ABSTRACT
Next-generation sequencing of DNA from nematode eggs has been utilised to give the first account of the equine ‘nemabiome’. In all equine faecal samples investigated, multiple species of Strongylidae were detected, ranging from 7.5 (SEM 0.79) with 99+% identity to sequences in the NCBI database to 13.3 (SEM 0.80) with 90+% identity. This range is typical of the number of species described previously in morphological studies using large quantities of digesta per animal. However, the current method is non-invasive; relies on DNA analysis, avoiding the need for specialist microscopy identification; and can be carried out with small samples, providing significant advantages over current methods.
Keywords: diversity, faecal egg, horse, nemabiome
Helminths of the digestive tract can pose problems for the animal they infect. Some helminths are unique to one host species, while others infect a range of hosts. Before studying changes to helminth diversity in the digestive tract, it is important to determine how many helminth species are likely to inhabit the animal’s tract. Recently, deep sequencing approaches using DNA have proposed the ruminant gastrointestinal ‘nemabiome’ [4] as a means of investigating this, as next-generation sequencing (NGS) of DNA has improved species specificity and detection sensitivity [14]. To date, the equine ‘nemabiome’ has not been investigated.
Strongylidae are among the most important helminth parasites in the equine gut, having been reported in horses, asses, zebras, and inter-species hybrids. To date 64 species, from 19 genera, have been described [10]. Estimates suggest around 5 to 10 species are present in individual horses [12, 15], but using larger samples, less abundant species were detected, meaning the number present is more likely to be more, with 20–29 species having been described [5].
These helminths have various ontogenic stages within their life cycle. Eggs within this cycle are defecated by the host animal, and detection of faecal eggs is the easiest way to observe these organisms. The relationship between numbers of intestinal worms and faecal eggs is not directly correlated [11] but provides an approximation of the worm burden, as higher faecal egg count (FECs) are assumed to be associated with higher numbers of adult worms within the digestive tract.
Two approaches are used to identify helminth species present in animals: (i) cultivating eggs in vitro and identifying larval and (ii) identifying worms extracted post mortem or during a digestive tract operation. Due to the effects co-infections in the host, such as faster immune modulation and susceptibility to infections [19] faster, non-invasive methods would make studies easier. The current work offers an alternative approach; extracting DNA from faecal eggs, followed by NGS to identify and quantify species numbers from horse faecal samples.
Materials and Methods
Fresh faecal samples were collected from 114 horses from 13 yards in West Wales between March and July 2015. As per Lester and Matthews [8], at least three faecal wafers were collected and homogenised to ensure even distribution of eggs. Total eggs per gram (EPG) in faeces were quantified using a modified Concentration McMaster Technique [17]. In each case a sub-sample was frozen and stored at −20°C for potential future DNA analysis.
DNA was extracted from twenty of the samples with the highest FECs using 50 g of sample stored at −20°C. DNA extraction was performed using thawed samples with a QIAamp® DNA Mini Stool Kit (Qiagen Ltd., West Sussex, U.K.) following the manufacturer’s standard protocol but increasing the initial incubation from 70 to 95°C for 5 min [6]. DNA purity and concentrations were assessed using A260 and A280 measurements on a BioTek Epoch Spectrophotometer System.
PCR was performed using the primers 5′-ACGTCTGGTTCAGGGTTGTT (NC1) and 5′-TTAGTTTCTTTTCCTCCGCT (NC2) [7], which have previously been shown to recognise and amplify the second internal transcribed spacer region (ITS-2) of the genomic DNA and recognise a range of internal parasites [3]. Reactions were performed in 25 µl volumes with a 4 µM final primer concentration, using the PCR buffer ImmoMix™ (Bioline, U.K.) at a 1 × reaction concentration. PCR was performed with a T100 Thermal Cycler (BioRad, Watford, U.K.) with the following temperatures: 95°C for 10 min followed by 40 cycles 95°C for 45 sec, 55°C for 45 sec, and 72°C for 45 sec and a final 5 min 72°C extension.
