There have been several advances in local and systemic mRNA delivery[1], especially to hepatocytes[2]. However, low dose mRNA delivery to other cell types within the liver microenvironment remains challenging. Using a high-throughput DNA barcode screen named JORDAN, we observed that in vitro nanoparticle delivery can be a poor predictor of in vivo delivery[3]. Since in vitro assays typically lack a complete immune system[4], variable blood flow[5], heterogeneous vasculature, off-target cells, and other factors that influence delivery in vivo[6], we reasoned that an ideal nanoparticle experiment would study LNPs directly in vivo. We therefore developed FIND[7] to quantify how over 100 LNPs deliver functional mRNA into the cytoplasm of target cells in a single mouse.
LNPs are formulated by mixing nucleic acids with ionizable lipids, PEG-lipids, phospholipids, and cholesterol. The diversity of available chemistries enables scientists to evaluate how LNP structure affects delivery. However, these studies are mostly performed in vitro[8, 9] and use LNPs formulated with unmodified cholesterol. Several lines of evidence led us to hypothesize that cholesterol chemical structure affected LNP mRNA delivery. First, cholesterol structure is modified via oxidation; this requires specific enzymes, demonstrating that cells regulate cholesterol structure[10]. Second, oxidized cholesterols signal differently than unmodified cholesterols, altering interactions with membrane and nuclear receptors that regulate gene expression[11]. Third, cholesterol is trafficked in lipoproteins using forward and reverse transport[12] and trafficking to endothelial cells, hepatocytes, and macrophages may change with cholesterol structure in vivo[13]. Fourth, LNPs and lipoproteins have similar size and composition. Finally, using JORDAN, we found that modified cholesterols alter LNP biodistribution[14], however this study measured LNP biodistribution; it did not measure functional mRNA delivery. Given that less than 4% of exogenously delivered RNA escapes endosomes[15] and endosomal escape changes with cell type[16], it is difficult to predict functional mRNA delivery using biodistribution.
To test the hypothesis that cholesterol structure affects LNP delivery of mRNA, we created a library of LNPs with nine cholesterol variants. We chose variants that were (i) produced naturally or synthetically and (ii) had oxidative modifications on sterol ring B, the hydrocarbon tail attached to sterol ring D, or both. We formulated LNPs to carry DNA barcodes and Cre mRNA[7], systemically injected them into Cre reporter mice, and analyzed the efficiency with which they delivered Cre mRNA in 28 cells types in vivo. We identified an LNP that delivers mRNA to Kupffer cells and hepatic endothelial cells five-fold more potently than to hepatocytes. Notably, the lead LNPs performed as predicted by the FIND screen, suggesting that FIND can rapidly identify LNPs for mRNA delivery.
