Abstract
Little is known about the spectrum of mitochondrial DNA (mtDNA) mutations across pediatric malignancies. In this study, we analyzed matched tumor and normal whole genome sequencing data from 616 pediatric patients with hematopoietic malignancies, solid tumors, and brain tumors. We identified 391 mtDNA mutations in 284 tumors including 45 loss-of-function mutations, which clustered at 4 statistically significant hotspots in MT-COX3, MT-ND4, and MT-ND5, and at a mutation hotspot in MT-tRNA-MET. A skewed ratio (4.83) of non-synonymous versus synonymous (dN/dS) mtDNA mutations with high statistical significance was identified based on Monte Carlo simulations in the tumors. By comparison, opposite ratios of 0.44 and 0.93 were observed in 616 matched normal tissues and in 249 blood samples from children without cancer, respectively. mtDNA mutations varied by cancer type and mtDNA haplogroup. Collectively, these results suggest that deleterious mtDNA mutations play a role in the development and progression of pediatric cancers.
Introduction
Cancer has long been considered a metabolic disease, following the discovery of the “Warburg effect”, or the elevated uptake of glucose in the majority of cancers [1, 2]. As noted by Porporato et al, Warburg himself attributed elevated glycolytic rates in malignant cells to primary mitochondrial defects, which incorrectly relegated mitochondria to a bystander role in the oncogenic process [3]. Mitochondrial metabolism plays an essential role in malignant transformation, tumor progression, metastasis, and response to treatment [3]. Many aspects of mitochondrial biology are important in cancer, including mitochondrial biogenesis and turnover, fission and fusion dynamics, oxidative stress, metabolism, bioenergetics, signaling, and variation in mitochondrial DNA (mtDNA) sequence and abundance [4]. All of these contribute to mitochondrial heterogeneity in different cells and microenvironments, which is essential for the considerable metabolic plasticity during tumorigenesis [3].
Cancer, especially pediatric cancer, is also considered a genetic disease. At least 8.5% of pediatric cancer patients have germline mutations in cancer predisposition genes [5]. Pan-cancer analyses of pediatric tumors reveal a spectrum of nuclear somatic DNA alterations that vary by tumor type [6, 7]. DNA mutations, chromosomal rearrangements, and copy number alterations are frequently distinct from those seen in adult tumors, even in similar cell types from the same anatomic locations. Epigenetic factors, such as chromatin remodeling and histone modifications, also appear to play a more critical role in pediatric compared to adult cancers.
Mitochondrial DNA mutations have a pathogenic role in complex, multi-system disorders affecting nearly every organ at every age [8, 9]. Despite extensive phenotypic heterogeneity, cancer is not commonly observed in patients with mitochondrial diseases. The importance of mtDNA mutations in cancer has been less well recognized [10-13], despite several large-scale studies showing significant numbers of mtDNA mutations in a variety of adult tumors [14, 15]. These mtDNA mutations, to a large extent, have been dismissed as passenger events during tumorigenesis. mtDNA mutations have been reported in a few pediatric cancer subtypes, e.g. medulloblastoma [16], glioma [17], acute myelogenous leukemia (AML) [18], and neuroblastoma [19], but little is known regarding the spectrum of mtDNA mutations across pediatric malignancies and its contribution to tumorigenesis.
Materials and Methods
Ethics statement
The pediatric cancer patients studied were originally consented to the St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project. We applied and were granted access to the data sets by the project’s data access committee. The use of control data from patients with non-cancer diagnosis in this study was approved by the Institutional Review Board of the Children’s Hospital Los Angeles for de-identified use in the study without patient consent (CCI #:12-00224). All investigations were conducted according to the principles expressed in the Declaration of Helsinki.
Mitochondrial DNA mutation and variant detection
Whole genome sequencing data were retrieved from European Genome Phenome archive under accession number EGAD00001001433 [5]. We extracted mtDNA reads from paired tumor-normal whole-genome sequencing data and aligned them to GRCh37 using the Edico DRAGEN Genome genome pipeline (version 2.3.1) (San Diego, CA).
Mitochondrial DNA somatic and germline calls were made by VarScan2 [20] and further filtered by our LUBA variant caller [21], which is a custom somatic variant caller that supports the pediatric genetic tests at our institution. VarScan2 was run in somatic mode with standard parameters which require at least 100x coverage in both the tumor and normal samples (--min-coverage 100), at least 2.5% of alternative allele frequency in the tumor to make a call (--min-var-freq 0.025) and minimum base quality of 20 at position (--min-avg-qual 20), and a strand bias filter (--strand filter 1). We removed known recurrent false positives [14] caused by misalignment in the polyC region (rCRS 302-315), the CAC repeat (rCRS 513-525), and the artifact deletion at position 3107 (3105-3110). Additionally, we applied a population frequency-based filter to remove any frequently recurrent calls (>10%) that are not reported in MitoMap (https://mitomap.org) [22]. Our custom LUBA variant caller generates a pile-up file from aligned reads, and base counts are adjusted depending on base qualities. Benjamini-Hochberg correction is applied to control the false discovery rate. Calls are then made for loci with sufficient coverage. After variants are called from the pile-up for specific positions, genotype qualities are assessed from haplotype analysis for indels and multi-allelic sites. To reduce false positives, the LUBA variant caller applies a number of filters, including filters based on a set of reference samples: genotype likelihood filter, strand bias filter, low complexity regions filter, variant occurrences filter which tags variants found in multiple reference samples, and strand-specific base variation filter which excludes loci with high average base variation across reference samples. Our LUBA variant caller was run with the same parameters as VarScan2 (stringent strand-bias filter, 2.5% AF and minimum 100x coverage) and was used to confirm calls made by VarScan2. LUBA supported 94% of tumor-only calls and 99% of germline calls. We determined this set of high confidence calls and used them for downstream analyses. In order to better describe potential functional impacts of these mutations, we calculated MutPred2 scores [23] for observed mutations in protein coding regions and MitoTIP scores [24] for tRNA mutations. Finally, we evaluated recurrence of observed mutations in publicly available database of mitochondrial variants MitoMap [22]. All tumor-only mtDNA mutations are listed in Supplementary Table 1.
Mitochondrial DNA genome sequencing of 249 blood samples of children without cancer
The 249 children were patients that we tested as part of our clinical exome sequencing test at the Children’s Hospital Los Angeles with no current or previous cancer diagnosis. DNA was extracted from peripheral blood using a commercially available kit (Promega Maxwell RSC DNA Extraction Kit; Fitchburg, WI). The WES library was generated using the Agilent SureSelect Human All Exon V6 plus, a custom mitochondrial genome capture kit (Santa Clara, CA), designed in collaboration with Agilent for consistent coverage across the mitochondrial genome]. Paired-end 2×100 basepair (bp) sequencing was performed using the Illumina (San Diego, CA) NextSeq 500 sequencing system.
Mitochondrial DNA genome recovery from single-cell ATACseq and heteroplasmy comparison
We employed the MTseeker package (https://doi.org/doi:10.18129/B9.bioc.MTseeker) to recover single-cell mitochondrial genomes from Corces & Buenrostro [25] and used the SNP-tolerant GSNAP variant caller [26] to characterize cells with mitochondrial read coverage of at least 20x (for error rates roughly equivalent to Sanger sequencing [27]). The subject’s cells had also been sequenced in bulk; we verified that haplogroup assignment and variant calls were consistent across single-cell and bulk runs.
Monte Carlo simulations
To assess the significance of departure from expected dN/dS ratios, we inferred parameters of the probability distribution of dN/dS ratio through Monte Carlo simulations. Since there are 12 classes of possible point substitutions (SC1, …, SC12), the probability of observing substitution SCx leading to a synonymous mutation in a set of observed mutations can be defined as
where P(syn∣SCx) is the frequency at which the substitution class SCx leads to a synonymous mutation and P(SCx) is the frequency of this substitution class in the data, i.e., mutational signature. Hence, the expected frequency of synonymous mutations in a set of observed mutations is a sum of probabilities across all substitution classes:
The calculated expected dN/dS ratios are listed in Supplementary Table 2, with detailed calculations included. For both pediatric and adult tumor-only mtDNA mutations, we created one million simulated datasets with the number of mutations and the mutational signature identical to the original empirical dataset, but with randomly assigned positions. For example, if we observed a m.4483C>T mutation in the real data, this mutation was randomly assigned to any position in a coding region where C>T mutation is possible in the simulated dataset.
We verified that dN/dS ratios observed in simulated datasets reasonably follow a normal distribution (D’Agostino and Pearson’s normality test P < 0.001) and we modeled the probability distribution function as a fit of the normal distribution to simulated data. Python code for Monte Carlo simulation can be provided upon request.
Meta-analysis of adult cancer datasets
For comparison of pediatric and adult cancer mtDNA somatic mutational profiles, we obtained reported calls from two large-scale studies from 1,675 and 567 individuals [14, 15]. The reported calls were extracted from the supplementary materials, filtered by basic QC criteria (minimum 20 reads covering position in both tumor and normal samples, minimum 5 reads supporting alternative allele) and annotated by our mtDNA pipeline. All adult cancer mutations are included in Supplementary Table 3.
