SUMMARY:
Next generation sequencing has provided information on actionable targets and biomarkers of response in oncology. In HCC, Wnt/CTNNB1 mutations characterize the immune excluded class (cold tumors) and might represent the biomarkers predicting resistance to immune checkpoint inhibitors. Large-scale validation of this data is needed to customize immunotherapy in advanced HCC.
In this issue of Clinical Cancer Research, Harding et al provide the first report of prospective genotyping of advanced HCC and the potential implications for clinical decision-making [1]. Tumor profiling using the MSK-IMPACT custom panel for targeted exome sequencing (468 genes) was used to inform the treating physician about the actionable genomic alterations harbored by each tumor in 127 patients with advanced HCC [1]. This first-in-class study provides three main results that warrant attention: a) Around 25% of advanced tumors harbored actionable mutations, although overall only 6% of patients received matched therapeutic interventions; b) Oncogenic PI3K-MTOR alterations might predict sorafenib resistance, as described in 81 HCC treated patients, and c) Previously reported immune exclusion HCC class, characterized by Wnt/CTNNB1 mutations [2,3], might predict innate primary resistance to immune checkpoint inhibitors, as explored in 31 patients receiving these therapies. Despite some limitations pointed out by the authors related to the small sample size and lack of analysis of confounding factors predicting response, the current data provides novel insights in defining biomarker-driven responses in advanced HCC that deserve confirmation in prospective large-scale clinical investigations.
In HCC, around 40% of patients are diagnosed at advanced stage when systemic therapies are indicated [2]. The multi-tyrosine kinase inhibitor (TKI) sorafenib was the first molecular targeted therapy that demonstrated survival benefit in advanced HCC cases [4]. More recently, additional TKIs (lenvatinib in first-line, and regorafenib, cabozantinib and ramucirumab in second-line) have demonstrated survival benefits in the setting of phase III trials. In addition, immune checkpoint inhibitors (nivolumab and pembrolizumab, both in second-line) obtained accelerated FDA approval based upon phase II positive clinical signals [2]. In spite of such advancements, median survival of these patients is still limited to ~12-13 months in front-line and ~9-11 months in second-line. No biomarkers are available for tailored treatments, except for AFP > 400 ng/mL in patients responding to ramucirumab. In this scenario, a deeper understanding of actionable targets in the clinical practice, mechanisms of response/resistance to identify biomarkers of response and synergies among treatments (immunotherapies (IOs) and TKIs/monoclonal antibodies (MAb)) are required to ultimately improve patients’ survival.
Deep-exome sequencing of HCC tumors has been reported in ~1,200 cases [2,5]. These studies, mostly conducted in resected tumors, have delineated that the most prevalent oncogenic drivers in HCC (TERT, TP53, CTNNB1, AXIN1, ARID1A and ARID2) are currently not actionable. As described for other solid tumor malignancies, in HCC there is also a “long tail” of mutations, amplifications or deletions at a prevalence of <10%, among which some are amenable for targeted therapies. In this sense, ~25% of such alterations have been defined as potentially targetable with the current drug armamentarium [5]. These include FGF19 and VEGFA amplifications, MET amplifications or mutations, mutations in PI3K-MTOR signaling (PI3KCA, PTEN, TSC1/2), among others. None of these potential biomarkers have reached a stage in clinical investigation to demonstrate survival benefits, although data with FGFR4 inhibitors in tumors overexpressing FGF19 is promising.
The current study demonstrates that a prospective genotyping approach in patients with advanced HCC is able to identify tumors (24%) with potentially actionable alterations. None of them could receive biomarker-based approved drugs for this indication (level of evidence 1-2A) and only 6% of patients received either drugs approved for another indication (level 2B) or agents in active clinical developments with appealing biomarkers predicting responses (level of evidence 3A and B). Specifically, 5 patients were treated in second-line with either everolimus (based upon TSC1/2 alterations) or monoclonal antibodies against VEGFR2 and MET (due to VEGFR and MET amplifications). Thus, this study, which is the first to report a prospective genotyping of HCC patients aimed to inform decision-making, describes 24% actionable mutations in advanced tumors, but ultimately only 6% of patients received matched therapeutics. Further approaches including large cohorts of patients are required to demonstrate the cost-effectiveness of this strategy.