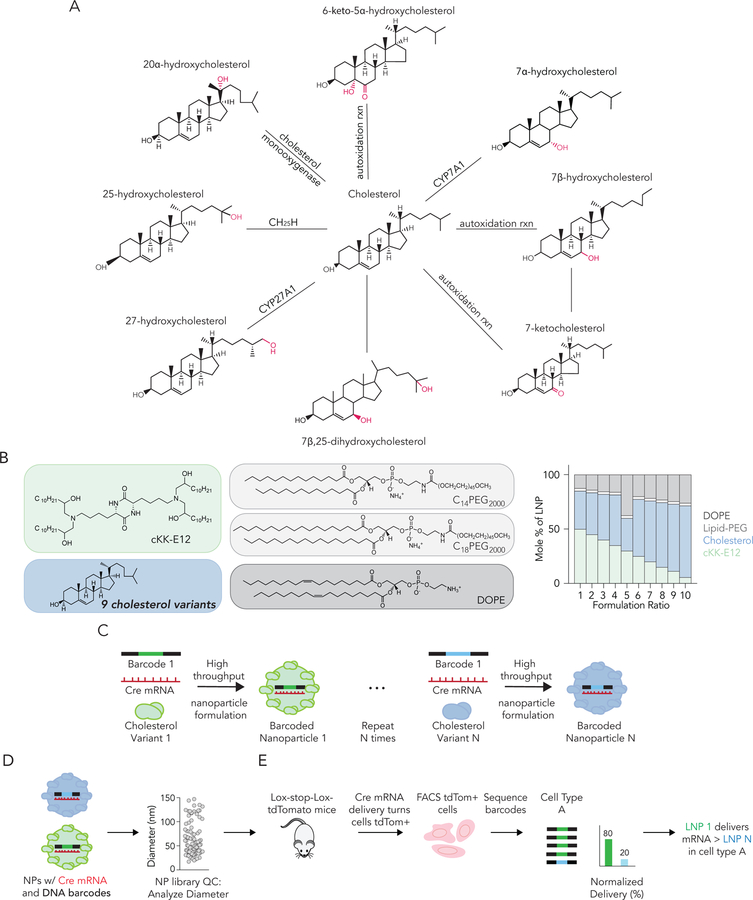
Unmodified cholesterol is acted upon by enzymes to form variants that are side-chain or ring oxidized (Figure 1A). To investigate whether these modifications altered LNP targeting, we formulated 125 FIND LNPs using microfluidics (Figure 1B). To minimize variation from components other than cholesterol, LNPs were made of the ionizable lipid-like material cKK-E12[8], two well validated PEG-lipids, the phospholipid 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), and one of nine different cholesterol variants (Figure 1B).
Figure 1.
Synthesizing a library of rationally designed nanoparticles containing oxidized cholesterols. (A) Cholesterol can be modified to form sterol variants that differentially act upon a number of biological pathways. Here we investigate whether cholesterol variants affected delivery. (B) We formulated 125 nanoparticles by combining the ionizable material cKK-E12, PEG-lipids, DOPE, and 9 different cholesterol variants. (C) These distinct nanoparticles were then barcoded and screened for functional mRNA delivery using FIND. (D) After formulation, barcoded nanoparticles are put through DLS-based quality control, pooled, and (E) intravenously administered to Ai14 mice. If Cre mRNA is delivered into the cytoplasm and translated into Cre protein that edits the genome, the cells become tdTomato+. tdTomato+ cells are isolated using FACS and barcodes are sequenced to identify nanoparticles that co-localize with cells transfected with Cre in vivo. This enables us to study how many LNPs deliver mRNA in a single mouse.
We formed LNPs by mixing the contents together in a microfluidic device[17]. Each LNP carried a unique DNA barcode, which acted as a tag for that LNP, as well as Cre mRNA, which signals functional mRNA delivery (Figure 1C). Stable LNPs were pooled together (Figure 1D) and administered to Ai14 mice at a total nucleic acid dose of 1.0 mg / kg. Ai14 mice contain a Lox-Stop-Lox-tdTomato construct under the control of a CAG promoter; as a result, cells in Ai14 mice become tdTomato+ if: (1) Cre mRNA is delivered into the cytoplasm, (2) Cre mRNA is translated into Cre protein, (3) Cre protein translocates from the cytoplasm into the nucleus, and (4) Cre protein edits the genome by removing the ‘Stop’ between Lox sites. Therefore, by isolating tdTomato+ cell types using fluorescence-activated cell sorting (FACS) and using next generation sequencing (NGS) to quantify barcodes within them, FIND identifies LNPs located in cells where functional mRNA delivery occurred (Figure 1E). We quantified NGS sequencing data as ‘normalized delivery’, analogous to counts per million in RNA-seq[7] (Figure 1E).