Somatic variant calling in samples with hypermutated mtDNA genome
In order to determine whether there is a link between the number of somatic mutations in mitochondrial and nuclear genomes, we performed tumor-normal paired somatic variant calling in these five mtDNA-hypermutated samples and 132 randomly selected samples (Supplementary Table 4). Tumor and normal WGS bam files were aligned using Edico DRAGEN pipeline version 2.3.1 (San Diego, CA). The same pipeline was used for the somatic variant calling pipeline with default settings and filters.
Results
Study subjects and mtDNA genome sequencing
The initial 621 matched tumor (somatic) and normal blood (germline) WGS data sets were generated from 23 pediatric cancer subtypes, including hematopoietic malignancies (49%), solid tumors (28%), and brain tumors (23%) (Table 1) (5). Due to the high cellular copy number of mtDNA genomes, the standard ~30X genome coverage provided much higher mtDNA genome coverage in both tumors (~7,130 ± 5,800X) (Supplementary Table 5) and matched blood (~4,100 ± 3,000X) (Supplementary Table 6) compared with the nuclear genome. Nine normal samples and four tumor samples yielded lower but acceptable coverage (~885 ± 210X) (Supplementary Table 7). mtDNA genome coverage was 99.9 ± 0.01% at 100X in all 621 pairs of matched tumor-germline samples, and 99.9 ± 3.5% at 800X in 1,226 of the 1,242 (98.7%) samples (Supplementary Tables 5 & 6). As pediatric controls, we also sequenced the mtDNA genomes from blood samples of 249 children with no history of cancer who were matched by age and gender, but not ethnicity, to a median depth of 6,930X (range 2,647 - 7,930X) (Supplementary Table 8) using Agilent custom mitochondrial probes [28]. mtDNA genome coverage at 100X was 99.99% and at 1,000X was 99.1% ± 0.9% (Supplementary Table 8). For comparison with adult cancers, we reanalyzed mtDNA genome mutations previously reported in 2,202 adult cancers [14, 15].
Table1.
Pediatric cancer subtypes studied and the number of samples of each subtype.
| Hematopoietic Malignancies | # of Samples | |
|---|---|---|
| AMLM | Acute megakaryoblastic leukemia | 4 |
| B-ALL | B-cell acute lymphoblastic leukemia | 45 |
| CBF | Core-binding factor acute lymphoblastic leukemia | 16 |
| EA | E2A-PBX1 B-lineage acute lymphoblastic leukemia | 33 |
| ERG | Acute lymphoblastic leukemia with alterations of ERG | 22 |
| ETV | ETV6-RUNX1 acute lymphoblastic leukemia | 49 |
| HYPER | Hyperdiploid B-cell acute lymphoblastic leukemia | 53 |
| HYPO | Hypodiploid B-cell acute lymphoblastic leukemia | 22 |
| INF | Infant B-cell acute lymphoblastic leukemia | 25 |
| TALL | T-cell acute lymphoblastic leukemia | 1 |
| PH-ALL | BCR-ABL1 acute lymphoblastic leukemia | 34 |
| Hematopoietic Malignancies (n) | 304 | |
| Solid Tumours | # of Samples | |
| ACC | Adrenocortical carcinoma | 20 |
| MEL | Melanoma | 5 |
| NBL | Neuroblastoma | 51 |
| OS | Osteosarcoma | 37 |
| RB | Retinoblastoma | 14 |
| RMS | Rhabdomyosarcoma | 13 |
| EWS | Ewing’s sarcoma | 35 |
| Solid Tumours (n) | 175 | |
| Brain Tumours | # of Samples | |
| CPC | Choroid plexus carcinoma | 4 |
| EPD | Ependymoma | 39 |
| HGG | High grade blioma | 31 |
| LGG | Low grade glioma | 36 |
| MB | Medulloblastoma | 32 |
| Brain Tumours (n) | 142 | |
| All subtypes (n) | 621 | |
mtDNA mutation analysis is complicated by heteroplasmy, which is the ratio of the number of copies of a mutated mtDNA genome to all mtDNA genomes in a cell or tissue [8, 9]. The high mtDNA coverage achieved in the next generation sequencing (NGS) data allowed us to accurately assess low-level heteroplasmy, measured as variant allele frequency (VAF), and minimize potential contamination of non-mtDNA genome reads from contaminating nuclear mtDNA transcripts (‘NuMTs’) [28].
Tumor-only mtDNA mutation profiles
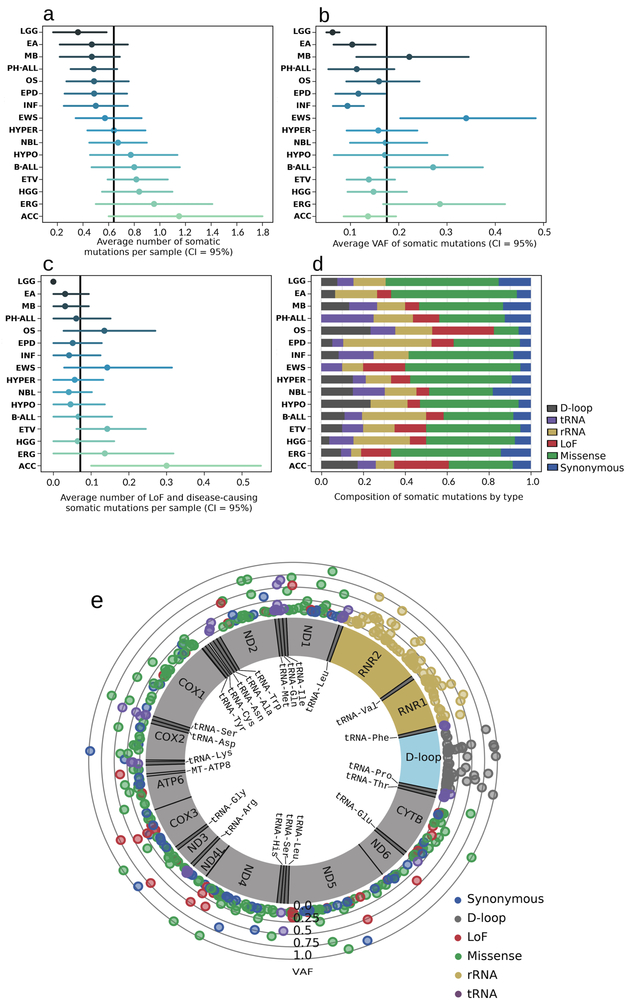
In the pediatric cancer data set, we excluded five mtDNA-hypermutated samples from downstream analyses. A separate analysis was performed as detailed below. Three hundred and ninety-one tumor-only mtDNA mutations were detected in 284 (45.7%) of the 616 remaining cancers (Supplementary Table 1). The mean number of tumor-only mtDNA mutations seen across all subtypes was 0.63 ± 0.84, ranging from a significantly lower number of 0.36 mtDNA mutations per tumor in low-grade gliomas (LGG) compared to other subtypes (p = 0.017, Mann-Whitney rank test) to 1.15 mtDNA mutations per tumor in adrenocortical carcinomas (ACC) (Fig. 1a). Nearly all tumor-only mtDNA mutations (99.2%) were present in a heteroplasmic state, with levels ranging from 2.5% to almost 100% (near-homoplasmy), but significantly skewed toward low-level heteroplasmy (skewness = 2.38, Pearson’s kurtosis = 5.23), with the average heteroplasmy at 17.7% (Fig. 1b). LGG mtDNA mutations again exhibited significantly lower levels of heteroplasmy compared to all other subtypes (Fig. 1b). A significant number of loss of function (LoF) and pathogenic (missense or nonsense) mutations were seen in all subtypes except LGG (Fig. 1c). The composition of mutation types also varied across tumor subtypes (Fig. 1d). In total, we identified 158 missense, 71 rRNA, 45 LoF, 43 D-loop, 36 synonymous, and 35 tRNA tumor-only mtDNA mutations with varied heteroplasmy levels (Fig. 1e). Only 119 of these mutations (30.4%) matched known variants in the general population, based on available data in the manually curated mtDNA mutation database, MitoMap (https://mitomap.org) (Supplementary Table 1) [22].
Figure 1. Distribution of tumor-only mutations across mtDNA genome and tumor subtypes. In panels a – d, only subtypes with more than 20 samples are shown.
a ∣ Average number of somatic mtDNA mutations in individual cancer subtypes. b ∣ Average heteroplasmy levels in individual cancer subtypes. c ∣ Average number of LoF and disease-causing mutations in individual cancer subtypes. d ∣ Composition of mutation types in individual cancer subtypes. e ∣ 391 mitochondrial somatic mutations detected in 616 pediatric cancers. Outer scale represents heteroplasmy level, color codes for type of variant.
