Abstract
To examine the association between metabolic deregulation and pancreatic cancer, we conducted a two-stage case-control targeted metabolomics study using pre-diagnostic sera collected one year before diagnosis in the Women’s Health Initiative study. We used the liquid chromatography–mass spectrometry to quantitate 470 metabolites in 30 matched case/control pairs. From 180 detectable metabolites, we selected 14 metabolites to be validated in additional 18 matched case/control pairs. We used the paired t-test to compare the concentrations of each metabolite between cases and controls and used the log fold change (FC) to indicate the magnitude of difference. False discovery rate (FDR) adjusted q-value < 0.25 was indicated statistically significant. Logistic regression model and receiver operating characteristic (ROC) curve analysis were used to evaluate the clinical utility of the metabolites. Among 30 case/control pairs, 1-methyl-L-tryptophan (L-1MT) was significantly lower in the cases than in the controls (log2 FC= − 0.35; q-value = 0.03). The Area under the ROC curve was 0.83 in the discrimination analysis based on the levels of L-1MT, acadesine, and aspartic acid. None of the metabolites was validated in additional independent 18 case/control pairs. No significant association was found between the examined metabolites and undiagnosed pancreatic cancer.
Keywords: biomarker, pancreatic cancer, early diagnosis, tryptophan, metabolomics
INTRODUCTION
Pancreatic cancer is currently the 3rd leading cause of cancer-related deaths in the United States and is projected to become the second most deadly cancer by 2030 (1, 2). More than 80% of patients with pancreatic cancer present with locally advanced or metastatic disease at diagnosis when a surgical resection is no longer a curative option. Consequently, pancreatic cancer has the lowest five-year all-cause survival rate among all malignancies (2). Early diagnosis of pancreatic cancer holds the promise for improving the prognosis of this disease. However, carbohydrate antigen 19-9 (CA19-9), the only US-FDA approved blood-based biomarker, is a poor screening tool because the antigen is non-specific (3). The sensitivity and specificity of the imaging modalities are also inappropriate to detect a tumor less than 2 cm (4). With the alarming increasing trend of pancreatic cancer incidence (1), there is an urgent need for non-invasive early diagnostic markers for pancreatic cancer.
Metabolomics is a powerful non-invasive technology capable of detecting low-molecular-weight metabolites in cells, tissues, and biofluids in combination with advanced bioinformatics approaches. Cancer (5), including pancreatic cancer (6), is known to have altered cellular metabolism (7). The subtle chemical change due to perturbed metabolism in cancer development can be detected by the sensitive gas chromatography or liquid chromatography-mass spectrometry (GC-MS or LC-MS) or nuclear magnetic resonance (NMR) (8). Prior studies have examined the metabolites using pancreatic tumor and normal adjacent tissues (9), urine (10, 11), serum (12–17), and plasma (18, 19) using MS or NMR. These studies concluded that the metabolites involved in the metabolism of the lipid, glucose, amino acid, choline, DNA synthesis, small organic acids, or muscle protein breakdown can discriminate pancreatic cancer from healthy controls and chronic pancreatitis (20, 21). However, whether the altered metabolite profile may indicate a subclinical early-stage disease has not been well investigated.
Several studies have used the metabolomics tool in pancreatic cancer early detection (22). One study found that the metabolite profiles are distinct among 50 pancreatic cancer patients, 50 diabetic patients, and 50 healthy controls (23). In another study of 30 pancreatic cancer patients and 30 patients with new-onset diabetes, N-succinyl-L-diaminopimelic acid and PE (18:2) were shown to have a high sensitivity (93.3%) and specificity (93.1%) in discriminating pancreatic cancer (24). The other study found that the metabolites in combination with CA19-9 can improve early detection rate of pancreatic cancer (25). However, most of the prior studies used blood collected after a pancreatic cancer diagnosis. It is unknown whether the observed metabolite changes were due to treatment or other post-diagnostic manipulations.
In this pilot study, we evaluated the metabolite change in pre-diagnostic sera of women who developed pancreatic cancer in the Women’s Health Initiative (WHI) Study. We hypothesized that a targeted metabolomics discovery platform would yield a panel of metabolites that can discriminate undiagnosed pancreatic cancers from non-cancer controls.
MATERIAL AND METHODS
Study population
We conducted a nested case-control study within the WHI Study. Details of the WHI Study (NCT00000611) design were previously published (26). Briefly, 161,808 postmenopausal women aged 50-79 years were recruited from 40 clinical centers throughout the United States between September 1993 and December 1998. A total of 68,132 women in the WHI clinical trials (CT) were randomized to three overlapping components: a hormone therapy (HT) trial, a dietary modification (DM) trial, and a calcium and vitamin D supplementation (CaD) trial. A total of 93,676 women were included in an Observational Study (WHI-OS). At baseline, blood samples were obtained following an overnight fast and stored at −70°C. We used serum sample that has never gone through a freeze-thaw cycle.
Study design
We performed a targeted profiling of 470 metabolites using the LC-MS in a two-stage nested case-control study. Both cases and controls had no cancer history at baseline and controls did not develop any cancer during follow-up. We identified 55 cases who were diagnosed during the first year of follow-up and 197,000 eligible controls. The controls were individually matched to the cases in a 1:1 ratio according to age (± 1 year), race/ethnicity, month of blood draw (± 12), time of blood draw (± 3 hours), season of blood draw (identical), diabetes status (yes or no), trial assignment, and body mass index (BMI) (± 5 kg/m2). Seven cases were not included in the study because no adequate controls could be identified. We, therefore, included 48 adjudicated cases with pancreatic cancer from OS and the placebo group of HT and CaD trials and 48 matched controls in this study. The matched pairs were randomly assigned to the discovery set (n = 30 pairs) or the validation set (n = 18 pairs).
