Abstract
When introduced to harsh conditions such as low pH, pathogenic Escherichia coli can secrete colanic acid to establish a protective barrier between the organism and the acidic environment. The colanic acid is made up of a six-sugar repeating unit polymer comprised of glucose, fucose, galactose, and glucuronic acid. The region of the E. coli genome that encodes for colanic acid biosynthesis has been reported, and the first enzyme in the biosynthesis pathway has been biochemically characterized. However, the specific roles of the remaining genes required for colanic acid biosynthesis have not been identified. Here we report the in vitro reconstitution of the next six steps in the assembly of the colanic acid repeating unit. To do this, we have cloned and overexpressed each gene within the colanic acid biosynthesis operon. We then tested the activity of the protein product of these genes using high performance liquid chromatography (HPLC) analysis and a fluorescent analogue of the isoprenoid anchor bactoprenyl diphospho-glucose as a starting substrate. To ensure that retention time changes were associated with varying sugar additions or modifications, we developed a liquid chromatography mass spectrometry (LC-MS) method for analysis of the products produced by each enzyme. We have identified the function of all but one encoded glycosyltransferase and have identified the function of two acetyltransferases. This work demonstrates the centrality of acetylation in the biosynthesis of colanic acid and provides insight into the activity of key proteins involved in the production of an important and highly conserved bacterial glycopolymer.
Graphical Abstract

Introduction
Virulent strains of Escherichia coli are responsible for as many as 70,000 infections, 2,000 hospitalizations and 60 deaths every year in the United States alone.1, 2 Pathogenic strains of the bacteria can exist harmlessly in the natural microbiome of the bovine digestive system. Therefore, it is not surprising that many virulent E. coli outbreaks are directly linked to cattle or indirectly through the crops that come into contact with their feces.3 Colanic acid is an exopolysaccharide secreted by E. coli and a number of other Enterobacteriaceae thought to promote biofilm formation and to protect the organism when exposed to harsh environments.4 When E. coli cannot synthesize colanic acid, it becomes highly susceptible to acidic pH. Therefore, it is thought that polymer production may allow the organism to survive in acidic foods.5
The colanic acid polymer is comprised of hexasaccharide repeat units consisting of glucose, two fucoses, two galactoses, and glucuronic acid (Figure 1).6–8 In addition, one fucose and one galactose may be acetylated to varying degrees, and the terminal galactose residue is pyruvylated. There is considerable discrepancy in the literature on the level of acetylation of colanic acid. Acetyl groups have been reported to be non-stoichiometric with the sugar polymer, and the repeat unit structure has been reported as mono-, di and tri-acetylated.6 It is not clear when these acetylations occur in the biosynthesis of the polymer.
Figure 1.
CA repeating unit consisting of six sugars; one glucose, two fucoses, two galactoses and a glucuronic acid. The repeating unit is assembled on a bactoprenyl phosphate anchor via a wzy-dependent pathway. Once the oligosaccharide repeat unit is formed it is flipped from facing the cytosol to the periplasm where it is polymerized and exported. Acetylation of fucose is non-stoichiometric with acetyl groups found at the 2 and 3 positions.
Interestingly, while there are three possible acetylations of colanic acid known, only two predicted acetyltransferases (wcaF, wcaB) are encoded by the biosynthesis operon, previously shown to encode all proteins required for production in E. coli.9 Other genes in the locus encode proteins homologous to an initiating glucose-1-phosphate transferase (wcaJ), five glycosyltransferases (wcaI, wcaE, wcaC, wcaL, wcaA), a presumed pyruvyltransferase (wcaK), a flippase (wzx), and a polymerase (wzy) as well as critical transport proteins.9 Generally, it is thought that colanic acid is biosynthesized in a Wzy-dependent pathway in which each sugar of the polymer repeat unit is added sequentially to the 55-carbon isoprenoid bactoprenyl phosphate (BP) (or undecaprenyl phosphate) (Figure 1). BP anchors the biosynthesis of the glycan repeating unit to the cytoplasmic face of the bacterial inner membrane. Once the repeat unit is complete, the isoprenoid-linked oligosaccharide is translocated across the inner membrane to face the periplasm and is polymerized by Wzy.8
Like many polysaccharide biosynthesis pathways, little is known about the precise biochemical roles of the genes associated with colanic acid production. The initiating glucose-1-phosphate transferase (WcaJ) has been biochemically identified and function has been reconstituted in vitro.10, 11 However, which reaction is catalyzed by each of the glycosyltransferases is unknown. There is a severe lack of information on bactoprenyl-dependent biosynthesis pathways, which hampers the ability to predict the functional role of these types of enzymes based on sequence alone.
Recently a new system was developed for characterizing the biosynthesis of complex bacterial polysaccharides that is based on a fluorescent reporter appended to the terminal isoprene of BP (Figure 2).12–14 A fluorescent 2-nitrileanilinobactoprenyl phosphate (2CNA-BP) has been used to elucidate each step in the biosynthesis of Capsular Polysaccharide A in Bacteroides fragilis.15 A similar probe was also functional in the early steps of N-linked oligosaccharide biosynthesis in Campylobacter jejuni.16 In this report, we have applied this probe to map the early steps of colanic acid production in vitro. In this work, the roles of the first four glycosyltransferases associated with the synthesis of the repeat unit have been identified, and function has been reconstituted in vitro. These studies highlight several important features of the CA biosynthesis pathway, including the role of, and centrality of, sugar acetylation in repeat unit production. The methods described here couple both fluorescence and mass spectrometry (MS) high performance liquid chromatography (HPLC) based detection for a powerful workflow to determine glycosyltransferase and acetyltransferase function.
Figure 2.
The fluorescent reporter 2-nitrileaniline (ex. 340 nm em. 390 nm) is appended to bactoprenyl phosphate (2CNA-B(Z)P) as a probe allowing simple HPLC analysis of glycan assembly enzymes.
Experimental Procedures:
Ligation-Independent Cloning (LIC)
A pET-24a vector (Novagen) was altered in two rounds of site-directed mutagenesis using PCR primers (Table S1) designed to sequentially replace a 5’ NdeI with a ScaI restriction site and a XhoI with MscI site. Mutagenesis reactions mixtures contained 0.5 μM each of forward and reverse primer, 4.0 ng of vector DNA, 0.2 mM dNTP mix, 1× Pfu Ultra DNA polymerase buffer (Agilent), and 1 unit of Pfu Ultra DNA Polymerase (Agilent). Mutagenesis was achieved with 25 cycles of 98°C for 10 s, 55°C for 30 s, and 72°C for 30 s. The PCR product was treated with 10 units of DpnI for 3 hrs at 37°C and transformed into chemically competent DH5α cells. Plasmid was isolated using a Fermentas GeneJET Plasmid Miniprep Kit and the process was repeated for the second mutation. ScaI and MscI restriction sites were confirmed with sequencing by Eurofins-Operon.