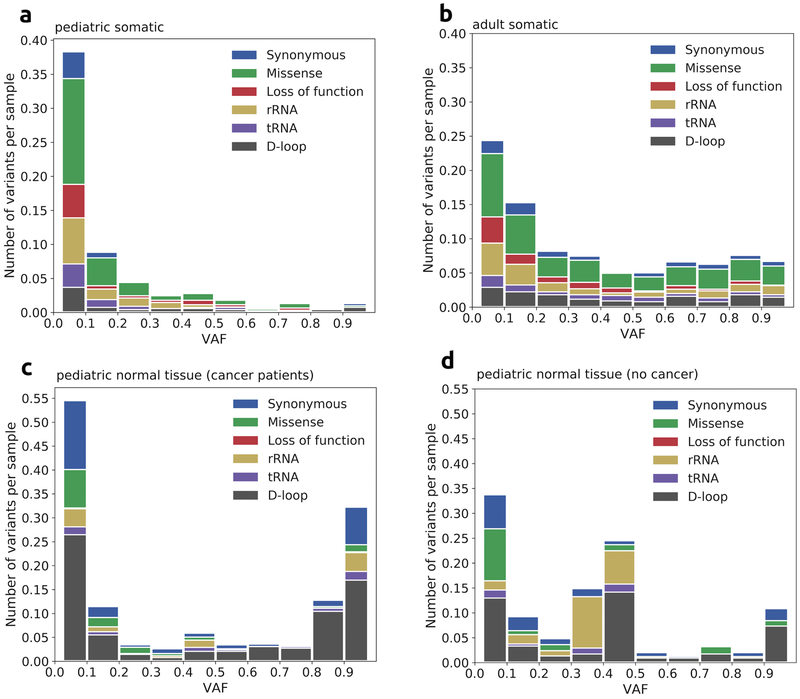
The mtDNA mutation profile we observed in pediatric cancers (Fig. 2a) was similar to what was previously reported in adult cancers (Fig. 2b), with missense mutations as the most frequent mutation type. However, the majority (68.4%) of pediatric, tumor-only mtDNA mutations were detected at heteroplasmy levels below 10% with a median heteroplasmy of 7.2% (Fig. 1b, Fig. 2a), while the distribution of adult tumor-only mtDNA mutations was more uniform across all heteroplasmy levels (Fig. 2b). We did not observe significant difference in heteroplasmy levels of different classes of tumor-only mtDNA mutations, in either pediatric cancers (Supplementary Figure 1a) or adult cancers (Supplementary Figure 1b). Tumor-only missense and LoF mtDNA mutations occurred at comparable or statistically significantly higher heteroplasmy levels than synonymous and D-loop mtDNA mutations, which are likely to be functionally neutral. We observed a significant number of missense and LoF mutations at high heteroplasmy in both pediatric and adult cancers, as well as low heteroplasmy tumor-only mtDNA mutations. The later was not excluded. As in some previous studies, as they could reflect intra-tumor heterogeneity, or subtle defects affecting the metabolic microenvironment..
Figure 2. Varied profiles of mtDNA mutations and variants.
a ∣ Composition of somatic mutations in 616 pediatric cancer tumor samples. Missense mutations were the most frequent type. The vast majority of mutations were present at low heteroplasmy levels (<10%). b ∣ Composition of somatic mtDNA mutations in 2,202 adult cancer tumor samples. Similarly to pediatric cancer, missense mutations were the most frequent type at all heteroplasmy levels. Distribution of mutations across heteroplasmy levels is more uniform compared to pediatric cancers. c and d ∣ Composition of heteroplasmic variants in normal tissues of (c) pediatric cancer patients and (d) control non-cancer pediatric patients. Most frequent types of variants were D-loop and synonymous variants. High-level heteroplasmic variants are almost entirely haplogroup-defining variants (Supplementary Tables 9 & 10).
Intra-tumor heterogeneity in mtDNA heteroplasmy
To test this hypothesis, we took advantage of the large fraction of mtDNA reads in single-cell ATACseq data, which are often treated as contamination as they are not useful for elucidating nuclear chromatin structure [29]. We found that single-cell ATACseq provides sufficient coverage to assess mtDNA variants in all cell types evaluated, and heteroplasmy of mtDNA variants could be readily assessed (Supplementary Figure 1). Reanalysis of data from Corces et al [25] revealed that tumor cells from the same patient harbored frameshift variants which, while rare in the bulk population, were nearly homoplasmic within some cells and absent in others. This intra-tumor variability, while apparent at single-cell level, was obscured in bulk samples; due to averaging of VAF, these mutations appear to have low heteroplasmy (Supplementary Figure 2). These results support our hypothesis that LoF mtDNA mutations can arise in distinct or individual tumor cells. Low observed heteroplasmy of mtDNA mutations in tumors can thus result from individual cells harboring different mtDNA mutations, suggesting that LoF mtDNA mutations in individual tumor cells may in fact have a functional role in tumor development or progression.
Heteroplasmic mtDNA variants in normal tissues
The composition of heteroplasmic mtDNA variants in matched normal blood from pediatric cancer patients and in our non-cancer pediatric control samples was dramatically different from the observed pediatric tumor-only mtDNA mutations. A chi-square test showed that the composition of the tumor-only mtDNA mutations was significantly different from that of the heteroplasmic mtDNA variants in non-cancer control samples (p-value < 2.2e-16). Specifically, 51.6% of all tumor-only mtDNA mutations were either missense or LoF, which is significantly higher than 6.2% found in the non-cancer control samples (Z-score = 16.4, p < 0.001, Z-test). In both the matched normal and non-cancer control datasets and in direct contrast to tumor-only mtDNA mutations, the most frequent heteroplasmic variants were within the D-loop region (43.8% in matched normal and 41.5% in non-cancer), followed by synonymous variants with uneven heteroplasmy levels (Fig. 2c, 2d). Nearly 85% of all heteroplasmic variants in both datasets were known to be benign, haplogroup-defining markers (Supplementary Tables 9 & 10), which demonstrated evidence of intensive purifying selection against deleterious variants in non-cancer cells. Missense variants were largely confined to low heteroplasmy levels and only two LoF variants were detected at very low heteroplasmy in matched normal tissues: m.12384T>TC, an MT-ND5 frameshift variant in a hyperdiploid B-cell leukemia patient at 4.8% heteroplasmy, and m.14527A>AC, an MT-ND6 frameshift variant, in an ependymoma patient at 3.1% heteroplasmy.
Known pathogenic mtDNA variants for mitochondrial disorders
Interestingly, four pediatric cancer patients harbored homoplasmic variants (ependymoma, m.14484T>C; Ewing sarcoma, m.1555A>G; hyperdiploid B-cell acute lymphoblastic leukemia (ALL), m.14484T>C; and neuroblastoma, m.11778G>A) in their matched tumor and normal samples that are causal of Leber’s Hereditary Optic Neuropathy (LHON) and hearing loss [22]. Three cancer patients’ matched tumor and normal samples had heteroplasmic mtDNA variants known to cause classic mitochondrial diseases: ACC, m.3243A>G (5.0%); B-cell ALL, m.9185T>C (4.9%); neuroblastoma, m.8363G>A (53.4%) (Supplementary Table 11) [22]. Compared to the low prevalence rate (1 in 5000) of mitochondrial disorders in the general population [30], these results are suggestive of increased cancer risk associated with known pathogenic mtDNA variants, contrary to an earlier report [31]. Clinical data for these patients were not available due to the de-identified nature of this dataset, so it is unclear whether these patients suffered from symptomatic mitochondrial disorders associated with these mtDNA variants.
Mitochondrial haplogroup analysis
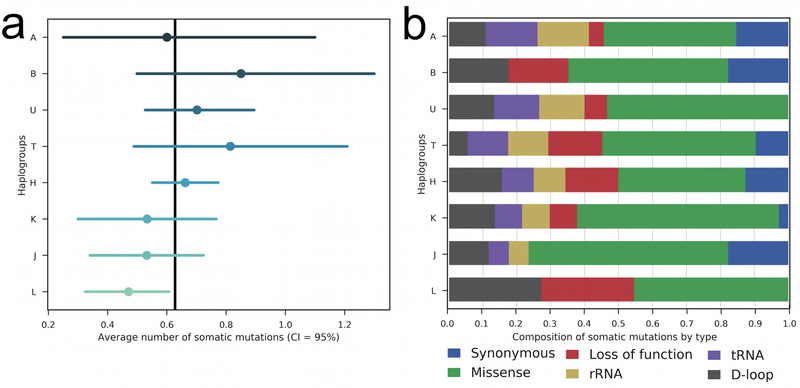
We determined the mitochondrial haplogroup for each of the 621 childhood cancer patients using Phy-Mer (Supplementary Table 12) [32], and summarized them into macro-haplogroups, A, B, C, D, E, H, HV, I, J, K, L, M, N, R, T, U, V, W, and X (Supplementary Table 13). For statistical power, we focused on macro-haplogroups with at least 20 patients: A (20), B (20), H (207), J (47), K (30), L (102), T (43) and U (57). While there appeared to be a greater representation of pediatric cancer patients in some macro-haplogroups, such as H, this may be a reflection of the local patient population. No inference can be made without an ethnicity-matched control population. However, we observed that the number of tumor-only mtDNA mutations per sample differed significantly between the two largest macro-haplogroups: 0.66 for H, which is mostly Eurasian-specific, and 0.47 for L, which is mostly African-specific (p = 0.03, Kruskal-Wallis test). The rates were not different between H and J (p = 0.44), H and U (p = 0.62), H and T (p = 0.82), H and K (p = 0.50), H and A (p = 0.46), or between H and B (p = 0.39) (Fig. 3a). The composition of mutation types in the L haplogroup was distinctly different from that in H and other haplogroups as well, lacking synonymous, rRNA, and tRNA mutations (Fig. 3b). We examined the number of patients belonging to each of the macro-haplogroups in different cancer subtypes and noticed an enrichment of certain subtypes in selected macro-haplogroups, such as hyperdiploid B-ALL in H (Supplementary Table 14). The Chi-Square test was highly significant (p-value < 0.001). We hypothesize that mitochondrial haplogroup background itself contributes to the varying risk for different pediatric cancers. However, patient ascertainment bias is a likely confounder, and cannot be ruled out.