All PCR products were checked by electrophoresis to ensure amplicons of the anticipated size. The ten amplicons with strongest signals were used for next-generation sequencing. PCR was performed for NGS with the adapter sequence CCATCTCATCCCTGCGTGTCTCCGACTCAG on the 5′ NC1 primer, with a sample-specific barcode between the adapter and the NC1 region. The following barcodes were used: CTAAGGTAACGT, TAAGGAGAACGT; AAGAGGATTCGT; TACCAAGATCGT; CAGAAGGAACGT; CTGCAAGTTCGT; TTCGTGATTCGT; TTCCGATAACGT; TGAGCGGAACGT and CTGACCGAACGT. PCR conditions were as described above. Prior to performing the NGS approach, all barcoded primers were checked to ensure that they gave comparable amplification efficiencies relative to that seen with the original primers (i.e., without the barcodes attached).
Two clean-up procedures (AMPure XP bead clean-up and E-Gel agarose gel electrophoresis) were carried out to remove short fragments. Sample concentrations were normalised (120 ng/µl), and equal quantities were pooled for all further stages.
Samples underwent emulsion PCR for Ion Torrent sequencing using the protocol described in the Ion PGM Template OT2 400 Kit User Guide (Life Technologies). The Ion OneTouch 2 Instrument (Life Technologies) was prepared following the User Guide’s instructions.
Initial quality filtering of DNA sequences was performed with standard settings on the Ion Torrent PGM platform, and final library outputs were filtered to ensure 100% identity to primers, checked for potential chimeric sequences, checked to ensure a maximum of one N and and homopolymers of ≤10, and checked to ensure that all sequences were ≥300 nucleotides in length. Individually barcoded files were merged for OTU clustering at five identity thresholds (99, 98, 97, 95 and 90%), with the CD-HIT-OTU program [9]. Sequences were clustered into groups sharing sequence similarity. Due to unequal numbers of sequences per sample, datasets were normalized to have the same number of sequences for statistical analyses using the script daisychopper.pl (http://www.genomics.ceh.ac.uk/GeneSwytch/Tools.html). All sequences generated were identified by BLASTn searches [1] using a set-set of data downloaded from the National Center for Biotechnology Information (NCBI) search engine (https://blast.ncbi.nlm.nih.gov/Blast.cgi), where only sequences which had been identified as either nematodes or Platyhelminthes were searched.
Results
Initially over 180,000 sequences were generated, with 73,534 remaining after meeting the filtering process (=7,353; SEM=920). All samples contained multiple sequences. The number of identifiable sequences varied depending on cut-off value (=7.5, SEM=0.79, at 99% identity; =13.3, SEM=0.80, at 90% identity). DNA database analyses identified 20 equid helminth species from 8 families (Table 1), 15 of which were at identity levels of ≥99+% relative to those in the database. Cyathostomum catinatum, Cylicocyclus nassatus, and Cylicostephanus longibursatus were found in all samples, at ≥99+% identity to database sequences. Sequences similar to Coronocyclus coronatus, Cyathostomum pateratum, Cylicocyclus ashworthi, Cylicocyclus elongates, Cylicocyclus radiates, and Cylicostephanus goldi were also observed in all samples, but often at lower identity levels.
Table 1. Percentage of sequences which could be identified at each cut-off threshold level following NGS, and the number of horses in which each species of helminth could be detected.
