Abstract
The cellular DNA replication stress response functions to stabilize DNA replication forks and inhibit genome instability and tumorigenesis induced by oncogenes. However, the specific proteins required for resolving oncogenic stress remain poorly understood. Here we report that Smarcal1 and Zranb3, closely related replication fork remodeling proteins, have non-redundant functions in resolving Myc-induced DNA replication stress. In Myc overexpressing primary cells, significant differences in replication fork stalling, collapse, and DNA damage were detected between cells deficient in Smarcal1 or Zranb3, leading to changes in proliferation and apoptosis. These differences were also reflected in Myc-induced lymphoma development; haploinsufficiency of Smarcal1 resulted in accelerated lymphomagenesis, whereas haploinsufficiency of Zranb3 inhibited lymphoma development. Complete loss of either protein resulted in disparate survival outcomes. Our results reveal that endogenous replication stress from Myc in primary cells requires both alleles of Smarcal1 and Zranb3 and demonstrate the requirement of both proteins to stabilize replication forks upon Myc dysregulation in a non-redundant manner.
Keywords: Smarcal1, Zranb3, Myc, DNA replication stress, lymphoma
Introduction
DNA replication stress leads to replisome stalling and potential replication fork collapse. It has been implicated in several human pathologies and in particular, is considered a significant contributor to cancer (1–6). Replication stress is present in pre-neoplastic lesions and is a hallmark of transformed cells (1,2,6). In cancer cells, dysregulated oncogenes, such as the transcription factor Myc, cause DNA replication stress by stimulating aberrant origin firing, driving premature S-phase entry, inducing transcriptional interference with the replisome, and modifying cellular metabolism (7). To suppress replication stress, which is believed to drive genomic instability and cellular transformation, cells activate a replication stress response (1–5).
The replication stress response is a highly complex, incompletely understood, and evolutionary conserved mechanism that functions to prevent replication fork collapse, safeguard DNA synthesis, and protect the genome (1–5). Critical aspects of the replication stress response include stabilization, repair, and restart of stalled replication forks. Although multiple proteins are reported to interact with stalled replisomes and facilitate fork repair and restart (8,9), much remains unknown. The vast majority of studies performed to-date have been conducted with yeast, were in vitro biochemical analyses, and/or utilized exogenous sources of replication stress to which mammalian cells are not normally exposed. Moreover, the contribution of specific replication stress response proteins in resolving different types of replication stress, whether there is functional redundancy between proteins, and the specific physiologically relevant cellular contexts (e.g., oncogenic stress, emergency hematopoiesis, etc.) that may require one protein over another remains largely unresolved.
One class of proteins involved in replication fork stability is the SNF2 family of DNA translocases. This family includes the closely related DNA fork remodeling proteins Smarcal1 and Zranb3 (10). Upon replication stress, both Smarcal1 and Zranb3 function similarly in vitro by binding DNA at stalled replication forks, re-annealing excessive single-stranded DNA, and promoting fork regression and remodeling (11–19). However, Smarcal1 and Zranb3 are recruited to stalled forks through different protein interactions (11–16,19,20) and have different substrate preferences (21). Smarcal1 also resolves replication stress at telomeres (22,23). While multiple studies have shown that these proteins functions to stabilize replication forks and promote replication restart upon treatment with genotoxic agents in cancer cell lines in vitro (11–13,15,16), the specific biological contexts requiring each protein remain poorly characterized. Although possessing similar biochemical functions (11–19), it is currently unknown whether Smarcal1 and Zranb3 can compensate for one another under physiological conditions in primary cells. Moreover, it is unknown whether endogenous sources of replication stress, such as Myc dysregulation in pre-neoplastic primary cells, require either (or both) protein for replication fork stability.
To address these outstanding questions, we utilized a transgenic mouse model (Eμ-myc) where Myc is overexpressed only in B lymphocytes causing B-cell lymphomagenesis (24). We observed profound differences in survival between Eμ-myc mice that were Smarcal1-deficient or Zranb3-deficient. These differences were due to alterations in replication fork stability, DNA damage, apoptosis, and proliferation of B cell, and the gene dosage of each protein. Our data identify Myc as the first endogenous source of replication stress that requires both Smarcal1 and Zranb3 for resolution, and establishes that there are unique, non-redundant biological functions of Smarcal1 and Zranb3 in vivo that influence Myc-induced tumor development.
Materials and Methods
Mice
C57Bl/6 Smarcal1+/Δ mice were previously provided by Dr. Cornelius Boerkoel (University of British Columbia). C57Bl/6 Zranb3+/− founder mice were purchased from the Texas Institute of Genomic Medicine (TIGM, Fort Worth, TX). Zranb3+/− mice were generated from murine embryonic stem cells retrovirally transduced with a vector encoding a gene trapping cassette. A unique gene trapping insertion occurred prior to exon 8 in the Zranb3 gene locus (Supplemental Fig. S1A). For survival studies, mice were monitored and sacrificed upon signs of tumor development and/or illness. The Smarcal1 Eμ-myc survival study was completed between 2010 and 2013. The Zranb3 Eμ-myc survival study was completed between 2015 and 2017. All spleen and bone marrow evaluated were removed from young mice prior to any lymphoma development. All experiments were performed with young male and female littermate-matched mice. All mouse studies were reviewed and approved by the Institutional Animal Care and Use Committee at either Vanderbilt University or Thomas Jefferson University.
Primary cell culture and retroviral infection
Short-term primary cell cultures of pro-B cells of each genotype were generated and passaged by culturing bone marrow cells on a bone marrow stromal feeder layer derived from the same mouse in RPMI 1640 with 20% FBS, 2 mM glutamine, 55 μM β-mercaptoethanol, penicillin/streptomycin, and 10 ng/mL interleukin-7. Pro-B cells were retrovirally infected with MSCV-MycER-IRES-GFP, as previously described (25), and GFP+ pro-B cells sorted by flow cytometry. Cell number and viability were determined (in triplicate) by trypan blue dye exclusion. Cell proliferation was evaluated by MTS assays (Promega, 490 nm) according to the manufacturer’s instructions.
B cell purification
Spleens or bone marrow from mice were processed into single cell suspensions. B cells were negatively selected by incubating cells with a biotinylated B cell enrichment antibody cocktail (CD43, CD4, Ter-119) followed by incubation with streptavidin magnetic particles and magnetization according to the manufacturer’s instructions (BD Biosciences).
