Abstract
Granulocytic myeloid infiltration and resultant enhanced neutrophil elastase (NE) activity is associated with poor outcomes in numerous malignancies. We recently showed that NE expression and activity from infiltrating myeloid cells was high in human prostate cancer xenografts and mouse Pten-null prostate tumors. We further demonstrated that NE directly stimulated human prostate cancer cells to proliferate, migrate, and invade, and inhibition of NE in vivo attenuated xenograft growth. Interestingly, reduced expression of SERPINB1, an endogenous NE inhibitor, also correlates with diminished survival in some cancers. Therefore, we sought to characterize the role of SERPINB1 in prostate cancer. We find that SERPINB1 expression is reduced in human metastatic and locally advanced disease and predicts poor outcome. SERPINB1 is also reduced in Pten-null mouse prostate tumors compared to wild type prostates, and treatment with sivelestat (SERPINB1 pharmacomimetic) attenuates tumor growth. Knockdown of highly expressed SERPINB1 in non-malignant prostatic epithelial cells (RWPE-1) increases proliferation, decreases apoptosis, and stimulates expression of epithelial-mesenchymal transition markers. In contrast, stable SERPINB1 expression in normally low-expressing prostate cancer cells (C4-2) reduces xenograft growth in vivo. Finally, EZH2-mediated histone (H3K27me3) methylation and DNMT-mediated DNA methylation suppress SERPINB1 expression in prostate cancer cells. Human TCGA analysis and pyrosequencing demonstrate hypermethylation of the SERPINB1 promoter in prostate cancer compared to normal tissue, and the extent of promoter methylation negatively correlates with SERPINB1 mRNA expression.
Introduction
Prostate cancer is the most common non-cutaneous cancer in men in the United States, with more than 160,000 new cases diagnosed each year. While most localized prostate cancers have an indolent course, it is essential to accurately assess which tumors will behave more aggressively [1]. The development of biomarkers has revolutionized oncology; yet few useful prostate cancer biomarkers have emerged in recent years. Biomarkers under investigation in prostate cancer include TMPRSS2:ERG rearrangement, PC antigen 3 (PCA3), and the androgen receptor splice variant-7 (ARv7) [2]. Moreover, several genomic, proteomic, and epigenetic tests such as the OncotypeDx, ProMark, and ConfirmMDx evaluate a panel of biomarkers and attempt to stratify patients by risk [2]. At this time however, clinical stage, Gleason grade, and serum prostate-specific antigen (PSA) remains the primary prognostic indicators used by clinicians to guide therapeutic decisions [2].
Growing evidence suggests that the tumor microenvironment contributes to cancer development and progression. Thus, understanding the complex interactions between tumor cells and surrounding reactive stroma may uncover novel biomarkers and therapeutic targets [3, 4]. While prostate cancer is defined as an adenocarcinoma, tumors in fact have a significant non-malignant cellular compartment consisting of fibroblasts, smooth muscle cells, endothelium, and immune cells [3, 4]. The tumor microenvironment is also rich in non-cellular components such as extracellular matrix (ECM), cytokines, growth factors, and proteases, which form fertile ground for cancer cell growth, migration, and invasion [5]. The cellular and non-cellular components evolve over the natural course of cancer. Specifically, tumor and stromal cells undergo genotypic and phenotypic changes, inflammatory cells infiltrate tumors, and the ECM becomes remodeled [3, 4]. Despite progress in other solid tumors, much remains to be learned about microenvironment changes in prostate cancer.
We recently showed that granulocytic myeloid cells (likely myeloid-derived suppressor cells or MDSCs) accumulate peripherally in blood and locally in tumors using a xenograft mouse model of human prostate cancer. These granulocytic myeloid cells directly promote tumor growth in the absence of adaptive immunity, since tumor size is significantly reduced upon sustained myeloid cell depletion in athymic mice [6]. Furthermore, we demonstrated the importance of myeloid-derived neutrophil elastase (NE) as a pro-tumorigenic factor for human prostate cancer cells, as NE promoted human prostate cancer cell line growth, migration, and invasion, while NE inhibition with the drug sivelestat attenuated human xenograft progression [6].
Although accumulation of NE-producing myeloid cells may be sufficient to explain enhanced NE activity within the tumor microenvironment, loss of endogenous NE inhibitors in tumor cells may further shift the balance [5]. There are several endogenous NE inhibitors that are expressed by numerous tissue types, including elafin (PI3), secretory leukocyte peptidase inhibitor (SLPI), and SERPINB1 [7-10]. We chose to focus our investigation on the most recently identified NE-specific inhibitor, SERPINB1. SERPINB1 is a 42-kD member of the Clade B family of serpins (serine protease inhibitors) that potently inhibits NE and neutrophil extracellular trap (NET) formation similar in mechanism to sivelestat [10-12]. SERPINB1 regulates neutrophil homeostasis within the bone marrow and promotes neutrophil survival via intracellular inhibition of neutrophil serine proteases [13-15]. Interestingly, SERPINB1 is expressed by many cell types beyond immune cells and detected both intra- and extra-cellularly [12, 16, 17]. While SERPINB1 lacks a classical signal peptide, it is secreted via an unconventional caspase-1 dependent secretory pathway that also mediates IL-1β and IL-18 secretion [17, 18]. The role of SERPINB1 in epithelial cells and cancer is not clear; however, limited screening studies demonstrate reduced expression in hepatocellular carcinoma, glioma, and melanoma compared to normal tissue counterparts, as well as possible roles in suppressing apoptosis and promoting tumor migration [19-21]. Microarray analysis of laser captured micro-dissected human prostate cancer specimens suggest that SERPINB1 mRNA expression is reduced early in the development of prostatic intraepithelial neoplasia (PIN) and remains low in prostate cancer [22]. 2D DIGE/MS proteomic analysis comparing prostate cancer to benign prostatic hyperplasia (BPH) also suggests lower SERPINB1 expression in tumors [23]. Finally, meta-analysis of epithelial-to-mesenchymal transition (EMT) encompassing several cancers, including prostate, identifies SERPINB1 as a commonly down-regulated gene [24]. However, all of the aforementioned studies simply classify SERPINB1 as a gene on a long list of other dysregulated genes, and neither a functional role for SERPINB1 loss nor mechanisms for its repression have been elucidated in prostate cancer.
Here we report that SERPINB1 expression is reduced in mouse Pten-null prostate tumors and treatment with the NE inhibitor sivelestat (SERPINB1 pharmacomimetic) attenuates tumor growth. We confirm that SERPINB1 is expressed in normal human prostatic epithelium but down-regulated in prostate cancer. We demonstrate that SERPINB1 is secreted by non-malignant human prostate cells and its expression is ERK1/2-dependent. Functionally, we show that SERPINB1 loss in non-malignant human prostate cells induces EMT and promotes a proliferative phenotype. In contrast, SERPINB1 overexpression in human prostate cancer cells suppresses xenograft progression. Finally, we find that SERPINB1 is epigenetically repressed in human prostate cancer cells by EZH2-mediated histone methylation (H3K27me) and DNMT-mediated DNA methylation. Overall, our data suggest a novel inhibitory role for SERPINB1 in prostate cancer progression, with potential application to both biomarker and therapeutic development.
Materials and Methods
Cell culture.
RWPE-1 cells (ATCC) were cultured in keratinocyte serum free media (K-SFM; Gibco) supplemented with 25 μg/mL bovine pituitary extract (BPE; Gibco), 5 ng/mL epidermal growth factor (EGF; Gibco), and 1% penicillin-streptomycin (P-S; Gibco). C4-2 (Ganesh Raj, UTSW), PC3 (ATCC), and LNCaP (ATCC) cells were cultured in RPMI-1640 media (Gibco) supplemented with 10% fetal bovine serum (FBS; Seradigm) and 1% P-S. BPH-1 cells (Donald DeFranco, UPitt) were cultured in 5% FBS and 1% P-S in RPMI-1640. VCaP, DU145, and CWR22Rv1 cells (Kent Nastiuk, UBuffalo) were cultured in 10% FBS and 1% P-S in DMEM (Gibco). LAPC-4 cells (Kent Nastiuk, UBuffalo) were cultured in 10% FBS and 1% P-S in RPMI-1640, 10 mM HEPES pH 7.4 (Gibco). Pten-Cap8 cells (Kent Nastiuk, UBuffalo) were cultured in 10% FBS, 25 μg/mL BPE, 6 ng/mL EGF, 5 μg/mL human recombinant insulin (Sigma), and 1% P-S in DMEM. All cells were maintained below passage 30 at 37°C, 95% air, 5% CO2.
Cell treatments and inhibitors.
Cells were serum starved for 24 hours and stimulated with 20 ng/mL EGF (Corning) in serum free media for 4 or 24 hours before subsequent lysis and analysis by quantitative PCR or Western, respectively. For inhibitor studies, cells were pre-incubated with 0.25 μM MEK inhibitor PD0325901 (Selleckchem) or 0.25 μM PI3K inhibitor LY294002 (Sigma) for 30 minutes before the addition of EGF. Cells were treated with 10 μM DNMT inhibitor 5-aza-2-deoxycytidine (Sigma) and 10 μM EZH2 inhibitor GSK343 (Selleckchem) in complete media for 72 hours.
SERPINB1 knockdown.