Figure 3. Distribution of tumor-only mtDNA mutations across 8 mitochondrial macro-haplogroups. In both panels, only macro-haplogroups with more than 20 samples are shown.
a ∣ Average number of somatic mtDNA mutations in patients belonging to individual mitochondrial macro-haplogroups. b ∣ Composition of mutation types in patients belonging to individual mitochondrial macro-haplogroups.
Skewed dN/dS ratio of tumor-only mtDNA mutations
We observed a highly skewed dN/dS ratio (4.83) of non-synonymous mutations (including missense, stop-gain and stop-loss) versus synonymous tumor-only mutations in pediatric cancers. If we include the LoF mutations in the non-synonymous category, the dN/dS ratio was even higher (5.64). The ratio remains largely unchanged within different heteroplasmy intervals (Supplementary Figure 3). Interestingly, the dN/dS ratio was higher for the tumor-only mtDNA mutations between 0.1 and 0.5 heteroplasmy (7.3) than at less than 0.1 heteroplasmy (5.2), although this is not statistically significant. This can partly be explained by the higher frequency of non-synonymous mutations among all possible point substitutions in mtDNA coding regions when mathematically calculated: out of 34,023 possible single nucleotide mtDNA changes, only 8,289 (24.4%) are silent. Furthermore, the mtDNA mutations are not random but affected by a strand-biased mutational signature that results from the replication-coupled mutation process [14, 15]. For these reasons, while Ju et al. and Stewart et al. reported skewed dN/dS ratios of 4.2 and 3.63 in adult cancers respectively, not including the LoF mutations, they dismissed their significance as reflecting the mtDNA mutational signature [14, 15].
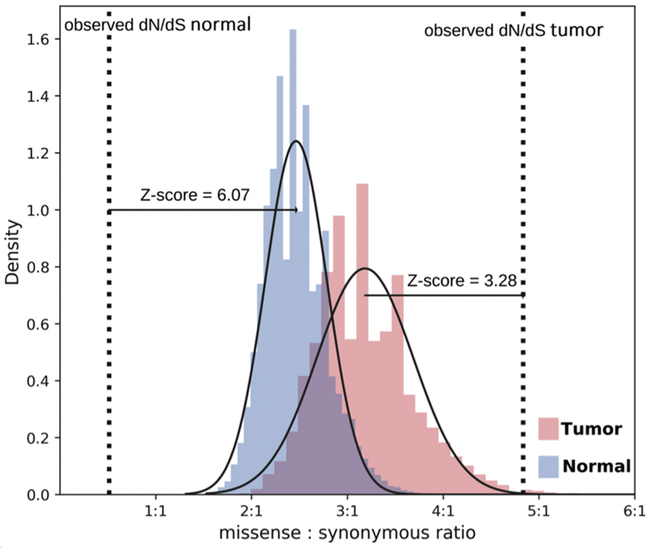
To determine whether this biased mutational signature could explain the skewed dN/dS ratio observed in pediatric tumors, we applied a Bayesian approach to determine the expected dN/dS ratio, with Monte Carlo simulations to calculate the probability density function (see Methods). Using the mutational signature shown for tumor-only mtDNA mutations in this study, we calculated an expected dN/dS ratio of 3.11 (Supplementary Table 2), which is significantly lower than the observed ratios of 4.83 (Z = 3.28, p = 5.19×10−4) (Fig. 4). In comparison, focusing solely on heteroplasmic variants, we extracted the mutational signature and determined the expected dN/dS ratios from matched normal samples of pediatric cancer patients to be 2.43 (Supplementary Table 2), which is significantly higher than the observed ratio of 0.44 (Fig. 4). Similarly, a lower dN/dS ratio of 0.93 was observed in the heteroplasmic mtDNA variants found in control samples with no history of cancer. In both matched normal and non-cancer groups, the expected ratio was much higher than what was observed (matched normal Z= 6.07, p = <1×10−5; non-cancer Z= 2.67, p = 3.79×10−3), which is in striking contrast to the pediatric cancer group. Combined, these analyses provide strong statistical evidence that non-synonymous tumor-only mtDNA mutations are under positive selection, hence enriched in tumors, but subjected to strong purifying selection, thus diminished, in non-tumor tissues (Fig. 4). We performed a similar analysis using only high-quality tumor-only mtDNA mutations found in adult cancers from the Ju et al. study [14]. Using the mutational signature of these calls, we determined an expected dN/dS ratio of 3.68 for adult tumors, which is also significantly lower than the calculated dN/dS ratio of 4.64 for these high-quality calls (Z = 3.02, p = 1.26×10−3). This indicates that the tumor-only mtDNA mutations in adult cancer are likely under similar strong positive selection.
Figure 4. Monte Carlo simulations of dN/dS ratio in pediatric tumors and normal samples.
Blue and pink histograms represent obtained dN/dS ratios in 1,000,000 simulated datasets in normal and tumor samples respectively, with fitted Gaussian curve. The dotted lines represent the observed dN/dS ratios in tumor and normal mtDNA mutations or variants detected. Significant difference between expected and observed dN/dS ratios in tumor and normal samples provide statistical evidence for opposite direction of evolutionary forces acting on non-synonymous mutations in tumor and normal tissues: positive selection is the dominant force in tumors while purifying selection dominates in normal tissues.
Loss-of-function mutations and cancer subtypes
Loss-of-function mutations are the most striking group of mtDNA mutations in tumor vs. control comparisons. We identified a total of 45 tumor-only mtDNA LoF (frameshift and nonsense) mutations, with heteroplasmy ranging from 2.5% to 73% (Table 2, Supplementary Table 1). Six LoF mutations were identified in 5 of 20 ACCs (25%). Similarly, LoF mutations were frequently observed in Ewing sarcomas (EWS) (11.4%) and osteosarcomas (OS) (13.5%). Combined, these three cancer subtypes had significantly more LoF mutations compared to other subtypes (p = 2.41×10−4). For example, we observed LoF mutations in only 2 of 45 neuroblastoma samples (4.4%), and in 5 of 157 CNS tumors combined (3.2%). In hematologic malignancies, 19 of 302 (6.3%) B-ALL and 2 out of 20 (10%) acute myeloid leukemia (AML) cases harbored a LoF mutation in the cancer sample. We did not find LoF mutations in retinoblastoma, choroid plexus carcinoma (CPC), LGG or T-cell ALL samples.
Table2.
Distribution of 45 loss-of-function somatic mtDNA mutations across mtDNA genes and tumor subtypes.
