Abstract
Although c-MYC and mTOR are frequently activated proteins in prostate cancer, any interaction between the two is largely untested. Here we characterize the functional crosstalk between FOXP3-c-MYC and TSC1-mTOR signaling during tumor progression. Deletion of Tsc1 in mouse embryonic fibroblasts (MEF) decreased phosphorylation of c-MYC at threonine 58 (pT58) and increased phosphorylation at serine 62 (pS62), an observation validated in prostate cancer cells. Conversely, inhibition of mTOR increased pT58 but decreased pS62. Loss of both Foxp3 and Tsc1 in prostate cancer cells synergistically enhanced c-MYC expression via regulation of c-Myc transcription and protein phosphorylation. This crosstalk between FOXP3 and TSC1 appeared to be mediated by both the mTOR-4EBP1-c-MYC and FOXP3-c-MYC pathways. In mice, Tsc1 and Foxp3 double-deletions in the prostate led to prostate carcinomas at an early age; this did not occur in these mice with an added c-Myc deletion. In addition, we observed synergistic anti-tumor effects of co-treating mice with inhibitors of mTOR and c-MYC in prostate cancer cells as well as in Foxp3 and Tsc1 double-mutant mice. In human prostate cancer, loss of nuclear FOXP3 is often accompanied by low expression of TSC1. Since loss of Foxp3 transcriptionally induces c-Myc expression and loss of Tsc1 activates mTOR signaling, these data suggest crosstalk between FOXP3-c-MYC and TSC1-mTOR signaling that converges on c-MYC to regulate tumor progression. Co-administration of c-Myc and mTOR inhibitors may overcome the resistance to mTOR inhibition so commonly observed prostate cancer cells.
Keywords: FOXP3, TSC1, prostate cancer, tumor progression, carcinogenesis
Introduction
FOXP3, localized on the X chromosome at Xp11.23, is a member of the forkhead-box/winged-helix transcription factor family. This gene functions as a master regulator in the development and function of regulatory T cells (Tregs). FOXP3 is also expressed in epithelial tissues of the breast, lung, and prostate (1). However, nuclear FOXP3 is lost in approximately 70% of human prostate cancers (2), which may be caused by epigenetic mechanisms. Of note, inactivation of FOXP3 contributes to the overexpression of c-MYC in human prostate cancer samples (2,3), and ectopic expression of wild-type (WT) FOXP3 induces growth inhibition and apoptosis of prostate cancer cells through downregulation of c-MYC (2,4), suggesting that FOXP3-mediated transcriptional repression of c-MYC is necessary to control c-MYC levels in prostate epithelial cells. Likewise, FOXP3 re-programs Treg cell metabolism by suppressing c-MYC expression, enhancing oxidative phosphorylation, and increasing nicotinamide adenine dinucleotide oxidation (5). Furthermore, lineage-specific ablation of Foxp3 in mouse prostate epithelial cells leads to mouse prostatic intraepithelial neoplasia (mPIN), as well as to increases in c-Myc mRNA and protein expression, indicating that loss of FOXP3 function is an early event in prostate carcinogenesis (2).
In 30–50% of prostate cancers, the PI3K/AKT/mTOR signaling pathway is upregulated, often through loss of PTEN suppressor function (6). In aggressive and metastatic prostate cancer, the most frequently altered genes are PIK3CA (4% mutation and 15–20% amplification) and PTEN (4% mutation and 30–39% deletion) (6). In prostate cancer cells, these mutated or deleted genes lead to constitutive activation of PI3K/AKT/mTOR signaling. Mice heterozygous for Pten deletion develop mPIN with 100% incidence, and homozygous deletion of Pten in the prostate induces invasive prostate cancer (7). The TSC1/2 complex is an essential component of the PI3K/AKT/mTOR signaling pathway. Either phosphorylation of the TSC1/2 complex by AKT or loss of TSC1/2 facilitates mTOR activation. Tsc1-deficient mice develop mPIN by 27 weeks of age, with increasing disease penetrance over time, and, in old mutant mice, mPIN progresses to prostate carcinomas (8). Thus, release of TSC1-dependent mTOR inhibition is sufficient to initiate prostate tumor progression. In addition, conditional Tsc1 deletion in Tregs impairs the suppressive activity and expression of Foxp3 and, under inflammatory conditions, results in increased IL-17 production (9,10). In angiosarcomas, Tsc1 deletion enhances mTOR complex 1 (mTORC1) activation through increased expressions of c-Myc and Hif1α (11). These data suggest potential functional correlations in the cells between Tsc1, Foxp3, and c-Myc expression.
Nuclear protein expression of c-MYC, present in 97% of human prostate cancers, positively correlates with the proliferation rate and negatively with apoptotic count (12). In prostate cancer, activation of c-MYC cooperates with PI3K/AKT/mTOR signaling (13-16), but the underlying molecular mechanisms remain unknown. Reductions in c-MYC increase Tsc2 expression, which further represses c-MYC expression (17,18), suggesting a feed-forward loop between c-MYC and the TSC1/2 complex. MYC binding to 4EBP1 induces translation (19), but eIF4E (a component of the eIF4F translation initiation complex) activity increases expression of c-MYC (20), suggesting a reciprocal induction of c-MYC and the mTOR-downstream 4EBP1. In addition, there is co-occurrence of c-MYC amplification and a PI3K/mTOR signaling alteration in human prostate cancers, raising the possibility that these two genetic hits cooperate to promote tumor progression. Mouse models show that this cooperation of c-MYC and PI3K/mTOR signaling pathways promotes progression of mPIN to invasive cancer and metastasis (14,16). Since Foxp3 deficiency leads the development of mPIN through transcriptional upregulation of c-Myc (2), and Tsc1 deficiency in aging mice promotes progression of mPIN to prostate carcinoma through constitutive mTOR activation (8), there may be a functional interaction between FOXP3-c-MYC and TSC1/2-mTOR axes during prostate cancer progression. Given the essential role of c-MYC in prostate cancer progression, we conducted the present study to determine if c-MYC is synergistically regulated by FOXP3 and TSC1 and to elucidate the mechanism for this regulation in prostate cancer cells, with the goal of developing a new therapeutic approach for prostate cancers with FOXP3 and TSC1 defects.
Methods and Materials
Cell lines, antibodies, and reagents
Prostate cancer cell lines PC3, DU145, and LNCaP were obtained from the American Type Culture Collection (Manassas, VA). Tsc2+/+ and Tsc1−/− mouse embryonic fibroblasts (MEFs) were generously provided by Dr. David Kwiatkowski (Harvard Medical School). Cells were cultured for less than 6 months, authenticated by examination of morphology and growth characteristics, and confirmed to be mycoplasma-free. A short tandem-repeat analysis for DNA fingerprinting was also used to verify the cell lines. The specific primary antibodies for Western blots and immunohistochemistry (IHC) are shown in Supplementary Table S1. TSC1, mTOR, and 4EBP1 siRNAs (IDT, Skokie, IL) were transfected into MEFs or prostate cancer cells. Torin1 (Cayman Chemical, Ann Arbor, MI), rapamycin (R8781, Sigma, St. Louis, MO), 10058-F4 (F3680, Sigma), and cycloheximide (CHX, C7698, Sigma) were used for treatment of cells and mice.
Human tissue specimens
For the formalin-fixed, paraffin-embedded (FFPE) tumor specimens, available information included patient age, race, prostate-specific antigen level, and pathologic stage. For gene copy number analysis, ten nuclear FOXP3+ and ten nuclear FOXP3− prostate tumors were Gleason sum-matched. These samples were collected from 20 patients with prostate carcinoma who underwent primary surgery between January 2012 and June 2017 at the University of Alabama at Birmingham (UAB) as presented in Supplementary Table S2. In addition, we used 151 FFPE tissue specimens of primary prostate cancer for IHC staining (Supplementary Table S3). All patients had histologically confirmed prostate carcinoma. The pathological staging was reassessed with the tumor-node-metastasis (TNM) staging system. Pathologic grading was based on specimens corresponding to Gleason scores of 2–6, 7, and 8–10, respectively. Written informed consent was obtained from all subjects in accordance with the requirements of the Institutional Review Board of UAB.
Laser capture microdissection of tumor tissues
Laser capture microdissection of tumor tissues was performed as described previously (2).