The present research was approved by the WHI and the combined Institutional Review Board of Baylor College of Medicine (BCM) and Michael E. DeBakey VA Medical Center. The research was conducted in accordance with the US. Common Rule. At baseline, all participants in the WHI study provided written informed consent for research.
Targeted metabolomics research
The Metabolomics core lab at BCM has established a validated pipeline for a targeted measurement of 433 metabolites that are involved in the metabolism of lipid, carbohydrate, nucleotide, amino acid, energy, CoA & vitamin, secondary compounds, xenobiotics, tricarboxylic acid, and urea cycles (27). In addition, we included 37 metabolites that had been associated with pancreatic cancer in previous studies (e.g., taurocholic acid, xylitol, and trimethylamine-N-Oxide) (13). Therefore, we included 470 metabolites in this study (Supplemental material 1).
Reagents
High-performance liquid chromatography (HPLC) grade acetonitrile, methanol, and water were purchased from Burdick & Jackson (Morristown, NJ). The mass spectrometry grade formic acid and internal standards, including N-acetyl L-aspartic acid-d3, L-tryptophan-15N2, sarcosine-d3, glutamic acid-d5, thymine-d4, gibberellic acid, trans-zeatin, jasmonic acid, 15N anthranilic acid, and testosterone-d3, were purchased from Sigma-Aldrich (St.Louis, MO). The ESI-L Low Concentration Tuning Mix (Agilent Technologies, Santa Clara, CA) was used to calibrate the mass spectrometer.
Serum preparation
Sample preparation was performed on ice. Enrichment of metabolites was achieved using a series of organic and aqueous extractions. Briefly, 50 μl of serum was sonicated in 1:4 ice cold water:methanol mixture containing an equimolar mixture of 10 standard compounds. This was followed by the sequential addition of ice-cold chloroform and water in 3:1 ratio and the separation of the organic (methanol and chloroform) and aqueous solvents (water:methanol:chloroform:water ratio 1:4:3:1). The aqueous extract was deproteinized using a 3 kDa molecular filter (Amicon Ultracel −3K Membrane, Millipore Corporation, Billerica, MA) and the filtrate containing metabolites was dried under vacuum (Genevac EZ-2plus, Gardiner, NY). The dried extract was resuspended in the identical volume of injection solvent composed of water:methanol (50:50).
Metabolomics experiment
A total of 10 μl of suspended samples was injected and analyzed using a 6495-triple quadrupole mass spectrometer coupled to a 1290 system (Agilent Technologies, Santa Clara, CA) via multiple reaction monitoring (MRM) of a total of 470 endogenous water-soluble metabolites. Source parameters were as follows: gas temperature was 250°C; gas flow was 14 L/min; Nebulizer pressure was 20 psi (Pounds per square inch); sheath gas temperature was 350°C; sheath gas flow was 12 L/min; capillary was 3000 V positive and 3000 V negative; Nozzle voltage was 1500 V positive and 1500 V negative. Approximately 8–11 data points were acquired per detected metabolite.
We used three chromatography methods to separate all the metabolites: Method 1: XBridge Amide columns (Waters, Milford, MA); mobile phase, A: 0.1% formic acid in HPLC grade water, B: 0.1% formic acid in HPLC grade acetonitrile. ESI positive mode. Method 2: Luna 3μ NH2 column 100A (Phenomenex, Torrance, CA); mobile phase, A: 20 mM ammonium acetate in HPLC grade water pH 9, B: HPLC grade acetonitrile. ESI positive mode. Method 3: Luna 3μ NH2 column 100A (Phenomenex); mobile phase, A: 20 mM ammonium acetate in HPLC grade water pH 9, B: HPLC grade acetonitrile. ESI negative mode. The gradients were run starting from 85% buffer B (HPLC grade acetonitrile or 0.1% formic acid in acetonitrile) to 35% buffer B for 0–3.5 minutes; 35% buffer B to 2% buffer B for 3.5–11.5 minutes; 2% buffer B was held for 11.5–16.5 minutes; 2% buffer B to 85% buffer B for 16.5–17.5 minutes; 85% buffer B was held for 7 minutes to re-equilibrate the column. The peak area for each metabolite was integrated using the MassHunter Workstation Software Quantitative Analysis Version B.06.00 (Agilent Technologies, Santa Clara, CA).
Quality control measures
The samples were run in random order and in triplicate. The matched case and control pairs were assayed consecutively in the same batch. The laboratory personnel performed blind sample testing. To check instrument response and exaction efficiency, two types of experiment controls were used. First, 20 μl of a matrix-free internal standard was reconstituted in 100 μl of methanol: water (50:50) and was analyzed by MRM. In addition, the process of metabolite extraction from serum was monitored using the pooled liver samples and spiked internal standards. Briefly, 100 mg of pooled liver lysate was extracted in tandem with serum using the buffer containing spiked internal standards. Once the metabolite of interest was identified, it was subjected to additional confirmation using selected reaction monitoring (SRM). The matrix-free internal standards and liver samples were analyzed twice daily. We included 3% blinded WHI samples in the study as the technical control.