RWPE-1 cells were transfected with 25 nM Silencer Select Negative Control #1 siRNA (#4390843, Ambion) or 25 nM Silencer Select SERPINB1 siRNA (#4392420, Ambion) using jetPRIME (Polyplus). Knockdown was carried out for 72-96 hours as specified.
SERPINB1 cloning and stable overexpression.
Human SERPINB1 was cloned via nested PCR. SERPINB1 cDNA was inserted into pcDNA3.1/Hygro(+) vector (Invitrogen). C4-2 cells were transfected with SERPINB1-pcDNA3.1/Hygro(+) or empty vector using X-tremeGENE 9 DNA Transfection Reagent (Sigma) and selected with 100 μg/mL hygromycin (Sigma). Monoclonal cell lines were established and maintained in 10% FBS and 1% P-S in RMPI-1640 containing 25 μg/mL hygromycin.
Conditioned media.
RWPE-1 and C4-2 cells were grown to 80% confluence in respective complete medias and then cultured without supplements for an additional 48 hours. Conditioned media were collected, centrifuged at 2000 rpm for 10 minutes at 4°C, and concentrated 5-fold using Amicon Ultra 0.5 mL 3 kDa centrifugal filter units (Millipore).
Gelatin zymography.
Conditioned media samples were diluted 1:1 with 2X non-reducing, non-denaturing sample buffer and separated on a 10% gel containing 1 mg/mL gelatin (Sigma). Zymograms were performed and analyzed as previously described [25, 26].
Western blot.
Cells were lysed in mammalian cell lysis buffer (Abcam) supplemented with 1X Halt protease and phosphatase inhibitor cocktail (ThermoFisher Scientific). Lysates or conditioned media were diluted 1:1 with 2X sample buffer containing 2-mercaptoethanol (Sigma) and denatured. Samples were processed for gel electrophoresis and blotted with mouse anti-SERPINB1 (1:2000, #TA800093, Origene), rabbit anti-GAPDH (1:2000, #2118, Cell Signaling), rabbit anti-phospho-ERK1/2 (1:1000, #9101, Cell Signaling), and rabbit anti-total-ERK1/2 (1:1000, #9102, Cell Signaling).
Quantitative PCR.
RNA was extracted using the E.Z.N.A. kit (Omega). Quantitative PCR (qPCR) was performed using the qScript XLT 1-Step RT-qPCR ToughMix kit (QuantaBio) and TaqMan primers (Applied Biosystems) for: human SERPINB1 (Hs00961948_m1), MMP9 (Hs00234579_m1), SNAI1 (Hs00195591_m1), TWIST1 (Hs01675818_s1), PI3 (Hs00160066_m1), SLPI (Hs00268204_m1), GAPDH (Hs03929097_g1). Expression was normalized to GAPDH using the ∆∆Ct method.
Chromatin Immunoprecipitation (ChIP)
ChIP for SERPINB1 was performed using the MAGnify Chromatin Immunoprecipitation System (Invitrogen) [27]. Briefly, chromatin fragments were immunoprecipitated with mouse monoclonal anti-H3K27me3 ChIP-grade antibody (#6002, Abcam) or mouse IgG. Quantitative PCR was performed using PerfeCTa SYBR Green SuperMix Reagent (QuantaBio) with primers directed against two regions of the human SERPINB1 promoter (P1-forward-5’ CGT GCG ATT CTA GAG ACG ATT T 3’, P1-reverse-5’ CGA GGA CAG GCA AAG AAG AA 3’; P2-primer – 5’ TCT GAG AGT GGA GAT CGA GAT G 3’, P2-primer-5’ GGT GTA GGA TGT GCC AGT TT 3’). Results were normalized by the fold enrichment method.
Apoptosis.
RWPE-1 cells were transfected with control and SERPINB1 siRNA for 72 hours. Cell pellets were collected with 0.05% trypsin-EDTA (Gibco), neutralized with 2% FBS in PBS, and washed with cold PBS. Apoptosis was assessed using the FITC Annexin V Apoptosis Detection Kit I (BD Pharmingen). Briefly, cells were resuspended in 1X Annexin Binding Buffer at 1×106 cells/mL, stained with PI and FITC Annexin V, collected on a LSRII flow cytometer (BD Biosciences), and analyzed with FCS Express 6 software.
Pyrosequencing.
Cells were treated with 10 μM 5-aza-2’-deoxycytidine for 72 hours and harvested for genomic DNA isolation with Blood and Cell Culture DNA Mini kit (Qiagen). 2 μg genomic DNA from each sample was used for bisulfite conversion with EpiTect Fast DNA Bisulfite kit (Qiagen). PyroMark PCR kit (Qiagen) was used for subsequent PCR reactions. Three pairs of primers amplifying three regions, each containing 2 CpG sites within 1500bp upstream of TSS, were designed with Qiagen PyroMark Assay Design Software 2.0. PyroMark Q24 sequencer (Qiagen) was used to analyze amplified PCR products, and methylation of CpG sites was determined with PyroMark Q24 Advanced software.
Proliferation assay.
RWPE-1 cells were transfected with control and SERPINB1 siRNA for 72 hours. Proliferation was assessed using the BrdU Cell Proliferation Assay Kit (Cell Signaling) [6].
Animal studies.
Experiments were performed in accordance with the guidelines for the Care and Use of Laboratory Animals and approved by the University Committee on Animal Resources at the University of Rochester. 6-8 week old male athymic nude mice were subcutaneously injected with 2×106 C4-2 cells (expressing SERPINB1 or vector) in 0.1 mL of a 1:1 mixture of Matrigel (Corning) and PBS. Tumor growth was monitored over 12 weeks. Once largest tumors reached end point (~2000 mm3), all tumors were harvested. Work with prostate-specific PbCre4/Ptenfl/fl mice in C57/BL6 background [28] was approved by the Roswell Park IACUC. Tumor volume was monitored using 3D ultrasound [29]. Mice bearing tumors of 300-500 mm3 were blindly randomized to vehicle (PBS) or sivelestat (Tocris; 5 mg/kg in PBS) treatment daily via intraperitoneal injection. Tumor volume was monitored weekly via ultrasound.
Flow cytometry for MDSCs.
Blood was collected from retro-orbital sinuses at indicated times and processed for CD11b, Ly6G, and Ly6C staining as previously described [6].
Immunohistochemistry.
Sections were de-paraffinized and rehydrated [6, 25]. Heat-mediated antigen retrieval was performed in 0.01 M Citrate pH 6 at 95°C. For human tissues, mouse anti-human SERPINB1 (#TA800093, Origene) was diluted 1:200 in antibody diluent (Thermo Scientific) and incubated overnight at 4°C. Biotinylated horse anti-mouse IgG (#BA-2000, Vector Laboratories) was diluted 1:200 in blocking serum (2.5% normal horse serum in PBS, Vector Laboratories). For mouse tissues, rabbit anti-mouse SERPINB1 (#TA340203, Origene) was diluted 1:200 in antibody diluent and incubated overnight at 4 °C. Biotinylated goat anti-rabbit IgG (#BA-1000, Vector Laboratories) was diluted 1:200 in blocking serum (2.5% normal goat serum in PBS, Vector Laboratories). Immunoreactivity was detected using the Vectastain Elite ABC and DAB peroxidase substrate kits (Vector Laboratories). Sections were counterstained with hematoxylin and mounted using Cytoseal 60 (ThermoFisher Scientific).
Immunofluorescence.
Primary antibodies were: biotin rat anti-mouse Ly6G (1:50, #127604, BioLegend) and goat anti-mouse proliferating cell nuclear antigen (PCNA; 1:50, #sc-9857, Santa Cruz Biotechnology). Primary antibodies were detected using streptavidin-Alexa 488 (1:200, #S11223, ThermoFisher Scientific), donkey anti-rat Alexa Fluor 488 (1:200, #A21208, ThermoFisher Scientific), and donkey anti-goat Alexa Fluor 568 (1:200, #A11057, ThermoFisher Scientific). Sections were counterstained and mounted using VECTASHIELD antifade mounting media with DAPI (Vector Laboratories).
NE imaging.
Mice received 4 nanomoles of Neutrophil Elastase 680 FAST probe (Perkin Elmer) in 0.1 mL PBS via tail-vein injection. Activity was measured on excised tumors using fluorescent microscopy [6, 25] and intensity was analyzed using ImageJ v1.48 software.
Gene expression profiling.
Microarray datasets of prostate adenocarcinoma and normal tissue were accessed through the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) and queried for SERPINB1 expression. Chandran et al (GEO accession number: GSE6919 [30]), Varambally et al (GEO accession number: GSE3325 [31]), Arredouani et al (GEO accession number: GSE55945 [32]), and Taylor et al (GEO accession number: GSE21036 [33]) expression datasets were analyzed. Kaplan–Meier analysis for recurrence-free survival was performed using the Taylor et al dataset through the open web interface Project Betastasis (http://www.betastasis.com). Analysis for DNA methylation at the SERPINB1 promoter and SERPINB1 expression in cancer and normal tissue was performed using The Cancer Genome Atlas (TCGA) dataset through MethHC (http://methhc.mbc.nctu.edu.tw/php/index.php) [34].
Statistical analysis.
Data are presented as mean ± standard error of the mean (SEM). Comparison between two groups was performed using two-tailed t-test, unless otherwise indicated. Comparison between more than two groups was performed using one-way ANOVA with appropriate post-hoc testing. Statistical analyses were performed using GraphPad Prism 7.0 software, and significance defined as p<0.05.