Quantification of gene copy numbers
Quantitative analysis of copy numbers was conducted by QIAGEN qBiomarker Copy Number PCR assays (QIAGEN, Germantown, MD) as described previously (21).
Quantitative real-time PCR (qPCR)
Relative levels of mRNA expression were analyzed by real-time PCR using the Applied Biosystems 7500 Real-Time PCR System, in accordance with the manufacturer’s protocol (Life Technologies, Grand Island, NY). Relative expression levels were determined by the comparative method (2−ΔCt) against endogenous Hprt controls. The primer sequences are listed in Supplementary Table S4.
Western blots
Western blotting was performed as described previously (2,22).
Experimental animals
Transgenic PB-Cre4 mice expressing Cre cDNA under the control of the probasin (PB) promoter and Tsc1 floxed mice were obtained from the National Cancer Institute Mouse Model Deposit (Frederick, MD). Male PB-Cre4 mice were crossed to female Foxp3flox/flox (2) or Tsc1flox/flox mice to generate prostate Foxp3, Tsc1, or both conditional knockout (cKO, PB-Cre4 × Foxp3flox/y, PB-Cre4 × Tsc1flox/flox, PB-Cre4 × Foxp3flox/y × Tsc1flox/flox) male mice. Cre-dependent c-Myc knock-in mice (23) that conditionally express c-MycWT or either the c-MycT58A or c-MycS62A phosphorylation mutant from the constitutively active Rosa26 locus were kindly provided by Dr. Rosalie C. Sears (Oregon Health & Sciences University). c-Myc floxed mice were from Dr. Andreas Trumpp (German Cancer Research Center). Pathologic evaluations were performed according to the Bar Harbor meeting guidelines (24). All animal experiments were conducted in accordance with accepted standards of animal care and approved by the Institutional Animal Care and Use Committee of UAB.
Magnetic resonance imaging (MRI) of prostates
Prostate size was measured by MRI as described previously (2).
Immunohistochemistry
Vectastain Elite ABC Kits (Vector Laboratories, Burlingame, CA) were used for immunostaining according to the manufacturer’s protocol as described previously (2,22). The percentage of positive tumor cells per slide (0% to 100%) was multiplied by the dominant intensity pattern of staining (1, weak; 2, moderate; 3, intense); therefore, the overall score ranged from 0 to 300 H-scores.
Treatments of Foxp3 and Tsc1 double cKO mice with c-MYC and mTOR inhibitors
To Foxp3/Tsc1 double-cKO mice at 18 weeks of age, we administered 100 μl of either vehicle, Torin1 (20 mg/kg body weight), 10058-F4 (12.5 mg/kg), or Torin1 (20 mg/kg) plus 10058-F4 (12.5 mg/kg) by intraperitoneal injection weekly for 25 weeks. Five mice from each group were sacrificed at 23, 27, 31, 35, 39, and 43 weeks of age. Histologic analysis was used to determine whether the administration of these drugs alone or in combination limited tumor formation and progression.
Statistical analyses
Differences of outcome between two groups were compared by use of a two-sided t test or the Mann-Whitney U test. One-way analysis of variance (ANOVA) was used to test for overall differences, followed by a protected least significant difference test for differences between groups. For survival analysis, Kaplan-Meier survival curves were assessed with a log-rank test for mice with different conditions during tumor development. All data were analyzed using Excel 2016 and SPSS (version 24; IBM, Armonk, NY).
Results
Identification of FOXP3 loss-related additional genetic hits in human prostate cancers
Although loss of nuclear FOXP3 is evident in most human prostate cancers, lineage-specific ablation of Foxp3 in mouse prostate epithelium leads only to mPIN (2). It is likely that inactivation of FOXP3 cooperates with additional genetic hits to cause tumor progression. Thus, we first investigated the somatic genomic alterations of FOXP3-related additional genetic hits in tumor progression. IHC was used to obtain 10 nuclear FOXP3+ and 10 nuclear FOXP3− prostate tumors for use in identifying additional genetic hits (Supplementary Figs. S1A-B and Table S2). Genomic DNA was extracted from micro-dissected prostate cancer cells and gene amplification or deletion was determined by gene copy number quantitative PCR. In 33 prostate-cancer related candidate genes in publically available datasets derived from large cohorts of patients with prostate cancer (6,25), there were increases in gene copy number of AKT1 and/or decreases in gene copy numbers of TSC1 and FOXP1 in FOXP3− tumors compared with FOXP3+ tumors (Supplementary Fig. 1B), raising the possibility that defects in these genes cooperate with FOXP3 defects to promote prostate cancer progression.
Prostate-specific deletion of Tsc1 accelerates prostate tumor progression in Foxp3 cKO mice through synergistic activation of c-MYC and mTOR signaling pathways
Prostate-specific deletion of the potential tumor suppressor gene Tsc1 was introduced into Foxp3 cKO mice with a C57BL/6 background (Supplementary Figs. S2A-B). Micro-dissected prostate epithelial tissues from 8-week-old mice showed approximately 90% deletions of the Tsc1 and Foxp3 loci (Fig. 1A). The mRNAs of Tsc1 and Foxp3 were reduced by 95% and 93%, respectively (Fig. 1B). Thus, in the mouse prostate epithelial cells, the deletions of Foxp3/Tsc1 were sufficient for the double KOs of both two genes. Furthermore, the double-deletions led to tumor progression starting at 22 weeks of age, and 100% of these mice developed prostate carcinomas by 43 weeks of age (Figs. 1C and Supplementary Fig. S3A). At high magnification, definitive alterations in nuclear and cytoplasmic features were evident in prostate tissue, including larger and more vesicular nuclei and more densely eosinophilic cytoplasm with larger cell size. In humans, similar cytologic alterations are characteristics of a variety of early invasive carcinomas (24). These features are similar to those of high-grade PIN and associated invasive carcinomas (Gleason pattern 3) in humans (24). As determined by IHC analysis, more Ki67+ cells (Fig. 1D) were present in the cancer lesions compared with adjacent normal tissues, indicating that the tumor cells were highly proliferative. In addition, prostate tumors developed in the double cKO mice by 43 weeks of age and showed losses of epithelium staining along with CK5 and p63 in prostate epithelial basal cells as compared to the WT and single cKO mice (Fig. 1E), suggesting a basal-to-luminal differentiation and potential invasive prostate cancer in the mice.
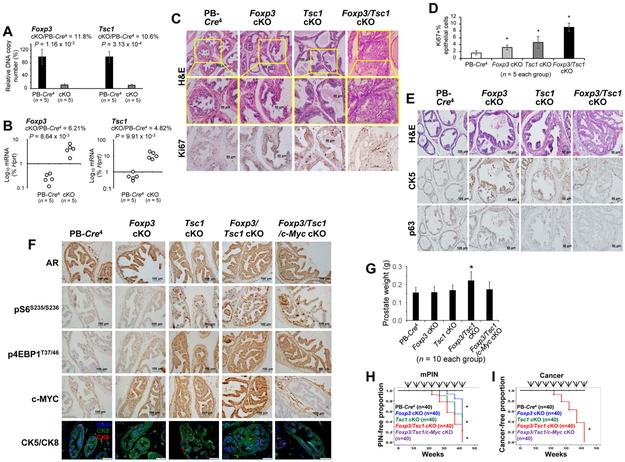
Figure 1. Prostate-specific deletions of Tsc1, Foxp3, and c-Myc and tumor progression in mouse prostate.