Metabolomics data analysis
The metabolites were identified from the preprocessed mass spectral data using the METLIN (Agilent Technologies, Santa Clara, CA) according to both mass and retention time. They were further processed via an in-house biostatistics pipeline to determine the metabolite levels and define differential compounds (28). The log2 transformed data were normalized using the internal standards (L-zeatin for water positive and luna positive; L-zeatin and gibberellic acid for luna negative). The log2 transformed data were centered by the median and scaled by its inter-quartile range. Following the normalization, the paired t-test was used to detect the significantly differentially present metabolites between cases and controls. In the discovery stage, we used the less stringent unadjusted P value < 0.10 in combination with the area under the receiver operating characteristics (ROC) curve (AUC) to identify the candidate metabolites to be validated. In the validation stage, the false discovery rate (FDR) corrected q value < 0.25 was considered statistically significant19.
The normalized data was further scaled for generation of the heatmap and boxplots for each comparison. We used the unsupervised principal component analysis (PCA) multivariate pattern recognition technique to discriminate cases from controls where each axis value explained the percentage of total variance contributed by each principal component.
Statistical construction of a diagnostic model
We used the unconditional logistic regression model to examine the diagnostic value of the metabolites identified from the discovery stage. The sensitivity and specificity of each and combined metabolite were evaluated using the ROC curve analysis. Age, ethnicity, smoking status, and BMI were included in the multivariable logistic regression model. The statistical analyses were conducted using R for Windows and SAS 9.4 (SAS Inc., Cary, NC). All the tests were two-sided.
Results
Study participants
Table 1 presents the baseline characteristics of the participants. Participants in the validation stage were more likely to be non-Hispanic white and never smokers, and have a lower BMI. A total of 42% cases had early or regionally localized cancers. The blinded quality control samples showed excellent lab performance with respect to the batch effect (Supplemental figure 1).
Table 1.
Baseline characteristics of cases and controls by study stages
| Characteristics | Discovery stage | Validation stage | ||
|---|---|---|---|---|
| Cases n=30 | Controls n=30 | Cases n=18 | Controls n=18 | |
| Age (year) | 68.9 (6.6) | 68.9 (6.7) | 68.5 (7.3) | 68.1 (7.4) |
| Race, n (%) | ||||
| Non-Hispanic White | 23 (76.7) | 23 (76.7) | 17 (94.4) | 17 (94.4) |
| Black or African-American | 5 (16.7) | 5 (16.7) | 1 (5.6) | 1 (5.6) |
| Hispanic/Latino | 1 (3.3) | 1 (3.3) | 0 | 0 |
| Other | 1 (3.3) | 1 (3.3) | 0 | 0 |
| BMI (kg/m2) | 28.9 (5.6) | 28.3 (5.6) | 25.7 (4.1) | 25.8 (3.9) |
| Smoking status, n (%) | ||||
| Current | 1 (3.3) | 3 (10.0) | 3 (16.7) | 0 |
| Never | 13 (43.4) | 10 (33.3) | 9 (50.0) | 11 (61.1) |
| Past | 15 (50.0) | 16 (53.4) | 6 (33.3) | 6 (33.3) |
| Missing | 1 (3.3) | 1 (3.3) | 0 | 1 (5.6) |
| Cancer stage, n (%) | ||||
| Distant | 9 (30.0) | 0 | 7 (38.9) | 0 |
| Localized | 2 (6.7) | 0 | 0 | 0 |
| Regional | 9 (30.0) | 0 | 9 (50.0) | 0 |
| Missing | 10 (33.3) | 0 | 2 (11.1) | 0 |
| Medical history (yes, n (%)) | ||||
| Hypertension | 14(46.7) | 13 (44.8) | 6 (33.3) | 3 (16.7) |
| Type 2 diabetes | 1 (3.3) | 1 (3.3) | 2 (11.1) | 2 (11.1) |
| Hormone therapy use | 14 (46.7) | 15 (50.0) | 4 (22.2) | 3 (16.7) |
| Ever used oral contraceptives | 10 (62.5) | 6 (20.0) | 3 (16.7) | 6 (33.3) |
Targeted metabolomics study – the discovery stage
Among 470 metabolites, the signal intensity of 290 metabolites was below the limit of detection (LOD) or had a low signal to noise ratio (< 10). The list of 180 metabolites that were successfully quantified in the discovery stage is shown in Supplemental table 1. In the discovery stage, 19 metabolites were differentially abundant in cases and in controls (P < 0.10) (Table 2). The difference in 1-methyl-L-tryptophan (L-1 MT, Aldrich: 447439), glycerol-3-phosphate, and acadesine remained statistically significant after adjustment for multiple comparisons (FDR q < 0.25) (Figure 1).
Table 2.