Results
Neutrophil elastase (NE) inhibition reduces prostate tumor growth in immunocompetent Pten-null mice
We recently demonstrated that NE inhibition with sivelestat reduces human prostate cancer xenograft growth in athymic mice. Furthermore, as in our xenograft models, we showed that granulocytic myeloid cells (likely myeloid-derived suppressor cells or MDSCs) are elevated in peripheral blood and infiltrate prostate tumors of immunocompetent probasin-driven Pten-null mice. Accordingly, Pten-null prostate tumors exhibit enhanced NE activity [6].
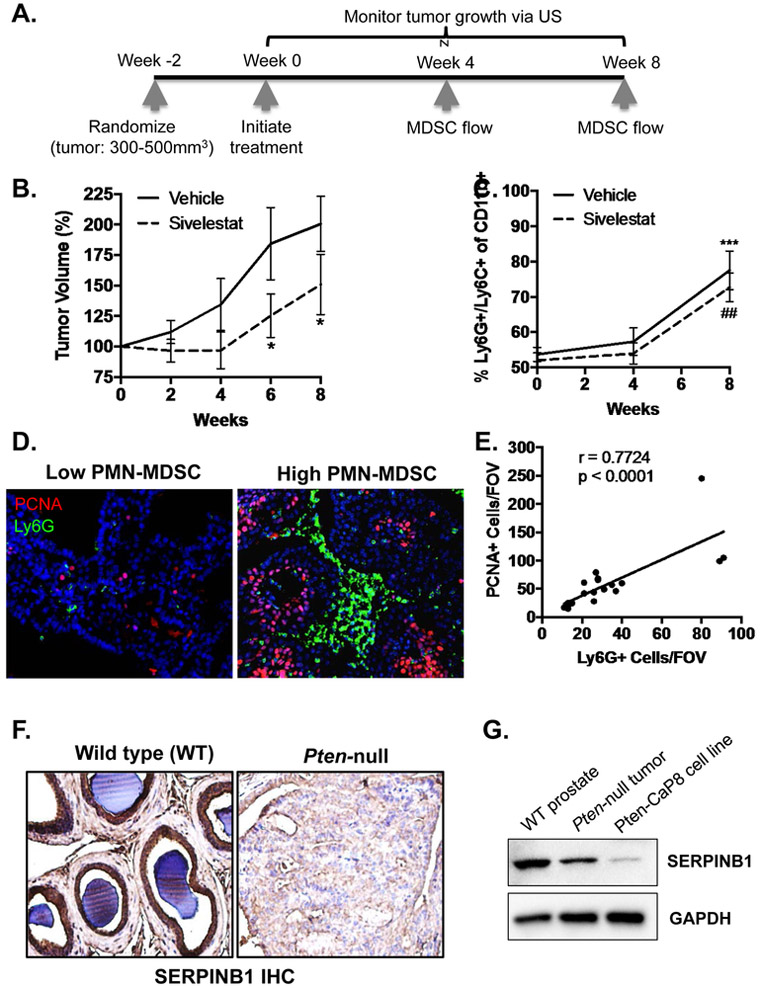
To determine whether NE is a pro-tumorigenic factor in Pten-null mice, we treated mice with sivelestat or vehicle and monitored tumor growth over eight weeks (Fig. 1A). Moreover, we assessed dynamics of peripheral blood MDSC accumulation via flow cytometry at zero, four, and eight weeks of treatment (Fig. 1A). We examined prostates by ultrasound to measure tumor volumes and initiated treatment at ~400 mm3. Tumors were smaller in sivelestat versus vehicle treated mice throughout the study, reaching statistical significance by the final two time points (Fig. 1B), suggesting that NE indeed promotes tumor growth in a prostate cancer mouse model with an intact immune system. Interestingly, we observed time dependent accumulation of MDSCs (CD11b+/Ly6G+/Ly6C+) in the blood of both sivelestat and vehicle treated animals (Fig. 1C), confirming that NE inhibition does not mitigate tumor induced MDSC production. Examination of MDSC infiltration within tumors via Ly6G immunofluorescence revealed that, while there was no difference between sivelestat and vehicle treated tumors (not shown), there was a strong positive correlation between infiltrating MDSCs and expression of proliferating cell nuclear antigen (PCNA) in prostate cells (Fig. 1D & E), supporting the hypothesis that MDSCs contribute to tumor proliferation.
Figure 1. Neutrophil elastase (NE) inhibition reduces prostate tumor growth in immunocompetent Pten-null mice.
A. Schematic illustrating important time points during the Pten-null experiment. Mice bearing tumors 300 – 500mm3 were randomized and treated daily with vehicle (PBS) or sivelestat (5mg/kg in PBS) via intraperitoneal (IP) injection. B. Ultrasound measurements were performed biweekly and are presented as percent volume increase from week 0. Tumor volume was compared using ANOVA with post-hoc Tukey HSD (n = 8 for vehicle, n = 8 for sivelestat; * p < 0.05). C. Peripheral blood MDSCs were assessed using flow cytometry at week 0, 4, and 8. The percent of Ly6G+/Ly6C+ cells at week 8 was compared to week 4 using ANOVA with post-hoc Tukey HSD (n = 8 for vehicle, n = 8 for sivelestat; *** p < 0.001 for vehicle ## p < 0.01 for sivelestat). D. Representative immunofluorescence stain for Ly6G positive infiltrating granulocytic MDSCs and PCNA positive proliferating epithelium in Pten-null prostates is shown. E. The number of PCNA positive cells versus the number of Ly6G positive cells per field of view (FOV) were plotted to examine correlation using two-tailed Pearson correlation analysis (n = 20; r = 0.7724, p < 0.0001). F. Representative immunohistochemistry stain for SERPINB1 in wildtype (WT) and Pten-null prostates is shown (n = 3 with identical results). G. Western blot for SERPBIN in wild-type prostate (WT), Pten-null prostates, and the Pten-null mouse derived CaP8 cell line.
Since sivelestat is a pharmacomimetic of the endogenous NE inhibitor SERPINB1 [35], we next examined SERPINB1 expression via immunohistochemistry in untreated Pten-null tumors and wild type mouse prostates. SERPINB1 strongly stained glandular epithelium of normal prostates but was reduced in the overgrown epithelium of Pten-null tumors (Fig. 1F & Supp. Fig. 1). We confirmed by Western blot that SERPINB1 was reduced in Pten-null tumors and the Pten-null prostate cancer cell line Pten-CaP8 compared to wild type prostate (Fig. 1G), suggesting that epithelial down-regulation of SERPINB1, along with increased MDSC infiltration, may contribute to the observed enhanced NE activity in Pten-null prostate tumors [6].
SERPINB1 mRNA expression is reduced and predicts poor prognosis in human prostate cancer
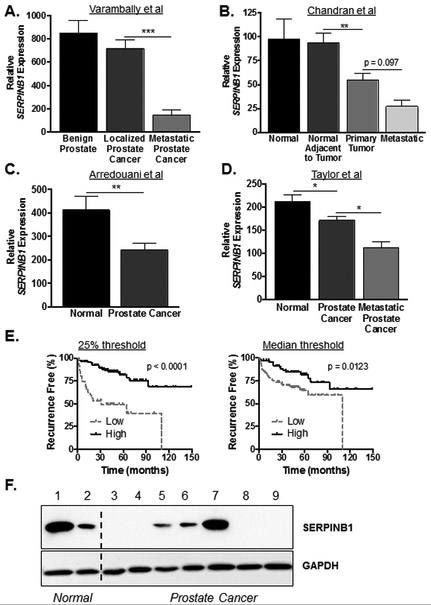
To determine whether SERPINB1 down-regulation occurs in human prostate cancer, we queried public datasets of prostate adenocarcinoma and normal tissue. Interestingly, SERPINB1 mRNA expression was reduced in prostate cancer compared to normal tissue in all four datasets, though the extent and pattern of reduction varied (Fig. 2A-D). Furthermore, SERPINB1 mRNA expression was lowest in metastatic prostate cancer (Fig. 2A, 2B, & 2D). Accordingly, using the Taylor et al dataset that contains clinical patient parameters, we found that low SERPINB1 mRNA expression significantly correlated with diminished recurrence free survival using both 50% and 25% cut-off thresholds (Fig. 2E).
Figure 2. SERPINB1 expression is reduced and predicts poor prognosis in human prostate cancer.
Human prostate cancer SERPINB1 mRNA expression data were obtained from (A) Varambally et al (Cancer Cell, 2005 [31]), (B) Chandran et al (BMC Cancer, 2007 [30]), (C) Arredouani et al (Clin Cancer Res, 2009 [32]), and (D) Taylor et al (Cancer Cell, 2010 [33]) through NCBI GEO. Differences between groups were assessed using ANOVA with post-hoc Holm-Sidak in A, B, and D. In C, difference was assessed using Student’s t-test. * p < 0.05, ** p < 0.01, *** p < 0.001. E. Kaplan-Meier plots for patients expressing SERPINB1 above (high) and below (low) the first quartile (left panel) and median (right panel) were constructed with Taylor et al data via the web interface Betastasis (http://www.betastasis.com ). Differences in recurrence free survival were assessed using the log-rank (Mantel-Cox) test. F. SERPINB1 expression in normal and prostate cancer cell lines were examined via Western blot; 1: RWPE-1, 2: BPH-1, 3: LNCaP, 4: C4-2, 5: DU145, 6: PC3, 7: 22Rv1, 8: VCaP, 9: LAPC-4.