A. Efficient deletions of the Foxp3 and Tsc1 loci in the PB-Cre4+Foxp3flox/yTsc1flox/flox (Foxp3/Tsc1 double cKO), but not the PB-Cre4+ transgenic control mice, at 12 weeks of age (p value by a two-tailed t test). Data shown are means and standard deviation (SD) of the ratio of product A/product B for Foxp3 or product C/product D for Tsc1 of DNA isolated from the micro-dissected prostate epithelium. B. Targeted deletions of the Foxp3 and Tsc1 loci caused by the reduction of Foxp3 and Tsc1 transcripts in prostate tissue (p value by a Mann-Whitney U test). The RNAs isolated from micro-dissected prostate epithelia were quantitated by real-time PCR. C. Tumor development and cell proliferation in the dorsal and lateral lobes of the prostates in Foxp3/Tsc1 double-cKO mice compared with that in Tsc1 single-cKO, Foxp3 single-cKO, and PB-Cre4+ control mice at 22 weeks of age under a C57BL/6 genetic background, identified by H&E and Ki67 staining. D. The percentage of Ki67+ cells as an indicator of proliferating cells among the mouse prostate or tumor tissues. For each mouse, at least five 40x fields were counted. Data are presented as means ± SD. * p < 0.05 by one-way ANOVA followed by protected least-significant difference test vs. PB-Cre4 control mice. E. H&E, CK5, and p63 staining in the dorsal and lateral lobes of the prostates in Foxp3/Tsc1 double-cKO mice compared with that in Tsc1 single-cKO, Foxp3 single-cKO, and PB-Cre4+ control mice at 43 weeks of age. F. Immunostaining for androgen receptor (AR), pS6S235/236, p4EBP1T37/46, c-MYC, and CK5/CK8 in the prostates of PB-Cre4 control, Foxp3 cKO, Tesc1 cKO, Foxp3/Tsc1 double cKO, and Foxp3/Tsc1/c-Myc triple cKO mice at 12 weeks of age. G. Weights of prostates in the mice at 12 weeks of age. Data are presented as the means and SD of the prostate weights. * p < 0.05, two-tailed t test vs. PB-Cre4 control mice. H and I. Kaplan–Meier curves of mPIN and cancer incidences in cKO and PB-Cre4 control mice (n = 40/group). At 7, 12, 17, 22, 27, 32, 37, and 43 weeks of age, 5 mice/per time point were sacrificed for pathologic analysis. Arrows indicate time points of animal sacrifice. * p < 0.05, log-rank test vs. PB-Cre4 control mice. cKO, prostate conditional knockout. All experiments were repeated two times.
For Foxp3/Tsc1 double, Foxp3 single, and Tsc1 single cKO and probasin (PB)-Cre4 control mice, no mPIN was present at 12 weeks of age. Expression of AR was evident in all prostates, but expressions of pS6S235/S236 and p4EBP1T37/46 were higher in Tsc1 single and Foxp3/Tsc1 double cKO prostates (Fig. 1F). Expression of c-MYC was higher in Foxp3 or Tsc1 single cKO prostate and enhanced in Foxp3/Tsc1 prostates with double cKO. Likewise, prostate weights were higher in Foxp3/Tsc1 double cKO mice, but this was not seen for Foxp3 or Tsc1 single cKO mice at 12 weeks of age (Fig. 1G). Furthermore, Foxp3/Tsc1 double, Foxp3 single, and Tsc1 single cKO mice developed mPIN after 22 weeks of age, but only double deletions of Foxp3/Tsc1 led to prostate carcinoma at 22 to 42 weeks of age (Figs. 1H-I and Supplementary Fig. S3B and Table S5), suggesting that the loss of both genes hastens tumor progression.
c-Myc is required for tumor initiation and progression of prostate cancers in Foxp3/Tsc1 double cKO mice
c-MYC is the most commonly overexpressed oncogene in prostate cancers (12). Thus, we deleted c-Myc in the mouse prostate by crossing PB-Cre4 mice with c-Mycflox/flox mice (Supplementary Fig. S4A). To determine whether c-Myc is required for Foxp3 and Tsc1-mediated tumor initiation and progression of prostate cancer, we generated prostate Foxp3/Tsc1/c-Myc triple-cKO mice on a C57BL/6 background. As determined with micro-dissected tissue samples of 8-week-old mice, c-Myc DNA and mRNA in c-Myc cKO prostate epithelial cells were lower by 85% and 92%, respectively (Supplementary Figs. S4B-C). The reduction of c-MYC protein in mouse prostate was validated by IHC staining (Supplementary Fig. S4D). Furthermore, there was no difference in prostate weights between PB-Cre4 control and c-Myc cKO mice at 12 weeks of age (Supplementary Fig. S4E), suggesting that c-Myc cKO does not influence prostate development. In addition, deletions of Foxp3/Tsc1 also elevated expressions of pS6S235/S236 and p4EBP1T37/46 in prostates of the Foxp3/Tsc1/c-Myc triple-cKO mice (Fig. 1E). Although mPIN was present in the Foxp3/Tsc1/c-Myc triple-cKO mice at 52 weeks of age (Supplementary Fig. S3B), cancer lesions did not occur at even one year of age. Thus in Foxp3/Tsc1 double cKO mice, c-Myc is most likely to be required for prostate tumor initiation and progression.
Prostate-specific double deletion of Foxp3 and Tsc1 upregulates c-MYC expression through post-translational modification of c-MYC in mutant mice
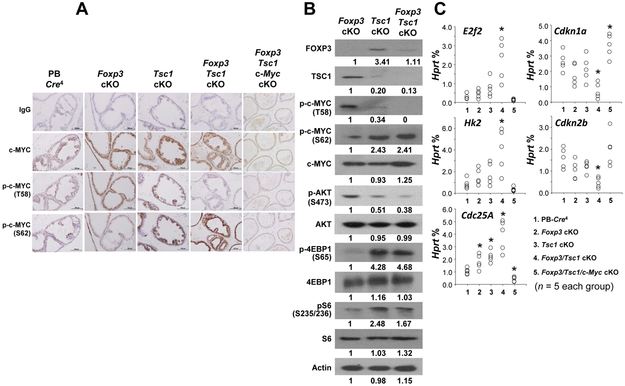
To determine if deletions of Foxp3/Tsc1 synergistically stabilize the c-MYC protein by regulating its phosphorylation at T58 and S62 in mouse prostate, we measured the levels of total c-MYC and serine 62 (pS62)/threonine 58 (pT58)-c-MYC protein in prostates of PB-Cre4 control, Foxp3, Tsc1, and Foxp3/Tsc1 double-cKO mice at 12 weeks of age were assessed by immunoblotting with specific antibodies. Total c-MYC protein was expressed in prostates of all mice (Fig. 2A). However, compared with PB-Cre4 control mice, the pT58-c-MYC protein was lightly expressed in prostates of Foxp3 and Tsc1 but not Foxp3/Tsc1 double-cKO mice, whereas pS62-c-MYC protein was more strongly expressed in prostates of Foxp3/Tsc1 double-cKO mice than in single-cKO mice (Fig. 2A). Likewise, Western blot analysis confirmed no expression of pT58-c-MYC protein but higher expression of the pS62-c-MYC protein in prostates of the Foxp3/Tsc1 double-cKO mice than in the single-cKO mice (Fig. 2B), suggesting that the upregulation of c-MYC is mediated by a post-translational regulation of c-MYC. Furthermore, lower expression of pAKTS473 but higher expressions of p4E-BP1S65 and pS6S235/236 were also present in prostates of the Tsc1 and Foxp3/Tsc1 double-cKO mice, suggesting an upregulation of mTORC1 activation but a downregulation of mTORC2 activation in these mice (Fig. 2B). In addition, c-Myc activity was evaluated in micro-dissected prostate epithelial cells by measuring, by use of qPCR, the mRNA expressions of c-MYC target genes, including E2f2, Hk2, Cdc25A, Cdkn1a and Cdkn2b. The expression levels of c-MYC target genes were greater in activated genes E2f2, Hk2, and Cdc25A but lower in repressed genes Cdkn1a and Cdkn2b in the prostate epithelial cells from double-cKO mice relative to that from single-cKO mice (Fig. 2C), indicating that tumor progression in the double-cKO mice is likely caused by elevated c-MYC activity.
Figure 2. Protein expression and activity of c-MYC in prostates of Foxp3/Tsc1 double-cKO mice.
A and B. Protein expressions and phosphorylation status of total and pT58/pS62-c-MYC and activation of mTOR as determined by IHC and Western blots of prostate tissues of Foxp3 cKO, Tsc1 cKO, or Foxp3/Tsc1 double cKO mice at 12 weeks of age. C. Relative mRNA levels of c-MYC target genes as a percentage of Hprt expression in micro-dissected prostate epithelial cells as determined by qPCR in the mice at 12 weeks of age. * p < 0.05 by one-way ANOVA followed by protected least-significant difference test between groups. T58: threonine 58; S62: serine 62; p: phosphorylation; cKO, prostate conditional knockout. All experiments were repeated two times.