Differentially presented metabolites in cases and controls in the discovery and validation stage
| Metabolites | Discovery stage (30 ca/co pairs) | Validation stage (18 ca/co pairs) | ||||
|---|---|---|---|---|---|---|
| Downregulated in cases | log FD | P valuea | q valuea | log FD | P valuea | q valuea |
| Indole | −0.70 | 0.09 | 0.93 | |||
| Ornithine | −0.36 | 0.07 | 0.93 | −0.05 | 0.53 | 0.69 |
| Ethylmalonate | −0.35 | 0.10 | 0.93 | −0.29 | 0.55 | 0.69 |
| 1-Methyl-tryptophan | −0.35 | 0.0004 | 0.03 | −0.02 | 0.78 | 0.90 |
| Glu-Glu | −0.32 | 0.07 | 0.93 | −0.03 | 0.74 | 0.74 |
| Glycerol-3-phosphate | −0.30 | 0.07 | 0.13 | −0.17 | 0.34 | 0.83 |
| Glycerophosphocholine | −0.30 | 0.01 | 0.75 | −0.07 | 0.38 | 0.69 |
| Citrulline | −0.25 | 0.05 | 0.77 | −0.16 | 0.31 | 0.58 |
| Ketoglutarate | −0.23 | 0.01 | 0.32 | −0.14 | 0.37 | 0.83 |
| Fucose | −0.56 | 0.03 | 0.39 | 0.23 | 0.45 | 0.83 |
| Upregulated in cases | ||||||
| Nicotinamide | 0.44 | 0.05 | 0.78 | 0.19 | 0.44 | 0.74 |
| Hypoxanthine | 0.41 | 0.03 | 0.77 | |||
| Acadesine | 0.38 | 0.003 | 0.14 | 0.13 | 0.56 | 0.75 |
| S-ribosyl-L-homocystein | 0.36 | 0.09 | 0.88 | |||
| Aspartic acid | 0.36 | 0.10 | 0.93 | 0.41 | 0.31 | 0.69 |
| Trehalose-6-phosphate | 0.34 | 0.06 | 0.40 | −0.01 | 0.97 | 0.97 |
| Myristoleic acid | 0.33 | 0.05 | 0.78 | |||
| TrimethyyamineN-oxide | 0.32 | 0.26 | 0.54 | 0.42 | 0.04 | 0.25 |
| S-methyl-5-thioadenosine | 0.25 | 0.09 | 0.93 | |||
Ca/co: case/control; FD: fold change.
P or q values for paired t test
Figure 1.
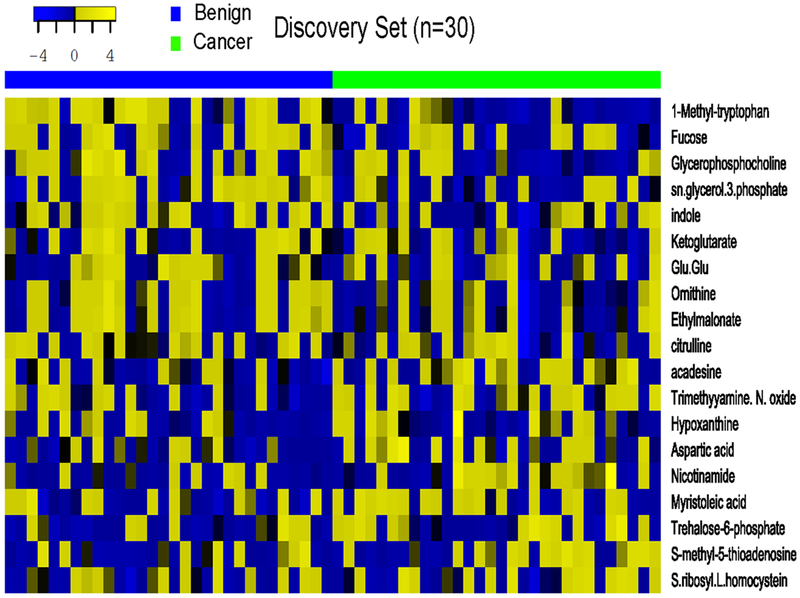
Heatmap of 19 metabolites in cases and controls in the discovery stage.
Targeted metabolomics study - the validation stage
Twelve out of 19 metabolites were further validated in 18 case/control pairs (Table 2, Figure 2). We included trimethylamine N-oxide (TMAO) in the validation stage because this gut microbiome derived metabolite has been shown to play a key role in chronic inflammatory diseases (29). We included aspartic acid in the validation stage because of the high AUC for this compound (AUC > 50%). However, after the FDR adjustment for multiple comparisons, none of the metabolites was significantly differentially present in cases versus controls. The direction of change for fucose and trehalose-6-phosphate was opposite in two stages. The PCA did not detect a distinct pattern between cases and controls (Supplemental figures 2 and 3).
Figure 2.
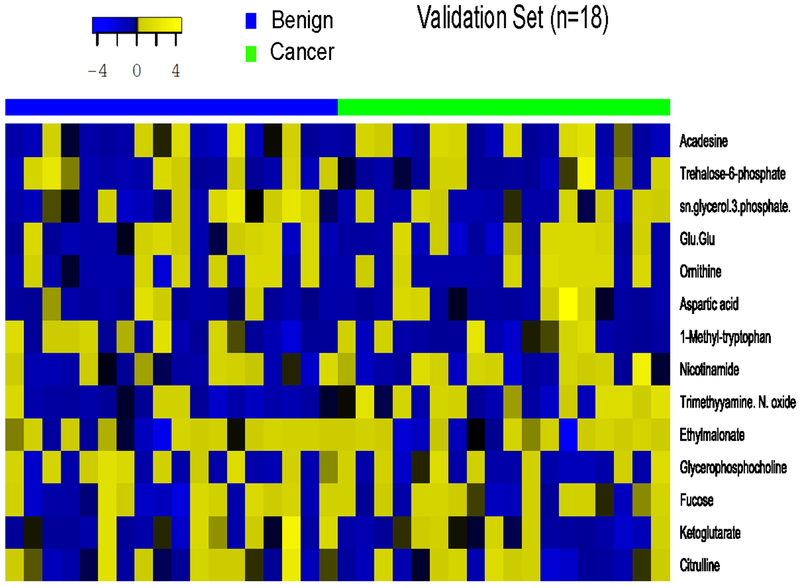
Heatmap of 14 metabolites in cases and controls in the validation stage.