Linear pET24a-LIC vector was generated by digestion with ScaI and MscI (New England BioLabs) in reaction mixtures containing vector DNA, 1× BSA and 20 units of ScaI and MscI and were incubated for 3 hr at 37°C. Linear plasmid DNA was isolated by gel electrophoresis. Gene inserts were amplified from E. coli K12 genomic DNA in 50 μL PCR reactions containing 1× HF buffer (Thermo Fisher Scientific), 0.2 mM dNTPs, 0.2 μM of each primer (Table S1), and 1 unit of Phusion High Fidelity DNA polymerase (Thermo Fisher Scientific). Amplification was performed with 30 cycles of 98 °C for 10 s, 64 °C for 30 s, and 72 °C for 30 s. Isolated insert and vector were treated with 1 unit of T4 DNA polymerase (Thermo Fisher Scientific) and 2.5 mM dCTP for vector DNA or 2.5 mM dGTP for the gene insert DNA.to produce 20–22 base single-stranded 5’ overhangs. All PCR products and T4 digested DNA were purified using the Promega Wizard SV Gel and PCR Clean Up Kit. T4 DNA polymerase treated insert and vector DNA were incubated together in 1:1 or 1:3 vector to insert ratios at 25°C for 30 min and subsequently transformed into chemically competent DH5α cells. After picking a single colony and isolation of vector, clones were confirmed by sequencing (Eurofins-Operon).
Induction and Overexpression of Cps2E
A pET-20b vector with cps2E gene inserted was previously transformed into BL21-A1 cells for protein expression by the Yother group.17 A 500 mL culture was prepared from this cell stock with shaking at 37 °C until an O.D. of at least 0.4 was reached. Arabinose was added to induce protein expression at a final concentration of 100 μg/mL, and the temperature was lowered to 16°C and allowed to shake overnight. Cells were pelleted for 15 minutes at 5,000 relative centrifugal force (RCF), resuspended in 0.9 % NaCl and pelleted again for storage at −80 °C. Cell envelope fractions with Cps2E protein were prepared as described below.
Expression of the CA Biosynthesis Proteins
Each pET-24a-LIC vector containing the CA biosynthesis genes was transformed into chemically competent C41 expression cells. A single colony was picked, grown overnight, and mixed 1:1 with glycerol then was stored at −80 °C. Overnight cultures of cells transformed with each vector were then used to inoculate 500 mL cultures, which were then incubated at 37°C with shaking until reaching an optical density (O.D.) of at least 0.4. Once an appropriate O.D. was reached, 0.5 mM IPTG was added to each culture flask and the samples were incubated at 30 °C with shaking. After 3 hours, cells were harvested by 15 minutes of centrifugation at 5,000 RCF. The supernatant was discarded and the cells were resuspended in 0.9 % NaCl. The resuspended cells were transferred to a 50 mL conical tube and spun for 15 minutes at 5,000 RCF. The supernatant was discarded and the pelleted cells were stored at −80°C.
Isolation and Purification of Soluble CA Biosynthesis Proteins
Pelleted cells were resuspended in lysis buffer (50 mM Tris (pH 8), 200 mM NaCl, 20 mM imidazole). The resuspended cells were lysed by sonication for four minutes (pulse: one second on, one second off). The lysed contents were spun for one hour at 90,140 RCF. Supernatant was poured over a Ni2+-NTA resin, volume 1 mL. After application of the supernatant, 12 mL of wash buffer (50 mM Tris (pH 8), 50 mM imidazole, 200 mM NaCl) was poured through the column and collected. Lastly, six 0.5 mL aliquots of elution buffer (50 mM Tris (pH 8), 500 mM imidazole, 200 mM NaCl) were added and fractions were collected. All fractions containing proteins, based on SDS-PAGE analysis, were combined then dialyzed (3×) at 4°C in 1000 mL of dialysis buffer (50 mM Tris, 300 mM NaCl). Purified protein was stored at −80° C. The concentration of the final purified protein was measured using the calculated extinction coefficients for each protein and UV absorbance (Table S2–S3). The difference between UV absorbance of denatured and folded protein was negligible.
Preparation of Membrane-Bound Cps2E, WcaA and WcaL
Cell envelope fractions containing Cps2E, WcaA and WcaL were prepared by first lysing overexpressing cells as described above followed by centrifugation for 30 minutes at 2,500 RCF. The pellet was discarded, and the remaining supernatant was centrifuged at 90,140 RCF for one hour. The pellet containing Cps2E, WcaA or WcaL was then homogenized in 1mL of 50 mM Tris (pH 8.0), 300 mM NaCl. Total protein was determined using a Bradford assay with BSA as the standard.
Cps2E Activity Assay
Reaction mixtures were prepared containing 100 mM Bicine (pH 8), 2.5 mM MgCl2, 7.5 mM sodium cholate, 25 μM 2CNA-B(5Z)P, 25 mM UDP-Glc, and 53 μg total protein Cps2E cef in a total volume of 1 mL for one hour. The reaction mixture was analyzed by HPLC on a C18 (Zorbax XDB-C18 3.5 μm. 4.6 mm × 50 mm) column with an isocratic mobile phase consisting of 42% n-propanol and 58% 100 mM ammonium bicarbonate. The 2CNA-B(5Z)PP-Glc was then isolated on a semipreparative C18 HPLC column (Eclipse XDB-C18 5 μm, 9.4 mm × 250 mm) with an isocratic method of 50:50 n-propanol, 100 mM ammonium bicarbonate at 2 mL/min. The isolated product was used for the remaining reactions. The HPLC used was an Agilent 1100 equipped with a fluorescence detector, autosampler, and quaternary pump.
General Glycosyltransferase Activity Assays
All reaction mixtures were prepared using a master mix of 10.5 μM 2CNA-B(5Z)P P-Glc, 100 mM Bicine (pH 8), 2.5 mM MgCl2, 7.5 mM sodium cholate, 37.5 μM GDP-Fuc, 3.8 mM Acetyl-CoA, 1 mM UDP-Gal, and 1 mM GlcA. Each CA pathway enzyme (WcaA/B/C/E/F/I/L) was added individually to the reactions at a final concentration of 4 μM, except for WcaA, which was added at 5 μg total protein as a cef. The total volume for each analytical reaction was 20 μL and reactions were carried out at room temperature for one hour except with WcaA, which was allowed to continue overnight. Next, products were analysed stepwise where complete conversion was monitored by HPLC with the preceding enzyme followed by addition of the remaining enzymes at either 4 μM (purified protein) or 5 ug total protein (WcaA or WcaL cef). For example, reactions were prepared to form disaccharide with WcaI, then analyzed by HPLC to ensure turnover before aliquoting the WcaI product into mixtures containing the above proteins. All glycosyltransferase and acetyltransferase reactions were analyzed by HPLC using a C18 (Zorbax XDB-C18 3.5 μm. 4.6 mm × 50 mm) column and an isocratic mobile phase consisting of 35% n-propanol and 65% 100 mM NH4HCO3.
Ammonium hydroxide/propanol HPLC transferase assays
General: MS and fluorescence HPLC analysis on reactions with known protein function, based on the above analyses, was performed on an Agilent 1260 Infinity II system equipped with autosampler, quaternary pump, fluorescence detector, and a single quadrapole electrospray ionization MS detector. The column used was the high pH stable Waters XBridge Peptide BEH C18 4.6 mm × 50 mm column with 300 Å pore size, and 2.5 μm particle size. Mobile phases were 100 mM ammonium hydroxide (A) and n-propanol (B). The gradient method started at 25% B which was raised to 40% B over 10 minutes at 1 mL/min. All LC-MS analysis was performed in selective ion mode (SIM) selecting for the [M-H]− ion of 2CNA-B(5Z)P (689.45), and the [M-H]− of the substrate and product of each protein unless noted otherwise. In addition, on each analysis a scan was performed from 500–1800 m/z. MS settings were negative ion mode, nebulizer pressure 50 psig, 1.2 sec/cycle, and peakwidth 0.15 min with a SIM dwell time of 215 msec. The scan mode threshold was 150, stepsize 0.1, and speed 2600 u/sec. A scan and three SIM ions were monitored for every run. Fragmentor value, optimized for 2CNA-BP detection, was set to 240 v for all analyses. Reported scans are averages of mass spectra at the SIM detected product peaks. All reported SIM data has been divided by 36 to give similar peak heights to the fluorescence values. Fluorescence HPLC injection volumes were adjusted to inject 7.7 pmoles total 2CNA in each run. SIM HPLC injection volumes were also adjusted to ensure product peak in the full spectrum could be observed above noise. The amount injected increased with increasing molecular weight. Reported SIM intensity normalizes signal to the total amount of 2CNA injected. All reactions contain 200 mM Bicine (pH=8.5), 7.5 mM cholate, 4 mM MgCl2, and 5% n-propanol (from 2CNA-BP resuspension buffer). When reaction mixtures are added to new protein mixtures, the new mixture always contains the above buffer components.