| 99% | 98% | 97% | 95% | 90% | |
|---|---|---|---|---|---|
| Percentage of sequences identified | 30 | 41 | 49 | 61 | 79 |
| Coronocyclus coronatus | 9 | 9 | 10 | 10 | 10 |
| Coronocyclus labiatus | 3 | 3 | 3 | 3 | 3 |
| Cyathostomum catinatum | 10 | 10 | 10 | 10 | 10 |
| Cyathostomum pateratum | 7 | 7 | 10 | 10 | 10 |
| Cylicocyclus ashworthi | 5 | 7 | 7 | 9 | 10 |
| Cylicocyclus auriculatus | - | - | 1 | 5 | 6 |
| Cylicocyclus brevicapsulatus | - | - | - | - | 2 |
| Cylicocyclus elongates | - | - | 4 | 9 | 10 |
| Cylicocyclus insigne | 3 | 4 | 5 | 6 | 8 |
| Cylicocyclus nassatus | 10 | 10 | 10 | 10 | 10 |
| Cylicocyclus radiates | 1 | 5 | 6 | 8 | 10 |
| Cylicocyclus ultrajectinus | - | 1 | 1 | 1 | 1 |
| Cylicodontophorus bicoronatus | 1 | 1 | 1 | 3 | 4 |
| Cylicodontophorus mettami | 1 | 1 | 1 | 1 | 1 |
| Cylicostephanus bidentatus | 3 | 3 | 3 | 3 | 3 |
| Cylicostephanus calicatus | 4 | 4 | 4 | 4 | 9 |
| Cylicostephanus goldi | 7 | 8 | 9 | 9 | 10 |
| Cylicostephanus longibursatus | 10 | 10 | 10 | 10 | 10 |
| Tridentoinfundibulum gobi | - | - | - | 1 | 5 |
| Triodontophorus serratus | 1 | 1 | 1 | 1 | 1 |
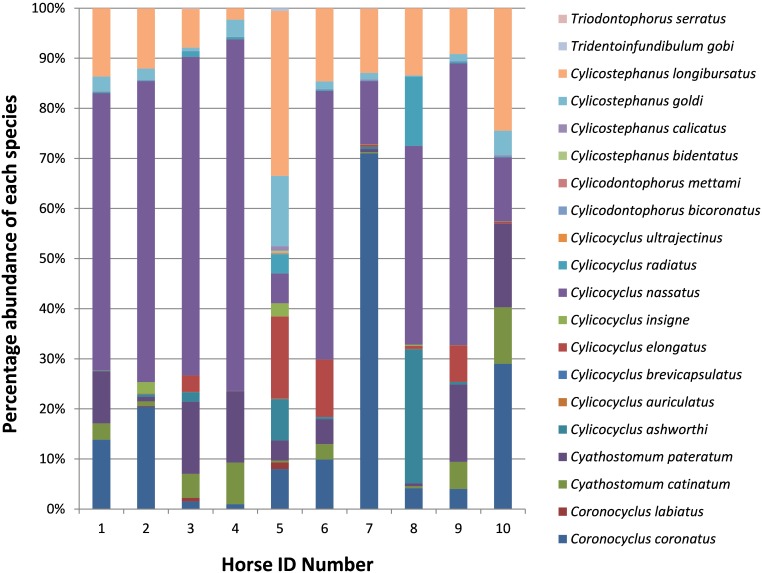
The relative distribution of species varied between samples (Fig. 1). For example, Coronocyclus coronatus, which was present in all samples, constituted ≥70% of identifiable sequences in horse 7 but ~1% of those in horse 4. Conversely, C. nassatus, which was also present in all samples, constituted ≥50% of the sequences detected in six of the animals (≥70% in horse 4) but only 12.6% of the sequences in horse 7. In addition, all horses contained at least one further sequence constituting ≥10% of its total, although the species varied from animal to animal.
Fig. 1.
The percentage of each species of gastrointestinal helminth found in faecal samples of ten different horses based on the total number of reads following NGS. The identification of each helminth species is based on the sequence identity relative to sequences present in the NCBI DNA database.
Samples also revealed sequences found at very low levels (≤ 0.1%) in individual animals, including Cylicocyclus auriculatus (6 horses), Cylicocyclus brevicapsulatus (horses 5 and 6), Cylicocyclus ultrajectinus (horse 5), and Cylicodontophorus bicoronatus (4 horses), although only at lower identity levels in some samples, so they may be sequences which have not been described previously.