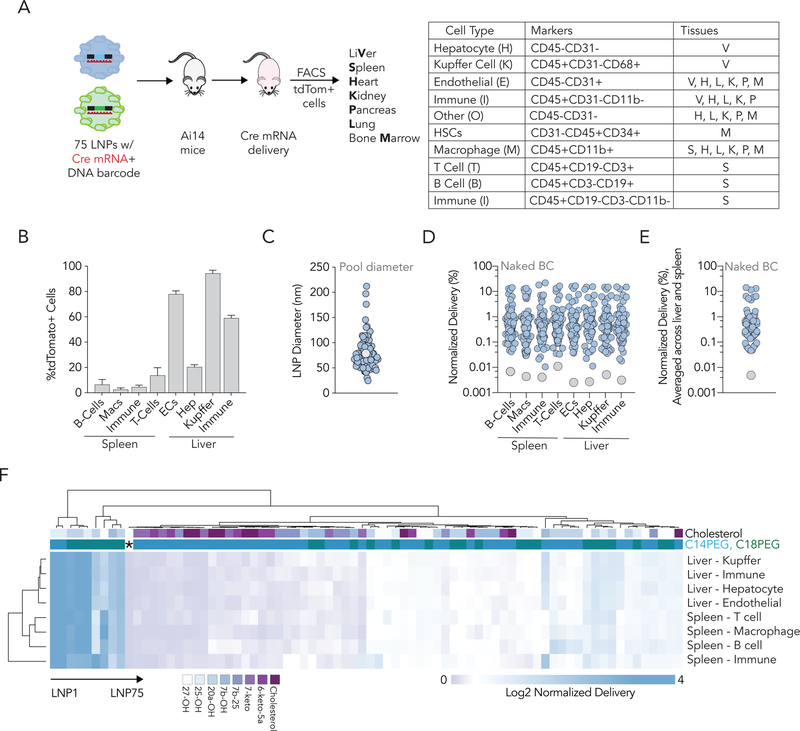
As a quality control, we analyzed the size of each individual LNP. LNPs were only pooled if their hydrodynamic diameter was between 20 and 215 nm and their autocorrelation curve contained one inflection point (SI Figure 1A,B). Eighty-six of the 125 LNPs we formulated met these criteria and were pooled. As a control, we compared the diameters of all 86 LNPs to the diameter of the pooled LNP solution, and found they were similar (Figure 2C). This suggests that the pooled LNPs did not aggregate after mixing; we have previously observed that pooled LNPs are stable[7]. We added a naked DNA barcode as a negative control[3] since naked DNA does not readily enter cells. After isolating cells and performing NGS, we found that - as expected - the naked DNA was delivered into cells less frequently than all the DNA barcodes delivered by LNPs (Figure 2D,E).Mice were sacrificed 72 hours after we injected the pooled LNPs[7]; this timepoint allows cells to express tdTomato after Cre mRNA delivery (Figure 2A). We isolated the liver, spleen, heart, kidney, pancreas, lung, and bone marrow. Using fluorescence activated cell sorting (FACS), we then isolated 28 different tdTomato+ cell types (Figure 2A). Comparing the percentage of tdTomato+ cell types in different organs, cells in the liver tended to be targeted more than cells in other organs (Figure 2B). The organ with the second-highest percentage of tdTomato+ cells was the spleen; the remaining five organs had negligible delivery (SI Figure 1C). We then clustered the barcode sequencing data using an unbiased Euclidean algorithm. This bioinformatics technique is regularly applied to gene expression data[18] and can analyze nanoparticle barcoding data[7]. Euclidean clustering revealed that the 4 liver cell types tended to ‘cluster’ together more closely than they did to splenic cell types (Figure 2F).
Figure 2.
Quantifying mRNA delivery mediated by 75 nanoparticles to 28 cell types in vivo. (A) After injecting mice with pooled nanoparticles carrying mRNA and DNA barcodes, we isolated 28 cell types from 7 tissues using FACS. (B) Functional Cre mRNA delivery, quantified as %tdTomato+ cells, in 8 cell types from the liver and spleen. (C) Hydrodynamic diameter of all administered LNPs; the diameter of the LNP pool is similar. (D) Normalized delivery of all LNPs, for all 8 cell types individually as well as (E) averaged for all 8 cell types. The naked barcode – negative control – falls out of every cell type. (F) Unbiased Euclidean clustering of 8 different cell types in 2 different tissues based on LNP delivery (%) clusters cell types according to tissue. The naked barcode is designated by an asterisk.