A 100 μL reaction mixture was prepared with 30.3 μM 2CNA-B(5Z)P, 1.3 mM UDP-Glc, and 0.6 mg/mL total protein Cps2E cef. After two hours the reaction mixture was extracted with 200 μL of n-butanol (2×), then was dried under a stream of air. The product was resuspended in 25 μL of a 1:1 mixture of 5 mM ammonium hydroxide and n-propanol, then was placed in a sonicating water bath for 3 min for use in other reactions. A 1:10 dilution of the extracted Cps2E product was analyzed by both fluorescence HPLC (0.64 μL: 7.7 pmoles total 2CNA) and SIM HPLC (5 μL, 60 pmoles). Ions selected were the 2CNA-B(5Z)P [M-H]− 689.45 (monitored for all reactions), and 931.46 for the [M-H]− of 2CNA-B(5Z)PP-Glc. This preparation of 2CNA-B(5Z)PP-Glc was used for all subsequent reactions.
For analysis of WcaI a solution was prepared with 12 μM 2CNA-B(5Z)PP-Glc, 0.15 μM WcaI, and 0.17 mM GDP-Fuc. Fluorescence HPLC analysis was performed on 0.64 μL and MS analysis was performed on 5 μL (60 pmoles). SIM ions included [M-H]− for substrate 931.46, and [M-H]− for expected product 2CNA-B(5Z)PP-Glc-Fuc (1077.52). To test WcaF, 50 μL of the WcaI solution was added to 50 μL of a solution containing 3.5 mM acetylCoA and 0.44 μM WcaF final concentrations. Fluorescence HPLC was performed on 1.3 μL, and MS analysis was performed on 12 μL (72 pmoles). SIM ions detected included [M-H]− of the WcaI product and [M-H]− of expected product 2CNA-B(5Z)PP-Glc-AcFuc (1119.53). For WcaE analysis, 70 μL of the WcaF reaction was added to a 10 μL mixture containing WcaE final concentration of 6.9 μM, and an additional 0.2 mM final concentration GDP-Fuc. Fluorescence HPLC was performed on 1.5 μL, and MS analysis was performed on 14 μL (74 pmoles). SIM ions detected were the substrate [M-H]−, and [M-H]− for 2CNA-B(5Z)PP-Glc-AcFuc-Fuc (1265.59). For fluorescence analysis of WcaC, 50 μL of the WcaE reaction was diluted into 10 μL of the appropriate buffer containing 2.3 mM final concentration UDP-Gal and WcaC at 1 μM final concentration. Fluorescence HPLC was performed on 1.8 μL. For MS analysis a new reaction mixture was prepared with 12 μM 2CNA-B(5Z)PP-Glc, 0.7 mM GDP-Fuc, 3.5 mM acetylCoA, and 2.8 mM UDP-Gal. This was then mixed with a solution containing 1.2 μM WcaC, 0.3 μM WcaI, 11 μM WcaE, and 0.9 μM WcaF final concentrations. After one hour, 20 μL (240 pmoles) was injected for HPLC-MS analysis. SIM ions detected included the WcaE product [M-H]− and expected WcaC product [M-H]− (1427.64). The WcaB reaction was prepared like the WcaC reaction with the addition of 0.5 μM WcaB. Fluorescence HPLC anlaysis was performed on 0.7 μL and MS analysis was performed on 10 μL (120 pmoles). To test WcaA, 20 μL of the WcaB reaction mixture was added to 10 μL of a WcaA mixture containing 0.2 mg/mL total protein final concentration WcaA, and 2 mM final concentration UDP-GlcA. Fluorescence HPLC analysis was performed on 1 μL, and MS analysis was performed on 18 μL (144 pmoles).
Results
Expression and isolation of CA biosynthesis proteins.
Our primary objective was to identify the function of each glycosyltransferase (wcaI, wcaE, wcaC, wcaA, wcaL) and acetyltransferase (wcaF, wcaB) gene located in the CA biosynthesis operon. Using ligation independent cloning, each gene was inserted into a modified pET-24a vector encoding a C-terminal hexahistidine tag and overexpressed in C41 E. coli cells. Using immobilized metal affinity chromatography, proteins were purified to homogeneity (Figure 3) and quantified by UV absorbance. Consistent with the lack of predicted transmembrane domains in the proteins based on sequence analysis (Figure S1), all proteins were soluble and surfactant was not required for isolation. However, we were unable to obtain large quantities of solubilized WcaA and WcaL due to the majority of the proteins remaining with lysed cell membranes (Figure S2). We attempted to extract the proteins from cell envelope fractions (cef) with poor recovery. This data suggested that WcaA and WcaL proteins either have transmembrane regions not detected by prediction software, interact with membranes peripherally, or form insoluble inclusion bodies when overexpressed.
Figure 3. SDS-PAGE and Western blot analysis of overexpressed CA biosynthesis gene products.
(Top panel) SDS-PAGE gel displaying the expected molecular weights and purities of all of the CA biosynthesis proteins consisting of the glycolsyltransferases and the sugar modifying enzymes. (Lower panel) Anti-His western blot of the same enzymes displaying the hexahistidine tag associated with the enzymes from the cloning of the genes for CA biosynthesis (lower panel). The lanes are: 1- Ladder (kDa), 2- WcaA (34 kDa). 3- WcaC (46 kDa), 4- WcaE (29 kDa), 5- WcaI (45 kDa), 6- WcaL (46 kDa), 7- ladder 8-WcaB (18 kDa), 9- WcaF (21 kDa), 10- WcaK (48 kDa), 11- WcaM (52 kDa).
Assembly of a fluorescent bactoprenyl diphosphate-linked glucose.
In previous work, Patel et al. demonstrated the in vitro activity of the initiating phosphoglycosyltransferase, WcaJ, which catalyzes the addition of glucose phosphate to BP.11, 18 Another protein, Cps2E from Streptococcus pneumoniae, was shown by Cartee and coworkers to catalyze an identical reaction.17 We chose to use Cps2E in this work to assemble the starting fluorescent 2CNA-BPP-glucose needed to assess the remaining proteins in the colanic acid biosynthesis pathway. We chose this protein in lieu of WcaJ because it was readily available and was known from preliminary work in our group to accept our 2CNA-BP as a substrate. Cps2E, like WcaJ, is a WbaP family protein with a similar predicted topological structure (Figure S3).10, 17 Cps2E was overexpressed in E. coli C41(DE3) cells and membrane fractions were prepared with a vector encoding the protein and with an empty vector control. Using methods previously described we chemoenzymatically synthesized the fluorescent analogue 2-nitrileanilinobactoprenyl phosphate with five Z-configuration isoprene units (2CNA-B(5Z)P).19 The 2CNA-B(5Z)P configuration analogue was then utilized by Cps2E to provide glucose-linked product (Figure 4A). HPLC analysis (Figure 4B) of the Cps2E reactions indicated the formation of a new product with decreased retention time (13.1 min to 7.6 min), consistent with the addition of glucose phosphate to the 2CNA-B(5Z)P. Optimization of the reaction conditions led to a method capable of consuming over 90% of the starting isoprenoid with Cps2E expressing cef, where no turnover was observed with cef from empty vector control cells.