Biomarkers predicting response to sorafenib are lacking in advanced HCC [2]. Companion studies of the SHARP trial using blood-based biomarkers showed a non-significant trend for sorafenib benefit in patients with high levels of c-Kit and low levels of HGF. Similarly, a recent study identified a gene signature able to predict sorafenib recurrence-free responders in the adjuvant setting after resection/local ablation, a result that warrants further validation [6]. Thus, after more than 10 years of research we lack biomarker guidance to identify sorafenib responders. Only clinical predictors of response, such as HCV-infected HCC and those with liver-only disease, have been associated with a greater benefit with this TKI. In this current study, Harding and colleagues report retrospectively that tumors harboring PI3K/MTOR mutations (PI3KCA, PTEN, TSC1/2) are more likely to be primary resistant to sorafenib (survival of 10.4mo vs 17.9mo in those without mutations). These mutational alterations accounted for ~15% of the tested patients, a figure above the 5-10% published by TCGA series or in a large meta-analysis [2]. Nonetheless, these results represent a positive signal that deserves further investigations.
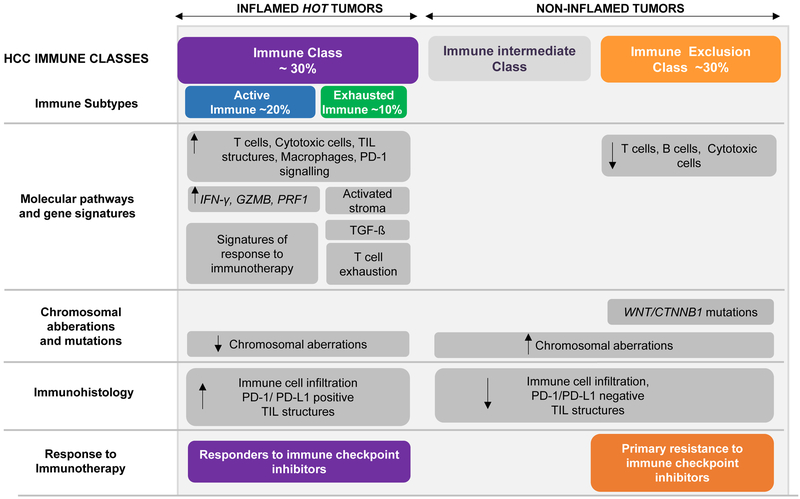
The most appealing data reported by Harding and colleagues refers to the first confirmation of the hypothesis that HCC ‘cold’ tumors -defined by Wnt/CTNNB1 mutations [3]- are those refractory to immune checkpoint inhibitors [2,3] (Figure 1). In order to identify biomarkers of response to IOs, the authors correlated outcome with the mutational profile of 27 patients receiving monoclonal antibodies anti-PD-1/PD-L1 (81%), or anti-CTLA-4, and combinations of anti-PD-1/PD-L1 with anti-CTLA-4, anti-KIR or anti-LAG3 (19%) [1]. Immunotherapies are more likely to be efficacious in ‘hot’ tumors, as defined by the presence of tumor-infiltrated lymphocytes (TILs). In parallel, tumors displaying immune exclusion, the so-called ‘cold’ tumors, are resistant to immunotherapies. In 2017 our group described a new molecular HCC classification based upon immune traits [2,5]. In brief, ~30% of HCCs were enriched by TILs and with molecular features resembling melanomas responsive to immunotherapies, and were defined as HCC Immune class (Figure 1) [2,5]. Conversely, we observed that ~30% of patients showed exclusion of TILs, enrichment in CTNNB1 mutations and known molecular fingerprints of primary resistance (HCC Exclusion class, Figure 1) [2,3]. Therefore, while tumors within the HCC Immune subtype may represent patients most likely to respond to immune checkpoint inhibitors, those within the HCC Exclusion class might represent those with innate resistance to anti-PD1/PD-L1 or similar therapies [2]. In their manuscript, Hardling et al. observed that 10 out of 27 patients (37%) treated with IOs exhibited Wnt/CTNNB1 mutations (7 CTNNB1, 3 AXIN1). Those 10 patients displaying an altered Wnt-β-catenin pathway were refractory to immune checkpoint blockers (all had progressive disease as best response), with a trend to poorer median survival (9.1mo vs 15.2 mo in those without mutations). These results are in line with the seminal study in melanoma demonstrating that non-T-cell-inflamed phenotype displayed altered b-catenin signaling [7]. This data is the first clinical signal, to our knowledge, to provide a potential biomarker defining primary resistance to checkpoint inhibitors in HCC. Certainly, whether mutations in CTNNB1 and AXIN1 are true biomarkers of innate resistance to immunotherapy needs to be validated in the setting of thorough phase II or III clinical trials. Confirmation of this preliminary data would be of utmost importance for the prospect of patient selection and trial enrichment in current and future investigations, and certainly for the precise decision-making in the management of advanced HCC cases.