To begin elucidating the functional significance of SERPINB1, we performed Western blots for SERPINB1 in human non-malignant and malignant cell lines. The most commonly used non-malignant prostatic epithelial cell line RWPE-1 exhibited strong SERPINB1 expression (Fig. 2F). The benign prostatic hyperplasia cell line BPH-1 expressed lesser amount of SERPINB1. Of the seven prostate cancer cell lines tested, four did not express detectable SERPINB1 (LNCaP, C4-2, VCaP, and LAPC-4), two expressed low levels of SERPINB1 (DU-145 and PC3), and one expressed equal amount of SERPINB1 (22Rv1), relative to RWPE-1.
SERPINB1 loss stimulates epithelial-to-mesenchymal (EMT) transition and proliferative phenotype in non-malignant prostate cells
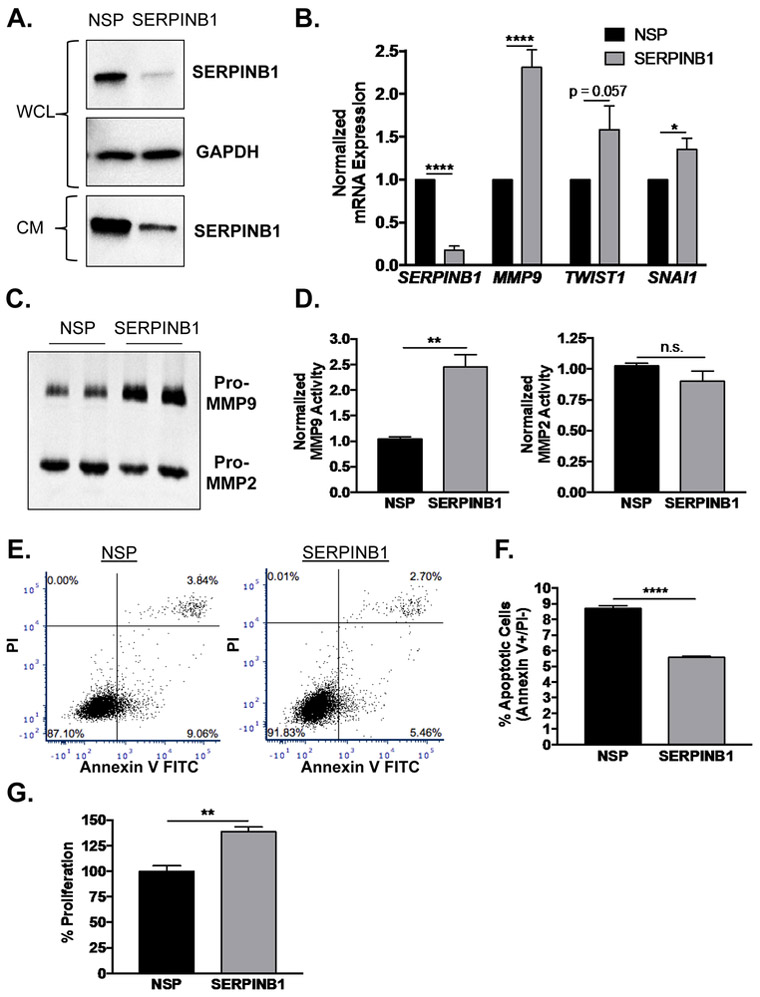
Since non-malignant RWPE-1 cells express abundant SERPINB1, we knocked down both cellular and secreted SERPINB1 expression by siRNA to determine how gene expression and physiology would be altered (Fig. 3A). Similar to gene arrays in other cancers [19, 20, 24], we found that mRNA expression of EMT-related genes MMP9, TWIST1, and SNAI1 were induced by SERPINB1 knockdown (Fig. 3B). Gelatin zymography on conditioned media collected from SERPINB1 and NSP siRNA treated RWPE-1 cells confirmed increased expression of pro-MMP9, but not pro-MMP2, activity (Fig. 3C, quantified in Fig. 3D). Since SERPINB1 has also been implicated in cell viability [12], we assessed effects of SERPINB1 loss on apoptosis via flow cytometry for PI and Annexin V. Apoptotic cells (Annexin V+/PI-) were significantly reduced by ~36% in response to SERPINB1 knockdown (Fig. 3E, quantified in Fig. 3F). Furthermore, proliferation by BrdU was increased by ~39% with SERPINB1 knockdown (Fig. 3G). Together, these data suggest that SERPINB1 loss contributes to EMT, enhanced proliferation, and reduced apoptosis in normal prostatic epithelial cells and may consequently drive malignant transformation.
Figure 3. SERPINB1 loss stimulates EMT and a proliferative phenotype in normal prostate cells.
A. RWPE-1 cells were transfected with non-specific (NSP) and SERPINB1-specific siRNA, and knockdown was verified using Western blot. GAPDH was used as a loading control. A representative blot is shown. WCL=whole cell lystate, CM=complete media (secreted SERPBINB1). B. mRNA expression of SERPINB1 and EMT markers MMP9, TWIST1, SNAI1 were determined in RWPE-1 cells after SERPINB1 knockdown using quantitative PCR and normalized to GAPDH. Data were normalized to NSP treated samples, and differences were assessed using Student’s t-test (n = 7; * p < 0.05, **** p < 0.0001). C. Expression and activity of MMP2 and MMP9 were determined in RWPE-1 cells after SERPINB1 knockdown using gelatin zymography. A representative gel is shown. D. Band densitometry of MMP9 and MMP2 was performed using ImageJ, and normalized differences were assessed using Student’s t-test (n = 3; ** p < 0.01, n.s. = not significant). E. Apoptosis was assessed in RWPE-1 cells after transfection with NSP and SERPINB1 siRNA via PI and Annexin V staining. Representative plots are shown. F. Difference in apoptosis (% Annexin V+/PI− cells) was determined using Student’s t-test (n = 3; **** p < 0.0001). G. Proliferation was assessed in RWPE-1 cells after transfection with NSP and SERPINB1 siRNA via colorimetric BrdU incorporation assay. Data were normalized to NSP treated samples, and difference was assessed using Student’s t-test (n = 3; ** p < 0.01).
SERPINB1 expression is induced via ERK1/2 signaling in normal prostate but not prostate cancer cells
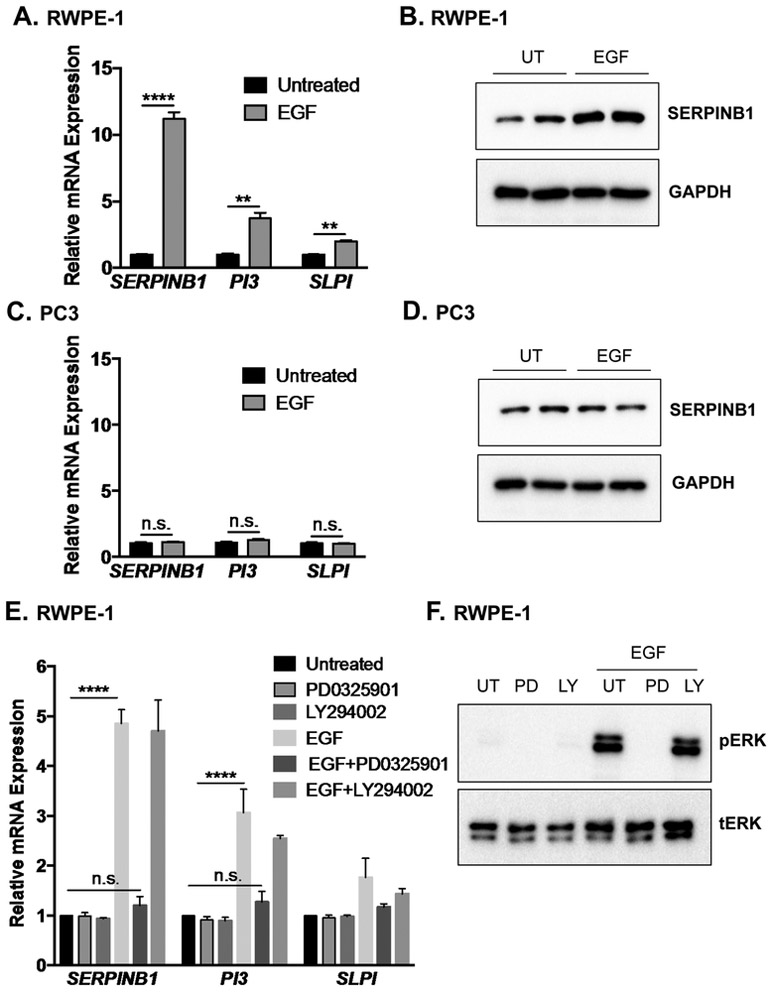
To determine how SERPINB1 is repressed in prostate cancer, we first examined regulation of SERPINB1 expression in non-malignant prostatic epithelial cells. We focused on epidermal growth factor (EGF) induced pathways, hypothesizing that enhanced growth factor signaling as seen in cancers might suppress SERPINB1 expression. Unexpectedly, EGF significantly induced SERPINB1 as measured by qPCR and Western blot in RWPE-1 cells (Fig. 4A & 4B). EGF induced expression of mRNAs encoding other endogenous NE inhibitors, PI3 (elafin) and SLPI (secretory leukocyte protease inhibitor), though less dramatically (Fig. 4A). In contrast, EGF did not increase SERPINB1 mRNA or SERPINB1 protein expression in PC3 (Fig. 4C & 4D) or DU145 prostate cancer cells (Supp. Fig. 2A & 2B), which express low but detectable SERPINB1 at baseline (Fig. 2F). Moreover, EGF was unable to induce SERPINB1 mRNA in LNCaP prostate cancer cells that do not express endogenous SERPINB1 (Supp. Fig. 2C). In RWPE-1 cells, MEK-inhibition by PD0325901 abrogated EGF-induced SERPINB1 transcription whereas the PI3K inhibitor had no effect (Fig. 4E), with similar but less dramatic results seen when examining PI3 and SLPI transcription (Fig. 4E). Thus, SERPINB1 expression in non-malignant RWPE-1 cells may be regulated in part by ERK1/2 signaling.