Figure 4. Production of a fluorescent bactoprenyl diphosphate-linked glucose.
(A) Cps2E product formation was analyzed by (B) reverse phase HPLC (42% n-propanol and 58% 100 mM ammonium bicarbonate isocratic) with UDP-Glc and 2CNA-B(5Z)P showing 90% turnover (based on the area under the curve (solid line) compared to the 2CNA-B(5Z)P) (broken line). Product was extracted into butanol, dried then resuspended in 1:1 water/propanol, which was then analyzed by (C) RP-HPLC (gradient 25% n-propanol →40% n-propanol over ten minutes with aqueous component 100 mM ammonium hydroxide) with fluorescence detection (bottom) or selective ion mode LC-MS detection (top) focused on 2CNA-B(5Z)PP-Glc expected mass/charge of 931.46 [M-H]− (solid line) and 689.45 [M-H]− 2CNA(5Z)P (broken line). Note that the ammonium hydroxide and ammonium bicarbonate HPLC peak sizes are comparable across all subsequent figures as they are normalized for total amount of material injected. In addition all SIM data intensities for this figure and all subsequent figures were divided by 36 to fit the data on the same scale as the fluorescence trace. (D) The product peak mass spectrum was collected as an average of all spectra at the retention time of the product peak indicating that the primary component of the product had an m/z of 931.3. Isotope ratios (inset): 931.3 (100%), 932.3 (42%), 933.3 (14 %), 934.2 (2.1 %) were consistent with expected 931.5 (100%), 932.5 (51%), 933.5 (16 %), 934.5 (2.7%).
LC-MS detection of a 2-nitrileaniline labeled Cps2E product.
To ensure that the product formed in the Cps2E reaction was 2CNA-B(5Z)PP-Glc the material was first isolated then analyzed by electrospray ionization mass spectrometry (ESI -MS) in the negative ion mode to confirm hexose-phosphate addition (data not shown). The process for isolating and confirming the formation of product was time consuming, and analysis without isolation would be much more favorable. Our standard HPLC conditions required 100 mM ammonium bicarbonate aqueous mobile phase with n-propanol. The relatively high concentration of ammonium bicarbonate was not amenable to mass spectral characterization, and lower concentrations of ammonium bicarbonate lead to severe peak broadening. As an alternative, we tested the ability to use ammonium hydroxide as our aqueous mobile phase with a high pH stable reverse phase column. The Cps2E reaction product was extracted into butanol, then dried, and resuspended in a 1:1 ammonium hydroxide/n-propanol mixture. Using a gradient elution profile optimized for analysis of oligosaccharides-linked to our fluorescent isoprenoid we were able to observe a clear separation between 2CNA-B(5Z)P remaining in the mixture and the Cps2E product (5.9 min to 5.5 min) (Figure 4C). Next, we tested the ability to detect the isoprenoids by LC-MS with selective ion mode (SIM) monitoring for the Cps2E starting material and product (Figure 4C). Both the starting 2CNA-B(5Z)P and Cps2E product were readily detected with relative abundances similar to what was observed by fluorescence, and each had similar retention times (6.1 to 5.7 min). The slight difference in retention time is likely due to differences in delay volume to the fluorescence and MS detector. On our instrument fluorescence and MS must be analyzed separately due to instrument limitations on the back pressure associated with the fluorescence detector flow cell. To analyze the major components of the product peak we averaged the mass spectra of species retained at the same time as the SIM product peak in the chromatogram (Figure 4D). The major ionizable component of the product peak had an m/z of 931.3, consistent with the expected 931.46 for the [M-H]− product of Cps2E. Although, the spectrometer is a low resolution instrument, we also inspected the isotopic ratios of the product peak to further confirm that we were observing the 2CNA-B(5Z)PP-Glc. The expected isotopic ratios of the product peak are 931.5 (100%), 932.5 (51%), 933.5 (16%), and 934.5 (2.7 %) We observed isotopes at 931.3 (100%), 932.3 (42%), 933.3 (14%), and 934.2 (2.1%). Overall, the retention time shift to a more polar species in both HPLC methods, the detection of specific ions at those retention times in selective ion mode, the identity of the major product peak in the mass spectrum, and isotopic abundance consistency together clearly demonstrate that the product is a 2CNA-B(5Z)PP-hexose, the expected product of Cps2E.
WcaI transfers unmodified fucose to BPP-Glc.
With two robust HPLC methods in hand, we were next interested in using the 2CNA-B(5Z)PP-Glc to analyze the remaining steps in CA biosynthesis. There were five proposed glycosyltransferases (WcaA, WcaC, WcaE, WcaI, and WcaL) associated with the CA biosynthesis gene cluster that could potentially promote the formation of a disaccharide with BPP-Glc (Figure 5A). To determine if one of these catalyzed the introduction of fucose in the structure, we mixed isolated 2CNA-B(5Z)PP-Glc with each glycosyltransferase and GDP-Fuc, then analyzed for product formation by HPLC (Figure 5B). Of the five glycosyltransferases, only incubation with WcaI altered the retention time of the substrate. The retention shift from 9.7 to 8.7 min was consistent with the addition of a single sugar moiety and suggested that WcaI was the next enzyme in the pathway. Another reaction was prepared with the extracted Cps2E product above then was analyzed using the high pH ammonium hydroxide HPLC conditions (Figure 5C). The product peak shifted from the Cps2E product retention time of 5.5 min to 5.3 min. The remaining 2CNA-B(5Z)P retention time remained at 5.9 min serving as an internal standard for the chromatography analysis. This was important because the high pH chromatography method was very sensitive to fluctuations in ammonium hydroxide, which would occur with aqueous solvent left overnight, or small differences in preparation of the solvent. This was not an issue for the ammonium bicarbonate-based HPLC analysis. As was observed with the Cps2E product, a slightly longer retention time was observed for the product by SIM relative to fluorescence (5.5 min), but no Cps2E product remained within our detection limits in the WcaI reaction. In all LC-MS work described here, the 2CNA-B(5Z)P [M-H]− ion was also monitored, and was consistent in intensity and retention time throughout (data not shown). The average of the spectra within the product peak confirmed the formation of a species with an m/z of 1077.4, consistent with the expected [M-H]− of 2CNA-B(5Z)PP-Glc-Fuc (1077.52) (Figure 5D). In addition, the isotopic ratios of the product: 1077.4(100%), 1078.3 (67%), 1079.5 (30%), 1080.3 (9.4%), were consistent with the expected 1077.5 (100%), 1078.5(58.4%), 1079.5(20%), 1080.5 (5%) for 2CNA-B(5Z)PP-Glc-Fuc.
Figure 5. Assembly of a disaccharide by the predicted glycosyltransferase WcaI.