DNA fiber labeling
Purified bone marrow B cells or bone marrow-derived pro-B cells were pulse labeled with 25 μM IdU (5-Iodo-2’-deoxyuridine; Sigma Aldrich) for 20 or 30 minutes, respectively, washed with 1× DPBS, and labeled with 250 μM CldU (5-Chloro-2’-deoxyuridine; Sigma Aldrich) for 20 or 30 minutes, respectively, both at 37°C. Cells were lysed with DNA spreading buffer (0.5% SDS, 200 mM Tris-HCl (pH 7.4), 50 mM EDTA) for 6 minutes and DNA spread across frosted microscope slides. Slides were air dried for 40 minutes and fixed with 3:1 methanol/acetic acid for 2 minutes. DNA was denatured by submerging slides in 2.5N HCl for 80 minutes. Slides were blocked for 1 hour with blocking buffer (10% normal goat serum, 0.1% Triton-X 100 in 1× DPBS) and incubated with rat anti-CldU antibody (Abcam) diluted 1:50 in blocking buffer for 1 hour. After washing 3× with 1× DPBS, slides were incubated with AlexaFluor-488 goat anti-rat secondary antibody (Invitrogen) diluted 1:200 in blocking buffer for 30 minutes. Slides were then washed 3× in 1× DPBS and incubated with mouse anti-IdU antibody (BD) diluted 1:50 in blocking buffer for 1 hour. Slides were washed 3× with 1× DPBS and incubated with Cy3 goat anti-mouse secondary antibody (Invitrogen) diluted 1:200 in blocking buffer for 30 minutes. All antibody incubations were done in a humidified chamber. Slides were washed 3× with 1× DPBS, air dried for 20 minutes and mounted with ProLong Gold (Invitrogen) and allowed to cure overnight. All incubations and washes were at room temperature. Images were captured (blinded) on a Nikon Eclipse Ni using a 100× oil objective (Nikon) and fibers were measured and analyzed using ImageJ software.
Flow cytometry analysis of immunophenotype, BrdU incorporation, cell cycle, and apoptosis
Bone marrow or spleens were harvested from mice and processed into single cells suspensions. Immunophenotyping was performed as we previously reported (26,27), using fluorochrome-linked antibodies specific for IgM, IgD, CD19, and B220 from Southern Biotech, CD43, Ly6A/E, and CD34 from BD Biosciences, and CD4 from eBiosciences. For BrdU experiments, mice were intraperitoneally injected with 1 mg BrdU and tissues harvested after 16 hours. BrdU incorporation was measured according to the manufacturer’s instructions (BD Biosciences). Cell cycle and subG1 DNA content was evaluated by propidium idodide (Sigma) staining of DNA, as we previously reported (28,29). Apoptosis was measured by Annexin-V/7AAD (BD BioSci) staining, as we previously reported (28,30). All samples were evaluated on an LSRII or BD Fortessa instrument (BD Biosciences) and analyzed using FlowJo software.
Western blotting
Whole cell lysates were Western blotted using antibodies specific for Rpa32, phospho-Rpa32 (Ser33), phospho-Rpa32 (Ser4/8), and Zranb3 from Bethyl Laboratories; Chk1, phospho-Chk1 (Ser345), and cleaved Caspase 3 from Cell signaling; Myc from EMD Millipore; and β-actin from Sigma Aldrich, as previously described (31). The Smarcal1 antibody was provided by Dr. David Cortez (Vanderbilt University).
γH2AX immunofluorescence
Quantification of γH2AX foci by immunofluorescence was performed as previously described (32). Images were captured on a Nikon C2 or A1R confocal microscope (Nikon) and analyzed using ImageJ. All samples were blinded for analysis and a minimum of 100 cells were analyzed per experiment.
Comet assay
Neutral comet assays were performed on pro-B cells or purified splenic B cells as previously described (33,34). Images were captured on a Nikon Eclipse Ni (Nikon) with a 10× objective and analyzed using CometScore software. All samples were blinded and a minimum of 75 cells per sample were scored.
Statistical analysis
Statistical analysis was determined using Student’s t-test (Figs. 1B, 1E, 1G, 6A–6D, 6F-6I), chi-square test (Figs. 1D, 1F, 3G, 3H, 3K, 3L, 4G, 4H, 4K, 4L), log-rank tests (Figs. 2A, 2B), or one-way ANOVA with a Bonferroni post-test (Figs. 3B, 3E, 3I, 3J, 3M, 3N, 4B, 4E, 4I, 4J, 4M, 4N, 5A–5F). All calculations were performed using GraphPad Prism software.
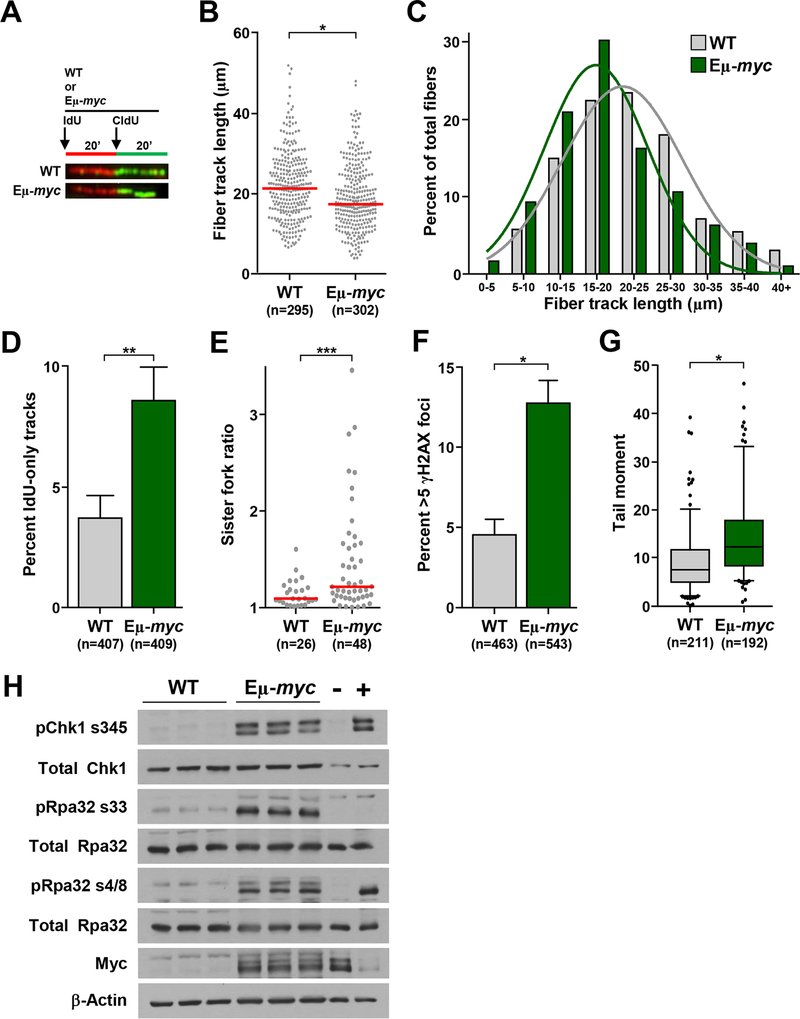
Figure 1. Myc overexpression leads to replication stress in primary B cells.