Diagnostic performance
In the discovery stage, the metabolites nicotinamide, acadesine, aspartic acid, L-1MT, and ketoglutarate were significantly associated with pancreatic cancer in a univariate unconditional logistic regression model (P value < 0.05). When including these metabolites and age, ethnicity, smoking status and BMI in the multivariable models, only L-1MT remained statistically significant (P = 0.004). The best diagnostic model included acadesine, aspartic acid, and L-1MT (area under the ROC curve = 0.83) (Figure 3). The validation stage did not show significant differences in metabolites between cases and controls.
Figure 3.
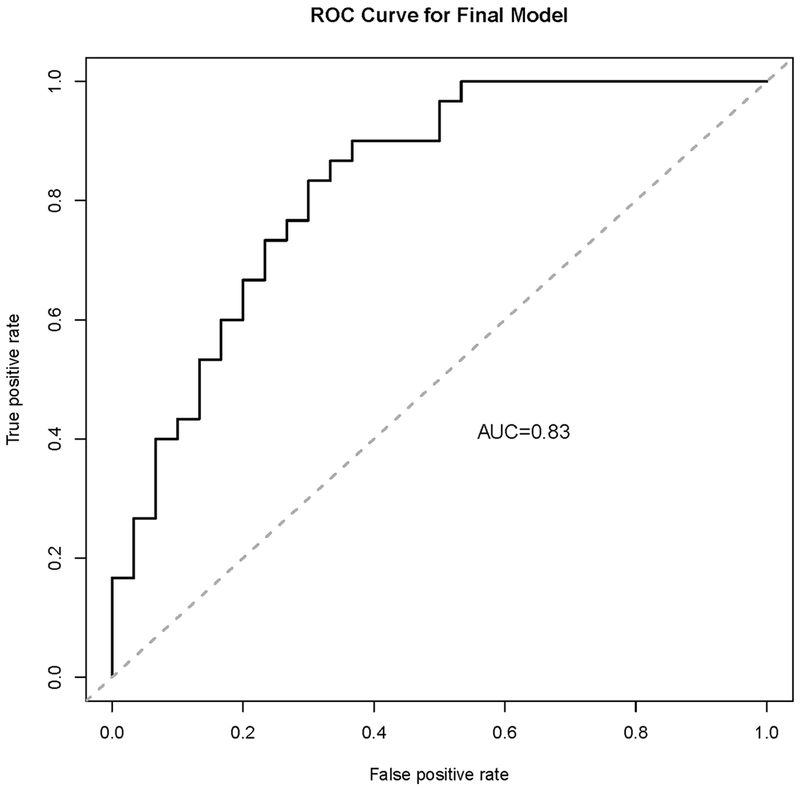
ROC curve of differentiation of cases and controls using acadesine, aspartic acid and L-1MT. The AUC was 0.83.
DISCUSSION
In this two-stage targeted metabolomics case-control study in the WHI Study, the abundance of metabolite did not differ significantly in cases with pancreatic cancer and in controls. Although we found the depletion of L-1MT in near-diagnostic blood of 30 pancreatic cancer cases in the discovery study, this finding was not validated in an independent study. Larger studies are required to further evaluate the value of metabolomics biomarkers in patient stratification or early detection of pancreatic cancer.
Several studies have investigated amino acid metabolites in pancreatic cancer (14, 30, 31). Like our study, one cross-sectional study of 360 patients and 8372 controls found that patients had significantly lower plasma levels of tryptophan and histidine but higher levels of serine than controls (31). However, another study of 32 cases and 32 controls showed the higher levels of urinary tryptophan and threonine in cases than in controls (10). Tryptophan metabolism has been increasingly recognized as a microenvironmental factor that suppresses antitumor responses in pancreatic cancer (32). The degradation of L-tryptophan to kynurenine in tumor cells induced by indoleamine-(2,3)-dioxygenase (IDO) is proposed as a mechanism of local immune suppression leading to tumor immune escape (33). A few metabolomics studies support the dysregulation of tryptophan metabolism in pancreatic cancer. One study found a panel of six metabolites including 5-hydroxytryptophan can distinguish benign pancreatic diseases from early-stage cancer with high accuracy (34). Another study showed that a metabolite panel that includes decreased indole-derivatives significantly increased the discriminating capacity of plasma protein biomarkers (35). In the present study, we found that the serum levels of indole showed the largest reduction in cases in comparison to controls. Indole is produced from dietary tryptophan by bacteria that express tryptophanase (36). It remains to be determined whether impaired tryptophan metabolism by both host and bacteria can serve as a surrogate marker for immunosuppression in patient stratification.