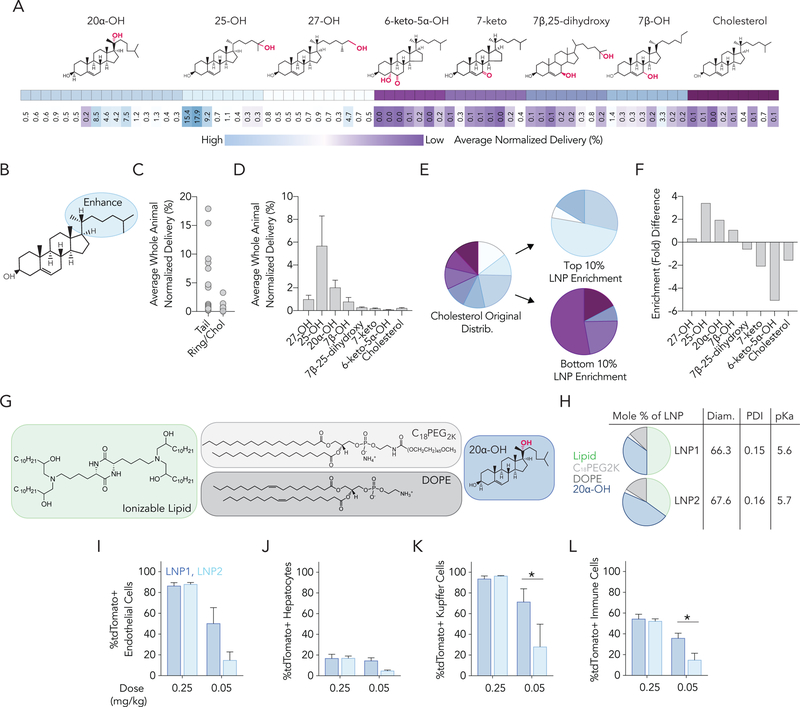
Unexpectedly, we found that the percentage of tdTomato+ hepatic endothelial cells, hepatic immune cells, and Kupffer cells was much higher than the percentage of tdTomato+ hepatocytes (Figure 2B). This result was surprising, however, the stability of the pooled LNPs, the poor performance of the naked barcode, the high percentage of tdTomato+ cells in the liver relative to other organs, and the Euclidean clustering convinced us the data were reliable enough to test our hypothesis: LNP cholesterol structure alters mRNA delivery in vivo. To quantify the impact of cholesterol structure on overall splenic and hepatic delivery, we calculated the normalized delivery across all 8 cell types we sequenced (4 in spleen and 4 in liver) (Figure 3A). We noted that side-chain oxidized cholesterol variants tended to enhance delivery relative to the other cholesterol variants (Figure 3B,C). In particular, LNPs formulated with 25-hydroxycholesterol (25-OH) and 20α-hydroxycholesterol (20α-OH) resulted in higher normalized delivery across all 8 cell types (Figure 3D). To complement these calculations, we assessed which cholesterol variants were enriched in the top 10% of LNPs (Figure 3E). We then calculated the enrichment in the bottom 10% of LNPs[19] and subtracted it from enrichment in the top 10% (Figure 3E,F). This identifies how likely it is that a given cholesterol variant is found in the best- and worst-performing LNPs. Enrichment calculations are detailed in SI Figure 2A. As an additional control, we performed the same two analyses – normalized delivery across all 8 cell types, and enrichment – for cholesterol mole percentage, ionizable lipid mole percentage, and phospholipid mole percentage (SI Figure 2B–D). We did not observe any significant trends.
Figure 3.