(A-E) B cells purified from bone marrow of Eμ-myc and littermate wild-type (WT) mice were subjected to single-molecule DNA fiber analysis. A) Design of DNA fiber analysis and representative fiber images. B) Quantification of total fiber length from two independent experiments; red line is median. C) Binned fiber length frequencies from (B). D) Quantification of fibers that only incorporated IdU from (B); SEM. E) Ratios of measured CldU tracks from left and right moving forks from the same replication origin; median indicated with red bar. F) Quantification of immunofluorescence of γH2AX foci in purified bone marrow B cells; SEM. G) Box-and-whisker plots of tail moments of purified bone marrow B cells. Box is the 25th and 75th percentiles, whiskers are 5th and 95th percentiles, line is the median. H) Whole cell lysates from spleens of littermate-matched Eμ-myc and WT mice were Western blotted for the indicated proteins. B cell lymphoma treated with etoposide or vehicle served as positive (+) and negative (–) controls, respectively, for the replication stress response. Total number (n) of fibers (B), replication structures (D, E), or cells (F, G) noted. Student’s t-tests (B, E, G) or chi-square tests (D, F); *p<0.0001, **p=0.0037, ***p=0.0052.
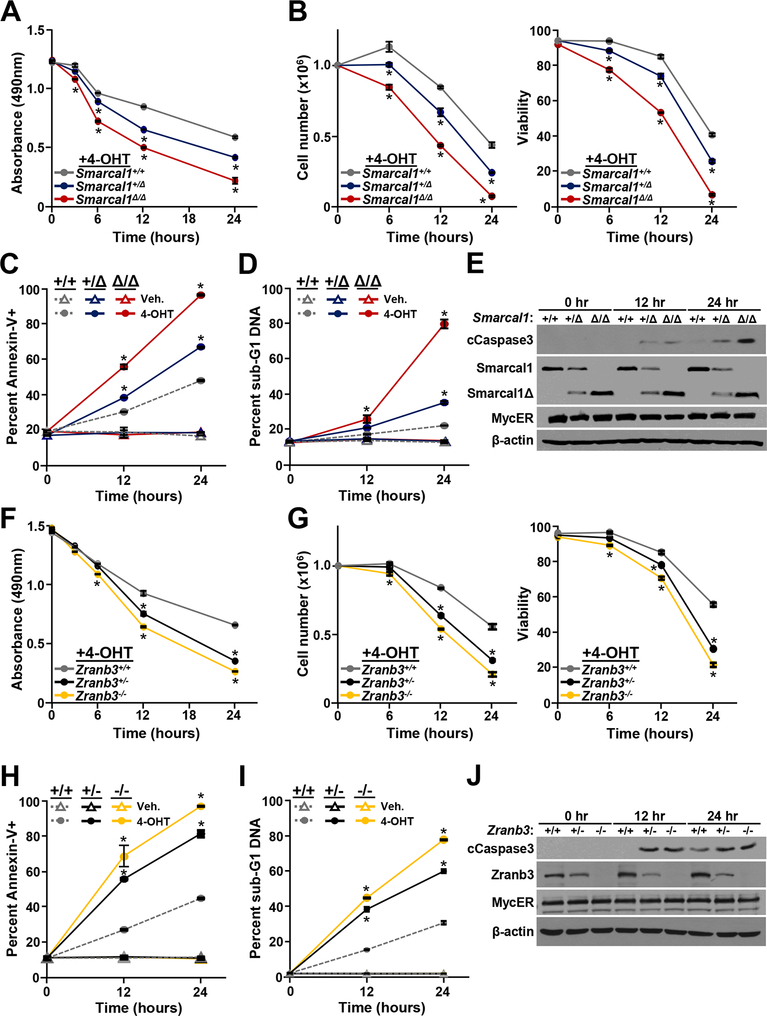
Figure 6. Loss of Smarcal1 or Zranb3 sensitizes B cells to Myc-induced apoptosis.
Bone marrow derived pro-B cells expressing MycER of the indicated genotypes. MycER was activated with 4-hydroxytamoxifen (4-OHT) and at intervals MTS assays (A, F), cell numbers and viability (B, G), Annexin-V positivity (C, H), and sub-G1 DNA content (D, I) were evaluated. Student’s t-tests determine significance, *p<0.0001. E, J) Western blots of whole cell lysates for the indicated proteins at intervals after MycER activation with 4-OHT.
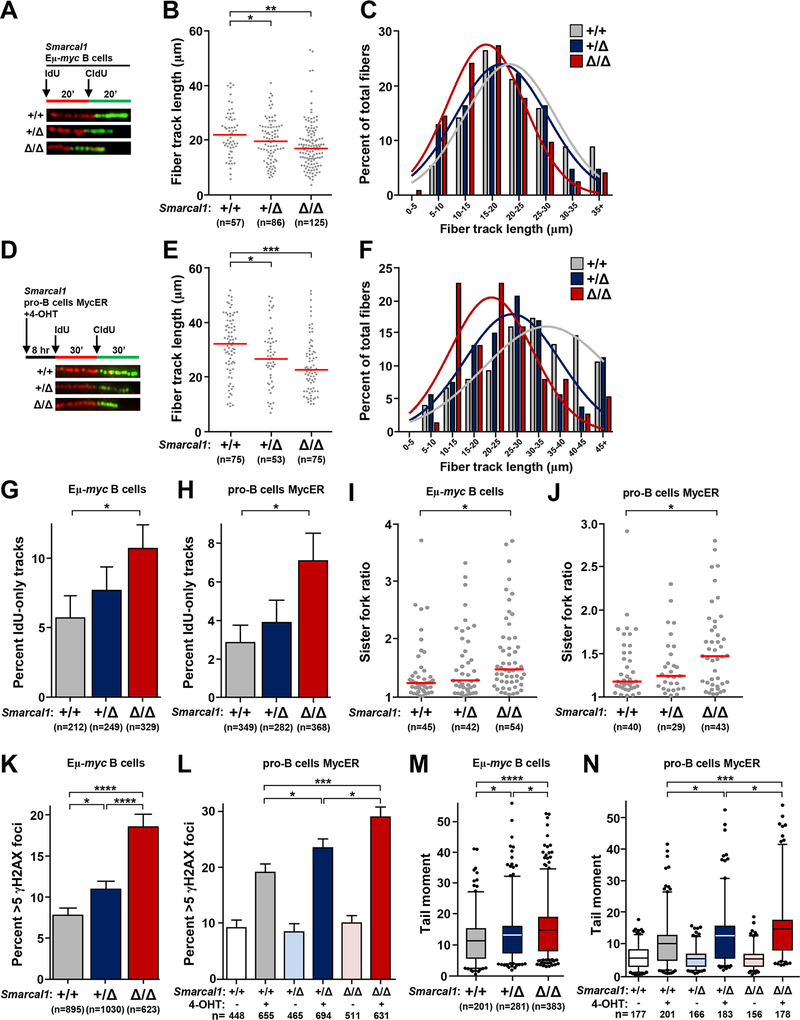
Figure 3. Smarcal1-deficient B cells have increased rates of replication fork collapse with Myc dysregulation.