| a. | |||
|
Mitochondrial region |
Gene |
Number of LoF mutations |
Cancer subtype |
| Complex I | MT-ND6 | 3 | B-ALL (2), EPD |
| MT-ND5 | 11 | ACC (5), B-ALL (3), EPD, EWS, NBL | |
| MT-ND4 | 7 | B-ALL (3), AML, EWS, OS, HGG | |
| MT-ND3 | 2 | B-ALL, HGG | |
| MT-ND2 | 3 | ACC, B-ALL, RMS | |
| MT-ND1 | 3 | B-ALL (2), MEL | |
| Complex III | MT-CYB | 6 | B-ALL (2), EWS (2), OS (2) |
| Complex IV | MT-CO3 | 6 | OS (2), B-ALL (2), MB, NBL |
| MT-CO1 | 2 | B-ALL | |
| ATP Synthase | MT-ATP8 | 1 | B-ALL |
| MT-ATP6 | 1 | AML | |
| b. | |||
|---|---|---|---|
| Major category | Cancer subtype | Mitochondrial mutations | |
| Brain Tumors | EPD | MT-ND5, MT-ND6 | |
| HGG | MT-ND3, MT-ND4 | ||
| MB | MT-CO3 | ||
| Solid Tumors | ACC | MT-ND5 (5), MT-ND2 | |
| OS | MT-CO3 (2), MT-CYB (2), MT-ND4 | ||
| EWS | MT-CYB (2), MT-ND4, MT-ND5 | ||
| NBL | MT-ND5, MT-CO3 | ||
| MEL | MT-ND1 | ||
| RMS | MT-ND2 | ||
| Hematopoietic Malignancies | B-ALL, NOS | MT-ND3, MT-ND4, MT-CO3 | |
| B-ALL, EA | MT-CO1 | ||
| B-ALL, ERG | MT-ND2, MT-ND6, MT-CO1 | ||
| B-ALL, ETV | MT-ND1, MT-ND4, MT-ND5, MT-ND6, MT-CYB, MT-ATP8 | ||
| B-ALL, HYPER | MT-ND4, MT-ND5, MT-CYB | ||
| B-ALL, HYPO | MT-ND1 | ||
| B-ALL, PHALL | MT-ND5, MT-CO3 | ||
| AML, CBF | MT-ND4, MT-ATP6 | ||
Clustering of loss-of-function somatic mtDNA mutations
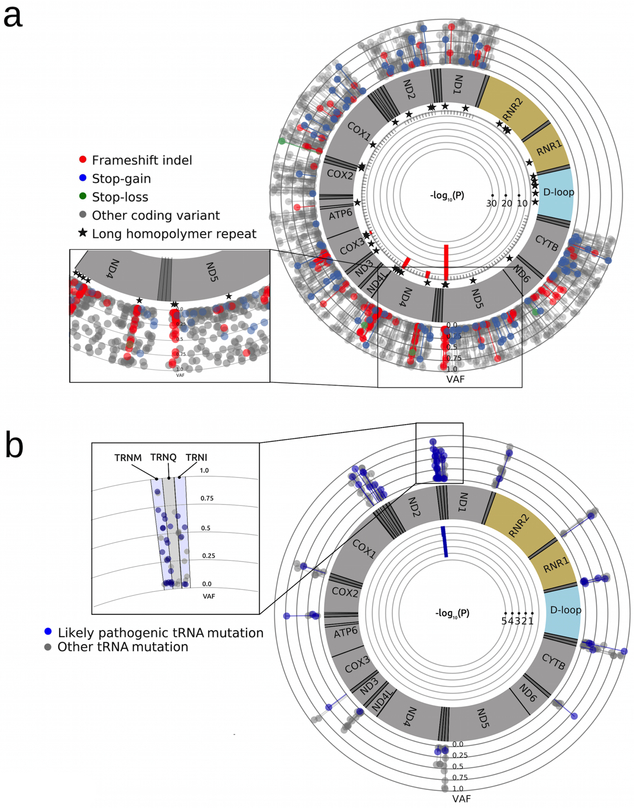
The majority of LoF mutations were within the mitochondrial complex I genes, particularly, with the most (11) in MT-ND5. We observed three recurrent tumor-only LoF mutations, including m.12417C>CA in MT-ND5 in four cases (ependymoma, Ewing sarcoma, ETV6-RUNX1 B-ALL, Ph-positive ALL), m.10191TC>T in MT-ND3 in two cases (B-ALL and high-grade glioma (HGG)), and m.9531A>AC in MT-CO3 in two cases (OS and Ph-positive ALL). To further explore possible mutational hotspots, we conducted a meta-analysis of pediatric tumor-only mutations, in combination with previously published tumor-only mtDNA mutations from 2,202 primarily adult cancers (Supplementary Table 3) [14, 15]. We searched for clustering of LoF mutations by counting LoF mutations in 100bp non-overlapping windows in coding regions, modeled as a Poisson distribution, and used the probability mass function to test whether a window departed from the model. Four windows (9445-9544, 10960-11059, 11768-11867, 12374-12473) within MT-ND4, MT-ND5 and MT-CO3 genes exhibited a substantial and significant enrichment of frameshift indels (p = 5.9×10−3 to 1.8×10−28, Bonferroni correction, Supplementary Table 15), indicating the presence of mutational hotspots (Fig 5a). Interestingly, the frameshift mutations within these windows were all located at homopolymer regions. Because indel mutations in repetitive regions may result from polymerase slippage [33], we tested for enrichment of frameshift indels among all long homopolymer regions. After restricting the analysis to 20 long homopolymers within coding regions, hotspots in MT-ND4 (polyA 11032-11038 and polyC 11867-11872) and MT-ND5 (polyA 12418-12425) remained significant (p = 3.1×10−3 - 9.2×10−11, Bonferroni correction, Supplementary Table 16). These data indicate that the presence of homopolymer repeats alone does not explain frameshift indel clustering in these hotspots. A selective advantage for these mutations is an attractive alternative explanation.
Figure 5. Meta-analysis of hotspots for pathogenic somatic mtDNA mutations in 2,818 adult and pediatric tumor samples.
Shown p-values are after Bonferroni correction. a ∣ LoF mtDNA mutations in 2,818 tumor samples. Frameshift indels exhibit high degree of clustering in four regions located at long homopolymer regions in MT-ND4, MT-ND5 and MT-CO3 genes. b ∣ Probable pathogenic mtDNA tRNA mutations (MitoTIP score >16.25) in 2,818 tumor samples. Pathogenic mtDNA tRNA mutations form a hotspot in the MT-TM gene.
Clustering of pathogenic tumor-only mtDNA tRNA mutations
We used the same approach to test for clustering of probable pathogenic tumor-only tRNA mutations (as defined by predicted MitoTIP score ≥ 16.25) [24]. One 20bp window located in MT-TM (TRNM), encoding the mitochondrial tRNA for methionine, was significantly enriched for pathogenic mutations (p = 2.361×10−5, Bonferroni correction, Supplementary Table 17). Identification of a MT-TM mutational hotspot is novel and may lead to important insights into tumor pathogenesis, since the mitochondrial tRNA genes are hotspots for pathogenic mutations that also cause mitochondrial disorders [34].
Predicted pathogenicity of missense and tRNA mutations
We used in-silico prediction algorithms MutPred2 [23] and MitoTIP [24] to estimate the pathogenicity of missense and tRNA mutations, respectively. We attempted to assess predicted pathogenicity for every gene separately in either the pediatric or the adult cancers (Supplementary Figure 4a-d). However, in most cases only the adult sample dataset provided enough data points for sufficient statistical power to investigate individual genes. We observed a statistically significant enrichment of highly pathogenic variants in the MT-TM gene in adult samples (p = 2.78×10−4, Welch’s t-test) compared to all other tRNA genes (Supplementary Figure 4d), which is in agreement with the presence of a hotspot for pathogenic mutations in this gene in both pediatric and adult cancers. Two genes in the coding region of Complex I, MT-ND4L and MT-ND6, had significantly higher MutPred scores (p = 2.02×10−3 and p = 3.28×10−4, respectively, Welch’s t-test, Supplementary Figure 4b). In contrast, missense mutations in MT-CYB had significantly lower pathogenicity levels (p = 1.97×10−12, Welch’s t-test), and this observation was also marginally significant in pediatric samples (p = 0.02).
mtDNA-hypermutated samples
Fifteen to 29 tumor-only mtDNA mutations per sample were found in 5 mtDNA-hypermutated samples (Supplementary Table 18). The mtDNA mutational profile of these samples was fundamentally different from the rest of the samples not only in the total number of mutations, but also with respect to low heteroplasmy levels (average 4.4%) and a high proportion of synonymous mutations (36.5%) (Supplementary Figure 5a&5b). The high proportion of synonymous mutations in these hypermutated samples, nevertheless, supports our hypothesis that the observed skewed ratio of non-synonymous to synonymous mutations in other samples is not a simple reflection of a generic mtDNA mutation bias. Almost all of the mutations (97.1%) found in these hypermutated samples have been described in the general population. Accordingly, predicted pathogenicity values for missense and tRNA mutations were very low and no LoF mutation was found. We performed tumor-normal paired nuclear somatic variant calling in the five mtDNA-hypermutated samples along with 134 other randomly selected samples (Supplementary Table 4). We observed a strong correlation between the number of missense mutations in the nuclear genome and the number of mutations in the mitochondrial genome (Pearson correlation coefficient = 0.43, p = 1.33×10−7), suggesting potentially common mechanisms underlying the increased number of nuclear and mtDNA tumor-only mutations. Of interest, three of the 5 mtDNA-hypermutated samples were from leukemia patients, and two were from neuroblastoma patients. There was no evidence for a mutation in the common DNA mismatch repair genes in the nuclear genome of these patients, including MSH1, MSH2, MSH6, PMS1, PMS2, MLH1, BRCA2, PALB, and MLH3.
Discussion
Tumor-only mtDNA mutations occur across diverse subtypes of childhood cancer.