Modified cholesterols can alter nanoparticle delivery in vivo. (A) Normalized delivery averaged across all 8 liver and splenic cell types for each LNP, sorted by cholesterol type. (B,C) We observed that cholesterols modified on the cholesterol tail tended to enhance delivery more so than modifications made to the B ring. (D) Normalized delivery averaged across all 8 liver and splenic cell types suggested that 25-OH and 20α-OH cholesterols promote delivery. (E) Enrichment of cholesterol variants in the top 10% and bottom 10% of nanoparticles. (F) Fold enrichment in the top 10% of LNPs, calculated by subtracting enrichment in the bottom 10% of LNPs from enrichment in the top 10% of LNPs. (G) Based on these results we selected LNP1 and LNP2; they contain cKK-E12, DOPE, C18PEG2k, and 20α-OH. (H) LNP mole %, diameter (nm), polydispersity index (PDI), and pKa. (I-L) As predicted by the FIND screen, LNP1 and LNP2 delivered Cre mRNA to (I) endothelial cells, (J) hepatocytes, (K) Kupffer cells, and (L) immune cells, at 0.25 mg / kg. LNP1 delivered Cre mRNA more efficiently than LNP2 at 0.05 mg / kg. (M) The efficiency of LNP1 carrying Cre mRNA was compared to a control LNP1 that contained cholesterol and a previously reported control LNP, all 3 of which carried Cre mRNA. %tdTomato+ cells with each LNP are shown for (N) endothelial cells, (O) hepatocytes, (P) Kupffer cells, and (Q) immune cells. P-values are represented as p-value < 0.0332 (*), <0.0021 (**), <0.0002 (***).
These data suggested that LNP cholesterol chemical composition was an important factor in LNP targeting. However, they did not take LNP size into account. We previously found no relationship between nanoparticle size and delivery for hydrodynamic diameters between ~20 and ~200 nm26,34. To investigate how size altered delivery in this experiment, we calculated whether normalized delivery varied with LNP size for all LNPs (SI Figure 2E), LNPs with tail oxidized cholesterols (SI Figure 2F), and LNPs with ring oxidized cholesterols (SI Figure 2G); we found no relationship. We then calculated the percentage of formulated LNPs that met our inclusion criteria as a function of cholesterol structure and the average size of stable LNPs based on cholesterol structure and found no significant differences (SI Figure 2H,I). We performed the same analyses as a function of cholesterol mole percentage, and reached the same conclusions (SI Figure 2J,K). Thus, we did not find evidence that size affected LNP delivery between 20 and 220 nm. However, we cannot exclude the possibility that size could affect delivery if very small or very large nanoparticles are analyzed.
Like all high-throughput screens, the utility of FIND is governed by its ability to make predictions that can be verified. To this end, we formulated our top 3 LNP candidates identified by FIND with Cre mRNA. Of the 3 LNPs formulated, one LNP – which contained 25-OH cholesterol - did not formulate consistently and was excluded. The remaining two LNPs – which contained 20α-OH cholesterol (Figure 3G,H, SI Figure 3F,G) – formed stable LNPs, and were administered to Ai14 mice at a total mRNA dose of 0.25 mg / kg. Encouragingly, we found that both LNPs recapitulated the results from the FIND screen (Figure 3I–L). Both LNP1 and LNP2 robustly targeted cells in the microenvironment after a 0.25 mg / kg injection; as predicted by the screen, hepatocytes were targeted far less efficiently. LNP1 did not cause mouse weight loss in any experiment; LNP2 did cause mice to lose weight at 0.25 mg / kg (SI Figure 4B). Encouraged by robust delivery at 0.25 mg / kg, we injected LNP1 and LNP2 at a dose of 0.05 mg / kg.