Losses of FOXP3 and TSC1 synergistically upregulate expression of c-MYC through a post-translational modification in MEFs
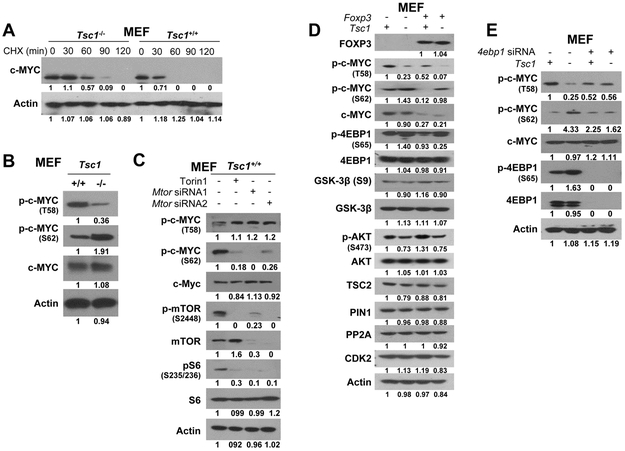
To determine if TSC1 is involved in c-MYC stability, the half-life of c-MYC was measured after treatment of mouse Tsc1−/− and Tsc1+/+ MEFs with the protein synthesis inhibitor CHX. Although c-MYC was degraded after 30 min of treatment with CHX, c-MYC degradation was more delayed in Tsc1−/− MEFs than in Tsc1+/+ MEFs (Fig. 3A), indicating that, in these cells, Tsc1-deficiency stabilized c-MYC. Furthermore, Tsc1 KO resulted in the simultaneous increase of pS62-c-MYC and decrease of pT58-c-MYC as determined by a comparison of Tsc1−/− and Tsc1+/+ MEFs (Fig. 3B), suggesting that the phosphorylation status of c-MYC is affected by Tsc1-deficiency. Since TSC1 is an essential component of the mTOR pathway, and loss of TSC1/2 facilitates mTOR activation, we investigated the effect of mTOR on the expression of pT58/pS62-c-MYC in MEFs. In Tsc1+/+ MEFs, there was a simultaneous increase of pT58-c-MYC and a decrease of pS62-c-MYC after inhibition of mTOR with Torin1, a mTOR inhibitor of both mTORC1 and mTORC2 (26,27), or silencing of mTOR with siRNAs (Fig. 3C). The inhibition of mTOR activation was validated by downregulation of pmTORS2448 and pS6S235/236. These data suggest a post-translational regulation of the phosphorylation and stabilization of c-MYC by TSC1-mTOR signaling.
Figure 3. Synergistic cross-talk between Foxp3 and Tsc1 in the regulation of c-MYC in MEFs.
A. c-MYC degradation in MEFs after exposure to cycloheximide (CHX, 100 μg/ml) for 2 hours. B. Expression levels of pT58/pS62-c-MYC in Tsc1+/+ MEFs and Tsc1−/− MEFs. C. Expression levels of pT58/pS62-c-MYC and mTOR activation in Tsc1−/− MEFs after inhibition of mTOR with Torin1 or silencing of mTOR with siRNAs. D. Phosphorylation and expression of c-MYC after Foxp3 transfection in Tsc1+/+ and Tsc1−/− MEFs. E. Effect of 4EBP1 on phosphorylation and expression of c-MYC in Tsc1+/+ MEFs and Tsc1−/− MEFs after 4ebp1 siRNA transfection for 48 hours. MEF, mouse embryonic fibroblasts; T58: threonine 58; S62: serine 62; p: phosphorylation; siRNA, small interfering RNA. Min, minute. All experiments were repeated three times.
To determine the role of Foxp3 in the Tsc1-dependent regulation of pT58/pS62-c-MYC, Foxp3 was transfected into Tsc1−/− and Tsc1+/+ MEFs. The exogenous Foxp3 reduced the increase of pS62-c-MYC by Tsc1-deficiency, but pT58-c-MYC was low in Tsc1−/− MEFs, regardless of Foxp3 transfection (Fig. 3D). However, in both Tsc1−/− and Tsc1+/+ MEFs, total c-MYC levels were reduced after Foxp3 transfection (Fig. 3D). These data suggest a synergistic effect of Foxp3 and Tsc1 on the expression of c-MYC in MEFs. Although pT58/pS62-c-MYC is controlled by TSC1-mTOR signaling (Figs. 3A-C), it remained unknown whether this regulation was direct or indirect. By use of siRNAs, we established that, in MEFs, silencing of either mTOR or 4EBP1 changed the Tsc1-dependent regulation of pT58/pS62-c-MYC (Figs. 3C and 3E). In Tsc1+/+ MEFs, silencing or inhibition of mTOR increased c-MYCT58 and decreased c-MYCS62, but did not change total c-MYC (Fig. 3C). In contrast, in Tsc1+/+ MEFs, silencing of 4ebp1 decreased c-MYCT58 and increased c-MYCS62, but did not change total c-MYC (Fig. 3E). Thus, depleting mTOR or 4EBP1 had the opposite effect on c-MYC phosphorylation (Figs. 3C and 3E). Of note, depleting 4EBP1 blocked the Tsc1-dependent changes in c-MYC phosphorylation; thus, Tsc1-dependent c-MYC phosphorylation is most likely to be through 4EBP1. In addition, we observed that, for Tsc1−/− MEFs, silencing of 4ebp1 increased c-MYCT58 and decreased c-MYCS62, but did not change total c-MYC (Fig. 3E). Although it is difficult to understand this observation, there may be other mechanisms in the regulation of c-MYC phosphorylation upon depleting both TSC1 and 4EBP1. However, it is still difficult to distinguish whether pT58/pS62-c-Myc levels are regulated by mTOR directly or through the 4E-BP1, which can also induce c-MYC (13,17-20) and may, therefore, have a role in c-MYC phosphorylation and subsequent activity. In addition, GSK3β, TSC2, PIN1, PP2A, and CDK2 (17,18,28-31) may be implicated in the Tsc1-dependent translational regulation of c-MYC. However, we did not find, in MEFs, any changes of these proteins in the absence or presence of Tsc1 and Foxp3 (Fig. 3D), supporting a direct regulation of c-MYC phosphorylation by mTOR or its substrates. However, there was lower expression of pAKTS473 in Tsc1−/− MEFs compared with in Tsc1+/+ MEFs but independent of Foxp3 transfection, suggesting a potential downregulation of mTORC2 activation due to a negative feedback loop of the mTORC1 pathway on AKT S473 (32).
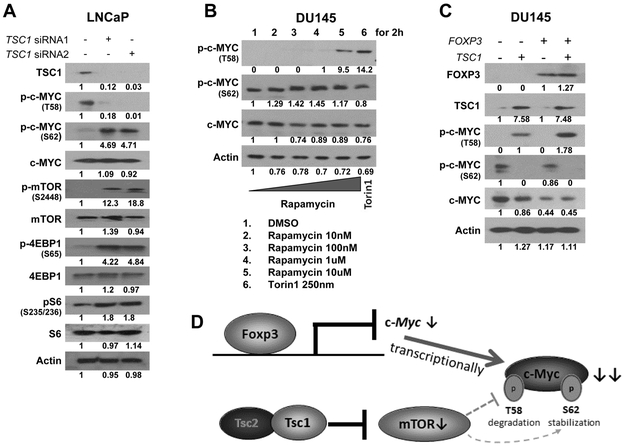
FOXP3 and TSC1 synergistically downregulate expression of c-MYC through a post-translational modification in prostate cancer cells
To confirm the involvement of TSC1 in regulating c-MYC in prostate cancer cells, siRNAs were used to silence human TSC1, which is normally expressed in the androgen-dependent prostate cancer cell line LNCaP. Although total c-MYC levels were not changed, a simultaneous increase of pS62-c-MYC and decrease of pT58-c-MYC were evident after silencing of TSC1 (Fig. 4A). TSC1-dependent mTOR activation was validated by increased expression of pmTORS2448, p4EBP1S65, and pS6S235/236 (Fig. 4A). Next, the androgen-independent prostate cancer cell line DU145 was treated with mTOR inhibitors. Treatment for 2 hours with rapamycin or Torin1 induced pT58-c-MYC and lightly reduced pS62-c-MYC levels (Fig. 4B), suggesting that, in prostate cancer cells, mTOR signaling regulates phosphorylation of c-MYC. There is a missense mutation (G1034C) of TSC1 in the prostate cancer cell line DU145 (www.mskcc.org), and mutated TSC1 is expressed at low levels in DU145 cells. To validate the cross-talk between FOXP3 and TSC1 in prostate cancer cells, WT TSC1 and FOXP3 were transfected into DU145 cells, and levels of c-MYC and mTOR and their phosphorylation status were compared. After TSC1 transfection, there was an increase in pT58-c-MYC and a decrease in pS62-c-MYC, and total c-MYC was reduced by transfections with both TSC1 and FOXP3 (Fig. 4C). In both normal and malignant prostate epithelial cells and tissues, FOXP3 is necessary and sufficient to repress c-MYC transcriptionally (2), indicating that, in prostate cancer cells, FOXP3 and TSC1 converge to inhibit the activity of c-MYC (Fig. 4D and Supplementary Fig. S5).