Figure 4. SERPINB1 expression is induced via ERK1/2 signaling in normal prostate but not prostate cancer cells.
A. RWPE-1 cells were serum starved and treated with epidermal growth factor (EGF; 20 ng/mL) for 4 hours. mRNA expression of SERPINB1, PI3, and SLPI were determined using quantitative PCR and normalized to GAPDH. Data were normalized to untreated samples, and differences were assessed using Student’s t-test (n = 3; ** p < 0.01, *** p < 0.001). B. RWPE-1 cells were serum starved and left untreated (UT) or treated with EGF (20 ng/mL) for 24 hours. SERPINB1 expression was determined using Western blot. GAPDH was used as a loading control. A representative blot with two of each treatment condition is shown. C. PC3 cells were serum starved and left untreated or treated with EGF (20 ng/mL) for 4 hours. mRNA expression of SERPINB1, PI3, and SLPI were determined using quantitative PCR and normalized to GAPDH. Data were normalized to untreated samples, and differences were assessed using Student’s t-test (n = 3, n.s. = not significant). D. PC3 cells were serum starved and treated with EGF (20 ng/mL) for 24 hours. SERPINB1 expression was determined using Western blot. GAPDH was used as a loading control. A representative blot with two of each treatment is shown. E. RWPE-1 cells were serum starved and treated with EGF (20 ng/mL) in the presence of MEK inhibitor PD0325901 (0.25 μM) or PI3K inhibitor LY294002 (0.25 μM) for 4 hours. mRNA expression of SERPINB1, PI3, and SLPI were determined using quantitative PCR and normalized to GAPDH. Data were normalized to untreated samples, and differences were assessed using ANOVA with post-hoc Tukey (**** p < 0.0001, n.s. = not significant). F. RWPE-1 cells were serum starved and treated with EGF (20 ng/mL) in the presence of PD0325901 (0.25 μM) or LY294002 (0.25 μM) for 30 minutes. pERK1/2 and tERK1/2 levels were examined via Western blot.
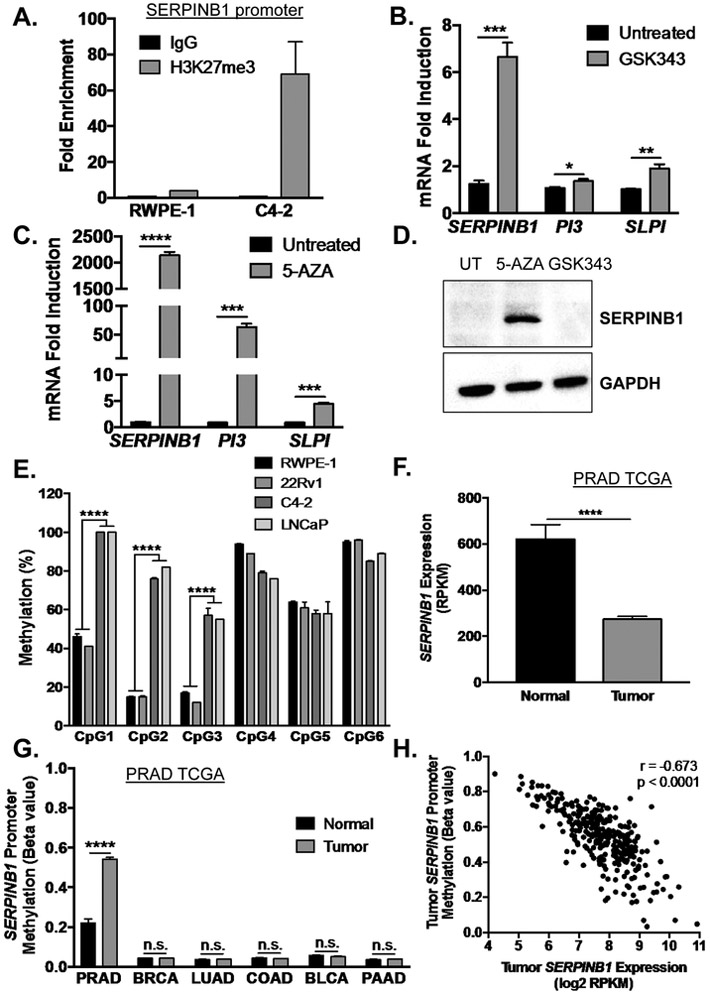
SERPINB1 is repressed by EZH2-mediated histone methylation and DNMT-mediated DNA methylation in prostate cancer
If EGF was not promoting SERPINB1 expression in tumor cells, then we hypothesized that some signal was preventing the EGF effects. Tumor suppressors are frequently turned off via epigenetic alterations such as histone and DNA methylation in cancers. Because EZH2 is up-regulated in and considered a driver of prostate cancer, we performed chromatin immunoprecipitation (ChIP) for histone 3 lysine 27 tri-methylation (H3K27me3) on the SERPINB1 promoter in non-malignant RWPE-1 versus castration resistant prostate cancer C4-2 cells. We observed marked enrichment of H3K27me3 in C4-2 compared to RWPE-1 cells (Fig. 5A). 72-hour treatment of C4-2 cells with an EZH2 inhibitor (GSK343) modestly induced expression of SERPINB1 mRNA, with minimal induction of PI3 and SLPI mRNA (Fig. 5B). Since this effect was relatively small, we examined the effects of DNA methyltransferase (DNMT) inhibition on SERPINB1 expression. 72-hour treatment of C4-2 cells with 5-aza-2-deoxycytidine (5-AZA) enhanced SERPINB1 mRNA expression by over 2,000-fold, comparable to transcript levels detected in RWPE-1 cells (Fig. 5C). Moreover, 5-AZA, but not GSK343, enhanced SERPINB1 protein expression in C4-2 cells (Fig. 5D). Similar effects on mRNA levels were observed in LNCaP prostate cancer cells (Supp. Fig. 3A & 3B).
Figure 5. SERPINB1 is epigenetically repressed by EZH2-mediated histone methylation and DNMT-mediated DNA methylation in prostate cancer.
A. Chromatin immunoprecipitation for H3K27me3 on the SERPINB1 promoter was performed in RWPE-1 and C4-2 cells. Data are presented as fold enrichment over IgG controls, and the average of two experiments is shown. B. C4-2 cells were treated with EZH2 inhibitor GSK343 (10 μM) for 72 hours. mRNA expression of SERPINB1, PI3, and SLPI were determined using quantitative PCR and normalized to GAPDH. Data were normalized to untreated samples, and differences were assessed using Student’s t-test (n = 3; * p < 0.05, ** p < 0.01, *** p <0.001). C. C4-2 cells were treated with DNMT inhibitor 5-aza-2-deoxycytidine (5-AZA, 10 μM) for 72 hours. mRNA expression of SERPINB1, PI3, and SLPI were determined using quantitative PCR and normalized to GAPDH. Data were normalized to untreated samples, and differences were assessed using Student’s t-test (n = 3; *** p < 0.001, **** p < 0.0001). D. C4-2 cells were treated with GSK343 and 5-AZA (10 μM) for 72 hours. SERPINB1 expression was determined using Western blot. GAPDH was used as a loading control. A representative blot is shown E. Promoter methylation at six CpG sites in C4-2, LNCaP, 22Rv1, and RWPE-1 cells was measured using pyrosequencing (CpG1 is the island closest to the transcription start site), and differences were determined using Student’s t-test (n = 4, *** p < 0.001). F. SERPINB1 expression in TCGA PRAD was assessed, and difference was determined using Student’s t-test (**** p < 0.0001). G. Analysis for DNA methylation at the SERPINB1 promoter in normal and cancer samples of various malignancies was performed using The Cancer Genome Atlas (TCGA) dataset through the open web interface MethHC (http://methhc.mbc.nctu.edu.tw/php/index.php) [34]. PRAD - prostate adenocarcinoma, BRCA - breast invasive carcinoma, LUAD - lung adenocarcinoma, COAD - colon adenocarcinoma, BLCA – bladder urothelial carcinoma, and PAAD - pancreatic adenocarcinoma. Differences were assessed using Student’s t-test (**** p < 0.0001, n.s. = not significant). H. SERPINB1 methylation versus expression were plotted to examine correlation using two-tailed Pearson correlation analysis (r = −0.673, p < 0.0001).