Once again, we found robust delivery (Figure 3I–L). Whereas LNP1 and LNP2 both performed equally well at 0.25 mg / kg by saturating our Cre-based system, LNP1 outperformed LNP2 at the lower dose of 0.05 mg / kg. We then investigated whether LNP1 formulated with unmodified cholesterol performed as effectively as LNP1 formulated with 20α-OH (Figure 3M). Three days after injecting mice with 0.05 mg / kg Cre mRNA, we found that 20α-OH improved delivery to the liver, across all hepatic cell types (Figure 3N–Q). As an additional control, we assessed the potency of LNP1 compared to a previously reported LNP with potency at 0.2 mg / kg (Figure 3M); LNP1 outperformed this positive control LNP (Figure 3N–Q) between 14- and 77-fold within the liver microenvironment. We then used QUANT[20], a highly sensitive biodistribution system. to assess LNP biodistribution, for LNP1, LNP2, and LNP1-Chol. We did not find a difference in LNP biodistribution within the liver microenvironment (SI Figure 3D,E).
Systemic delivery of therapeutic RNA to hepatocytes has led to an FDA approved drug[21]. Delivery to non-hepatocytes has remained more challenging. Thus, an unbiased, high throughput method to study how LNPs deliver RNA in vivo could accelerate the discovery of nanoparticles with new tropisms. Here we report that FIND can predict delivery of LNPs to the liver microenvironment. If this holds true for other cell types, then FIND could reduce the time and resources needed to identify clinically relevant LNPs, relative to traditional in vitro screening. Future studies will also be needed to identify therapeutic mRNA that – when delivered to Kupffer cells – will alleviate disease.
We found that LNPs containing oxidized cholesterol can deliver mRNA to cells in the liver microenvironment more potently than to hepatocytes. Notably, robust delivery occurred at 0.05 mg / kg, which is below the dose regime used for siRNA therapies in humans[21]. Given the importance of liver endothelial cells[22] and Kupffer cells[23] in disease, these data suggest that additional advances in delivery could eventually result in protein replacement therapies within the liver microenvironment. We observed that oxidative modifications made to the hydrocarbon tail were more well tolerated than those made to the B cholesterol ring. These results need to be repeated with more ionizable lipids before they can be considered a general statement. However, in the context of these experiments, cholesterol structure impacts functional delivery. We find it feasible that different protein coronas may adsorb onto LNPs based on cholesterol structure, thereby altering targeting. In this paper, one limitation is that we did not identify the mechanism; although future work is needed to substantiate this hypothesis, we believe these data are an exciting first step to understanding how cholesterol structure can be rationally altered to change LNP tropism.
Experimental Section
Nanoparticles were formulated in a microfluidic device by mixing Cre mRNA, DNA, and LNP components[17]. The nucleic acid was diluted in citrate buffer (Teknova). Nanoparticle materials were diluted in ethanol. The phases were mixed together via microfluidics. Each LNP was formulated to carry a distinct barcode; LNP1 carried Cre mRNA and DNA barcode 1, whereas LNP2 carried Cre mRNA and DNA barcode 2. LNP hydrodynamic diameter was measured using dynamic light scattering. All animal experiments were performed in accordance with the Georgia Tech IACUC. C57BL/6J mice were purchased from Jackson Laboratory. Mice were aged 5–8 weeks, and N = 3–4 mice per group were injected intravenously. Sequencing was performed on MiniSeq™ using Illumina protocols.