Discussion
Previous studies based on adult worm morphology have shown that horses may carry more than one species of worm [12, 15], although larger faecal samples were needed to detect less common species [5]. The current work reiterates this observation regarding multiple helminth species by an alternative method. Moreover, NGS from faecal samples has the additional benefit of being non-invasive as opposed to that with samples from more proximal areas of the digestive tract. The range of species identified here by NGS is in keeping with the numbers of species observed when large digesta samples were analysed [5], but in the current work, the species could be detected using much smaller sample quantities.
Based on DNA sequence identity, it is unclear where to define the cut-off for discriminating between helminth species, as genetic isolation is generally used to define species boundaries rather than DNA differences. Hence the current analysis used a number of cut-off thresholds (99, 98, 97, 95 and 90%) to investigate this approach. Because of this, it may mean that for those identities of ~90%, the sequences may actually be from species for which the sequence had not been determined previously. However this is not equivalent to identifying a novel species, as not all known species of gastrointestinal helminths have had their ITS sequence determined, and it is possible that these sequences correspond to organisms reported morphologically but as yet not characterised at the level of the DNA. Nevertheless, it shows the methodology’s ability to recognise helminth sequences over and above those described in previous work for relatively small samples [12, 15].
Generally, studies to investigate worm burdens in the digestive tract are carried out by FEC, using EPG to give approximations of worms producing these eggs within the tract. However, this method does not allow identification of individual species and relies on analysing collective numbers. NGS allows a more detailed approach and has been used to uncover the ruminant ‘nemabiome’ [4].
It is also worth noting that not all species previously described in horses were detected within the current work. This may be a reflection of seasonal variation in the numbers of different helminths, which is known to be a factor [16] in terms of the species observed. Likewise, the absence of any ruminants on the pastures grazed by these animals may also have helped to influence which parasites were detected. For example, Trichostrongylus axei, which was not detected, is known to be capable of infecting ruminants as well as horses [13]. Moreover, it is important to highlight that these data are based on a single time point and one geographical location. It is already known that these factors can play an influential role in the diversity of helminths detected [10].
Identification of individual species of helminths causing infection is likely to become progressively important in veterinary management practices. All parasitic helminths, irrespective of species, are potentially a burden on the host animal. Therefore, it is viewed as advantageous to treat all animals which have a high faecal egg count with an anthelmintic to control gastrointestinal helminths [9, 18]. There has been a call for reliable diagnostic tools to help combat infections [2] for improved veterinary management, particularly for treatments which offer scope for targeted approaches to treatment of specific organisms. If knowledge of the species present can be obtained prior to treatment, this may in turn allow a more targeted approach to dosing for specific infecting organisms. Based on the work presented here, next-generation sequencing offers a relatively rapid approach towards species identification, uncovering the equid ‘nemabiome’. Moreover, with the cost of NGS continuing to drop and the potential for barcoding of samples to allow multiple analyses to be performed in single runs, NGS offers potential for provision of treatment selection methods in the future.
In conclusion, the current work presents an alternative method of studying helminth diversity in the horse. It has a high level of sensitivity, as evidenced by the number of sequences detected per sample, and reiterates the point that earlier estimates of the scale of diversity of the equine nemabiome may have been underestimations.
Acknowledgments
This work was part-funded by an Access to Masters Scholarship awarded to CJM as part of the European Social Fund (ESF) through the European Union’s Convergence programme administered by the Welsh Government.