Accordingly, pyrosequencing analysis on the SERPINB1 promoters revealed that CpG1, CpG2, and CpG3, the islands closest to the transcription start site, had markedly elevated DNA methylation in C4-2 cells compared to non-malignant RWPE-1 cells (Fig. 5E). 22Rv1 cancer cells that expressed comparable level of SERPINB1 as RWPE-1 also exhibited a similar pattern of CpG methylation as RWPE-1 (Fig. 5E). Conversely, LNCaP cancer cells that lacked endogenous SERPINB1 had a methylation pattern that closely resembled C4-2 cells (Fig. 5E). Furthermore, in assessing DNA methylation at the SERPINB1 promoter in various cancers using TCGA data available through the open web interface MethHC, only prostate adenocarcinoma (PRAD) exhibited increased methylation relative to adjacent normal tissue (Fig. 5G). Similar to individual prostate cancer expression datasets (Fig. 2A-D), SERPINB1 mRNA expression was significantly reduced in tumor compared to normal samples using the PRAD TCGA (Fig. 5F), with SERPINB1 promoter methylation negatively correlating with SERPINB1 mRNA expression in tumors (Fig. 5H). These data suggest that DNA methylation may silence SERPINB1 expression specifically in human prostate cancer versus other cancers.
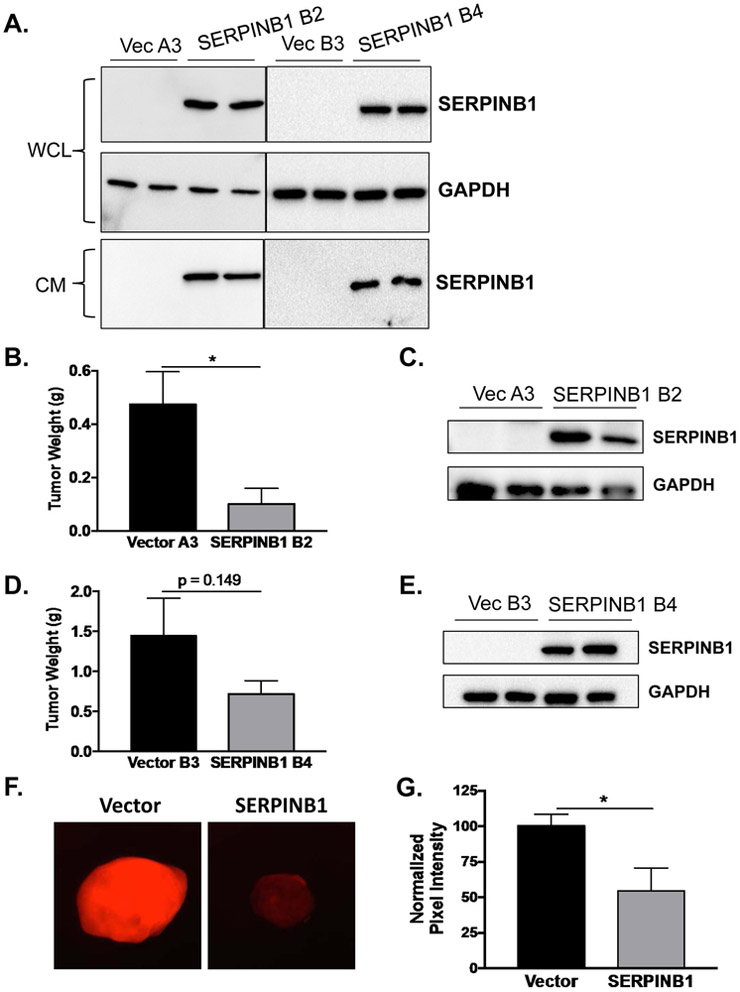
SERPINB1 overexpression reduces prostate cancer xenograft growth in vivo
We next determined whether rescuing SERPINB1 expression in normally low-expressing prostate cancer cells would diminish tumor burden. We therefore generated two stable monoclonal cell lines overexpressing SERPINB1 or vector control in C4-2 cells (Fig. 6A). Notably, these cells expressed and secreted SERPINB1 just like RWPE-1 (Fig. 6A). Xenografts expressing SERPINB1 were markedly smaller than vector control xenografts after 12 weeks of growth in athymic nude mice (Fig. 6B & D). Stable SERPINB1 overexpression was verified in tumors of both clonal cell lines collected at the end of the experiment (Fig. 6C & E). Using a NE activity probe [6], we observed reduced NE activity in SERPINB1 overexpressing tumors compared to vector controls (Fig. 6F and 6G, quantified), suggesting that SERPINB1 inhibits local NE activity to attenuate prostate cancer xenograft growth.
Figure 6. SERPINB1 overexpression reduces prostate cancer xenograft growth in vivo.
A. SERPINB1 was detected in whole cell lysates (WCL) and concentrated conditioned media (CM) collected over 24 hours in C4-2 cells stably expressing SERPINB1 (SERPINB1 clones B2 and B4) and vector controls (Vector clones A3 and B3). GAPDH was used as a loading control. Representative blots are shown. B. Tumor weight was compared between Vector A3 and SERPINB1 B2 xenografts injected subcutaneously into the flanks of athymic nude mice using Student’s t-test (n = 10 for Vector, n = 3 for SERPINB1; * p < 0.05). C. SERPINB1 was detected in harvested tumors at the end of the experiment using Western blot. A representative blot is shown. D. Tumor weight was compared between Vector B3 and SERPINB1 B4 xenografts injected subcutaneously into the flanks of athymic nude mice using Student’s t-test (n = 10 for Vector, n = 11 for SERPINB1; p = 0.149). SERPINB1 was detected in harvested tumors at the end of the experiment using Western blot. A representative blot is shown. F. Intra-tumoral neutrophil elastase activity was measured ex vivo using an NE specific optical probe. G. Intra-tumoral NE activity was quantified using ImageJ and normalized to Vector controls. Difference was assessed using Student’s t-test (n = 8 for Vector, n = 4 for SERPINB1; * p < 0.05).
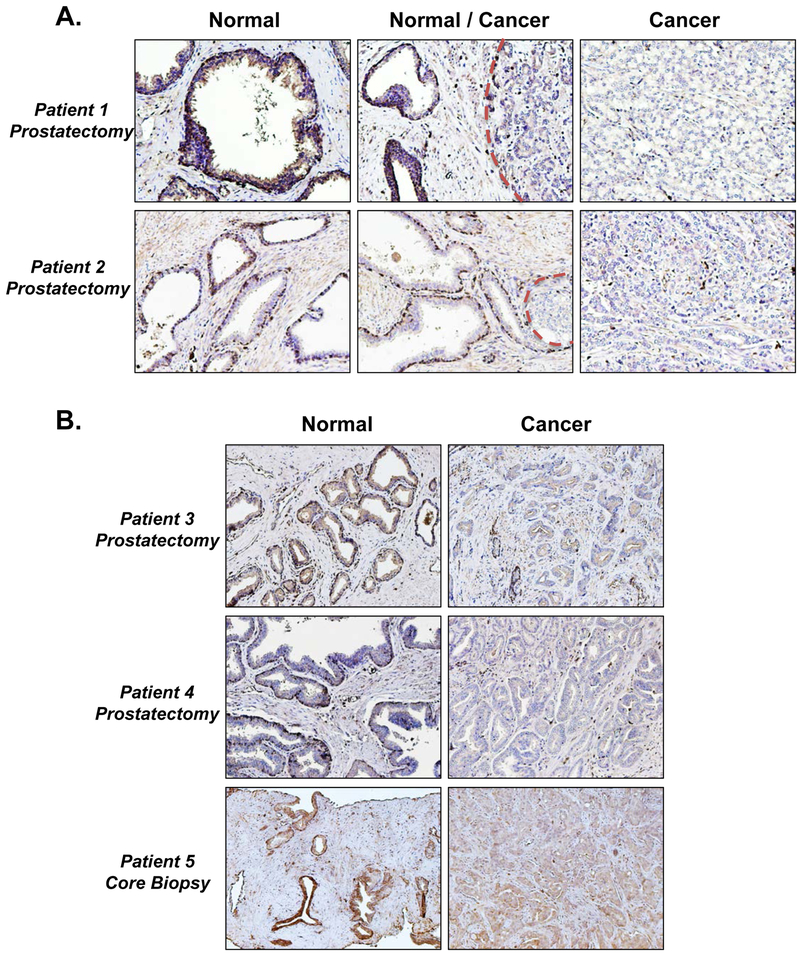
SERPINB1 is expressed by normal human prostate glands but reduced in prostate cancer
Finally, we confirmed loss of SERPINB1 expression in prostate cancer by performing immunohistochemistry on prostatectomy and core biopsy specimens that contain regions of both normal and malignant tissue. As expected, SERPINB1 was localized to cells lining normal prostate glands and exhibited diffuse stromal staining, both of which were largely absent in cancer (Fig. 7). Scattered SERPINB1 positive cells present in cancer regions were likely infiltrating immune cells expressing SERPINB1. Therefore, epithelial and/or stromal down-regulation of SERPINB1 may serve as a novel biomarker for prostate cancer progression.
Figure 7. SERPINB1 is expressed by normal human prostate glands but reduced in prostate cancer.
A. Representative immunohistochemistry stains for SERPINB1 in human prostatectomy specimens, showing regions of normal, normal and cancer, and cancer only. Dotted red lines outline cancer foci adjacent to normal prostate glands. B. Representative immunohistochemistry stains for SERPINB1 in human prostatectomy and core biopsy specimens, showing regions of normal and cancer.