Figure 4. Functional cross-talk between FOXP3 and TSC1 in the regulation of c-MYC in prostate cancer cells.
A. The effect of TSC1 on c-MYC phosphorylation and mTOR activation in LNCaP cells afterTSC1 siRNA transfection for 48 hours. B. Phosphorylation and expression of c-MYC in DU145 cells after treatment with rapamycin and Torin1 for 2 hours. C. Phosphorylation and expression of c-MYC in DU145 cells after FOXP3, TSC1, or both transfections for 48 hours. D. Diagram of the synergistic action of FOXP3 and TSC1 in the regulation of transcription and phosphorylation of c-MYC in prostate cancer cells. Since FOXP3 transcriptionally inhibits c-Myc expression (2), and TSC1 regulates the phosphorylation status of c-MYC in prostate cancer cells, dysfunction of both FOXP3 and TSC1 synergistically promotes tumor progression through regulation of transcription, phosphorylation, and stabilization of c-MYC. T58: threonine 58; S62: serine 62; p: phosphorylation; siRNA, small interfering RNA; DMSO, dimethyl sulfoxide. All experiments were repeated three times.
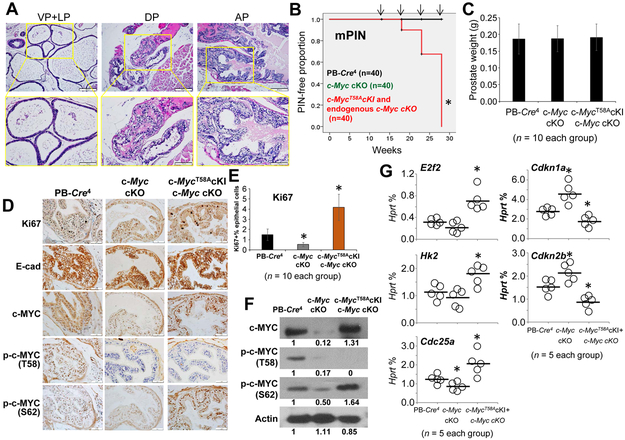
Mutagenesis of pT58A-c-Myc causes precancerous lesions in mouse prostate
Phosphorylation of c-MYC regulates its protein stability, and elevated ratios of pS62/pT58 c-MYC are present in human cancer cells (33,34). In vitro functional analysis shows that mutation of S62 to alanine (S62A) reduces the transforming activity of c-MYC, whereas mutation of T58 to alanine (T58A) enhances its transforming activity, suggesting that phosphorylation changes at these sites affect c-MYC function (31). To determine the role of c-MYC phosphorylation in tumor progression, we generated a mouse model of prostate c-MycT58A knock-in (cKI) under endogenous c-Myc cKO and observed the histologic changes in prostates of the c-MycT58A cKI mice with aging. In a small fraction of the anterior prostate lobes, c-MycT58A cKI mice at 18 weeks of age displayed signs of early mPIN characterized by atypical epithelial cells with clear cell cribriform hyperplasia: crowded glands with pale clear cytoplasm, round uniform nuclei, and prominent basal layers (Figs. 5A-B and Supplementary Figs. S3B and S6). By 28 weeks of age, all c-MycT58A cKI mice had early mPIN lesions in all prostate lobes (Figs. 5A-B and Supplementary Fig. S6). However, at this time, there was no difference in prostate weights between c-MycT58A cKI, c-Myc cKO, and PB-Cre4 control mice (Fig. 5C). IHC analysis revealed high proliferation in the epithelial cells of c-MycT58A cKI mice compared with other mice at 18 weeks of age (Figs. 5D-E). In the prostate epithelial cells of c-MycT58A cKI mice at 18 weeks of age, there was high c-MYC and pS62-c-MYC and low pT58-c-MYC, but no change in E-cadherin expression compared with cells from other mice (Fig. 5D). Western blot analysis confirmed these observations (Fig. 5F). In addition, there were high expressions of E2f2, Hk2, and Cdc25A, but low expressions of Cdkn1a and Cdkn2b in prostates of the c-MycT58A cKI mice at 18 weeks of age (Fig. 5G), suggesting elevated c-MYC activity by pT58A mutagenesis. This is the first direct evidence for an association between c-MYC pT58 phosphorylation and prostate tumorigenesis in vivo, which may also give insight into the mechanism of tumor initiation in Tsc1 cKO mice. We also generated a mouse model of prostate c-MycS62A cKI under endogenous c-Myc cKO, but, with aging, there were no apparent histologic changes in prostates of the c-MycS62A cKI mice, although pS62 is involved in cell proliferation, growth, and survival (23).
Figure 5. Effects of c-Myc T58A mutagenesis in mouse prostate.
A. Histologic analysis in the ventral (VP), lateral (LP), dorsal (DP), and anterior (AP) lobes of the prostates from 28-week-old prostate c-MycT58A knock-in (cKI) mice under endogenous c-Myc cKO (PB-Cre4+c-Mycflox/floxc-MycT58AcKI), identified by H&E staining. B. Kaplan-Meier curve of mPIN in PB-Cre4+ control, c-Myc cKO, and c-MycT58A cKI + c-Myc cKO mice. Arrows indicate time points of animal sacrifice (n=10 mice/per time point). * p < 0.05, log-rank test vs. PB-Cre4 control mice. C. Weights of prostates in the mice at 18 weeks of age. Data are presented as the means and SD. D. Representative IHC staining of Ki67, E-cadherin, c-MYC, pT58-c-MYC, and pS62-c-MYC in the anterior prostates of 18-month-old PB-Cre4+ control, c-Myc cKO, and c-MycT58A cKI + c-Myc cKO mice. E. The percentage of Ki67+ prostate epithelial cells among the mouse prostate tissues. Data are presented as the means and SD. * p < 0.05, two-tailed t test vs. PB-Cre4 control mice. F. Protein expression levels of c-MYC, pT58-c-MYC, and pS62-c-MYC in the prostates of PB-Cre4+ control, c-Myc cKO, and c-MycT58A cKI + c-Myc cKO mice at 18 weeks of age as determined by Western blots. G. Relative mRNA levels of c-MYC target genes as a percentage of Hprt expression in micro-dissected prostate epithelial cells as determined by qPCR in the mice at 18 weeks of age. Horizontal lines represent the average values. * p < 0.05 by one-way ANOVA followed by protected least-significant difference test vs. the PB-Cre4+ control group. All experiments were repeated two times.