FIGURE 1.
Molecular classification of HCC based upon immune status. Around 30% of HCCs belong to the ‘Immune class’, with high levels of immune cell infiltration, expression of PD-1 and/or programmed cell death 1 ligand 1 (PD- L1), activation of IFNγ signaling, and markers of cytolytic activity (such as granzyme B and perforin 1). This HCC tumors are more likely to respond to immune checkpoint inhibitors. On the other hand, the ‘Immune Exclusion class’ accounting for ~30% of HCCs, is characterized by T cell exclusion from the TME and CTNNB1 mutations. Tumors classified within the HCC Exclusion class would represent those with innate resistance to anti-PD-1/PD-L1 inhibitors. Non-inflamed tumors with wild-type CTNNB1 and intermediate levels of immune infiltration (the so called ‘Immune intermediate class’) needs to be further characterized to be able to anticipate the response that these patients would have when treated with immunotherapies. Figure partly modified from Llovet et al. (2).
Acknowledgments
FUNDING:
JML is supported by National Cancer Institute (P30-CA196521), U.S. Department of Defense (CA150272P3), European Commission (EC)/Horizon 2020 Program (HEPCAR, Ref. 667273-2), EIT Health (CRISH2, Ref. 18053), Accelerator Award (CRUCK, AECC, AIRC) (HUNTER, Ref. C9380/A26813), Samuel Waxman Cancer Research Foundation, Spanish National Health Institute (SAF2016-76390) and the Generalitat de Catalunya/AGAUR (SGR-1358). RP is funded by EC and EIT Health. DS is supported by the Gilead Sciences Research Scholar Program in Liver Disease.
Footnotes
CONFLICT OF INTEREST:
D. Sia and J. M. Llovet are inventors of a patent on immune classifiers to detect immune class. J. M. Llovet reports receiving commercial research grants from Bayer Healthcare Pharmaceuticals, Eisai, Bristol Myers-Squibb, Ipsen, Blueprint, and Incyte and is a consultant/advisory board member for Eli Lilly, Bayer Healthcare Pharmaceuticals, Bristol Myers-Squibb, Eisai Inc., Celsion, Exelixis, Merck, Blueprint, Ipsen, Glycotest, Navigant, Leerink Swann LLC., Midatech Ltd., Fortress Biotech Inc., Spring Bank Pharmaceuticals, Nucleix, and Can-Fite Biopharma. No potential conflicts of interest were disclosed by the other author.
REFERENCES:
- 1.Harding JJ, Nandakumar S, Armenia J, Khalil DN, Albano M, Ly M, et al. Prospective Genotyping of Hepatocellular Carcinoma: Clinical Implications of Next Generation Sequencing for Matching Patients to Targeted and Immune Therapies. Clin Cancer Res. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, Castro de Moura M, et al. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology. 2017;153:812–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, Cosme A, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten T, Galle P, Seitz JF, Borbath I, Häussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D and Bruix J, for the SHARP Investigators Study Group. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378–90 [DOI] [PubMed] [Google Scholar]
- 5.Schulze K, Imbeaud S, Letouzé E Alexandrov L, Calderaro J,Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud AL, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc JF, Mazzaferro V, Calvo F, Villanueva A, Nault JC, Bioulac-Sage P, Stratton M, Llovet JM, Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nature Genetics 2015;47:505–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinyol R, Montal R, Bassaganyas L, Sia D, Takayama T, Chau G-Y, et al. Molecular predictors of prevention of recurrence in HCC with sorafenib as adjuvant treatment and prognostic factors in the phase 3 STORM trial. Gut. 2018; gutjnl-2018-316408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–5. [DOI] [PubMed] [Google Scholar]