Discussion
Granulocytic myeloid cells or MDSCs are increased in the circulation, peripheral organs, and primary tumors in mouse models of numerous malignancies, including prostate cancer [36, 37]. MDSCs contribute to cancer progression and metastasis indirectly via T cell immunosuppression and directly via release of pro-inflammatory cytokines, growth factors, and proteases such as matrix metalloproteinases (MMPs) and neutrophil elastase (NE). Consequently, targeting MDSC production and recruitment leads to decelerated tumor growth and reduced metastases [5, 36]. We reported that myeloid-derived NE directly promotes human prostate cancer growth in athymic mice, and that sivelestat (NE inhibitor) reduces tumor burden to the same extent as anti-Gr-1 MDSC depletion [6]. We now demonstrate that targeting NE activity with sivelestat in the immunocompetent Pten-null mouse model of prostate cancer similarly attenuates tumor progression without altering levels of circulating or infiltrating MDSCs. Therefore, tumors continue to recruit pro-tumorigenic MDSCs despite reduced NE activity, which likely explains why the tumor inhibitory effect of sivelestat is somewhat diminished by the later time points when the MDSC burden is too great. MDSC depletion via antibodies against CXCL5 and Gr-1 or inhibitors targeting CXCR2 similarly result in modestly decelerated prostate tumor growth in Pten-null mouse models [38-40]. Unfortunately, MDSC levels rebound after pro-longed antibody-mediated depletion, thus the translational potential of such therapy is uncertain [36]. Therefore, we postulate that perhaps dual targeting of MDSCs and NE activity will lead to synergistic growth inhibitory effects.
Local MDSC recruitment likely drives the preponderance of immune-derived proteases in prostate and other cancers, and in fact we find that, in Pten-null mice, prostate cancer cell proliferation directly correlates with the density of surrounding MDSCs. Alternatively, while inflammatory protease levels are increasing in the tumor microenvironment, expression of endogenous protease inhibitors by epithelial cells is often suppressed, favoring enhanced proteolytic activity and resultant cancer cell proliferation, migration, and invasion [41]. For instance, loss of Timp3, an endogenous MMP inhibitor, accelerates tumor growth and invasion of Pten-null prostate tumors via up-regulated MMP activity [42]. Here, we are first to demonstrate that SERPINB1, an endogenous NE inhibitor, is reduced in Pten-null mouse tumors, which might further explain their heightened intra-tumoral NE activity [6].
SERPINB1 is a potent regulator of neutrophil serine proteases and neutrophil extracellular trap (NET) formation [13]. Consequently, SERPINB1 contributes to the resolution of both acute infections and chronic inflammatory diseases, similar to the therapeutic effects of its pharmacomimetic sivelestat [12]. SERPINB1 is widely expressed in various cell types, and several studies suggest a role in cancer progression separate from its NE inhibitory function. In hepatocellular carcinoma (HCC), glioma, and melanoma, SERPINB1 down-regulation in tumors is associated with poor patient prognosis [19-21]. Intriguingly, two exploratory studies identify SERPINB1 as a down-regulated gene, among many others, in prostate cancer compared to non-malignant prostate [22, 23]. We now demonstrate that SERPINB1 is in fact reduced in human prostate cancer compared to normal prostatic epithelium using a variety of transcriptional and translational approaches. SERPINB1 is further reduced in metastatic disease, and low expression predicts diminished recurrence free survival in patients. Immunohistochemistry staining reveals SERPINB1 is strongly expressed by the basal layer in normal prostate glands, which is largely absent in regions of cancer. Interestingly, there is strong evidence for the basal progenitor as the cell type of origin in both mouse models and human prostate cancer [43, 44]. Moreover, loss of tumor suppressor Pten in basal cells promotes basal-to-luminal differentiation and development of invasive prostate cancer in mice [45]. Accordingly, we observe decreased SERPINB1 expression in mouse Pten-null prostates as well as a Pten-null prostate cell line. It is therefore possible that SERPINB1 loss also plays a critical role during prostate tumorigenesis, though this remains to be carefully dissected.
The functional role of SERPINB1 in cancer at this time is unclear, though limited studies suggest an involvement in regulation of cell motility. For instance, SERPINB1 reduction in hepatocellular carcinoma (HCC) and glioma cells enhances migration and invasion in vitro, potentially through increased MMP-2 expression [19, 20]. Conversely, SERPINB1 overexpression in lung and breast cancer cells decreases migratory and invasive capacity [46]. Here we demonstrate that SERPINB1 loss in non-malignant prostatic epithelial cells induces expression of EMT markers, including MMP-9, TWIST1, and SNAI1. EMT is the process by which polarized epithelial cells acquire a mesenchymal, pro-migratory, and pro-invasive phenotype, and activation of an EMT program is considered a critical step in malignant transformation [47, 48]. Therefore, as mentioned above, loss of SERPINB1 may be an early event during tumorigenesis. Accordingly, a meta-analysis of 18 independent gene expression datasets of EMT identified SERPINB1 as a commonly down-regulated gene in various cancers [24].
In addition to EMT, two hallmarks of cancer are the ability of tumor cells to resist cell death and sustain proliferative signaling [48]. Here, we report that SERPINB1 loss impedes apoptosis and stimulates proliferation in normal prostatic epithelial cells. SERPINB1 is proposed to control cell viability via interaction with several proteins involved in apoptosis, including PARP-1, BCL-2, and apoptosis inducing factor (AIF), among others [12]. Moreover, SERPINB1 loss induces proliferation in keratinocytes by promoting G1/S cell cycle transition. [49]. Nonetheless, the effect of ectopic SERPINB1 overexpression on tumor growth in vivo has not been previously examined. Here, we demonstrate that stable ectopic expression of SERPINB1 in prostate cancer cells with low endogenous SERPINB1 expression reduces xenograft growth and NE activity in vivo. We also report SERPINB1 secretion by both normal prostatic epithelial cells and ectopically expressing SERPINB1 prostate cancer cells. Together, these data suggest that extracellular SERPINB1 may act as a functional NE inhibitor in vivo. Consequently, down-regulation of SERPINB1 by cancer cells may permit NE activation within the tumor microenvironment to enhance tumor progression.
Still, how SERPINB1 is regulated in normal tissues and cancer is not well understood. In this study, we demonstrate that SERPINB1 is strongly induced by epidermal growth factor (EGF) signaling through the ERK1/2 pathway in non-malignant but not malignant prostatic epithelial cells. Furthermore, we demonstrate a novel mechanism of SERPINB1 repression in prostate cancer cells via EZH2-mediated histone methylation (H3K27me) and DNA methyltransferase (DNMT)-mediated DNA methylation. While EZH2 inhibition modestly induces SERPINB1 expression, DNMT inhibition induces SERPINB1 expression by over 2,000-fold and rescues protein expression from previously undetectable levels. EZH2 may facilitate gene silencing directly through histone methylation and indirectly by serving as a platform for DNMT complex recruitment [50]. Thus, the two epigenetic repression systems may be functionally linked, and both are likely responsible for SERPINB1 silencing during the complex process of tumor initiation and progression. Accordingly, EZH2 and various DNMTs (i.e. DNMT1, DNMT3A, DNMT3B) are overexpressed in mouse and human prostate cancers [51, 52].
Remarkably, SERPINB1 promoter methylation appears to be specific to prostate cancer, as no other human cancers examined through the TCGA exhibit such a stark difference compared to their normal tissue counterparts. Not surprisingly, we observe that promoter methylation and SERPINB1 expression are tightly correlated in prostate cancer. Therefore, promoter methylation may serve as a surrogate marker of SERPINB1 expression and have diagnostic and prognostic potential. Furthermore, SERPINB1 down-regulation, combined with elevated markers of inflammation such as neutrophil to lymphocyte ratio (NLR), may identify prostate cancer patients who will best respond to MDSC or NE-targeting therapy. Better mechanistic understanding of the methyltransferases responsible for SERPINB1 repression will be advantageous for future drug and diagnostic test development. Analogously, GSTP1 is a tumor suppressor silenced via promoter methylation in prostate cancer, and this epigenetic alteration may be evaluated clinically through the ConfirmMDx assay [53].
In summary, our findings demonstrate that SERPINB1 is a novel tumor suppressor that is epigenetically silenced in prostate cancer. While SERPINB1 loss may permit NE-driven prostate cancer progression, it also exhibits significant NE-independent tumor promoting effects. Importantly, SERPINB1 expression and promoter methylation status may serve as a potential biomarker to guide therapeutic decisions.
Supplementary Material
Implications:
Our findings suggest that the balance between SERPINB1 and NE is physiologically important within the prostate and may serve as a biomarker and therapeutic target in prostate cancer.
Acknowledgements
S. Hammes was supported by NIH grants R01GM101709 and R01CA193583-01A1. I. Lerman was supported by NIH grant F30CA203517.