B cells from Smarcal1+/+ Eμ-myc, Smarcal1+/Δ Eμ-myc, and Smarcal1Δ/Δ Eμ-myc mice were purified from bone marrow (A-C, G, I) or spleen (K, M). Pro-B cell cultures from Smarcal1+/+, Smarcal1+/Δ, and Smarcal1Δ/Δ mice expressing MycER (D-F, H, J, L, N) with MycER activated by the addition of 4-hydroxytamoxifen (4-OHT) for 8 hours. (A, D) Design of DNA fiber labeling experiments and images of representative fiber tracks. B, E) Quantification of total fiber length in purified Eμ-myc B cells (B) and MycER pro-B cells (E); median indicated with red bar. C, F) Fiber length frequencies from (B) and (E), respectively. G, H) Quantification of DNA fibers that only incorporated IdU from Eμ-myc B cells (G) and MycER pro-B cells (H); SEM. I, J) Ratios of measured CldU track lengths from left and right moving forks from the same replication origin from Eμ-myc B cells (I) and MycER pro-B cells (J); median indicated with red bar. K, L) Quantification of immunofluorescence of γH2AX foci in Eμ-myc B cells (K) and MycER pro-B cells (L); SEM. M, N) Box-and-whisker plots of tail moments of Eμ-myc B cells (M) and MycER pro-B cells (N). Line is the median, boxes are the 25th and 75th percentiles, whiskers are the 5th and 95th percentiles. The total number (n) of fibers (B, E), replication structures (G-J), or cells (K-N) analyzed is indicated. All data are from 2–3 independent experiments. One-way ANOVA with Bonferroni correction (B, E, M, N, I, J) or chi-square test (G, H, K, L); *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
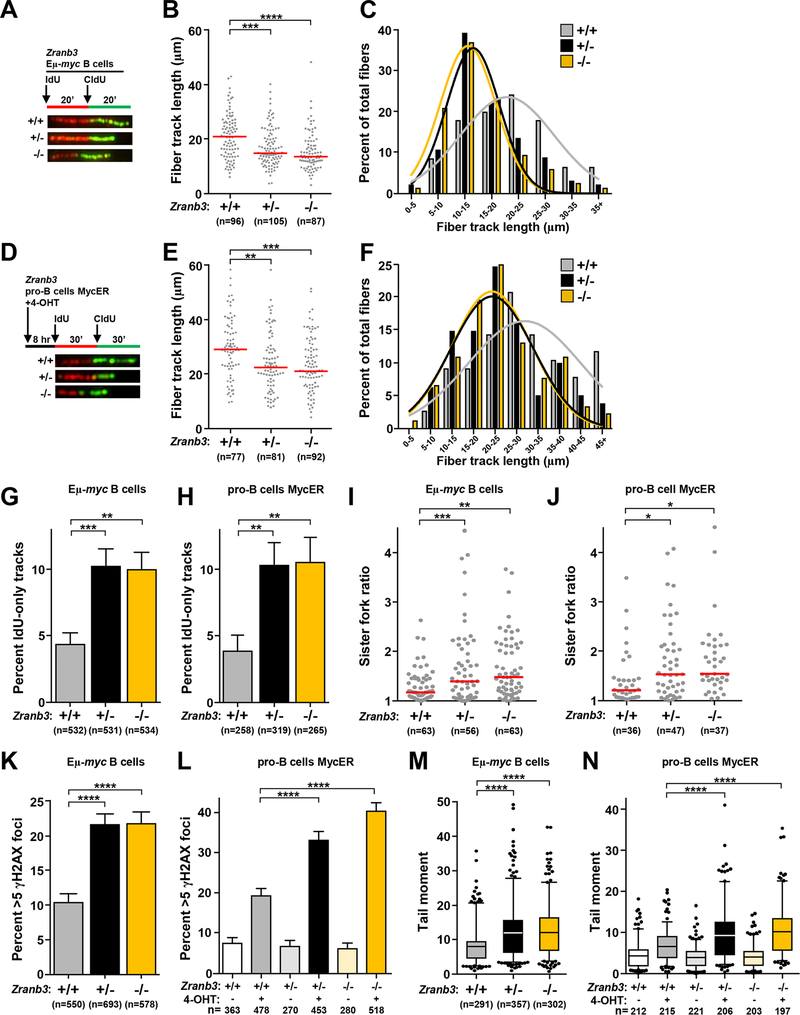
Figure 4. Zranb3 protects replication forks during Myc-induced replication stress.
B cells from Zranb3+/+ Eμ-myc, Zranb3+/− Eμ-myc, and Zranb3−/− Eμ-myc mice were purified from bone marrow (A-C, G, I, K, M). Pro-B cells from Zranb3+/+, Zranb3+/− and Zranb3−/− mice expressing MycER (D-F, H, J, L, N) with MycER activated by the addition of 4-hydroxytamoxifen (4-OHT) for 8 hours. (A, D) Design of DNA fiber labeling experiments and images of representative fiber tracks. B, E) Quantification of total fiber length in purified Eμ-myc B cells (B) and MycER pro-B cells (E); median indicated with red bar. C, F) Fiber length frequencies from (B) and (E), respectively. G, H) Quantification of DNA fibers that only incorporated IdU from Eμ-myc B cells (G) or pro-B cells (H); SEM. I, J) Ratios of measured CldU track lengths from left and right moving forks from the same replication origin from Eμ-myc B cells (I) and MycER pro-B cells (J); median indicated with red bar. K, L) Quantification of immunofluorescence of γH2AX foci in Eμ-myc B cells (K) and MycER pro-B cells (L); SEM. M, N) Box-and-whisker plots of tail moments of Eμ-myc B cells (M) and MycER pro-B cells (N). Line is the median, boxes are the 25th and 75th percentiles, whiskers are the 5th and 95th percentiles. The total number (n) of fibers (B, E), replication structures (G-J), or cells (K-N) analyzed is indicated. All data are from 2–3 independent experiments. One-way ANOVA with Bonferroni correction (B, E, M, N, I, J) or chi-square test (G, H, K, L); *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
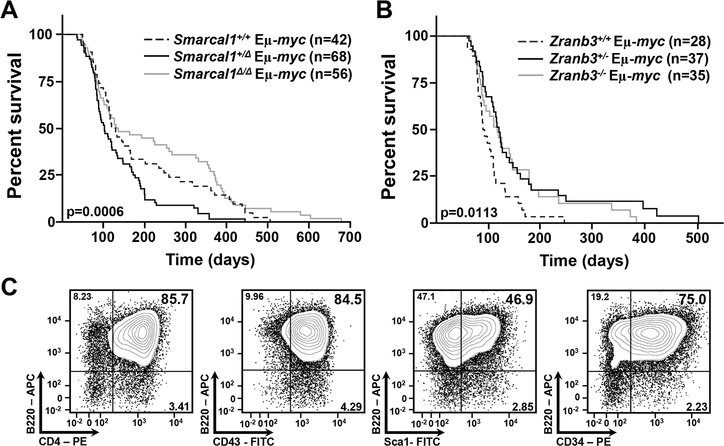
Figure 2. Deficiency of Smarcal1 or Zranb3 significantly alters Myc-induced lymphomagenesis.