Supplementary Material
References
- [1].Hajj KA, Whitehead KA, Nature Reviews Materials 2017, 2, 17056. [Google Scholar]
- [2].Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, Zhu H, Siegwart DJ, Angew Chem Int Ed Engl 2017, 56, 1059; C. Jiang, M. Mei, B. Li, X. Zhu, W. Zu, Y. Tian, Q. Wang, Y. Guo, Y. Dong, X. Tan, Cell research 2017, 27, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Paunovska K, Sago CD, Monaco CM, Hudson WH, Castro MG, Rudoltz TG, Kalathoor S, Vanover DA, Santangelo PJ, Ahmed R, Bryksin AV, Dahlman JE, Nano Lett 2018, 18, 2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jones SW, Roberts RA, Robbins GR, Perry JL, Kai MP, Chen K, Bo T, Napier ME, Ting JP, Desimone JM, Bear JE, J Clin Invest 2013, 123, 3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tsoi KM, MacParland SA, Ma XZ, Spetzler VN, Echeverri J, Ouyang B, Fadel SM, Sykes EA, Goldaracena N, Kaths JM, Conneely JB, Alman BA, Selzner M, Ostrowski MA, Adeyi OA, Zilman A, McGilvray ID, Chan WC, Nat Mater 2016, 15, 1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cheng CJ, Tietjen GT, Saucier-Sawyer JK, Saltzman WM, Nat Rev Drug Discov 2015, 14, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sago CD, Lokugamage MP, Paunovska K, Vanover DA, Monaco CM, Shah NN, Gamboa Castro M, Anderson SE, Rudoltz TG, Lando GN, Mummilal Tiwari P, Kirschman JL, Willett N, Jang YC, Santangelo PJ, Bryksin AV, Dahlman JE, Proceedings of the National Academy of Sciences 2018. [DOI] [PMC free article] [PubMed]
- [8].Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, Yin H, Chen Y, Vegas AJ, Alabi CA, Sahay G, Olejnik KT, Wang W, Schroeder A, Lytton-Jean AK, Siegwart DJ, Akinc A, Barnes C, Barros SA, Carioto M, Fitzgerald K, Hettinger J, Kumar V, Novobrantseva TI, Qin J, Querbes W, Koteliansky V, Langer R, Anderson DG, Proceedings of the National Academy of Sciences of the United States of America 2014, 111, 3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hao J, Kos P, Zhou K, Miller JB, Xue L, Yan Y, Xiong H, Elkassih S, Siegwart DJ, J Am Chem Soc 2015, 29, 9206. [DOI] [PubMed] [Google Scholar]
- [10].Mutemberezi V, Guillemot-Legris O, Muccioli GG, Progress in lipid research 2016, 64, 152; A. J. Brown, W. Jessup, Molecular aspects of medicine 2009, 30, 111. [DOI] [PubMed] [Google Scholar]
- [11].Kulig W, Cwiklik L, Jurkiewicz P, Rog T, Vattulainen I, Chemistry and physics of lipids 2016, 199, 144. [DOI] [PubMed] [Google Scholar]
- [12].Mahley RW, Science 1988, 240, 622. [DOI] [PubMed] [Google Scholar]
- [13].Ikonen E, Nature reviews. Molecular cell biology 2008, 9, 125. [DOI] [PubMed] [Google Scholar]
- [14].Paunovska K, Gil CJ, Lokugamage MP, Sago CD, Sato M, Lando GN, Gamboa Castro M, Bryksin AV, Dahlman JE, ACS nano 2018, 12, 8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M, Nat Biotechnol 2013, 31, 638. [DOI] [PubMed] [Google Scholar]
- [16].Palm W, Thompson CB, Nature 2017, 546, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, Jeon A, Dong Y, Whitehead KA, Anderson DG, J Am Chem Soc 2012, 134, 6948. [DOI] [PubMed] [Google Scholar]
- [18].Ronan T, Qi Z, Naegle KM, Science signaling 2016, 9, re6. [DOI] [PubMed] [Google Scholar]
- [19].Mui BL, Tam YK, Jayaraman M, Ansell SM, Du X, Tam YY, Lin PJ, Chen S, Narayanannair JK, Rajeev KG, Manoharan M, Akinc A, Maier MA, Cullis P, Madden TD, Hope MJ, Mol Ther Nucleic Acids 2013, 2, e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sago CD, Lokugamage MP, Lando GN, Djeddar N, Shah NN, Syed C, Bryksin AV, Dahlman JE, Nano Lett 2018. [DOI] [PMC free article] [PubMed]
- [21].Rizk M, Tuzmen S, Pharmacogenomics and personalized medicine 2017, 10, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Poisson J, Lemoinne S, Boulanger C, Durand F, Moreau R, Valla D, Rautou PE, Journal of hepatology 2017, 66, 212. [DOI] [PubMed] [Google Scholar]
- [23].Kolios G, Valatas V, Kouroumalis E, World journal of gastroenterology 2006, 12, 7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.