(A) Expected reaction catalyzed by WcaI, R=2CNA-B(5Z)PP. (B) Reverse phase HPLC analysis (isocratic 35% n-propanol and 65% 100 mM NH4HCO3) of reactions of 2CNA-B(5Z)PP-Glc with each of the glycolsyltransferases WcaA, WcaC, WcaE, WcaI and WcaL along with the acetyltransferases WcaF and WcaB. (C) Product of a WcaI reaction was analyzed by HPLC using the gradient described for LC-MS in figure 4 with fluorescence (bottom) and selective ion detection (top). Ions targeted were those of 2CNA-B(5Z)PP-Glc-Fuc ([M-H]− 1077.52, solid line) and 2CNA-B(5Z)PP-Glc ([M-H]− 931.4,: broken line). (D) The average mass spectrum was collected on the product peak giving an m/z of 1077.4 for the product. Isotope ratios (inset): 1077.4(100%), 1078.3 (67%), 1079.5 (30%), 1080.3 (9.4%) were consistent with the expected: 1077.5 (100%), 1078.5(58.4%), 1079.5(20%), 1080.5 (5%)
WcaF acetylates isoprenoid-linked disaccharide.
There was ambiguity in the next step in the biosynthesis of CA; the fucose residue in 2CNA-B(5Z)PP-Glc-Fuc, could either be acetylated then a second fucose added to form a trisaccharide, or the unmodified fucose could be the acceptor for the addition of a second fucose (Figure 6A). To distinguish between these possibilities we prepared 2CNA-B(5Z)PP-Glc-Fuc then added each of the remaining glycosyltransferases and the acetyltransferases in separate reactions with additional GDP-Fuc or acetyl-CoA. We did not observe a shift in retention time with any of the glycosyltransferases under these conditions (Figure 6B), suggesting that the sugar may need to be acetylated prior to the next step in the pathway. We found that in the presence of the acetyltransferase WcaF, and not WcaB, that there was a slight retention time change from 8.7 min to 8.8 min, which was consistent with the slight increase in hydrophobicity associated with acetylation of the disaccharide. Neither WcaF nor WcaB had any influence on the retention of GDP-fucose alone (data not shown) suggesting that the fucose residue had to be associated with 2CNA-B(5Z)PP-Glc before modification. Analysis of the isoprenoid-linked product using the high pH HPLC method indicated a product with retention time 5.3 min, which was the same as the retention time of the WcaI product (Figure 6C). However, we did see a broadening of the peak shape with WcaF relative to the WcaI product. SIM-based HPLC-MS analysis indicated that a product with m/z 1119.53 ([M-H]− for 2CNA-B(5Z)PP-Glc-AcFuc) was detected at 5.6 min, and no peak was observed for the [M-H]− ion of the WcaI product. The major component of that peak was a species with an m/z of 1119.3 (Figure 6D), which was consistent with the [M-H]− of 2CNA-B(5Z)PP-Glc-AcFuc. An additional peak was observed for an [M-H+Na]− adduct at 1141.2. The [M-H]− isotope abundance was also consistent with the formation of the acetylated trisaccharide (Figure 6D).
Figure 6. Assembly of an acetylated disaccharide by the acetyltransferase WcaF.
(A) The expected reaction catalyzed by the protein encoded by the wcaF gene identified as a disaccharide acetyltransferase R=2CNA-B(5Z)PP-Glc. Acetylation can be at either of the underlined positions, but not both at the same time. (B) Reverse phase HPLC analysis (isocratic 35% n-propanol and 65% 100 mM NH4HCO3) of reactions with 2CNA-B(5Z)PP-Glc-Fuc product of WcaI and WcaA, WcaC, WcaE, WcaL, WcaF, or WcaB. (C) Fluorescence (bottom) and selective ion mode (SIM, top) ammonium hydroxide based HPLC analysis of a WcaF reaction product. SIM ions were those of 2CNA-B(5Z)PP-Glc-AcFuc ([M-H]− 1119.53, solid line) and WcaF substrate 2CNA-B(5Z)PP-Glc-Fuc ([M-H]− 1077.52, broken line). (D) The average of mass spectra across the product peak was obtained and the major component had an m/z of 1119.3 m/z. Isotope ratios (inset): 1119.3(100%), 1120.4(61%), 1121.5(35%), 1122.3 (12.4%) were consistent with expected 1119.5(100%), 1120.5(61%), 1121.5(23%), 1122.5 (4.8%)
Acetylation is required for WcaE activity.
Since WcaF was able to catalyze the transfer of the acetyl group to the B(5Z)PP-Glc-Fuc disaccharide, and none of the glycosyltransferases would utilize the un-acetylated material, it was likely that this modification was required before the formation of the trisaccharide (Figure 7A). To test this, reactions were prepared with 2CNA-B(5Z)PP-Glc, WcaI, GDP-Fuc, WcaF, and acetyl-CoA, then were monitored for completion by HPLC. We then took aliquots of the reaction and added them to mixtures containing the remaining glycosyltransferase enzymes (WcaA, WcaC, WcaE, and WcaL) in separate tubes. Only the addition of WcaE protein led to an 8.8 to 7.6 min shift in retention time from the WcaF product (Figure 7B). Further HPLC analysis of a WcaE reaction product using the ammonium hydroxide based method indicated a new fluorescent peak at 5.2 min, which was a significant shift relative to the 5.3 min retention time of the WcaF product (Figure 7C). SIM analysis of the product indicated that again, there was no substrate leftover in the reaction mixture. Analysis of the product peak indicated that the major component was an ion at 1265.3 m/z which was similar to the [M-H]− of the 2CNA-B(5Z)PP-Glc-AcFuc-Fuc (Figure 7D).
Figure 7. Assembly of isoprenoid-linked trisaccharide by WcaE.
(A) Expected reaction catalyzed by WcaE, R=2CNA-B(5Z)PP-Glc. Acetylation at either of the underlined positions. (B) Reverse phase HPLC analysis of reactions (isocratic 35% n-propanol and 65% 100 mM NH4HCO3) of 2CNA-B(5Z)PP-Glc-AcFuc with each of the glycolsyltransferases WcaA, WcaC, WcaE and WcaL along with the acetyltransferase WcaB. (C) Fluorescence (bottom) and SIM (top) analysis of WcaE product. Ions analyzed were 1265.59 (2CNA-B(5Z)PP-Glc-AcFuc-Fuc [M-H]−, solid line), and 1119.53 (2CNA-B(5Z)PP-Glc-AcFuc [M-H]−, broken line). (D) Average of mass spectra across the product peak giving an m/z of 1265.3 as the major component. Isotope ratios (inset): 1265.3(100%), 1266.4(63%), 1267.4(30%), 1268.4(20%), were consistent with expected 1265.6(100%), 1266.6(68%), 1267.6 (26%), 1268.6(7.7%).
WcaC catalyzes the formation of an acetylated tetrasaccharide.