A, B) Kaplan-Meier survival curves of mice of the indicated genotypes. Overall P values in figures and p=0.0142 (+/+ vs. +/Δ), p=0.0002 (+/Δ vs. Δ/Δ), p=0.3224 (+/+ vs. Δ/Δ) (B), and p=0.0066 (+/+ vs. +/−), p=0.0199 (+/+ vs. −/−), log-rank tests. Number (n) of mice indicated. C) Representative flow cytometry contour plots of the early precursor B cell lymphomas arising in a fraction of the Smarcal1+/Δ Eμ-myc, Zranb3+/− Eμ-myc, and Zranb3−/− Eμ-myc mice (also see Table 1) using fluorochrome-linked antibodies specific for B cell surface markers (indicated).
Figure 5. Loss of Smarcal1 reduces cell cycle progression of B cells, whereas loss of Zranb3 does not when Myc is overexpressed.
A, C) Quantification of total splenic leukocytes from litters of the indicated genotypes. B, D) Representative contour plots and quantification of mature (B220+/IgM+) B splenocytes from litters of the indicated genotypes. E) Quantification of BrdU+ splenic B cells (CD19+; left) and the percentage of purified B cells in each phase of the cell cycle (right) from litters of the Eμ-myc mice of the indicated Smarcal1 genotype. Cell cycle was determined using Dean-Jett-Fox analysis. F) Quantification of BrdU+ splenic B cells (CD19+) from litters of the Eμ-myc mice of the indicated Zranb3 genotype. Total number (n) of mice indicated; data are from 3–7 independent litters. Error bars are SEM; *p<0.05, one-way ANOVA with Bonferroni post-test.
Results
Myc overexpression generates replication stress in primary B cells.
Myc dysregulation is a source of endogenous DNA replication stress, which is thought to contribute to tumorigenesis (35–38). Currently, there is a paucity of knowledge on the molecular consequences of this endogenous stress at the replication fork. To evaluate the effects of a physiological source of DNA replication stress, Eμ-myc transgenic mice that overexpress Myc only in B cells and develop B cells lymphomas (24) were utilized. We performed single-molecule DNA fiber analysis on purified B cells from Eμ-myc transgenic mice prior to lymphoma development and littermate-matched wild-type controls (Fig. 1A). Myc-overexpressing B cells showed a significant reduction in fiber length, which is indicative of impaired DNA replication (Figs. 1B and 1C). Fibers that only incorporate the first nucleotide analog (IdU), but not the second (CldU), represent stalled forks or terminated replication. These IdU-only fibers were significantly increased in Eμ-myc B cells compared to wild-type B cells (Fig. 1D). To determine whether Myc overexpression caused fork stalling, we evaluated the incidence of sister replication fork asymmetry by comparing the relative CldU track lengths of replication forks originating from the same origin. B cells overexpressing Myc had significantly higher rates of asymmetric sister forks (Fig. 1E). Thus, Myc overexpression results in fork stalling and impaired replication fork progression in primary murine B cells.
Prolonged fork stalling can lead to replication fork collapse resulting in double-stranded DNA breaks (5). Evaluation of DNA damage in B cells showed Eμ-myc mice had a significantly higher percentage of B cells with γH2AX foci, a marker of double-stranded DNA breaks (Fig. 1F), and increased DNA breaks (Fig. 1G) compared to wild-type littermates. Additionally, B cells from Eμ-myc mice had increased activation of the replication stress response compared to wild-type littermates. There were elevated levels of phosphorylated Chk1 (Ser345) and Rpa32 (Ser33), as well as phospho-Rpa32 (Ser4/8), a marker of DNA damage (Fig. 1H). Therefore, Myc overexpression generates replication stress, inducing a robust replication stress response in primary B lymphocytes prior to cellular transformation.
Loss of Smarcal1 or Zranb3 significantly alters Myc-driven lymphoma development
Smarcal1 and Zranb3 have been implicated in resolving replication stress caused by drugs that induce fork stalling in vitro (11–16), but their role in responding to replication stress under physiological conditions is incompletely understood. To assess in an unbiased, biologically relevant system the contribution of Smarcal1 and Zranb3 in the replication stress response to Myc overexpression, we utilized mouse models. We previously reported Smarcal1 loss-of-function mice, which express an N-terminal truncated, non-functional Smarcal1 protein, Smarcal1Δ (32). Zranb3 knockout mice contain a gene trapping cassette in the Zranb3 gene locus and do not express Zranb3 protein (Supplemental Fig. S1A and B). Similar to Smarcal1-deficient mice, Zranb3+/− and Zranb3−/− mice displayed no overt abnormalities.
We crossed the Smarcal1Δ and Zranb3 knockout mice to Eμ-myc transgenic mice and monitored lymphoma development. Previously, using a γ-radiation replication stress-induced model of T-cell lymphomagenesis, we reported loss of one or two alleles of Smarcal1 inhibited lymphomagenesis and increased overall survival (32). However, with Myc-induced replication stress, there was a significant acceleration in B cell lymphoma development and decreased survival of Smarcal1+/Δ Eμ-myc mice compared to Smarcal1+/+ Eμ-myc mice (Fig. 2A) with mean survivals of 135 days and 187 days, respectively. Unexpectedly, Smarcal1Δ/Δ Eμ-myc mice showed no statistically significant difference in overall survival compared to wild-type Eμ-myc mice (p=0.3224) and had a mean survival of 224 days. However, Smarcal1Δ/Δ Eμ-myc mice showed significantly increased survival compared to Smarcal1+/Δ Eμ-myc mice (p=0.0002, Fig. 2A), revealing a Smarcal1 gene dosage effect on Myc-induced lymphomagenesis.