We have demonstrated the spectrum of the mtDNA mutations that are both shared by and unique to individual pediatric cancer subtypes. While different pediatric cancer subtypes showed different mean numbers of tumor-only mtDNA mutations, the types of mtDNA mutations seen also varied among different subtypes. For example, LGGs had the fewest number of tumor-only mtDNA mutations, lacked any LoF mutations, and any mtDNA mutations present in these tumors occurred at very low heteroplasmy levels. These data suggest that mtDNA mutations do not play a pathogenic role in the development of pediatric LGG. In contrast, a total of six LoF mutations were identified in 5 of 20 (25%) ACC samples, which is significantly higher than in any other subtype of pediatric malignancy. Thirteen percent of OS samples had LoF mtDNA mutations. 15-70% of ACCs and the vast majority of osteosarcomas have inactivating TP53 alterations [35-39]. Singh et al. previously reported that MT-ND5 somatic mutations down regulate mitochondrial electron transport chain complex I enzyme activity and increase reactive oxygen species, also epigenetically upregulate nuclear anti-apoptotic genes in p53 deficient cells [40]. Based on this finding we initially postulated that a link between TP53 mutations and complex I activity could explain why LoF mutations in ACC and OS were observed at such high frequency relative to the rest of the pediatric tumors studied. However, only 2 of the 20 ACC tumors in our dataset had a nuclear TP53 mutation. Similarly, while CPCs also harbor TP53 mutations, no LoF mutations were observed in the four CPC samples that we studied. Thus, while increased mtDNA mutation frequency occurs in ACC, it is not clear that this is influenced by co-occurrence with nuclear TP53 mutations. The high mtDNA mutation rate may be related to another functional alteration in p53 or a distinct DNA repair pathway.
Increased non-synonymous tumor-only mtDNA mutations occur in pediatric cancers.
Ju et al. and Stewart et al. both observed a skewed dN/dS ratio (indicating an increase in non-synonymous tumor-only mtDNA mutations compared to synonymous mtDNA mutations) in adult cancers [14, 15]. The authors suggested that this was due to a replication-coupled mutational process. Through Monte Carlo simulations, we demonstrated computationally that although it does clearly contribute to the skewed ratio, the biased mtDNA mutational signature does not fully explain the highly skewed dN/dS ratio observed in both pediatric and adult cancers. Sequencing and analyzing the mtDNA genome in a large number of children without cancer provided further support for this conclusion. Our analysis clearly demonstrated that non-synonymous or deleterious mtDNA mutations were under strong negative selection in normal cells, but under strong positive selection in tumor cells. Thus, we have demonstrated for the first time that the highly skewed dN/dS ratio of tumor-only mtDNA mutations is a strong indicator that mtDNA mutations have broad contributions to tumor development not only in pediatric cancers but also in adult cancers. We postulate that a significant fraction of the observed non-synonymous or deleterious mtDNA mutations provide the tumor cells with a significant selective advantage. Evidence from functional studies is required to support this hypothesis.
Clustering of loss-of-function (LoF) and pathogenic tRNA tumor-only mtDNA mutations occurs across pediatric and adult cancers.
While LoF tumor-only mtDNA mutations in a variety of cancers have previously been reported, including several recurrent mutations, we discovered clustering of LoF mtDNA mutations in MT-CO3, MT-ND4, and MT-ND5, predominantly in the Complex I (ND) subunit genes (Fig. 5a). Our analysis provides strong statistical evidence that this finding did not result from stochastic mutation drift. Rather, these hotspot mtDNA mutations were shared by diverse childhood and adult cancers as demonstrated by our meta-analysis. While the observed LoF mutations were spread across all 13 mtDNA protein-coding genes, the existence of hotspots of LoF mtDNA mutations in childhood and adult cancers indicates that defects in specific mitochondrial genes, function, and related pathways such as mitochondrial apoptosis may play a more important role than previously appreciated for tumorigenesis of childhood and adult cancers. This is supported by the high predicted pathogenicity scores of tumor-only mutations in some of the mtDNA genes.
The importance of individual LoF mtDNA mutations to tumor development, especially those occurring within complex I genes, is supported by previous studies. For example, Sharma et al. experimentally demonstrated that mitochondrial complex I dysfunction, caused by a heteroplasmic nonsense MT-ND5 mutation, promoted tumorigenesis [41]. This result is similar to that reported by Ishikawa et al., who demonstrated complex I deficiencies caused by a MT-ND6 frameshift mutation contributed to tumor progression and metastasis [42]. Recently, Gopal et al reported that LoF mutations in the complex I genes lead to rewiring of glutathione metabolism in renal oncocytoma and are early genetic events that contribute to tumor formation [43]. Notably, 13 and 8 of these 45 LoF mutations that we found were above 15% and 40% heteroplasmy, respectively, with the highest observed at 73% in a neuroblastoma tumor (Supplementary Table 1). High heteroplasmy levels support a contributing role for a mtDNA mutation in tumorigenesis, consistent with the Park et al study demonstrating that a LoF MT-ND5 mutation promoted tumorigenesis at high-level heteroplasmy but not at homoplasmy [44]. Conversely, tumor heterogeneity may lead to skewed lower heteroplasmy levels observed for other LoF mutations, as we illustrated in the single-cell ATACseq data from two acute myeloid leukemia patients. The functional significance of low heteroplasmic mtDNA mutations should therefore not be dismissed, and will need to be addressed in future studies.
We have demonstrated clustering of tumor-only mtDNA mutations in the MT-TM gene encoding the tRNA for methionine (Fig. 5b). A heteroplasmic pathogenic MT-TM mutation was first reported in a patient with splenic lymphoma [45], with deficient mitochondrial respiratory chain enzyme activities and Complex IV COX2 subunit expression. However, our pan-cancer analyses suggest that pathogenic MT-TM mutations do not occur sporadically. Of note, the 22 mitochondrial tRNA genes are also hotspots for pathogenic mutations that cause classical mitochondrial disorders [34].
mtDNA haplogroup background influences the development of specific tumor types
Mitochondrial haplogroups, as defined by common fixed (homoplasmic) mtDNA polymorphisms, have long been found to be associated with varied risks for diverse cancers [46]. For example, the mtDNA haplogroup M7B2 was found to be a risk factor for leukemia [47], while haplogroup U was found to increase the risk for both renal and prostate cancers [48]. Indeed, we observed an enrichment of certain cancer types in patients with specific mtDNA haplogroup backgrounds, for example, hyperdiploid B-ALL in haplogroup H. Patient ascertainment bias is a significant confounder in these studies; therefore larger data sets are required to allow us to formally determine the risk of cancer within haplogroups. However, we observed significantly fewer tumor-only mtDNA mutations occurring in patients belonging to the L (African) macro-haplogroup compared to the H (Eurasian macro-haplogroup). To our knowledge, this is the first report of such a finding.
Functional significance of the tumor-only mtDNA mutations
The highlights of our findings are the clustering of hotspot LoF mutations. Naturally, the next-step will be to validate their functional significance experimentally. Because of the nature of these mutations (loss-of-function), there is less uncertainty of their functional significance in terms of protein function. However, determining their potential functional significance in terms of tumorigenesis is challenging since mitochondrial metabolism plays an essential role in all aspects of tumorigenesis, namely malignant transformation, tumor progression, metastasis, and response to treatment [3]. There is significant mitochondrial heterogeneity in different cells and microenvironments which, while highlighting the tumor extraordinary metabolic plasticity during tumorigenesis, makes it extremely challenging to select a single representative or appropriate assay to assess the functional significance of the candidate mtDNA mutation. A thorough characterization of the likely functional significance of a mtDNA mutation would likely require a suite of assays targeting all aspects of tumorigenesis.
Heteroplasmy levels of the tumor-only mtDNA mutations
Determining the functional significance of tumor-only mtDNA mutations is complicated by heteroplasmy and intra-tumor heterogeneity and, in some cases, tumor percentage. A tumor-only mtDNA mutation may be observed at low VAF from next-generation sequencing of a tumor sample. In previous studies a pathogenic role for mtDNA mutations was dismissed because of their low heteroplasmy. However it is highly possible that such mtDNA mutations may be present at high heteroplasmy in some tumor cells but not others. The observed low heteroplasmy may be an artifact of pooling and sequencing DNA from heterogeneous groups of tumor (and normal) cells. Kang et al sequenced iPS cell lines derived from individual skin cells and demonstrated somatic mutations at various frequencies in different cell lines [49]. Similarly, our analysis of the single-cell ATACseq data from two acute myeloid leukemia patients also supports this hypothesis, since individual tumor cells had near-homoplasmy levels of some LoF mtDNA mutations that were observed at low heteroplasmy at bulk cell populations.
If the near homoplasmy state of LoF mutations in single cells translates to a selective disadvantage, these tumor cells are expected to be purified away, so that they would represent lower fractions of all mtDNA mutations at higher heteroplasmy levels when the bulk tumor cells are sequenced. However, our data showed that the likely functionally deleterious tumor-only mtDNA mutations, including the LoF mutations, occurred at comparable if not higher heteroplasmy levels than the likely functionally neutral tumor-only mtDNA mutations, such as the synonymous and D-loop region mutations (Supplementary Figure 6). This is true for tumor-only mtDNA mutations found in both pediatric and adult cancers. In each case, there were also a significant number of LoF mtDNA mutations at high heteroplasmy (Supplementary Figure 6a & 6b). Furthermore, although most of the tumor-only mtDNA mutations were seen at low heteroplasmy, the composition of these mutations across different heteroplasmy levels remained similar (Figure 2a &2b, Supplementary Figure 6), which argues that the deleterious mtDNA mutations were not selected against. Combined with single-cell ATACseq data, we should not dismiss the likely functional significance of a tumor-only mtDNA mutation because of low heteroplasmy.