The diagnostic value of metabolites has been evaluated using post-diagnostic blood (22, 30, 37, 38). Three studies showed > 0.85 AUC when the metabolites were used in combination with CA19-9 (31, 39, 40). The diagnostic performance of any single metabolite in our study was encouraging with the sensitivity ranged from 63% to 80%. The highest AUC was achieved with the combination of L-1MT, acadesine, and aspartic acid in the discovery study. Future study is needed to test the metabolite changes in longitudinally collected samples yearly before cancer diagnosis to identify early detection markers.
A recent cell-line study using metabolite profiling in concert with gene expression identified three distinct metabolic subtypes of pancreatic cancer, including reduced proliferative capacity, glycolysis, and lipogenic (41). However, the clinical studies on circulating metabolite biomarkers have less agreement. For example, we found non-significantly higher levels of TMAO in cases than in controls, whereas an earlier study found the opposite (15). Our study also could not validate the markers that were identified by prior studies, including glutamate, choline, 1,5-anhydro-d-glucitol, betaine, and methyl-guanidine (13, 38). One meta-analysis research tested 18 plasma metabolites (3-hydroxybutyrate, alanine, asparagine, choline, glutamate, histidine, isoleucine, lactate, leucine, lysine, lysolPC (18:2), methionine, palmitic acid, threonine, tyrosine, valine, and phenylalanine) in 59 pancreatic cancer patients, 48 normal controls, 66 patients with colorectal cancer, and 19 with type 2 diabetes. The downregulation of alanine, threonine, and tyrosine in pancreatic cancer was consistently shown in all published studies (22). These three amino acids were non-significantly downregulated in cases in our research.
Three prospective studies have conducted the metabolomics research using pre-diagnostic blood. One study of 234 pancreatic cancer cases in China found that glycerophospholipids were dysregulated in pancreatic cancer (42). In the same line, although not validated, we found that the levels of glycerol-3-phosphate were significantly lower in cases than in controls (FDR q < 0.25). The other two studies, one in Japanese (43) and one in European descendants (44), showed that the levels of branched-chain amino acid including isoleucine, leucine, and valine were significantly higher in cases than in controls. However, we did not observe this association.
Our study had multiple uniqueness and strengths. Unlike most previous studies, we used near-diagnostic fasting blood and the cases and controls were strictly individually matched by factors that may confound the observations, such as BMI and diabetes. These stringent criteria minimized the non-specific inter-individual variation of metabolites and maximized the signal specific to pancreatic cancer. In addition to our rigorous study design, the blinded QC samples showed a highly reproducible performance across different batches. We used the targeted approach that allowed for an absolute quantification. However, it is likely that untested metabolites could have been missed. The non-targeted approach should be complemented with the targeted approach for a comprehensive discovery of biomarker for early detection.
Our study was a pilot study by nature. None of the metabolites was validated in the small validation study. Although the study participants were randomly assigned to the discovery or the validation stage, we found the distribution of ethnicity and BMI differed by the study stage. This imbalance also likely contributed to an unsuccessful validation. Only 180 out of 470 targeted metabolites were reportedly detected in this project. For some of the low-abundant compounds, the signal was below LOD probably due to the matrix effect, sample preparation method, or small sample volume (45). The normal phase chromatography columns used in this study can detect most metabolites, but we might have missed some non-polar metabolites if the experimental condition was not optimized. Some compounds with the large fold change were not promoted to validation, such as indole and hypoxanthine. Lastly, we did not quantitate CA19-9 as a gold standard for differentiating pancreatic cancer cases from controls.
In summary, we found no significant association between examined metabolites and undiagnosed pancreatic cancer. Additional studies are required to provide more insight into the translational value of metabolomics in managing pancreatic cancer in combination with other omics data and CA19-9 (46). Standardized and optimized protocol for sample collection, processing, measurement, and bioinformatics should be developed and incorporated in future research.
Supplementary Material
Acknowledgments
We acknowledge the dedicated effort of the investigators and staff at the Women’s Health Initiative (WHI) clinical centers, the WHI Clinical Coordinating Center, and the National Heart, Lung and Blood program office (listing available at http://www.whi.org). We also recognize the WHI participants for their commitment to the WHI program. For a list of all the investigators who have contributed to WHI science, please visit: http://www.whiscience.org/publications/WHI_investigators_longlist.pdf.
Financial support
This work and the Women’s Health Initiative program are funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, US Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221 as a pilot grant (L.J). Gillson Longenbaugh foundation (L.J), Golfers Against Cancer Organization (L.J). The Center for Innovations in Quality, Effectiveness and Safety, Michael E. DeBakey Veterans Affairs Medical Center (CIN13-413) also fund the research. The work was further supported by the Global Center for Mass Spectrometry Excellence supported by Agilent Technologies at Baylor College of Medicine (BCM), ACS 127430-RSG-15-105-01-CNE (N.P), and R01CA220297 (N.P), Metabolomics core at BCM received funding from the NIH (P30 CA125123), Cancer Prevention and Research Institute of Texas (CPRIT) Proteomics and Metabolomics Core Facility (RP170005) in Dan L Duncan Cancer Center at BCM.
Footnotes
Conflict of interest statement: The authors declare no conflict of interest.