In contrast to Smarcal1-deficient Eμ-myc mice, lymphoma development was significantly inhibited in both Zranb3+/− and Zranb3−/− Eμ-myc mice compared to wild-type littermates (Fig. 2B), with mean survivals of 149, 138 and 104 days, respectively. Thus, for proteins thought to be very similar biochemically, our data demonstrate a difference in function in responding to Myc-induced replication stress in vivo.
Eμ-myc mice typically develop pre-B and/or B cell lymphomas (24,27,39), but a subset of Smarcal1+/Δ, Zranb3+/−, and Zranb3−/− Eμ-myc mice developed early progenitor B cell lymphomas (Fig. 2C and Table 1), suggesting Smarcal1 and Zranb3 loss influenced the tumor cell of origin. Taken together, our data indicate both Smarcal1 and Zranb3 are critical non-redundant proteins in the cellular response to Myc-induced replication stress. Moreover, the expression levels of these proteins profoundly affect Myc-induced lymphoma development and survival.
Table 1.
Precursor B cell lymphomas develop in the Smarcal1- and Zranb3-deficient Eμ-myc cohorts
| Smarcal1 | Zranb3 | |||||
|---|---|---|---|---|---|---|
| Phenotype | +/+ | +/ Δ | Δ/Δ | +/+ | +/− | −/− |
| B220+ CD19+ IgM+/− IgD+/− | 9 (100%) | 9 (81.8%) | 9 (100%) | 8 (100%) | 9 (75.0%) | 6 (85.7%) |
| B220+ CD19+/− IgMlo Sca1+/lo CD34+ CD4+ |
0 (0%) | 2 (18.2%) | 0 (0%) | 0 (0%) | 3 (25.0%) | 1 (14.3%) |
Smarcal1 loss results in replication fork collapse upon Myc overexpression.
To begin to determine the cause(s) of the differences in survival of the Smarcal1- and Zranb3-deficient Eμ-myc mice, we evaluated the consequences of Smarcal1 loss at the replication fork in B cells overexpressing Myc. Single-molecule DNA fiber analysis was performed on purified bone marrow B cells from Smarcal1 Eμ-myc littermates (Fig. 3A). Loss of both alleles of Smarcal1 led to a significant reduction in mean fiber length and loss of a single Smarcal1 allele led to an intermediate, but statistically significant reduction in fiber length (Figs. 3B and 3C). To independently validate these results, we retrovirally expressed a 4-hydroxytamoxifen (4-OHT)-inducible form of Myc, MycER (40), in pro-B cells from wild-type, Smarcal1+/Δ and Smarcal1Δ/Δ mice. We performed fiber analysis in these cells 8 hours after activation of MycER with 4-OHT addition (Fig. 3D), a point when MycER has induced S-phase entry, but prior to significant Myc-induced apoptosis. With MycER induction, we observed a significant reduction in mean fiber length in the Smarcal1Δ/Δ cells and an intermediate reduction in the Smarcal1+/Δ cells (Figs. 3E and 3F). The differences in fiber lengths were not due to Smarcal1 loss alone, as Smarcal1-deficient pro-B cells with normal Myc levels, did not show changes in fiber length (Supplemental Figs. S2A–C). Therefore, two independent approaches with primary B cells demonstrate loss of Smarcal1 function at replication forks in the presence of oncogene overexpression impairs DNA replication.
Since the fork reversal/remodeling function of Smarcal1 stabilizes stalled forks and promotes replication restart (11,12,15,21), we sought to determine whether fibers from Smarcal1-deficient cells displayed evidence of increased fork stalling/collapse. In both the Smarcal1Δ/Δ Eμ-myc bone marrow B cells and the Smarcal1Δ/Δ pro-B cells with activated MycER, there was a significant increase in the percentage of fibers that had only incorporated IdU (Figs. 3G and 3H). Moderate differences were observed with loss of a single Smarcal1 allele, but it did not reach statistical significance. There were no differences in IdU-only fibers in B cells between the genotypes that did not overexpress Myc (Supplemental Fig. S2D). However, in both Eμ-myc and MycER activated Smarcal1Δ/Δ B cells, we observed increased rates of fork asymmetry (Figs. 3I and 3J), indicating loss of Smarcal1 results in replication fork stalling when B cells are under oncogenic stress.
Evaluation of DNA damage in Eμ-myc B cells and pro-B cells following MycER activation showed a significant increase in the percentage of Smarcal1-deficient B cells with γH2AX foci and DNA breaks (Figs. 3K–3N). Myc overexpression in Smarcal1Δ/Δ B cells had higher levels of DNA damage than Smarcal1 heterozygous B cells, which had a small, but significant increase in damaged DNA (Figs. 3K–3N). This DNA damage was not due to Smarcal1 loss alone, as B cells from non-Myc overexpressing Smarcal1-deficient mice showed no increase in DNA damage (Supplemental Figs. S2E and F). Thus, Smarcal1 stabilizes replication forks in response to oncogene dysregulation in a gene dosage-dependent manner, preventing fork collapse.
Zranb3 is required for replication fork stability when Myc is overexpressed.
To determine effects of Zranb3 loss on replication fork stability in B cells with Myc dysregulation, and to compare with Smarcal1 loss, we evaluated DNA fiber length, fork collapse, and DNA damage. In both Eμ-myc bone marrow B cells (Figs. 4A–4C) and MycER activation in pro-B cells (Figs. 4D–4F), there was a significant analogous reduction in mean DNA fiber length in Zranb3+/− and Zranb3−/− cells. Evaluation of B cells for fork collapse showed significantly higher rates of IdU-only fibers and asymmetric sister forks in Zranb3+/− and Zranb3−/− Eμ-myc B cells and MycER activated pro-B cells compared to wild-type controls (Figs. 4G–4J). However, unlike Smarcal1, loss of a single Zranb3 allele induced the same level of fork stalling as that in Zranb3−/− B cells, indicating a loss of both Zranb3 alleles was no more deleterious to forks than Zranb3 haploinsufficiency. The increase in fork collapse in Zranb3-deficient B cells led to significantly increased rates of γH2AX foci and DNA breaks in both the Eμ-myc B cells and MycER activated pro-B cells (Figs. 4K–4N). The differences in DNA fiber length, fork collapse, and DNA damage/breaks with loss of Zranb3 were dependent on Myc overexpression, as B cells lacking one or both Zranb3 alleles with normal Myc levels did not show differences compared to wild-type controls (Supplemental Fig. S3A–F). Together, these data demonstrate Zranb3 is crucial for maintaining replication fork stability in the presence of dysregulated Myc. Moreover, unlike Smarcal1, loss of a single Zranb3 allele results in effects equally as severe as observed in Zranb3-null cells, suggesting a more critical biological function for Zranb3 in resolving oncogene-induced replication stress. Furthermore, these data demonstrate that Smarcal1 and Zranb3 are unable to biologically compensate for one another, as loss of either protein led to significant replication defects.
