. F, Each dot represents the frequency of the indicated MDSC subpopulation (G-MDSC: CD11b+Ly6Ghigh; M-MDSC: CD11b+Ly6Chigh) in the colonic mucosa from individual mice in the indicated groups. One-way ANOVA and Tukey’s Multiple Comparison Test was used to determine significance. Data are from one experiment showing the means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001; NS, not significant.
Discussion
A previous report demonstrated that AOM/DSS-treated GF mice develop more CAC compared to SPF mice and linked this phenotype to impaired inflammatory responses and delayed repair processes (21). Our complementary findings show that exGF mice had an altered microbiota, enhanced chemokine expression and G-MDSCs in the colon, and were hyper-susceptible to CAC. These data indicate that early-life exposure to microbiota induced enduring microbial and host changes that predisposed mice to CAC.
The microbiota differences in exGF mice were characterized by increases in Verrucomicrobiaceae, Rikenellaceae, and Erysipelotrichaceae. These results are consistent with reports that Akkermansia, Bacteroidales, and Erysipelotrichaceae correlate with increased CAC (22,23). Both Bacteroides and Akkermansia are mucin degraders, and an overabundance of these bacteria may impair mucosal barrier defenses leading to increased inflammation and tumorigenesis in exGF mice. Alternatively, Dehalobacteriaceae, Peptococcaceae, and Ruminococcaceae, which have been linked to protection from CAC (24–26), were significantly reduced in exGF mice. These findings provide insight into how the altered microbiota in exGF mice may lead to enhanced CAC.
Collectively, our results suggest that CAC susceptibility in adulthood may be imprinted in the prenatal and/or early postnatal period by maternal-derived microbiota (27). Additional insights into how early-life exposure to microbiota regulates intestinal homeostasis, inflammation, and tumorigenesis may lead to opportunities for targeted interventions (28,29).
Supplementary Material
Acknowledgements
This work was supported by: a National Institutes of Health grant DK097256 (to T.L.D.); a Crohn’s and Colitis Foundation Research Fellowship Award (to A.H.) and a Career Development Award (to E.V.); Japanese Society for the Promotion of Science KAKENHI Grants 17H07013 and 18K15128 (to A.H.); and a Senior Research Career Scientist Award BX00447601 from the Veterans Health Administration (to D.M.).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interests were disclosed by any of the authors.
References
- 1.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 2008;6(10):776–88 doi 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol 2013;14(7):668–75 doi 10.1038/ni.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature 2016;535(7610):75–84 doi 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 4.Hasegawa M, Osaka T, Tawaratsumida K, Yamazaki T, Tada H, Chen GY, et al. Transitions in oral and intestinal microflora composition and innate immune receptor-dependent stimulation during mouse development. Infect Immun 2010;78(2):639–50 doi 10.1128/IAI.01043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol 2007;5(7):e177 doi 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science 2016;351(6279):1296–302 doi 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 7.Gagliani N, Hu B, Huber S, Elinav E, Flavell RA. The fire within: microbes inflame tumors. Cell 2014;157(4):776–83 doi 10.1016/j.cell.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Viennois E, Merlin D, Gewirtz AT, Chassaing B. Dietary Emulsifier-Induced Low-Grade Inflammation Promotes Colon Carcinogenesis. Cancer Res 2017;77(1):27–40 doi 10.1158/0008-5472.CAN-16-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehrad B, Strieter RM, Moore TA, Tsai WC, Lira SA, Standiford TJ. CXC chemokine receptor-2 ligands are necessary components of neutrophil-mediated host defense in invasive pulmonary aspergillosis. J Immunol 1999;163(11):6086–94. [PubMed] [Google Scholar]
- 10.Harusato A, Flannigan KL, Geem D, Denning TL. Phenotypic and functional profiling of mouse intestinal antigen presenting cells. J Immunol Methods 2015;421:20–6 doi 10.1016/j.jim.2015.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015;519(7541):92–6 doi 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7(5):335–6 doi 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26(19):2460–1 doi 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 14.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6(3):610–8 doi 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 2009;26(7):1641–50 doi 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12(6):R60 doi 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen CC, Pekow J, Llado V, Kanneganti M, Lau CW, Mizoguchi A, et al. Chitinase 3-like-1 expression in colonic epithelial cells as a potentially novel marker for colitis-associated neoplasia. Am J Pathol 2011;179(3):1494–503 doi 10.1016/j.ajpath.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harusato A, Abo H, Ngo VL, Yi SW, Mitsutake K, Osuka S, et al. IL-36gamma signaling controls the induced regulatory T cell-Th9 cell balance via NFkappaB activation and STAT transcription factors. Mucosal Immunol 2017;10(6):1455–67 doi 10.1038/mi.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh H, Wang D, Daikoku T, Sun H, Dey SK, Dubois RN. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell 2013;24(5):631–44 doi 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol 2018;19(2):108–19 doi 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhan Y, Chen PJ, Sadler WD, Wang F, Poe S, Nunez G, et al. Gut microbiota protects against gastrointestinal tumorigenesis caused by epithelial injury. Cancer Res 2013;73(24):7199–210 doi 10.1158/0008-5472.CAN-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baxter NT, Zackular JP, Chen GY, Schloss PD. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome 2014;2:20 doi 10.1186/2049-2618-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Wilson JE, Koenigsknecht MJ, Chou WC, Montgomery SA, Truax AD, et al. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat Immunol 2017;18(5):541–51 doi 10.1038/ni.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richard ML, Liguori G, Lamas B, Brandi G, da Costa G, Hoffmann TW, et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes 2018;9(2):131–42 doi 10.1080/19490976.2017.1379637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez J, Moreno FJ, Olano A, Clemente A, Villar CJ, Lombo F. A Galacto-Oligosaccharides Preparation Derived From Lactulose Protects Against Colorectal Cancer Development in an Animal Model. Front Microbiol 2018;9:2004 doi 10.3389/fmicb.2018.02004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alteber Z, Sharbi-Yunger A, Pevsner-Fischer M, Blat D, Roitman L, Tzehoval E, et al. The anti-inflammatory IFITM genes ameliorate colitis and partially protect from tumorigenesis by changing immunity and microbiota. Immunol Cell Biol 2018;96(3):284–97 doi 10.1111/imcb.12000. [DOI] [PubMed] [Google Scholar]
- 27.Macpherson AJ, de Aguero MG, Ganal-Vonarburg SC. How nutrition and the maternal microbiota shape the neonatal immune system. Nat Rev Immunol 2017;17(8):508–17 doi 10.1038/nri.2017.58. [DOI] [PubMed] [Google Scholar]
- 28.Torow N, Hornef MW. The Neonatal Window of Opportunity: Setting the Stage for Life-Long Host-Microbial Interaction and Immune Homeostasis. J Immunol 2017;198(2):557–63 doi 10.4049/jimmunol.1601253. [DOI] [PubMed] [Google Scholar]
- 29.Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med 2016;22(7):713–22 doi 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.