Abbreviations:
- MDSC
myeloid derived suppressor cell
- NE
neutrophil elastase
Footnotes
COI: The authors declare no potential conflicts of interest
References
- 1.Litwin MS and Tan HJ, The Diagnosis and Treatment of Prostate Cancer: A Review. JAMA, 2017. 317(24): p. 2532–2542. [DOI] [PubMed] [Google Scholar]
- 2.Gaudreau PO, et al. , The Present and Future of Biomarkers in Prostate Cancer: Proteomics, Genomics, and Immunology Advancements. Biomark Cancer, 2016. 8(Suppl 2): p. 15–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strasner A and Karin M, Immune Infiltration and Prostate Cancer. Front Oncol, 2015. 5: p. 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barron DA and Rowley DR, The reactive stroma microenvironment and prostate cancer progression. Endocr Relat Cancer, 2012. 19(6): p. R187–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lerman I and Hammes SR, Neutrophil elastase in the tumor microenvironment. Steroids, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lerman I, et al. , Infiltrating Myeloid Cells Exert Protumorigenic Actions via Neutrophil Elastase. Mol Cancer Res, 2017. 15(9): p. 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan AL, et al. , Neutrophil elastase reduces secretion of secretory leukoproteinase inhibitor (SLPI) by lung epithelial cells: role of charge of the proteinase-inhibitor complex. Respir Res, 2008. 9: p. 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Wetering S, et al. , Regulation of secretory leukocyte proteinase inhibitor (SLPI) production by human bronchial epithelial cells: increase of cell-associated SLPI by neutrophil elastase. J Investig Med, 2000. 48(5): p. 359–66. [PubMed] [Google Scholar]
- 9.Hunt KK, et al. , Elafin, an inhibitor of elastase, is a prognostic indicator in breast cancer. Breast Cancer Res, 2013. 15(1): p. R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benarafa C, et al. , Characterization of four murine homologs of the human ov-serpin monocyte neutrophil elastase inhibitor MNEI (SERPINB1). J Biol Chem, 2002. 277(44): p. 42028–33. [DOI] [PubMed] [Google Scholar]
- 11.Farley K, et al. , A serpinB1 regulatory mechanism is essential for restricting neutrophil extracellular trap generation. J Immunol, 2012. 189(9): p. 4574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torriglia A, Martin E, and Jaadane I, The hidden side of SERPINB1/Leukocyte Elastase Inhibitor. Semin Cell Dev Biol, 2017. 62: p. 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benarafa C, The SerpinB1 knockout mouse a model for studying neutrophil protease regulation in homeostasis and inflammation. Methods Enzymol, 2011. 499: p. 135–48. [DOI] [PubMed] [Google Scholar]
- 14.Benarafa C, et al. , SerpinB1 protects the mature neutrophil reserve in the bone marrow. J Leukoc Biol, 2011. 90(1): p. 21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heit C, et al. , Update of the human and mouse SERPIN gene superfamily. Hum Genomics, 2013. 7: p. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooley J, et al. , SerpinB1 in cystic fibrosis airway fluids: quantity, molecular form and mechanism of elastase inhibition. Eur Respir J, 2011. 37(5): p. 1083–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Ouaamari A, et al. , SerpinB1 Promotes Pancreatic beta Cell Proliferation. Cell Metab, 2016. 23(1): p. 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller M, et al. , Active caspase-1 is a regulator of unconventional protein secretion. Cell, 2008. 132(5): p. 818–31. [DOI] [PubMed] [Google Scholar]
- 19.Cui X, et al. , Decreased expression of SERPINB1 correlates with tumor invasion and poor prognosis in hepatocellular carcinoma. J Mol Histol, 2014. 45(1): p. 59–68. [DOI] [PubMed] [Google Scholar]
- 20.Huasong G, et al. , Serine protease inhibitor (SERPIN) B1 suppresses cell migration and invasion in glioma cells. Brain Res, 2015. 1600: p. 59–69. [DOI] [PubMed] [Google Scholar]
- 21.Willmes C, et al. , SERPINB1 expression is predictive for sensitivity and outcome of cisplatin-based chemotherapy in melanoma. Oncotarget, 2016. 7(9): p. 10117–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashida S, et al. , Molecular features of the transition from prostatic intraepithelial neoplasia (PIN) to prostate cancer: genome-wide gene-expression profiles of prostate cancers and PINs. Cancer Res, 2004. 64(17): p. 5963–72. [DOI] [PubMed] [Google Scholar]
- 23.Davalieva K, et al. , Proteomics analysis of malignant and benign prostate tissue by 2D DIGE/MS reveals new insights into proteins involved in prostate cancer. Prostate, 2015. 75(14): p. 1586–600. [DOI] [PubMed] [Google Scholar]
- 24.Groger CJ, et al. , Meta-analysis of gene expression signatures defining the epithelial to mesenchymal transition during cancer progression. PLoS One, 2012. 7(12): p. e51136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prizant H, et al. , Estrogen maintains myometrial tumors in a lymphangioleiomyomatosis model. Endocr Relat Cancer, 2016. 23(4): p. 265–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Light A and Hammes SR, LH-Induced Steroidogenesis in the Mouse Ovary, but Not Testis, Requires Matrix Metalloproteinase 2- and 9-Mediated Cleavage of Upregulated EGF Receptor Ligands. Biol Reprod, 2015. 93(3): p. 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma X, et al. , Androgens Regulate Ovarian Gene Expression Through Modulation of Ezh2 Expression and Activity. Endocrinology, 2017. 158(9): p. 2944–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia AJ, et al. , Pten null prostate epithelium promotes localized myeloid-derived suppressor cell expansion and immune suppression during tumor initiation and progression. Mol Cell Biol, 2014. 34(11): p. 2017–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh S, et al. , Ultrasound enhanced enzymatic hydrolysis of Parthenium hysterophorus: A mechanistic investigation. Bioresour Technol, 2015. 192: p. 636–45. [DOI] [PubMed] [Google Scholar]
- 30.Chandran UR, et al. , Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer, 2007. 7: p. 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varambally S, et al. , Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell, 2005. 8(5): p. 393–406. [DOI] [PubMed] [Google Scholar]
- 32.Arredouani MS, et al. , Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res, 2009. 15(18): p. 5794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor BS, et al. , Integrative genomic profiling of human prostate cancer. Cancer Cell, 2010. 18(1): p. 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang WY, et al. , MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res, 2015. 43(Database issue): p. D856–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turk B, Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov, 2006. 5(9): p. 785–99. [DOI] [PubMed] [Google Scholar]
- 36.Marvel D and Gabrilovich DI, Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Invest, 2015. 125(9): p. 3356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hossain DM, et al. , TLR9-Targeted STAT3 Silencing Abrogates Immunosuppressive Activity of Myeloid-Derived Suppressor Cells from Prostate Cancer Patients. Clin Cancer Res, 2015. 21(16): p. 3771–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bezzi M, et al. , Diverse genetic-driven immune landscapes dictate tumor progression through distinct mechanisms. Nat Med, 2018. 24(2): p. 165–175. [DOI] [PubMed] [Google Scholar]
- 39.Di Mitri D, et al. , Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature, 2014. 515(7525): p. 134–7. [DOI] [PubMed] [Google Scholar]
- 40.Wang G, et al. , Targeting YAP-Dependent MDSC Infiltration Impairs Tumor Progression. Cancer Discov, 2016. 6(1): p. 80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caruso JA, et al. , The serine protease inhibitor elafin maintains normal growth control by opposing the mitogenic effects of neutrophil elastase. Oncogene, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adissu HA, et al. , Timp3 loss accelerates tumour invasion and increases prostate inflammation in a mouse model of prostate cancer. Prostate, 2015. 75(16): p. 1831–43. [DOI] [PubMed] [Google Scholar]
- 43.Packer JR and Maitland NJ, The molecular and cellular origin of human prostate cancer. Biochim Biophys Acta, 2016. 1863(6 Pt A): p. 1238–60. [DOI] [PubMed] [Google Scholar]
- 44.Stoyanova T, et al. , Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc Natl Acad Sci U S A, 2013. 110(50): p. 20111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu TL, et al. , Conditionally ablated Pten in prostate basal cells promotes basal-to-luminal differentiation and causes invasive prostate cancer in mice. Am J Pathol, 2013. 182(3): p. 975–91. [DOI] [PubMed] [Google Scholar]
- 46.Chou RH, et al. , Suppression of the invasion and migration of cancer cells by SERPINB family genes and their derived peptides. Oncol Rep, 2012. 27(1): p. 238–45. [DOI] [PubMed] [Google Scholar]
- 47.Kalluri R and Weinberg RA, The basics of epithelial-mesenchymal transition. J Clin Invest, 2009. 119(6): p. 1420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanahan D and Weinberg RA, Hallmarks of cancer: the next generation. Cell, 2011. 144(5): p. 646–74. [DOI] [PubMed] [Google Scholar]
- 49.Hui L, et al. , Calcitriol inhibits keratinocyte proliferation by upregulating leukocyte elastase inhibitor (serpin B1). J Dermatol, 2014. 41(5): p. 393–8. [DOI] [PubMed] [Google Scholar]
- 50.Simon JA and Lange CA, Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res, 2008. 647(1-2): p. 21–9. [DOI] [PubMed] [Google Scholar]
- 51.Yang YA and Yu J, EZH2, an epigenetic driver of prostate cancer. Protein Cell, 2013. 4(5): p. 331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi Y, et al. , DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer. Genome Res, 2011. 21(7): p. 1017–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martignano F, et al. , GSTP1 Methylation and Protein Expression in Prostate Cancer: Diagnostic Implications. Dis Markers, 2016. 2016: p. 4358292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.