REFERENCES
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913–2921. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Cancer Facts & Figures 2017 Atlanta; 2017. [Google Scholar]
- 3.Ballehaninna UK, Chamberlain RS. Serum CA 19-9 as a Biomarker for Pancreatic Cancer-A Comprehensive Review. Indian J. Surg. Oncol 2011;2:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee MX, Saif MW. Screening for early pancreatic ductal adenocarcinoma: an urgent call! JOP 2009;10:104–108. [PubMed] [Google Scholar]
- 5.Zhou W, Liotta LA, Petricoin EF. Cancer metabolism and mass spectrometry-based proteomics. Cancer Lett 2015;356:176–183. [DOI] [PubMed] [Google Scholar]
- 6.Sousa CM, Kimmelman AC. The complex landscape of pancreatic cancer metabolism. Carcinogenesis 2014;35:1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardie RA, van Dam E, Cowley M, Han TL, Balaban S, Pajic M, Pinese M, et al. Mitochondrial mutations and metabolic adaptation in pancreatic cancer. Cancer Metab 2017;5:2. doi: 10.1186/s40170-017-0164-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YS, Maruvada P, Milner JA. Metabolomics in biomarker discovery: future uses for cancer prevention. Future. Oncol 2008;4:93–102. [DOI] [PubMed] [Google Scholar]
- 9.Zhang G, He P, Tan H, Budhu A, Gaedcke J, Ghadimi BM, Ried T, et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin. Cancer Res 2013;19:4983–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis VW, Schiller DE, Eurich D, Bathe OF, Sawyer MB. Pancreatic ductal adenocarcinoma is associated with a distinct urinary metabolomic signature. Ann. Surg. Oncol 2013;20 Suppl 3:S415–S423. [DOI] [PubMed] [Google Scholar]
- 11.Napoli C, Sperandio N, Lawlor RT, Scarpa A, Molinari H, Assfalg M. Urine metabolic signature of pancreatic ductal adenocarcinoma by (1)h nuclear magnetic resonance: identification, mapping, and evolution. J. Proteome. Res 2012;11:1274–1283. [DOI] [PubMed] [Google Scholar]
- 12.Bathe OF, Shaykhutdinov R, Kopciuk K, Weljie AM, McKay A, Sutherland FR, Dixon E, et al. Feasibility of identifying pancreatic cancer based on serum metabolomics. Cancer Epidemiol. Biomarkers Prev 2011;20:140–147. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi T, Nishiumi S, Ikeda A, Yoshie T, Sakai A, Matsubara A, Izumi Y, et al. A novel serum metabolomics-based diagnostic approach to pancreatic cancer. Cancer Epidemiol. Biomarkers Prev 2013;22:571–579. [DOI] [PubMed] [Google Scholar]
- 14.Leichtle AB, Ceglarek U, Weinert P, Nakas CT, Nuoffer JM, Kase J, Conrad T, et al. Pancreatic carcinoma, pancreatitis, and healthy controls: metabolite models in a three-class diagnostic dilemma. Metabolomics 2013;9:677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.OuYang D, Xu J, Huang H, Chen Z. Metabolomic profiling of serum from human pancreatic cancer patients using 1H NMR spectroscopy and principal component analysis. Appl. Biochem. Biotechnol 2011;165:148–154. [DOI] [PubMed] [Google Scholar]
- 16.Ritchie SA, Akita H, Takemasa I, Eguchi H, Pastural E, Nagano H, Monden M, et al. Metabolic system alterations in pancreatic cancer patient serum: potential for early detection. BMC. Cancer 2013;13:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tesiram YA, Lerner M, Stewart C, Njoku C, Brackett DJ. Utility of nuclear magnetic resonance spectroscopy for pancreatic cancer studies. Pancreas 2012;41:474–480. [DOI] [PubMed] [Google Scholar]
- 18.Urayama S, Zou W, Brooks K, Tolstikov V. Comprehensive mass spectrometry based metabolic profiling of blood plasma reveals potent discriminatory classifiers of pancreatic cancer. Rapid Commun. Mass Spectrom 2010;24:613–620. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Jin H, Guo X, Yang Z, Zhao L, Tang S, Mo P, et al. Distinguishing pancreatic cancer from chronic pancreatitis and healthy individuals by (1)H nuclear magnetic resonance-based metabonomic profiles. Clin. Biochem 2012;45:1064–1069. [DOI] [PubMed] [Google Scholar]
- 20.Marengo E, Robotti E. Biomarkers for pancreatic cancer: recent achievements in proteomics and genomics through classical and multivariate statistical methods. World J Gastroenterol 2014;20:13325–13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen VHS; Ayloo S; Molinari M Advances in pancreatic cancer: the role of metabolomics. JOP. J Pancreas 2015;16:244–248. [Google Scholar]
- 22.Mehta KY, Wu HJ, Menon SS, Fallah Y, Zhong X, Rizk N, Unger K, et al. Metabolomic biomarkers of pancreatic cancer: a meta-analysis study. Oncotarget 2017;8:68899–68915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lou YB, Fan FX, Mu YC, Dong X. The implication of diabetes metabolomics in the early diagnosis and pathogenesis of pancreatic cancer. J Biol Regul Homeost Agents 2018;32:75–82. [PubMed] [Google Scholar]
- 24.He X, Zhong J, Wang S, Zhou Y, Wang L, Zhang Y, Yuan Y. Serum metabolomics differentiating pancreatic cancer from new-onset diabetes. Oncotarget 2017;8:29116–29124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirata Y, Kobayashi T, Nishiumi S, Yamanaka K, Nakagawa T, Fujigaki S, Iemoto T, et al. Identification of highly sensitive biomarkers that can aid the early detection of pancreatic cancer using GC/MS/MS-based targeted metabolomics. Clin Chim Acta 2017;468:98–104. [DOI] [PubMed] [Google Scholar]