Pediatric cancer patients have an increased incidence of germline pathogenic mtDNA variants.
The most intriguing observation from this study was our novel finding that four childhood cancer patients harbored homoplasmic or heteroplasmic mtDNA germline variants that are known to cause classical mitochondrial disorders, which have not been previously linked to a cancer phenotype. A previous epidemiological study, as well as our own experience, have not shown that patients with mitochondrial dysfunction have an increased risk of cancer [31]. These findings are intriguing, however it is not known whether these patients exhibited other mitochondrial disease-related symptoms. Park et al reported that homoplasmic MT-ND5 mutations prevents tumor development [44]. Some residual mitochondrial function may be present in cells with these homoplasmic pathogenic mtDNA variants or they would be lethal instead of mitochondrial disease-causing. The remaining mitochondrial function may be sufficient or even promote tumorigenesis, which is consistent with findings from Park et al [44]. Prospective investigation is needed to clarify the tumorigenic potential of these mutations.
It is also intriguing that there are four pediatric cancer patients with homoplasmic pathogenic variants that cause primary mitochondrial diseases, yet, it was shown by Park et al that the homoplasmic MT-ND5 mutations prevents tumor development [44]. To explain this, we speculate that there is residual mitochondrial function in cells with these homoplasmic pathogenic mtDNA variants or they would cause death or miscarriage instead of mitochondrial diseases only. The remaining mitochondrial function may be sufficient or even promote tumorigenesis, which is consistent with findings from Park et al [44].
mtDNA-hypermutation occurs in a small subset of pediatric cancers
We found five samples that were mtDNA-hypermutated, with significantly more tumor-only mtDNA mutations than other samples (15-29 vs. 0.63 ± 0.84). Although we did not find mutations in the DNA mismatch repair genes in these samples, we did observe a strong correlation between the number of somatic nuclear DNA mutations and the number of tumor-only mtDNA mutations. It is possible that the same underlying mechanism causes somatic nuclear DNA and mtDNA mutations in cancer. It was shown previously that defective mitochondrial respiratory chain activity could lead to nuclear DNA damage and hypermutagenesis [50, 51]. These data provide further support for tumor-only mtDNA mutations contributing to diverse cancer development.
In conclusion, the present study establishes the broad landscape of germline and tumor-only mtDNA genome mutations across numerous subtypes of childhood leukemias, brain tumors, and solid tumors. We utilized a highly powered public dataset, combined with additional data from pediatric controls with no history of cancer, to demonstrate that deleterious mtDNA mutations contribute significantly to the tumorigenesis of pediatric cancers. The highly skewed dN/dS ratio clearly indicates non-synonymous mtDNA mutations in general, rather than a few isolated mutations, provide tumor cells a selective advantage. Our findings provide novel insights into the nature of mtDNA heteroplasmic mutations that alter normal mitochondrial electron transport chain function, and likely make substantial contributions to tumorigenesis in diverse pediatric cancers. This is highlighted by the discoveries of hotspots for mtDNA LoF and likely pathogenic tRNA mutations shared between pediatric and adult cancers, which suggests that certain mitochondrial genes and pathways may play a bigger role in tumorigenesis than others. Future functional studies are required to determine the effect of specific mutations on mitochondrial function and their role in cancer.
Supplementary Material
Statement of Significance: This pan-cancer mtDNA study establishes the landscape of germline and tumor mtDNA mutations and identifies hotspots of tumor mtDNA mutations to pinpoint key mitochondrial functions in pediatric malignancies.
Acknowledgments
This data makes use of data generated by the St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project. We are thankful to members of the Children’s Hospital Los Angeles Center for Personalized Medicine for their help during this study and particularly, Dennis Maglinte, Ananthanarayanan Govindarajan, Moiz Bootwalla, Alex Ryutov, and Andy Garcia. This study was supported in part by the Unravel Childhood Cancer foundation (to J.A. Biegel), by a grant from the United Mitochondrial Disease Foundation (UMDF) (to X. Gai & M.J. Falk.), and by grants from the National Institutes of Health: 5T32CA009656-22T32 (K. Kaneva), U54-NS078059 (M.J. Falk), U41-HG006834 (M.J. Falk), and U24 HD093483-01 (to M.J. Falk & X. Gai). The Meijer Foundation supported N. Sohail.’s work at Van Andel Research Institute (VARI). T.J. Triche’s work on this project was supported by both the VARI and the Michelle Lunn Hope Foundation.
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
References.
- 1.Heiden MG, Cantley LC, & Thompson CB (2009). Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science, 324(5930), 1029–1033. doi: 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warburg O (1927). The Metabolism Of Tumors In The Body. The Journal of General Physiology, 8(6), 519–530. doi: 10.1085/jgp.8.6.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porporato PE, Filigheddu N, Pedro JM, Kroemer G, & Galluzzi L (2017). Mitochondrial metabolism and cancer. Cell Research, 28(3), 265–280. doi: 10.1038/cr.2017.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vyas S, Zaganjor E, & Haigis MC (2016). Mitochondria and Cancer. Cell, 166(3), 555–566. doi: 10.1016/j.cell.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Walsh MF, Wu G, Edmonson MN, Gruber TA, Easton J, … Downing JR (2015). Germline Mutations in Predisposition Genes in Pediatric Cancer. New England Journal of Medicine, 373(24), 2336–2346. doi: 10.1056/nejmoa1508054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, … Zhang J (2018). Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature, 555(7696), 371–376. doi: 10.1038/nature25795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, … Pfister SM (2018). The landscape of genomic alterations across childhood cancers. Nature, 555(7696), 321–327. doi: 10.1038/nature25480 [DOI] [PubMed] [Google Scholar]
- 8.Wallace DC, & Chalkia D (2013). Mitochondrial DNA Genetics and the Heteroplasmy Conundrum in Evolution and Disease. Cold Spring Harbor Perspectives in Biology, 5(11). doi: 10.1101/cshperspect.a021220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muraresku CC, Mccormick EM, & Falk MJ (2018). Mitochondrial Disease: Advances in Clinical Diagnosis, Management, Therapeutic Development, and Preventative Strategies. Current Genetic Medicine Reports, 6(2), 62–72. doi: 10.1007/s40142-018-0138-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandon M, Baldi P, & Wallace DC (2006). Mitochondrial mutations in cancer. Oncogene, 25(34), 4647–4662. doi: 10.1038/sj.onc.1209607 [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee A, Mambo E, & Sidransky D (2006). Mitochondrial DNA mutations in human cancer. Oncogene, 25(34), 4663–4674. doi: 10.1038/sj.onc.1209604 [DOI] [PubMed] [Google Scholar]
- 12.Lu J, Sharma LK, & Bai Y (2009). Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Research, 19(7), 802–815. doi: 10.1038/cr.2009.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace DC (2012). Mitochondria and cancer. Nature Reviews Cancer, 12(10), 685–698. doi: 10.1038/nrc3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ju YS, Alexandrov LB, Gerstung M, Martincorena I, Nik-Zainal S, Ramakrishna M, … Campbell PJ (2014). Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. ELife, 3. doi: 10.7554/elife.02935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart JB, Alaei-Mahabadi B, Sabarinathan R, Samuelsson T, Gorodkin J, Gustafsson CM, & Larsson E (2015). Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers. PLOS Genetics, 11(6). doi: 10.1371/journal.pgen.1005333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lueth M, Deimling AV, Pietsch T, Wong L, Kurtz A, Henze G, & Driever PH (2010). Medulloblastoma Harbor Somatic Mitochondrial DNA Mutations in the D-loop Region. Journal of Pediatric Hematology/Oncology, 32(2), 156–159. doi: 10.1097/mph.0b013e3181c97c3f [DOI] [PubMed] [Google Scholar]
- 17.Luna B, Bhatia S, Yoo C, Felty Q, Sandberg DI, Duchowny M, … Roy D (2014). Proteomic and Mitochondrial Genomic Analyses of Pediatric Brain Tumors. Molecular Neurobiology, 52(3), 1341–1363. doi: 10.1007/s12035-014-8930-3 [DOI] [PubMed] [Google Scholar]