Dual targeting of c-MYC and mTOR signaling pathways blocks tumor progression in prostates of Foxp3/Tsc1 double-cKO mice
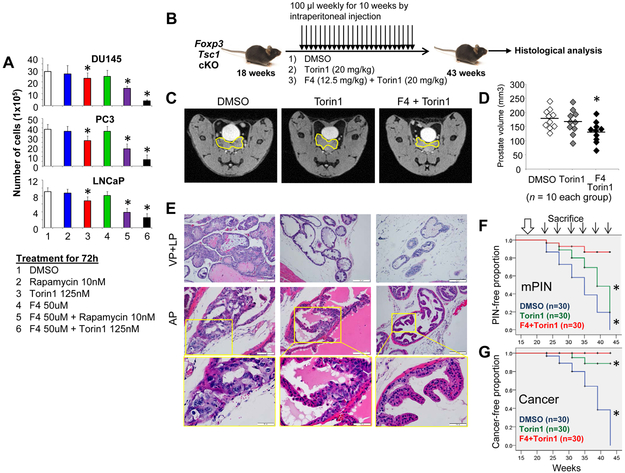
Clinical trials show that tumor resistance to rapamycin limits the efficacy of rapamycin or its analogues in inhibiting prostate cancer progression; however, cells lacking c-MYC are sensitive to rapamycin (17,35,36), indicating that c-MYC controls tumor resistance to mTOR inhibitors. Functional analysis suggests that c-MYC eliminates sensitivity to rapamycin through regulation of the 4E-BP1, but its capacity to confer rapamycin-resistance is not fully understood (13,35,37). Thus, inhibition of c-MYC expression may be an effective means of alleviating resistance to mTOR inhibitors. Here, we tested for synergistic action by blocking mTOR and c-MYC activity with inhibitors of both proteins in three commonly used prostate cancer cell lines, DU145, PC3, and LNCaP (Fig. 6A). Torin1 is an ATP-competitive mTOR inhibitor of both mTORC1 and mTORC2 and impairs cell growth and proliferation to a far greater degree than rapamycin (26,27). At 250 nM, Torin1 completely inhibits proliferation and causes a G1/S cell cycle arrest in MEFs (38). 10058-F4 is a c-Myc inhibitor that prevents the binding of c-Myc/Max dimers to DNA targets, downregulates c-Myc expression, and induces cell-cycle arrest and apoptosis (39-41). Treatment with 100 μM 10058-F4 inhibits the growth of acute myeloid leukemia cells (39). To observe the synergistic action of mTOR and c-Myc inhibitors, we applied only half of the reported effective doses of each drug to the cells. Several combinations of these inhibitors reduced cell growth, but, for all three cell lines, the combination of Torin1 and 10058-F4 inhibited cell growth most substantially (Fig. 6A).
Figure 6. Effects of dual targeting c-MYC and mTOR on prostate tumor progression in Foxp3 and Tsc1 double cKO mice.
A. Cells were exposed to DMSO, rapamycin, Torin1, 10058-F4, or their combinations. Data are presented as the means and SD of the cell numbers. * p<0.05, by one-way ANOVA followed by protected least-significant difference test vs. DMSO group. Rap: rapamycin; F4: 10058-F4; DMSO: dimethyl sulfoxide. B. Timeline of the therapeutic delivery of drugs. Foxp3 and Tsc1 double cKO littermates were dosed intraperitoneally with DMSO or Torin1 or Torin1+F4 weekly (black arrows). C. Representative MRI images of the prostates (outlined in yellow) of 40-week-old mice. D. Prostate volumes of individual mice. * p<0.05, by one-way ANOVA followed by protected least-significant difference test vs. the group dosed with DMSO. Horizontal lines represent the average values. E. Histologic analysis in the ventral (VP), lateral (LP), and anterior (AP) lobes of prostates from Foxp3 and Tsc1 double cKO mice after treatment, identified by H&E staining. F and G, Kaplan–Meier curves of mPIN and cancer incidences in the mice at 43 weeks of age after treatments. Large white arrow, starting time of treatment. Thin arrows, time points of mouse sacrifice (n=5 mice/per time point). * p<0.05, log-rank test vs. the DMSO group. All experiments were repeated two times.
In mice, Torin1 represses downstream effectors of mTOR at a dose of 20 mg/kg (27,42), and 10058-F4 inhibits tumor progression at a dose of 25 mg/kg (43). Next, we assessed the therapeutic potential of combined treatment with Torin1 and 10058-F4 against tumor progression in mice. To reduce the toxic side effects of the c-Myc inhibitor, only half of the reported effective dose of 10058-F4 was administered. Since Foxp3/Tsc1 double-cKO mice develop primary prostate tumors at 22 to 43 weeks of age (Fig. 1C and Supplementary Fig. S3), treatment was initiated at 18 weeks of age to correspond with cancer transition. DMSO, Torin1, or Torin1 plus 10058-F4 was administered into Foxp3/Tsc1 double-cKO mice by intraperitoneal injection weekly for 25 weeks (Fig. 6B). As determined by MRI images of mice at 40 weeks of age, prostate volumes were reduced in those treated with Torin1+10058-F4 compared with those treated with Torin1 or DMSO (Figs. 6C-D). Histologic analysis of prostates from mice at 43 weeks of age revealed that administration of Torin1 or Torin1+10058-F4 limited tumor formation and progression (Fig. 6E). Survival analysis revealed mPIN in 100% (5/5) of DMSO-, 60% (3/5) of Torin1-, and 0% (0/5) of Torin1+10058-F4-treated mice, and cancer in 100% (5/5) of DMSO-, 0% (0/5) of Torin1-, and 0% (0/5) of Torin1+10058-F4-treated mice (Figs. 6F-G), suggesting a synergistic inhibition of prostate tumor formation and progression by blocking the c-MYC and mTOR signaling pathways in Foxp3/Tsc1 double-cKO mice.
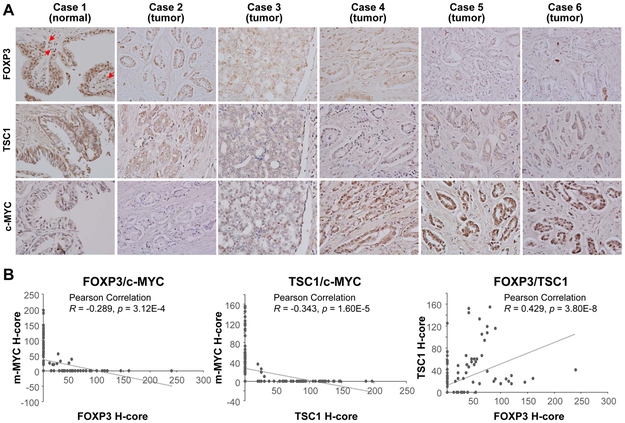
Co-downregulation of FOXP3 and TSC1 with upregulation of c-MYC in human prostate cancers
We next evaluated, by IHC, the protein expressions of FOXP3, TSC1, and c-MYC and their relationship in 151 prostate cancer tissues (Supplementary Table S3). Only 32.5% (49/151) of the prostate cancer samples expressed nuclear FOXP3 (Fig. 7A and Supplementary Table S6). All samples expressed cytoplasmic TSC1, but only 39.1% (59/151) had high expression of TSC1. All samples expressed nuclear c-MYC in at least one tumor cell, but only 35.8% (54/151) had high levels of nuclear c-MYC; 56.3% (85/151) of the samples showed cytoplasmic staining of c-MYC. Loss of nuclear FOXP3 was frequently accompanied by a low expression of TSC1 (56.3%, 85/151) or high expression of nuclear c-MYC (29.8%, 45/151). In addition, loss of nuclear FOXP3 and low expression of TSC1 were conversely associated with tumor stages (Supplementary Table S7). Finally, our quantitative analysis of the protein levels by H-score of all human samples showed statistically inverse correlations between c-MYC and FOXP3 or TSC1 but a positive correlation between FOXP3 and TSC1 (Fig. 7B). These data suggest frequent co-downregulation of FOXP3 and TSC1 with upregulation of c-MYC in human prostate cancers.
Figure 7. Protein expression levels of FOXP3, TSC1, and c-MYC in human primary prostate cancers.
A, IHC analyses with specific antibodies against human FOXP3, TSC1, and c-MYC were performed for 151 primary prostate cancer samples. Red arrows indicate Treg cells. This experiment was repeated two times. B, Correlation of the H-scores of FOXP3, TSC1, and c-MYC staining in the human prostate cancer samples. All experiments were repeated two times.
Discussion
Ours is the first effort to identify the synergy between the FOXP3 and TSC1 pathways and to investigate their role in the transcriptional and post-translational regulation of c-MYC during the progression of prostate cancer. The data showed that loss of nuclear FOXP3 is often accompanied by low expression of TSC1 in human prostate cancers. Prostate-specific deletions of Foxp3/Tsc1 in mice led to the progression of mPIN to cancer at an early age. Functional analysis suggested that TSC1-mTOR signaling affects the protein stability of c-MYC through T58/S62 phosphorylation in prostate cancer cells. Since FOXP3 represses the transcriptional activity of c-Myc (2), we found that cross-talk between FOXP3 and TSC1 regulates the expression and protein stability of c-MYC through a synergistic transcriptional and post-translational regulation to progress mPIN to prostate cancer. Notably, c-MycT58A cKI mice developed mPIN but not cancer. On the other hand, dual targeting of c-MYC and mTOR by their inhibitors blocked progression of mPIN to cancer in Foxp3/Tsc1 double-cKO mice, indicating a role of cross-talk between c-MYC and mTOR signaling pathways in tumor progression.