Smarcal1- and Zranb3-deficient Eμ-myc mice have reduced numbers of B cells.
To determine the biological consequences of increased fork collapse and DNA breaks in Smarcal1-deficient and Zranb3-deficient Myc overexpressing B cells, we evaluated B cell populations in both colonies. We first evaluated hematopoietic organs in non-Eμ-myc transgenic mice to determine if loss of either protein alone altered B cell populations. Smarcal1-deficient and Zranb3-deficient mice displayed no overt B cell phenotypes in either the spleen or the bone marrow. Numbers of splenic leukocytes (Fig. 5A) and percentages of splenic B cells (Fig. 5B) were unchanged between all non-Eμ-myc transgenic genotypes. Similarly, both the percentages and total numbers of developing B cell populations in the bone marrow were analogous in both colonies (Supplemental Fig. S4A and B). Additionally, loss of Zranb3 or Smarcal1 did not affect the ability of bone marrow cells to differentiate into pro-B cells ex vivo (Supplemental Fig. S5A and B). Therefore, our data indicate loss of either Smarcal1 or Zranb3 alone is insufficient to alter B cell development or total B cell numbers in young, unstressed animals.
We next evaluated B cell populations in Myc overexpressing Smarcal1-deficient and Zranb3-deficient mice. There were no significant differences in developing B cell populations in the bone marrow of Smarcal1-deficient or Zranb3-deficient Eμ-myc mice (Supplemental Fig. S6A and B). In Smarcal1Δ/Δ Eμ-myc mice there was a trend towards decreased splenic leukocyte numbers that did not reach statistical significance (Fig. 5C), but they had 22–28% fewer mature B cells compared to either Smarcal1+/Δ or wild-type Eμ-myc littermates (Fig. 5D). Zranb3+/− and Zranb3−/− Eμ-myc mice had a significant reduction (18–22%) in splenic leukocytes (Fig. 5C) and mature B cells (Fig. 5D) compared to wild-type Eμ-myc mice. These data suggest complete loss of Smarcal1 and loss of only a single Zranb3 allele is sufficient to sensitize B cells to Myc overexpression, resulting in a reduction in B cells in Eμ-myc mice.
Smarcal1-deficient but not Zranb3-deficient Eμ-myc mice have fewer proliferating B cells.
Due to the function of Smarcal1 and Zranb3 at DNA replication forks, we investigated whether a deficiency in Smarcal1 or Zranb3 would alter cell cycle, particularly S-phase, with Myc overexpression. Evaluation of S-phase revealed that B cells from Smarcal1Δ/Δ Eμ-myc mice incorporated significantly less BrdU compared to wild-type and Smarcal1+/Δ Eμ-myc littermates (Fig. 5E). Investigation of cell cycle with propidium iodide also showed a reduction in the percentage of B cells in S-phase with a concomitant increase in the percentage in G0/G1 in Smarcal1Δ/Δ Eμ-myc mice compared to wild-type Eμ-myc littermates (Fig. 5E). In contrast, neither Zranb3+/− Eμ-myc nor Zranb3−/− Eμ-myc B cells had significant differences in BrdU incorporation compared to wild-type Eμ-myc littermates (Figs. 5F). Lack of either Smarcal1 or Zranb3 alone without Myc overexpression had no effect on B cell proliferation or BrdU incorporation (Supplemental Fig. S7A–F). Thus, loss of both Smarcal1 alleles, but not Zranb3 leads to decreased cell cycle progression in B cells overexpressing Myc.
Loss of Smarcal1 or Zranb3 sensitizes B cells to Myc-induced apoptosis.
Myc overexpression induces apoptosis in normal cells, which serves as a barrier to transformation (41). We evaluated susceptibility to Myc-induced apoptosis of Smarcal1 and Zranb3-deficient B cells. We assessed pro-B cells ex vivo with MycER, because apoptotic B cells in vivo are cleared quickly. Following MycER activation with 4-OHT in Smarcal1+/Δ and Smarcal1Δ/Δ B cells, there was a significant decrease in cell expansion (Fig 6A) that was attributable to reduced cell number and viability (Fig. 6B) and increased numbers of B cells that were Annexin-V positive (Figs. 6C), contained sub-G1 DNA (Figs. 6D), and had cleaved caspase 3 (Figs. 6E). The effects of Myc activation in Smarcal1+/Δ B cells was intermediate between wild-type and Smarcal1Δ/Δ B cells, indicating a degree of retained Smarcal1 function in the heterozygous cells. Loss of one or both alleles of Zranb3 resulted in significantly decreased B cell expansion (Fig. 6F), cell number, and viability (Fig. 6G) and increased Annexin-V positivity (Fig. 6H), sub-G1 DNA (Figs. 6I), and cleaved Caspase 3 (Fig. 6J) following MycER activation. In contrast to Smarcal1, the effects of MycER activation in Zranb3+/− B cells were similar to that in Zranb3−/− B cells. These results provide additional evidence that loss of a single Zranb3 allele is profoundly deleterious to B cells overexpressing Myc, whereas Myc overexpressing B cells are impacted, but can tolerate loss of one allele of Smarcal1 providing fertile ground for transformation.
Discussion
The DNA replication stress response is an essential mechanism employed by cells to protect replication forks and facilitate successful DNA synthesis (1–3,5). However, while significant strides have been made in understanding the biochemical aspects of this process (21,42–45), the biological importance and consequences of specific proteins that mediate fork reversal, particularly under physiological conditions, remained unknown. In this study, utilizing primary cells and mouse models, our data reveal that both Smarcal1 and Zranb3 are essential and non-redundant for responding to DNA replication stress and stabilizing replication forks during oncogene dysregulation. Biologically, we demonstrate in mice the consequence of loss of either Smarcal1 or Zranb3 profoundly altered Myc-driven B cell lymphomagenesis and revealed that gene dosage had a significant impact. Zranb3 haploinsufficiency inhibited Myc-induced lymphoma development and increased survival, whereas loss of one Smarcal1 allele accelerated lymphomagenesis and decreased survival. Our data reveal levels of Smarcal1 and Zranb3 have unique roles in stabilizing forks and preventing DNA breaks and apoptosis during oncogenic replication stress in vivo that was not previously known.