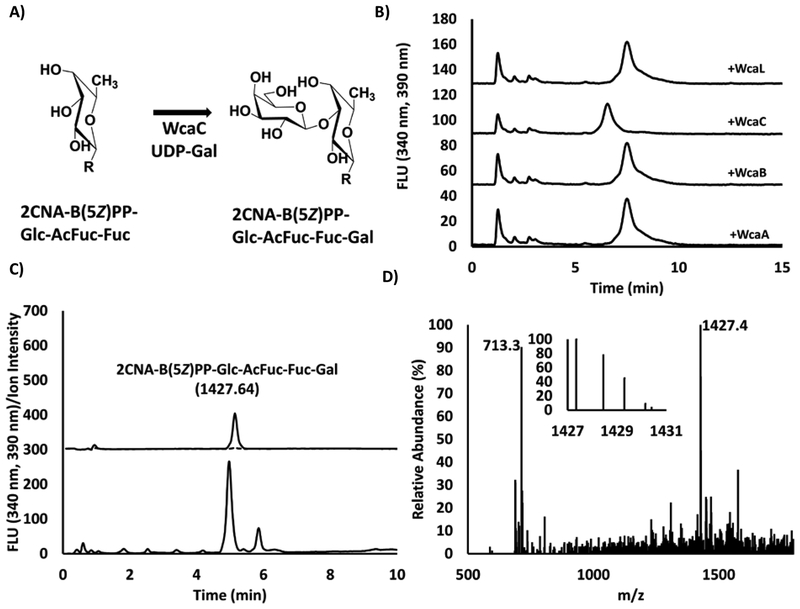
With the formation of the trisaccharide by WcaE the next step was presumably the introduction of galactose into the repeat unit precursor (Figure 8A). We prepared a reaction mixture containing 2CNA-B(5Z)PP-Glc with all components up to WcaE and tested the product of WcaE with the remaining glycosyltransferases (WcaA, WcaC, and WcaL) in separate reactions. Again, we found that only one of the remaining glycosyltransferases (WcaC) altered the retention of WcaE product from 7.6 min to 6.6 min (Figure 8B). HPLC analysis in the high pH conditions indicated that there was a peak shift with the WcaC product to 5.0 min from 5.2 min (WcaE product) providing further confirmation that a sugar was appended. Up to this point reactions had been diluted with the addition of each new enzyme for analysis by HPLC-MS. We compensated for this by increasing injection volumes. However, with the WcaC product there was a substantial decrease in signal from SIM analysis with the expected 2CNA-B(5Z)-PP-Glc-AcFuc-Fuc-Gal [M-H]− m/z of 1427.64. While the increase in injection required for fluorescence analysis had little effect on detection, the noise associated with the SIM analysis and obtaining the mass spectrum was problematic. To get around this issue, a new reaction was prepared with an identical 2CNA-B(5Z)PP-Glc concentration as was used for the WcaI reaction analysis. In this assay the isoprenoid-linked glucose was mixed with surfactant, buffer, MgCl2, all nucleotide-linked sugar donors, and acetylCoA. Separately the enzymes WcaI, WcaF, WcaE, and WcaC were combined then added to the new reaction mixture. Within one hour all isoprenoid-linked monosaccharide was converted to product based on HPLC (data not shown). SIM analysis of the product (Figure 8C) indicated that there were no intermediate materials left in the mixture, only the 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal. Analysis of the product peak mass spectrum indicated two major chemical species (Figure 8D). The species had an m/z of 1427.4 and 713.3, which are consistent with the [M-H]− and [M-2H]2- ions of 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal. Closer inspection of the isotope abundance of the [M-H]− species indicated the expected ratios of isotopes from the products. However, the abundance of the [M-2H]2- species isotopes were not as clear. This difficulty could be related to the low resolution analysis by this instrument, or limitations with the current configuration of the instrument required to monitor three ions and a scan for every run. It is not clear why only the WcaC product thus far had a major −2 charge ion, but it is likely that this was why detection by SIM required so much more material.
Figure 8. WcaC catalyzed formation of isoprenoid-linked tetrasaccharide.
(A) Expected reaction catalyzed by WcaC. R=2CNA-B(5Z)PP-Glc-AcFuc (B) Reverse phase HPLC ana lysis of 2CNA-B(5Z)PP-Glc-AcFuc-Fuc product of WcaE with each of the glycolsyltransferases WcaA, WcaC and WcaL along with the acetyltransferase WcaB. (C) As in figure 4–7 HPLC fluorescence (bottom) and SIM detection (top) analysis was performed on the WcaC product with a focus on 1427.64 (2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal [M-H]− solid line) and 1265.59 (2CNA-B(5Z)PP-Glc-AcFuc-Fuc [M-H]− broken line). (D) Average spectra across the product peak indicated the presence of two major ions: 1427.4 and 713.3 representing the [M-H]− and [M-2H]2- of 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal. Isotope ratios (inset) for [M-H]− 1427.4(100%), 1428.5 (77%), 1429.3 (45%), 1430.2 (9%) were consistent with expected 1427.64(100%), 1428.7 (74.7%), 1429.65 (32%), 1430.65 (9.4%).
WcaB catalyzes the addition of an acetyl group to the isoprenoid-linked tetrasaccharide.
With the formation of the tetrasaccharide 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal by WcaC, the introduction of glucuronic acid into the repeat unit precursor was the expected next step (Figure 9A). We prepared 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal and tested it for turnover with WcaA or WcaL in the presence of UDP-GlcA. With solubilized protein, we observed no easily reproducible product formation. However, when we used the cef of WcaA overexpressing cells, but not WcaL, we observed a retention time shift from 6.6 min to 4.3 min, consistent with the addition of the more polar glucuronic acid sugar (Figure S4). Limited product formation was observed with WcaA cef alone and we were unable to force the reaction to go to completion. Additionally, the use of an empty vector cef control in C41 cells did not modify the 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal product with UDP-GlcA (data not shown).
Figure 9. Assembly of a diacetylated acidic pentasaccharide.
(A)Expected reaction catalyzed by WcaB and WcaA. R=2CNA-B(5Z)PP-Glc-AcFuc-Fuc (B) Reverse phase HPLC analysis (isocratic 35% n-propanol and 65% 100 mM NH4HCO3) of 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal with each of the glycosyltransferases WcaA and WcaL, and the acetyltransferase WcaB. (C) HPLC analysis as in figures 4–8 with fluorescence (bottom) and SIM (top) detection of [M-H]− ions associated with 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-AcGal (1469.65, solid line) and 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal (1427.64, broken line). (D) Average of MS spectra across the product peak indicating a major component with m/z 1469.4. Additional peaks are observed with m/z of 688.3, 702.4, and 713.4. These are consistent with a 2CNA-B(5Z)P [M-H]−, acetylated tetrasaccharide [M]−, and unacetylated 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal [M-2H]2- respectively. WcaB product expected isotope ratios: 1469.65(100%), 1470.66 (76.9%), 1471.66(33.8%), 1472.66(10.3%) Actual (inset): isotope ratios: 1469.4(100%), 1470.5 (82.4%), 1471.5 (21%), 1472.3 (8.7%). (E) HPLC analysis of WcaA product with fluorescence (bottom) and SIM (top) detection with a focus on the [M-2H]2- 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-AcGal-GlcA (822.34, solid line) and WcaB product ([M-H]−:1427.64, broken line). (F) Product mass spectra average indicated that the major ionizable species had an m/z of 822.5 consistent with the expected product. Isotope abundance (inset) was inconsistent with expected values most likely due to instrument resolution limitations where the expected distribution was 822.34 (100%), 822.84 (82.2%), 823.34 (33.3%), 823.84(8.1%). The observed was 822.5(100%) 822.7 (29.5%), 823.3(9.2%), 823.8(13%). However, isotopic differences suggested a −2 charged species.
WcaB was then tested for modification of 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal in the presence of acetyl-CoA. A reproducible retention time shift was not readily observed that would be associated with the acetylation of the 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal. Interestingly, when WcaA and UDP-GlcA were added to the WcaB reaction we observed complete conversion of 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-Gal to form the presumed di-acetylated pentasaccharide (Figure 9B). As described for WcaC above, a new reaction mixture was prepared with 2CNA-B(5Z)PP-Glc, all nucleotide-linked sugars, buffer, surfactant, and acetylCoA. A mixture of WcaI, WcaF, WcaE, WcaC, and WcaB was then prepared and added to the reaction mixture. Within one hour we observed complete turnover to a product with a retention time consistent with the WcaC product (Figure 9C). SIM analysis of the product indicated that the major product was a diacetylated 2CNA-B(5Z)PP-linked tetrasaccharide with an m/z of 1469.65. We also detected a limited amount of monoacetylated tetrasaccharide WcaC product. It was not clear whether the WcaC product detected was unreacted material or an ionization fragment. Analysis of the spectrum of this peak indicated that the major species was an ion with an m/z consistent with the [M-H]− of a 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-AcGal. The isotopes associated with the [M-H]− diacetylated product were consistent with the expected isotopic ratios of the 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-AcGal.