During tumor progression, FOXP3 and TSC1 signaling pathways converge on c-MYC (Fig. 4D). First, deletion of Foxp3 in mouse prostate leads to mPIN through transcriptionally overexpressed c-Myc (2), suggesting that the FOXP3-c-MYC axis regulates tumor initiation. Second, introduction of a prostate-specific deletion of Tsc1 into Foxp3 cKO mice leads to prostate carcinoma at an early age, but this does not occur when these mice are crossed with c-Myc-cKO mice, suggesting that the deletion of Tsc1 accelerates tumor progression of Foxp3-related mPIN to cancer through c-Myc. Third, FOXP3 transcriptionally inhibits c-Myc expression (2), and TSC1 regulates the phosphorylation status of c-MYC in prostate cancer cells. Thus, in tumor progression, c-MYC is likely to be a bridge for the cross-talk between FOXP3 and TSC1. Therefore, in prostate cancers, dysfunction of both FOXP3 and TSC1 synergistically promotes tumor progression through regulation of transcription, phosphorylation, and stabilization of c-MYC (Fig. 4D). In addition, increases in gene copy number of AKT1 were found in FOXP3− tumors compared with FOXP3+ tumors (Supplementary Fig. 1B), raising the possibility that AKT1 cooperates with FOXP3 defects to promote prostate cancer progression. However, in the Foxp3 cKO crossed with Akt1 transgenic (MPAKT, rPb-myr-HA-Akt1) (15) mice, we did not observe accelerated tumor progression in the mice with aging until the age of 12 months. Although we cannot reject an interaction of FOXP3 with AKT1 in mouse prostate, AKT1, in our mouse models, is unlikely to promote the FOXP3-mediated tumor progression.
Although there is no known, direct connection between TSC1 and FOXP3 or c-MYC, the role of TSC1 opposes that of c-MYC in cell growth and proliferation (15,17,18,44). Expression of c-MYC is affected by phosphorylation at the conserved residues T58 and S62. Phosphorylation at S62 stabilizes c-MYC, but phosphorylation at T58 promotes c-MYC ubiquitination and degradation (45-47). Although both TSC1/2 and 4EBP1 and GSK3β, TSC2, PIN1, PP2A, and CDK2 (17,18,28-31) can affect the post-translation effects of c-MYC (13,17-20,48), our data support the concept that, in prostate cancer cells, TSC1-mTOR signaling regulates phosphorylation of c-MYC at T58/S62 through 4EBP1. Recent studies have identified new phosphorylation peptides of c-MYC in mTORC1 substrates (26,49), but these peptides appear to be correlated with S77, not with T58/S62. Thus, it is of interest to determine, in future studies, if mTORC1 regulates c-MYC function through S77 phosphorylation. In addition, in cancer cells, post-translational regulation of c-MYC is balanced by USP13-mediated deubiquitination and FBXL14-mediated ubiquitination (50), but no reports show a connection of them with the TSC1-mTOR signaling pathway.
Numerous studies investigating therapies for prostate cancer have focused on mTOR and c-Myc inhibitors. However, clinical trials with these drugs have failed to demonstrate clinical activity. The efficacy of mTOR inhibitors is limited by drug resistance due to insufficient blocking of mTOR or interference from other signaling pathways, such as c-Myc (17,35,36). For example, c-MYC disrupts the elimination of mPIN lesions in young Akt mutant mice treated with mTOR inhibitors (14), although the mechanism is not fully understood (13). Use of c-Myc inhibitors is limited by toxic side effects. In the present study, we tested the joint administration of an mTOR inhibitor (e.g., Torin1) with a c-MYC inhibitor (e.g., 10058-F4) for effects on prostate cancer cells. Although rapamycin was ineffective in inhibition of cancer cell proliferation, a combination of rapamycin with 10058-F4, especially Torin1, overcomes this resistance in cultured cells. Furthermore, we developed a new therapeutic approach by blocking both c-Myc and mTOR to inhibit tumor progression of prostate cancer. In Foxp3/Tsc1 double-cKO mice, we used a more effective mTOR inhibitor, Torin1, with a low dose of a c-Myc inhibitor, 10058-F4, to block progression of mPIN to cancer, which may overcome tumor resistance to the mTOR inhibitor. Since both mTOR and c-MYC are frequently activated in prostate cancer (6,12,25,51), dual targeting strategies may be an effective therapeutic approach for blocking tumor progression and alleviating resistance to mTOR inhibitors.
In addition, we performed a genetic analysis of FOXP3, TSC1, AKT1, and FOXP1 in prostate adenocarcinoma with the two most commonly used public datasets. As shown in Supplementary Figs. S7A-B, in the TCGA dataset, except for FOXP1 gene deletion in 8% of all cases, few genetic alterations were present in FOXP3, TSC1, and AKT1. Likewise, in the MSKCC/DFCI dataset with more than 1,000 cases, there were no genetic alterations for FOXP3 and few gene deletions, gene amplifications, or both for TSC1, AKT1, and FOXP1, suggesting that loss of FOXP3 protein is not a genomic loss but may be caused by an epigenetic mechanism, such as DNA methylation. In the present study, we used the genomic DNA from micro-dissected prostate cancer cells for the IHC analysis of nFOXP3+ and FOXP3− prostate tumors. The results suggest the presence of genetic differences of TSC1, AKT1, and FOXP1 between nFOXP3+ and FOXP3− prostate tumors (Supplementary Figs. S1A-B and Table S8). However, due to the small size in our sample cohort, these differences may be from an infrequent event and should be considered with caution.
In summary, we characterized the cross-talk between the signaling pathways of FOXP3-c-MYC and TSC1-mTOR in the regulation of c-MYC expression in prostate tumor progression. These results will help in understanding how c-MYC is regulated and its role in tumor progression of prostate cancer. Dual targeting c-MYC and mTOR signaling provides an effective intervention against tumor progression and offers potential benefits for patients with prostate tumors resistant to mTOR inhibitors. Future studies are required to identify a direct regulatory mechanism of pT58/pS62 phosphorylation of c-MYC by mTOR signaling.
Supplementary Material
Significance:
Our results establish the principle of a synergistic action of TSC1 and FOXP3 during prostate cancer progression and provide new therapeutic targets for patients who have prostate cancer with two signaling defects.
Acknowledgment
We thank Dr. Donald L Hill for editorial assistance in this manuscript, Dr. Andreas Trumpp for the c-Myc floxed mice, and Dr. Rosalie C. Sears for the c-MycWT, c-MycT58A, and c-MycS62A knock-in mice.
Grant support
Research reported in this publication was supported by the National Institutes of Health (CA164688, CA179282, and CA118948 for L.W.), the Department of Defense (BC160808, PC130594, and PC140308 for L.W. and R.L.), the Mike Slive Foundation for Prostate Cancer Research (L.W.), a Mercer University Seed Grant (W.H.Y.), a Rubye Smith Research Grant (W.H.Y.), and a Larsen Research Endowment Fellowship Program Grant (W.H.Y.).
Footnotes
No potential conflicts of interest were disclosed.