- 26.Hays J, Hunt JR, Hubbell FA, Anderson GL, Limacher M, Allen C, Rossouw JE. The Women’s Health Initiative recruitment methods and results. Ann Epidemiol 2003;13:S18–77. [DOI] [PubMed] [Google Scholar]
- 27.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc 2012;7:872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Putluri N, Shojaie A, Vasu VT, Vareed SK, Nalluri S, Putluri V, Thangjam GS, et al. Metabolomic profiling reveals potential markers and bioprocesses altered in bladder cancer progression. Cancer Res 2011;71:7376–7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown JM, Hazen SL. Targeting of microbe-derived metabolites to improve human health: The next frontier for drug discovery. J Biol Chem 2017;292:8560–8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tumas J, Kvederaviciute K, Petrulionis M, Kurlinkus B, Rimkus A, Sakalauskaite G, Cicenas J, et al. Metabolomics in pancreatic cancer biomarkers research. Med Oncol 2016;33:133. [DOI] [PubMed] [Google Scholar]
- 31.Fukutake N, Ueno M, Hiraoka N, Shimada K, Shiraishi K, Saruki N, Ito T, et al. A Novel Multivariate Index for Pancreatic Cancer Detection Based On the Plasma Free Amino Acid Profile. PLoS One 2015;10:e0132223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leja-Szpak A, Pierzchalski P, Goralska M, Nawrot-Porabka K, Bonior J, Link-Lenczowski P, Jastrzebska M, et al. Kynuramines induce overexpression of heat shock proteins in pancreatic cancer cells via 5-hydroxytryptamine and MT1/MT2 receptors. J Physiol Pharmacol 2015;66:711–718. [PubMed] [Google Scholar]
- 33.Witkiewicz A, Williams TK, Cozzitorto J, Durkan B, Showalter SL, Yeo CJ, Brody JR. Expression of indoleamine 2,3-dioxygenase in metastatic pancreatic ductal adenocarcinoma recruits regulatory T cells to avoid immune detection. J. Am. Coll. Surg 2008;206:849–854. [DOI] [PubMed] [Google Scholar]
- 34.Unger K, Mehta KY, Kaur P, Wang Y, Menon SS, Jain SK, Moonjelly RA, et al. Metabolomics based predictive classifier for early detection of pancreatic ductal adenocarcinoma. Oncotarget 2018;9:23078–23090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fahrmann JF, Bantis LE, Capello M, Scelo G, Dennison JB, Patel N, Murage E, et al. A Plasma-Derived Protein-Metabolite Multiplexed Panel for Early-Stage Pancreatic Cancer. J Natl Cancer Inst 2018. doi: 10.1093/jnci/djy126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang LS, Davies SS. Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome Med 2016;8:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siminska E, Koba M. Amino acid profiling as a method of discovering biomarkers for early diagnosis of cancer. Amino Acids 2016;48:1339–1345. [DOI] [PubMed] [Google Scholar]
- 38.Xie G, Lu L, Qiu Y, Ni Q, Zhang W, Gao YT, Risch HA, et al. Plasma metabolite biomarkers for the detection of pancreatic cancer. J Proteome Res 2015;14:1195–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayerle J, Kalthoff H, Reszka R, Kamlage B, Peter E, Schniewind B, Gonzalez Maldonado S, et al. Metabolic biomarker signature to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Gut 2017; 67:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakai A, Suzuki M, Kobayashi T, Nishiumi S, Yamanaka K, Hirata Y, Nakagawa T, et al. Pancreatic cancer screening using a multiplatform human serum metabolomics system. Biomark Med 2016;10:577–586. [DOI] [PubMed] [Google Scholar]
- 41.Daemen A, Peterson D, Sahu N, McCord R, Du X, Liu B, Kowanetz K, et al. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc Natl Acad Sci U S A 2015;112:E4410–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shu X, Zheng W, Yu D, Lan Li H, Lan Q, Yang G, Cai H, et al. Prospective metabolomics study identifies potential novel blood metabolites associated with pancreatic cancer risk. Int J Cancer 2018; 143:2161–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katagiri R, Goto A, Nakagawa T, Nishiumi S, Kobayashi T, Hidaka A, Budhathoki S, et al. Increased Levels of Branched-chain Amino Acid Associated With Increased Risk of Pancreatic Cancer in a Prospective Case-Control Study of a Large Cohort. Gastroenterology 2018; 155:1474–1482. [DOI] [PubMed] [Google Scholar]
- 44.Mayers JR, Wu C, Clish CB, Kraft P, Torrence ME, Fiske BP, Yuan C, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med 2014;20:1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sitnikov DG, Monnin CS, Vuckovic D. Systematic Assessment of Seven Solvent and Solid-Phase Extraction Methods for Metabolomics Analysis of Human Plasma by LC-MS. Sci Rep 2016;6:38885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liesenfeld DB, Habermann N, Owen RW, Scalbert A, Ulrich CM. Review of mass spectrometry-based metabolomics in cancer research. Cancer Epidemiol. Biomarkers Prev 2013;22:2182–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