WcaA catalyzes the addition of glucuronic acid to form a diacetylated pentasaccharide.
Since WcaB appeared to catalyze the addition of an acetyl group to the tetrasaccharide, and helped to promote catalysis by WcaA, we presumed that WcaA catalyzed the next step in the colanic acid biosynthesis pathway (Figure 9A). The WcaB reaction mixture above was added to a solution containing UDP-GlcA and WcaA cef. A large shift in retention time was observed for the product relative to the 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-AcGal (5.0 min to 3.1 min) when analyzed by fluorescence detection using the high pH method (Figure 9E). We suspected, based on the pka of the glucuronic acid residue and previous experience with the other compounds, that the [M-2H]2- ion (822.34) would be the easiest to detect by SIM-MS. Indeed, a large peak associated with the expected [M-2H]2- ion of 2CNA-B(5Z)PP-Glc-AcFuc-Fuc-AcGal-GlcA was detected with a similar retention time to the new fluorescent peak (Figure 9E). When the [M-H]- ion was used no product was detected (data not shown). No WcaB product was detected. In this assay some peaks that may be associated with intermediate products could be detected by fluorescence. We also noted that the WcaA SIM product peak was larger than the previous WcaB and WcaC peaks. Analysis of the mass spectrum of the product peak indicated that there was only one dominant species with an m/z of 822.5 (Figure 9F). Isotope abundance analysis of the major peak was inconsistent with the expected abundance, much like we saw with the WcaC [M-2H]2-. However, the ions associated with the isotopes indicated that this was a −2 charged species with isotopes at 822.5, 822.7, 823.3 and 823.8. Due to the poor performance of solubilized WcaA there was concern that earlier tests of the protein would be invalid because the functional form of the protein appeared to be associated with the cef. We retested all of the prior reactions with cef preparations of both WcaA and WcaL and still found no activity with the small amounts of solubilized protein or cef preparations (data not shown).
Discussion
The goal of this work was to identify the transfer function of each gene involved in colanic acid repeat unit biosynthesis in E. coli, and to establish the order in which the gene products function. Here we have demonstrated the function of six of the eight colanic acid biosynthesis cluster genes thought to be responsible for the assembly of the colanic acid repeat unit (Figure 10). The only two steps that remain would be the transfer of a galactose residue followed by pyruvylation of that galactose. The two assembly enzymes that remain uncharacterized are predicted, based on sequence alignments, to be a glycosyltransferase (WcaL) and a pyruvyltransferase (WcaK). However, we have thus far been unable to reconstitute the function of these last two proteins. Why we have been unable to reconstitute the next step in the pathway remains unclear. WcaL was one of the poorest soluble expressing proteins of the set, and the problem may simply be the ability to overproduce the protein in a functional form. Alternatively, other proteins may be involved in promoting the activity of WcaL, or the conditions for effective transfer by the first six proteins may not be amenable to this protein in vitro. WcaA for instance appears to be fully functional when still associated with membrane components. This suggests that WcaA is only properly folded in those conditions, or there are other components of the membrane that help to promote its function. It would be interesting in future work to prepare soluble constructs of both WcaL and WcaA to help reconstitute their function in vitro.
Figure 10. Biosynthesis pathway to colanic acid repeating unit.
Data from figures 5–9 were collated to provide a model for the pathway that leads to colanic acid biosynthesis in E. coli. Only two steps (presumably WcaL and WcaK) remain that have not been addressed in this report. However, WcaL is the only remaining predicted glycosyltransferase in the biosynthesis gene cluster and WcaK is the only predicted pyruvyl transferase.
It has been reported that CA has non-stoichiometric acetylations at the 2 and 3 positions of the disaccharide fucose residue.20–22 Since this was non-stoichiometric, we initially thought that these modifications would not be required for the formation of the polymer repeat unit, and may occur after repeat unit formation or upon polymer formation. However, our data demonstrate that the activity of the acetyltransferase WcaF on the polyprenyl-linked disaccharide is critical to the formation of the repeat unit. Since WcaF was importnat to the formation of the oligosaccharide, we wonder if it is selective for either the 2 or the 3 position. It may be possible that incorporation of the acetyl group in either position could lead to formation of a WcaE competent substrate. Therefore, the non-stoichiometric nature of this modification may be due to the acetyl group being split between these two positions. Our MS data suggests only one modification per fucose, and we have no evidence of multiple acetylations at this stage of the pathway. However, we cannot rule out that a second acetylation takes place after formation of the oligosaccharide or polymer. The importance of WcaF is consistent with studies showing that this protein is critical to the formation of colanic acid in vivo and more recently, the importance of that gene for biofilm formation.23, 24 We have also shown that WcaB, the other encoded acetyltransferase, had no influence on the disaccharide, but increases the ability of WcaA to add the fifth sugar onto the CA repeating unit, and also appears to be a critical protein in the assembly of the oligosaccharide repeat unit.
The identification of the functional roles of proteins involved in complex polysaccharide biosynthesis in bacteria remains a major challenge. Radiolabel based methods are most often used, but when the target protein is involved in the incorporation of a second or third sugar, or a sugar modification such as acetylation is involved, these methods become more complex. This report highlights the use of fluorescent polyisoprenoids as replacements for traditional radiolabel techniques. Importantly, this work has shown that these probes can be used for assessing the acetylation of sugars, in addition to glycosylation reactions, and has allowed us to map nearly the entire biosynthesis pathway of the glycopolymer in single pot reactions.
The LC-MS and fluorescent methods described in this report are excellent complementary techniques where each have advantages and disadvantages. One of the major values of the fluorescent probe is the ease with which it can be prepared then quantified spectrophotometrically. There is also very little change in fluorescent peak integral with the 2-ntirileaniline fluorophore upon sugar addition, allowing for the potential simple measurement of reaction rates. However, the problem with the HPLC analysis of the fluorescent isoprenoids is that in some cases, like acetylation or other sugar modifications, there may not be an easily resolved retention time shift. For example, here we see a small shift with WcaF, but no reproducible shift with WcaB. This is one place where coupling MS analysis is invaluable. Here we show that WcaB catalyzes acetylation of the isoprenoid-linked tetrasaccharide using LC-MS coupled with the analysis by fluorescence. The LC-MS method also provides rapid characterization of the product mass spectrum, which would otherwise require isolation of the product followed by direct injection mass spectrometry. The polyisoprenoid-linked compounds can be sensitive to conditions used for removing HPLC solvents where the phosphoanhydride and acetylations are relatively easily hydrolyzed. The problem with LC-MS alone is that, as observed with the proteins analyzed in this report, the efficiency of the mass detection differs depending on the particular molecular species. In general, as molecular weight goes up, if there is not an increase in charge density (WcaA), the efficiency of the mass spectrometric detection decreases. In addition, in our hands, obtaining a clear mass spectrum of a product peak required 1–2 orders of magnitude more material than detection by fluorescence. Coupling the two methods provides a powerful characterization system for these biosynthesis pathways that could further be developed for structure analysis including MS/MS fragmentation and methylation analysis.