- 18.Kang M, Kim Y, Lee JH, Szardenings M, Baek H, Kook H, … Shin M (2016). Clinicopathological Implications of Mitochondrial Genome Alterations in Pediatric Acute Myeloid Leukemia. Annals of Laboratory Medicine, 36(2), 101. doi: 10.3343/alm.2016.36.2.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riehl LM, Schulte JH, Mulaw MA, Dahlhaus M, Fischer M, Schramm A, … Beltinger C (2015). The mitochondrial genetic landscape in neuroblastoma from tumor initiation to relapse. Oncotarget, 7(6). doi: 10.18632/oncotarget.6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koboldt DC, Zhang Q, Larson DE, Shen D, Mclellan MD, Lin L, … Wilson RK (2012). VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Research, 22(3), 568–576. doi: 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryutov A, Bootwalla M, Govindarajan A, Buckley J, Maglinte D, Ji J, Gai X (2017). LUBA: A software toolbox for efficiently manipulating and analyzing NGS data. The American Society of Human Genetics Annual Conference. [Google Scholar]
- 22.Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, … Wallace DC (2013). MtDNA Variation and Analysis Using Mitomap and Mitomaster. Current Protocols in Bioinformatics. doi: 10.1002/0471250953.bi0123s44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pejaver V, Urresti J, Lugo-Martinez J, Pagel KA, Lin GN, Nam H, … Radivojac P (2017). MutPred2: Inferring the molecular and phenotypic impact of amino acid variants. bioRxiv 134981; doi: 10.1101/134981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonney S, Leipzig J, Lott MT, Zhang S, Procaccio V, Wallace DC, & Sondheimer N (2017). Predicting the pathogenicity of novel variants in mitochondrial tRNA with MitoTIP. PLOS Computational Biology, 13(12). doi: 10.1371/journal.pcbi.1005867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, …Chang HY (2016). Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu TD, & Nacu S (2010). Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics,26(7), 873–881. doi: 10.1093/bioinformatics/btq057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffin HR, Pyle A, Blakely EL, Alston CL, Duff J, Hudson G, … Chinnery PF (2014). Accurate mitochondrial DNA sequencing using off-target reads provides a single test to identify pathogenic point mutations. Genetics in Medicine,16(12), 962–971. doi: 10.1038/gim.2014.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falk MJ, Pierce EA, Consugar M, Xie MH, Guadalupe M, Hardy O, … Gai X (2012). Mitochondrial disease genetic diagnostics: optimized whole-exome analysis for all MitoCarta nuclear genes and the mitochondrial genome. Discovery Medicine, 14, 389–399. [PMC free article] [PubMed] [Google Scholar]
- 29.Montefiori L, Hernandez L, Zhang Z, Gilad Y, Ober C, Crawford G, … Sakabe NJ (2017). Reducing mitochondrial reads in ATAC-seq using CRISPR/Cas9. Scientific Reports,7(1). doi: 10.1038/s41598-017-02547-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorman GS, Schaefer AM, Ng Y, Gomez N, Blakely EL, Alston CL, … Mcfarland R (2015). Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Annals of Neurology, 77(5), 753–759. doi: 10.1002/ana.24362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lund M, Melbye M, Diaz LJ, Duno M, Wohlfahrt J, & Vissing J (2015). Mitochondrial dysfunction and risk of cancer. British Journal of Cancer, 112(6), 1134–1140. doi: 10.1038/bjc.2015.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarro-Gomez D, Leipzig J, Shen L, Lott M, Stassen AP, Wallace DC, … Gai X (2014). Phy-Mer: A novel alignment-free and reference-independent mitochondrial haplogroup classifier. Bioinformatics, 31(8), 1310–1312. doi: 10.1093/bioinformatics/btu825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madsen CS, Ghivizzani SC, & Hauswirth WW (1993). In vivo and in vitro evidence for slipped mispairing in mammalian mitochondria. Proceedings of the National Academy of Sciences, 90(16), 7671–7675. doi: 10.1073/pnas.90.16.7671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yarham JW, Elson JL, Blakely EL, Mcfarland R, & Taylor RW (2010). Mitochondrial tRNA mutations and disease. Wiley Interdisciplinary Reviews: RNA, 1(2), 304–324. doi: 10.1002/wrna.27 [DOI] [PubMed] [Google Scholar]
- 35.Barzon L, Chilosi M, Fallo F, Martignoni G, Montagna L, Palu G, & Boscaro M (2001). Molecular analysis of CDKN1C and TP53 in sporadic adrenal tumors. European Journal of Endocrinology, 145(2), 207–212. doi: 10.1530/eje.0.1450207 [DOI] [PubMed] [Google Scholar]
- 36.Amadou A, Achatz MI, & Hainaut P (2018). Revisiting tumor patterns and penetrance in germline TP53 mutation carriers. Current Opinion in Oncology, 30(1), 23–29. doi: 10.1097/cco.0000000000000423 [DOI] [PubMed] [Google Scholar]
- 37.Ragazzon B, Libe R, Gaujoux S, Assie G, Fratticci A, Launay P, … Bertherat J (2010). Transcriptome Analysis Reveals that p53 and -Catenin Alterations Occur in a Group of Aggressive Adrenocortical Cancers. Cancer Research, 70(21), 8276–8281. doi: 10.1158/0008-5472.can-10-2014 [DOI] [PubMed] [Google Scholar]
- 38.De MM, Al GA, Do CC, Assie G, Scoazec J, Leboulleux S, … Baudin E (2013). Molecular screening for personalized treatment approach in advanced adrenocortical cancer. Endocrine Abstracts. doi: 10.1530/endoabs.32.p11 [DOI] [PubMed] [Google Scholar]
- 39.Szyszka P, Grossman AB, Diaz-Cano S, Sworczak K, & Dworakowska D (2016). Molecular pathways of human adrenocortical carcinoma - translating cell signalling knowledge into diagnostic and treatment options. Endokrynologia Polska, 67, 427–450. Doi: 10.5603/EP.a2016.0054 [DOI] [PubMed] [Google Scholar]
- 40.Singh RK, Saini S, Verma D, Kalaiarasan P, & Bamezai RN (2017). Mitochondrial ND5 mutation mediated elevated ROS regulates apoptotic pathway epigenetically in a P53 dependent manner for generating pro-cancerous phenotypes. Mitochondrion, 35, 35–43. [DOI] [PubMed] [Google Scholar]
- 41.Sharma LK, Fang H, Liu J, Vartak R, Deng J, & Bai Y (2011). Mitochondrial respiratory complex I dysfunction promotes tumorigenesis through ROS alteration and AKT activation. Human Molecular Genetics, 20(23), 4605–4616. doi: 10.1093/hmg/ddr395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, … Hayashi J (2008). ROS-Generating Mitochondrial DNA Mutations Can Regulate Tumor Cell Metastasis. Science, 320(5876), 661–664. doi: 10.1126/science.1156906 [DOI] [PubMed] [Google Scholar]
- 43.Gopal RK, Calvo SE, Shih AR, Chaves FL, Mcguone D, Mick E, … Mootha VK (2018). Early loss of mitochondrial complex I and rewiring of glutathione metabolism in renal oncocytoma. Proceedings of the National Academy of Sciences, 201711888. doi: 10.1073/pnas.1711888115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park JS, Sharma LK, Li H, Xiang R, Holstein D, Wu J, … Bai Y (2009). A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Human Molecular Genetics, 18(9), 1578–1589. doi: 10.1093/hmg/ddp069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lombès A, Bories D, Girodon E, Fraction P, Ngo M, Breton-Gorius J, … Goossens M (1998). The first pathogenic mitochondrial methionine tRNA point mutation is discovered in splenic lymphoma. Human Mutation, 11(S1). doi: 10.1002/humu.1380110158 [DOI] [PubMed] [Google Scholar]
- 46.Singh KK, & Kulawiec M (2009). Mitochondrial DNA Polymorphism and Risk of Cancer. Methods in Molecular Biology Cancer Epidemiology, 291–303. doi: 10.1007/978-1-59745-416-2_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verma M, Naviaux RK, Tanaka M, Kumar D, Franceschi C, & Singh KK (2007). Meeting Report: Mitochondrial DNA and Cancer Epidemiology. Cancer Research, 67(2), 437–439. doi: 10.1158/0008-5472.can-06-4119 [DOI] [PubMed] [Google Scholar]
- 48.Canter JA, Kallianpur AR, & Fowke JH (2006). Re: North American White Mitochondrial Haplogroups in Prostate and Renal Cancer. The Journal of Urology, 176(5), 2308–2309. doi: 10.1016/j.juro.2006.07.067 [DOI] [PubMed] [Google Scholar]
- 49.Kang E, Wang X, Tippner-Hedges R, Ma H, Folmes C, Gutierrez N, … Mitalipov S (2016). Age-Related Accumulation of Somatic Mitochondrial DNA Mutations in Adult-Derived Human iPSCs. Cell Stem Cell, 18(5), 625–636. doi: 10.1016/j.stem.2016.02.005 [DOI] [PubMed] [Google Scholar]
- 50.Singh KK, Kulawiec M, Still I, Desouki MM, Geradts J, & Matsui S (2005). Inter-genomic cross talk between mitochondria and the nucleus plays an important role in tumorigenesis. Gene, 354, 140–146. doi: 10.1016/j.gene.2005.03.027 [DOI] [PubMed] [Google Scholar]
- 51.Singh K (2004). Mitochondrial damage checkpoint in apoptosis and genome stability. FEMS Yeast Research. doi: 10.1016/s1567-1356(04)00056-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.