LITERATURE CITED
- 1.Chen GY, Chen C, Wang L, Chang X, Zheng P, Liu Y. Cutting edge: Broad expression of the FoxP3 locus in epithelial cells: a caution against early interpretation of fatal inflammatory diseases following in vivo depletion of FoxP3-expressing cells. J Immunol 2008;180:5163–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, Liu R, Li W, Chen C, Katoh H, Chen GY, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell 2009;16:336–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katoh H, Qin ZS, Liu R, Wang L, Li W, Li X, et al. FOXP3 orchestrates H4K16 acetylation and H3K4 trimethylation for activation of multiple genes by recruiting MOF and causing displacement of PLU-1. Mol Cell 2011;44:770–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu R, Yi B, Wei S, Yang WH, Hart KM, Chauhan P, et al. FOXP3-microRNA-146-NF-kappaB axis and therapy for precancerous lesions in prostate. Cancer research 2015;75:1714–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angelin A, Gil-de-Gomez L, Dahiya S, Jiao J, Guo L, Levine MH, et al. Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments. Cell Metab 2017;25:1282–93 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 2003;4:209–21 [DOI] [PubMed] [Google Scholar]
- 8.Kladney RD, Cardiff RD, Kwiatkowski DJ, Chiang GG, Weber JD, Arbeit JM, et al. Tuberous sclerosis complex 1: an epithelial tumor suppressor essential to prevent spontaneous prostate cancer in aged mice. Cancer Res 2010;70:8937–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park Y, Jin HS, Lopez J, Elly C, Kim G, Murai M, et al. TSC1 regulates the balance between effector and regulatory T cells. J Clin Invest 2013;123:5165–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Zhang L, Zhang H, Xiao Y, Shao L, Li H, et al. Disruption of TSC1/2 signaling complex reveals a checkpoint governing thymic CD4+ CD25+ Foxp3+ regulatory T-cell development in mice. FASEB J 2013;27:3979–90 [DOI] [PubMed] [Google Scholar]
- 11.Sun S, Chen S, Liu F, Wu H, McHugh J, Bergin IL, et al. Constitutive Activation of mTORC1 in Endothelial Cells Leads to the Development and Progression of Lymphangiosarcoma through VEGF Autocrine Signaling. Cancer Cell 2015;28:758–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pettersson A, Gerke T, Penney KL, Lis RT, Stack EC, Pertega-Gomes N, et al. MYC Overexpression at the Protein and mRNA Level and Cancer Outcomes among Men Treated with Radical Prostatectomy for Prostate Cancer. Cancer Epidemiol Biomarkers Prev 2018;27:201–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balakumaran BS, Porrello A, Hsu DS, Glover W, Foye A, Leung JY, et al. MYC activity mitigates response to rapamycin in prostate cancer through eukaryotic initiation factor 4E-binding protein 1-mediated inhibition of autophagy. Cancer Res 2009;69:7803–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clegg NJ, Couto SS, Wongvipat J, Hieronymus H, Carver BS, Taylor BS, et al. MYC cooperates with AKT in prostate tumorigenesis and alters sensitivity to mTOR inhibitors. PLoS One 2011;6:e17449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Kobayashi T, Floc’h N, Kinkade CW, Aytes A, Dankort D, et al. B-Raf activation cooperates with PTEN loss to drive c-Myc expression in advanced prostate cancer. Cancer Res 2012;72:4765–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbard GK, Mutton LN, Khalili M, McMullin RP, Hicks JL, Bianchi-Frias D, et al. Combined MYC Activation and Pten Loss Are Sufficient to Create Genomic Instability and Lethal Metastatic Prostate Cancer. Cancer Res 2016;76:283–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt EV, Ravitz MJ, Chen L, Lynch M. Growth controls connect: interactions between c-myc and the tuberous sclerosis complex-mTOR pathway. Cell Cycle 2009;8:1344–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravitz MJ, Chen L, Lynch M, Schmidt EV. c-myc Repression of TSC2 contributes to control of translation initiation and Myc-induced transformation. Cancer Res 2007;67:11209–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenwald IB, Rhoads DB, Callanan LD, Isselbacher KJ, Schmidt EV. Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2 alpha in response to growth induction by c-myc. Proc Natl Acad Sci U S A 1993;90:6175–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin CJ, Cencic R, Mills JR, Robert F, Pelletier J. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res 2008;68:5326–34 [DOI] [PubMed] [Google Scholar]
- 21.Luo Y, Li B, Zhang G, He Y, Bae JH, Hu F, et al. Integrated Oncogenomic Profiling of Copy Numbers and Gene Expression in Lung Adenocarcinomas without EGFR Mutations or ALK Fusion. J Cancer 2018;9:1096–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu R, Yi B, Wei S, Yang WH, Hart KM, Chauhan P, et al. FOXP3-miR-146-NF-kappaB Axis and Therapy for Precancerous Lesions in Prostate. Cancer Res 2015;75:1714–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Cunningham M, Zhang X, Tokarz S, Laraway B, Troxell M, et al. Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res 2011;71:925–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, Rubin MA, et al. Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res 2004;64:2270–305 [DOI] [PubMed] [Google Scholar]
- 25.Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 2017;541:359–64 [DOI] [PubMed] [Google Scholar]
- 26.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 2011;332:1317–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, Chang JW, Wang J, Kang SA, Thoreen CC, Markhard A, et al. Discovery of 1-(4-(4-propionylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3-yl)benz o[h][1,6]naphthyridin-2(1H)-one as a highly potent, selective mammalian target of rapamycin (mTOR) inhibitor for the treatment of cancer. J Med Chem 2010;53:7146–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farrell AS, Pelz C, Wang X, Daniel CJ, Wang Z, Su Y, et al. Pin1 Regulates the Dynamics of c-Myc DNA Binding To Facilitate Target Gene Regulation and Oncogenesis. Mol Cell Biol 2013;33:2930–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hydbring P, Bahram F, Su Y, Tronnersjo S, Hogstrand K, von der Lehr N, et al. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci U S A 2010;107:58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol 2004;6:308–18 [DOI] [PubMed] [Google Scholar]
- 31.Vervoorts J, Luscher-Firzlaff J, Luscher B. The ins and outs of MYC regulation by posttranslational mechanisms. J Biol Chem 2006;281:34725–9 [DOI] [PubMed] [Google Scholar]
- 32.Albert S, Serova M, Dreyer C, Sablin MP, Faivre S, Raymond E. New inhibitors of the mammalian target of rapamycin signaling pathway for cancer. Expert Opin Investig Drugs 2010;19:919–30 [DOI] [PubMed] [Google Scholar]
- 33.Arnold HK, Zhang X, Daniel CJ, Tibbitts D, Escamilla-Powers J, Farrell A, et al. The Axin1 scaffold protein promotes formation of a degradation complex for c-Myc. EMBO J 2009;28:500–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malempati S, Tibbitts D, Cunningham M, Akkari Y, Olson S, Fan G, et al. Aberrant stabilization of c-Myc protein in some lymphoblastic leukemias. Leukemia 2006;20:1572–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci U S A 2011;108:E699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muellner MK, Uras IZ, Gapp BV, Kerzendorfer C, Smida M, Lechtermann H, et al. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat Chem Biol 2011;7:787–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Abronzo LS, Bose S, Crapuchettes ME, Beggs RE, Vinall RL, Tepper CG, et al. The androgen receptor is a negative regulator of eIF4E phosphorylation at S209: implications for the use of mTOR inhibitors in advanced prostate cancer. Oncogene 2017;36:6359–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 2009;284:8023–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang MJ, Cheng YC, Liu CR, Lin S, Liu HE. A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp Hematol 2006;34:1480–9 [DOI] [PubMed] [Google Scholar]
- 40.Mo H, Henriksson M. Identification of small molecules that induce apoptosis in a Myc-dependent manner and inhibit Myc-driven transformation. Proc Natl Acad Sci U S A 2006;103:6344–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Hammoudeh DI, Follis AV, Reese BE, Lazo JS, Metallo SJ, et al. Improved low molecular weight Myc-Max inhibitors. Mol Cancer Ther 2007;6:2399–408 [DOI] [PubMed] [Google Scholar]
- 42.Liu Q, Kang SA, Thoreen CC, Hur W, Wang J, Chang JW, et al. Development of ATP-competitive mTOR inhibitors. Methods Mol Biol 2012;821:447–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zirath H, Frenzel A, Oliynyk G, Segerstrom L, Westermark UK, Larsson K, et al. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc Natl Acad Sci U S A 2013;110:10258–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosner M, Hofer K, Kubista M, Hengstschlager M. Cell size regulation by the human TSC tumor suppressor proteins depends on PI3K and FKBP38. Oncogene 2003;22:4786–98 [DOI] [PubMed] [Google Scholar]
- 45.Lutterbach B, Hann SR. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol 1994;14:5510–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell 1999;3:169–79 [DOI] [PubMed] [Google Scholar]
- 47.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 2000;14:2501–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt EV. The role of c-myc in cellular growth control. Oncogene 1999;18:2988–96 [DOI] [PubMed] [Google Scholar]
- 49.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011;332:1322–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang X, Zhou W, Wu Q, Huang Z, Shi Y, Yang K, et al. Deubiquitinase USP13 maintains glioblastoma stem cells by antagonizing FBXL14-mediated Myc ubiquitination. J Exp Med 2017;214:245–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012;485:55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.