References
- 1.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 2.Andersen U.V., Howe D.K., Olsen S.N., Nielsen M.K. 2013. Recent advances in diagnosing pathogenic equine gastrointestinal helminths: the challenge of prepatent detection. Vet. Parasitol. 192: 1–9. [DOI] [PubMed] [Google Scholar]
- 3.Archie E.A., Ezenwa V.O. 2011. Population genetic structure and history of a generalist parasite infecting multiple sympatric host species. Int. J. Parasitol. 41: 89–98. [DOI] [PubMed] [Google Scholar]
- 4.Avramenko R.W., Redman E.M., Lewis R., Yazwinski T.A., Wasmuth J.D., Gilleard J.S. 2015. Exploring the gastrointestinal “Nemabiome”: deep amplicon sequencing to quantify the species composition of parasitic nematode communities. PLoS One 10: e0143559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman M.R., Kearney M.T., Klei T.R. 2003. Equine cyathostome populations: accuracy of species composition estimations. Vet. Parasitol. 116: 15–21. [DOI] [PubMed] [Google Scholar]
- 6.Edmunds J.L., Worgan H.J., Dougal K., Girdwood S.E., Douglas J.L., McEwan N.R. 2016. In vitro analysis of the effect of supplementation with activated charcoal on the equine hindgut. J. Equine Sci. 27: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasser R.B., Chilton N.B., Hoste H., Beveridge I. 1993. Rapid sequencing of rDNA from single worms and eggs of parasitic helminths. Nucleic Acids Res. 21: 2525–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lester H.E., Matthews J.B. 2014. Faecal worm egg count analysis for targeting anthelmintic treatment in horses: points to consider. Equine Vet. J. 46: 139–145. [DOI] [PubMed] [Google Scholar]
- 9.Li W., Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- 10.Lichtenfels J.R., Kharchenko V.A., Dvojnos G.M. 2008. Illustrated identification keys to strongylid parasites (Strongylidae: Nematoda) of horses, zebras and asses (Equidae). Vet. Parasitol. 156: 4–161. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen M.K., Baptiste K.E., Tolliver S.C., Collins S.S., Lyons E.T. 2010. Analysis of multiyear studies in horses in Kentucky to ascertain whether counts of eggs and larvae per gram of feces are reliable indicators of numbers of strongyles and ascarids present. Vet. Parasitol. 174: 77–84. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen M.K., Reinemeyer C.R., Donecker J.M., Leathwick D.M., Marchiondo A.A., Kaplan R.M. 2014. Anthelmintic resistance in equine parasites—current evidence and knowledge gaps. Vet. Parasitol. 204: 55–63. [DOI] [PubMed] [Google Scholar]
- 13.Palcy C., Silvestre A., Sauve C., Cortet J., Cabaret J. 2010. Benzimidazole resistance in Trichostrongylus axei in sheep: long-term monitoring of affected sheep and genotypic evaluation of the parasite. Vet. J. 183: 68–74. [DOI] [PubMed] [Google Scholar]
- 14.Pilotte N., Papaiakovou M., Grant J.R., Bierwert L.A., Llewellyn S., McCarthy J.S., Williams S.A. 2016. Improved PCR-based detection of soil transmitted helminth infections using a next-generation sequencing approach to assay design. PLoS Negl. Trop. Dis. 10: e0004578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proudman C., Matthews J. 2000. Control of intestinal parasites in horses. In Pract. 22: 90–97. [Google Scholar]
- 16.Rehbein S., Visser M., Winter R. 2013. Prevalence, intensity and seasonality of gastrointestinal parasites in abattoir horses in Germany. Parasitol. Res. 112: 407–413. [DOI] [PubMed] [Google Scholar]
- 17.Roepstorff A., Nansen P. 1998. Epidemiology, diagnosis and control of helminth parasites of swine. FAO Animal Health Manual: Number 3, Food and Agriculture Organization of the United Nations. [Google Scholar]
- 18.Stratford C.H., Lester H.E., Pickles K.J., McGorum B.C., Matthews J.B. 2014. An investigation of anthelmintic efficacy against strongyles on equine yards in Scotland. Equine Vet. J. 46: 17–24. [DOI] [PubMed] [Google Scholar]
- 19.Supali T., Verweij J.J., Wiria A.E., Djuardi Y., Hamid F., Kaisar M.M., Wammes L.J., van Lieshout L., Luty A.J.F., Sartono E., Yazdanbakhsh M. 2010. Polyparasitism and its impact on the immune system. Int. J. Parasitol. 40: 1171–1176. [DOI] [PubMed] [Google Scholar]