The methodology here is a complement for current methods available for the high-resolution NMR-based structural characterization of complex glycans isolated from the surface of microorganisms in substantially larger quantities. One of the primary difficulties in using a chemoenzymatic approach for NMR studies would be the expense of the individual sugar donors. However, with unlimited resources scale up would not be unreasonable. This report has intentionally focused solely on mapping the biosynthesis pathway to the colanic acid repeat unit. The biosynthesis pathway map provided here is consistent with the most recent structural characterization of colanic acid fragments bound to lipopolysaccharide molecules in E. coli.20
Supplementary Material
ACKNOWLEDGMENTS
We thank Janet Yother (University of Alabama at Birmingham) for clones of cps2E expressing vector. We thank Amanda Reid (UNC-Charlotte) for help producing fluorescent bactoprenyl phosphates, and we thank Beth Scarbrough (UNC-Charlotte) for help modifying the ligation independent cloning vector.
Uniprot Accession Codes: Cps2E: Q9ZII5, WcaI: P32057, WcaE: P71239, WcaC: P71237, WcaA: P77414, WcaL: P71243, WcaF: P0ACD2, WcaB: P0ACC9, WcaK: P71242, WcaM: P71244
Funding Sources
Funding supporting this work was received through grants from the National Institutes of Health General Medical Sciences grant numbers: R01GM123251 and R15GM114773.
Footnotes
Supporting Information
Figures S1–S4, Table S1–S3 which include topological predictions, SDS-PAGE analysis of membrane fraction proteins, HPLC analysis of WecA reactions without WecB, primer sequences, protein extinction coefficients, and protein sequences are available in supporting information available free of charge.
References
- [1].Rangel JM, Sparling PH, Crowe C, Griffin PM, and Swerdlow DL (2005) Epidemiology of Escherichia coli O157 : H7 outbreaks, United States, 1982–2002, Emerg Infect Dis 11, 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Raum E, Lietzau S, von Baum H, Marre R, and Brenner H (2008) Changes in Escherichia coli resistance patterns during and after antibiotic therapy: a longitudinal study among outpatients in Germany, Clin Microbiol Infec 14, 41–48. [DOI] [PubMed] [Google Scholar]
- [3].Nguyen Y, and Sperandio V (2012) Enterohemorrhagic E. coli (EHEC) pathogenesis, Front Cell Infect Mi 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mao Y, Doyle MP, and Chen JR (2001) Insertion mutagenesis of wca reduces acid and heat tolerance of enterohemorrhagic Escherichia coli O157 : H7, J Bacteriol 183, 3811–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mao Y, Doyle MP, and Chen J (2006) Role of colanic acid exopolysaccharide in the survival of enterohaemorrhagic Escherichia coli O157 : H7 in simulated gastrointestinal fluids, Lett Appl Microbiol 42, 642–647. [DOI] [PubMed] [Google Scholar]
- [6].Meredith TC, Mamat U, Kaczynski Z, Lindner B, Holst O, and Woodard RW (2007) Modification of lipopolysaccharide with colanic acid (M-antigen) repeats in Escherichia coli, J Biol Chem 282, 7790–7798. [DOI] [PubMed] [Google Scholar]
- [7].Sutherland IW (1969) Structural Studies on Colanic Acid, Common Exopolysaccharide Found in Enterobacteriaceae, by Partial Acid Hydrolysis - Oligosaccharides from Colanic Acid, Biochem J 115, 935–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Whitfield C (2006) Biosynthesis and assembly of capsular polysaccharides in Escherichia coli, Annu Rev Biochem 75, 39–68. [DOI] [PubMed] [Google Scholar]
- [9].Stevenson G, Andrianopoulos K, Hobbs M, and Reeves PR (1996) Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid, J Bacteriol 178, 4885–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Furlong SE, Ford A, Albarnez-Rodriguez L, and Valvano MA (2015) Topological analysis of the Escherichia coli WcaJ protein reveals a new conserved configuration for the polyisoprenyl-phosphate hexose-1-phosphate transferase family, Sci Rep-Uk 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Patel KB, Toh E, Fernandez XB, Hanuszkiewicz A, Hardy GG, Brun YV, Bernards MA, and Valvano MA (2012) Functional Characterization of UDP-Glucose:Undecaprenyl-Phosphate Glucose-1-Phosphate Transferases of Escherichia coli and Caulobacter crescentus, J Bacteriol 194, 2646–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Aragao-Leoneti V, Campo VL, Gomes AS, Field RA, and Carvalho I (2010) Application of copper(I)-catalysed azide/alkyne cycloaddition (CuAAC) ‘click chemistry’ in carbohydrate drug and neoglycopolymer synthesis, Tetrahedron 66, 9475–9492. [Google Scholar]
- [13].Chehade KAH, Andres DA, Morimoto H, and Spielmann HP (2000) Design and synthesis of a transferable farnesyl pyrophosphate analogue to Ras by protein farnesyltransferase, Journal of Organic Chemistry 65, 3027–3033. [DOI] [PubMed] [Google Scholar]
- [14].Lujan DK, Stanziale JA, Mostafavi AZ, Sharma S, and Troutman JM (2012) Chemoenzymatic synthesis of an isoprenoid phosphate tool for the analysis of complex bacterial oligosaccharide biosynthesis, Carbohyd Res 359, 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sharma S, Erickson KM, and Troutman JM (2017) Complete Tetrasaccharide Repeat Unit Biosynthesis of the Immunomodulatory Bacteroides fragilis Capsular Polysaccharide A, Acs Chem Biol 12, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Glover KJ, Weerapana E, and Imperiali B (2005) In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation, Proc Natl Acad Sci U S A 102, 14255–14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cartee RT, Forsee WT, Bender MH, Ambrose KD, and Yother J (2005) CpsE from type 2 Streptococcus pneumoniae catalyzes the reversible addition of glucose-1-phosphate to a polyprenyl phosphate acceptor, initiating type 2 capsule repeat unit formation, J Bacteriol 187, 7425–7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Saldias MS, Patel K, Marolda CL, Bittner M, Contreras I, and Valvano MA (2008) Distinct functional domains of the Salmonella enterica WbaP transferase that is involved in the initiation reaction for synthesis of the O antigen subunit, Microbiol-Sgm 154, 440–453. [DOI] [PubMed] [Google Scholar]
- [19].Troutman JM, Erickson KM, Scott PM, Hazel JM, Martinez CD, and Dodbele S (2015) Tuning the production of variable length, fluorescent polyisoprenoids using surfactant-controlled enzymatic synthesis, Biochemistry 54, 2817–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Meredith TC, Mamat U, Kaczynski Z, Lindner B, Holst O, and Woodard RW (2007) Modification of lipopolysaccharide with colanic acid (M-antigen) repeats in Escherichia coli, J Biol Chem 282, 7790–7798. [DOI] [PubMed] [Google Scholar]
- [21].Sutherland IW (1969) Structural studies on colanic acid, the common exopolysaccharide found in the enterobacteriaceae, by partial acid hydrolysis. Oligosaccharides from colanic acid, Biochem J 115, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Whitfield C (2006) Biosynthesis and assembly of capsular polysaccharides in Escherichia coli, Annu Rev Biochem 75, 39–68. [DOI] [PubMed] [Google Scholar]
- [23].Ranjit DK, and Young KD (2016) Colanic Acid Intermediates Prevent De Novo Shape Recovery of Escherichia coli Spheroplasts, Calling into Question Biological Roles Previously Attributed to Colanic Acid, J Bacteriol 198, 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang JY, and Poh CL (2018) Regulating exopolysaccharide gene wcaF allows control of Escherichia coli biofilm formation, Sci Rep-Uk 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.