Prior to this current study, no endogenous sources of replication stress had been reported to require Zranb3, and telomeric sequences were the only known source of endogenous stress requiring Smarcal1 (22,23). Here, we identified Myc overexpression as an endogenous source of replication stress that required both Smarcal1 and Zranb3 for replication fork stability. Notably, our data indicate their functions are not redundant in that neither Smarcal1 nor Zranb3 could compensate for the other when one was lost in the presence of Myc overexpression. Moreover, the different biological results that were observed in Eμ-myc mice and B cells from them with a Smarcal1-deficiency compared to a Zranb3-deficiency indicate distinct functions for these proteins in resolving the type of replication stress caused by an oncogene. Thus, while loss of Smarcal1 or Zranb3 negatively impacted fork stability during oncogenic stress, they appear to have unique functions in promoting fork repair and restart during this particular type of stress that are essential to prevent DNA damage leading to tumorigenesis or apoptosis.
Previously, SMARCAL1 was shown to be required for replication fork restart and cell viability when human cancer cell lines are treated with genotoxic agents in vitro (11,12,15). However, our data reveal there are different requirements for Smarcal1 with different replication stresses in vivo. Specifically, we reported that loss of one or both Smarcal1 alleles inhibited gamma radiation-induced DNA replication stress-mediated T-cell lymphoma development (32). In this model, radiation stimulates a burst of hematopoietic stem and progenitor cell proliferation, acutely generating replication stress in these cells, resulting in T-cell lymphoma (46,47). In contrast, here we show in Eμ-myc mice, which have a constitutively increased level of Myc (3–4 fold) in B cells causing chronic replication stress in B cells, an acceleration of B-cell lymphomagenesis with loss of one Smarcal1 allele and no change in the rate of B cell lymphoma development in Smarcal1-null Eμ-myc mice. In the radiation model, loss of single allele of Smarcal1 was sufficient to confer profound sensitivity to hematopoietic cells to this acute replication stress, causing significant apoptosis of cycling cells (32). However, with Myc overexpression, Smarcal1 haploinsufficiency resulted in a mild cellular phenotype in B cells with a modest increase in apoptosis and small changes in replication fork stability and DNA damage, which resulted in increased B cell transformation. The significant differences in the biological effects in these two studies uncover DNA replication stress-specific and/or cellular context differences of Smarcal1. Future studies investigating the distinctions between acute and chronic replication stress would further define the precise physiological settings that require Smarcal1 for fork protection and begin to characterize the differences Zranb3 has in relationship to Smarcal1.
Studies investigating the contribution of Zranb3 in responding to different types of stress under various conditions in vitro have not provided a clear understanding of Zranb3. Previous studies showed in human cancer cell lines knockdown of ZRANB3 had increased rates of fork stalling and impaired fork restart upon hydroxyurea treatment and withdrawal, respectively (13,16). In contrast, knockout of ZRANB3 in a human cancer cell line led to increased rates of fork progression due to the abolishment of Zranb3-mediated of fork reversal following treatment with genotoxic agents (20). Our data in primary B cells show Myc-overexpressing Zranb3-null had impaired fork progression due to fork collapse and increased DNA damage. Notably, loss of a single Zranb3 allele was as deleterious to B cells as loss of both alleles when Myc was overexpressed. Thus, future studies are needed to determine whether specific cellular contexts, sources, and duration of replication stress require Zranb3 differently.
Overall, our data significantly advance understanding of the closely related proteins, Smarcal1 and Zranb3, by revealing their essential, non-redundant in vivo function in replication fork stability during oncogenic stress and their contribution to Myc-driven tumor development. In addition, our data suggest that Smarcal1 and Zranb3 are likely to be important in other Myc-driven malignancies, and could provide a therapeutic opportunity. Since Myc is overexpressed in at least 70% of human cancers, these malignancies may rely on Smarcal1 and Zranb3 to stabilize forks and complete replication. Developing inhibitors against these proteins, particularly Zranb3, could provide therapeutic benefit for Myc-driven malignancies. Additionally, synthetic lethal approaches using targeted inhibitors combined with replication-stress inducing drugs may be useful to treat Myc-driven malignancies. Further studies investigating these approaches are needed to determine the therapeutic efficacy of drugging fork remodelers in Myc-driven cancers.
Supplementary Material
Significance.
Smarcal1 and Zranb3 are essential, but non-redundant, for responding to DNA replication stress and stabilizing replication forks following MYC overexpression.
Acknowledgements
The authors thank Dr. David Cortez for helpful discussion and for the Smarcal1 antibody, Tiffany Aleman for assistance with mouse husbandry and genotyping, Shanequia Jackson for technical assistance, and members of the Eischen lab for helpful discussions. This work was supported by F30CA189433 (MVP), the Vanderbilt MSTP Training Grant T32GM007347 (MVP), R01CA226432 (CME), and the National Cancer Institute Cancer Center Grant P30CA056036 for supporting the Bioimaging, Flow Cytometry, and Lab Animals core facilities.
Financial Support: This work was supported by F30CA189433 (M. V. Puccetti), the Vanderbilt MSTP Training Grant T32GM007347 (M. V. Puccetti), R01CA226432 (C. M. Eischen), and the National Cancer Institute Cancer Center Grant P30CA056036 for supporting C. M. Eischen and the Bioimaging, Flow Cytometry, and Lab Animals core facilities.
Footnotes
Conflict of Interest Statement: The authors have no conflicts of interest.
References
- 1.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 2005;434:864–70 [DOI] [PubMed] [Google Scholar]
- 2.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 2005;434:907–13 [DOI] [PubMed] [Google Scholar]
- 3.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science 2008;319:1352–5 [DOI] [PubMed] [Google Scholar]
- 4.Macheret M, Halazonetis TD. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 2018;555:112–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol 2014;16:2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaillard H, Garcia-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer 2015;15:276–89 [DOI] [PubMed] [Google Scholar]
- 7.Kotsantis P, Petermann E, Boulton SJ. Mechanisms of Oncogene-Induced Replication Stress: Jigsaw Falling into Place. Cancer Discov 2018;8:537–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 2010;11:683–7 [DOI] [PubMed] [Google Scholar]
- 9.Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol 2013;5:a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poole LA, Cortez D. Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit Rev Biochem Mol Biol 2017;52:696–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev 2009;23:2405–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, et al. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev 2009;23:2415–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, et al. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell 2012;47:396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Postow L, Woo EM, Chait BT, Funabiki H. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem 2009;284:35951–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev 2009;23:2394–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan J, Ghosal G, Chen J. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol Cell 2012;47:410–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science 2008;322:748–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yusufzai T, Kadonaga JT. Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif. Proc Natl Acad Sci U S A 2010;107:20970–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yusufzai T, Kong X, Yokomori K, Kadonaga JT. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev 2009;23:2400–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vujanovic M, Krietsch J, Raso MC, Terraneo N, Zellweger R, Schmid JA, et al. Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity. Mol Cell 2017;67:882–90e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betous R, Couch FB, Mason AC, Eichman BF, Manosas M, Cortez D. Substrate-selective repair and restart of replication forks by DNA translocases. Cell Rep 2013;3:1958–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox KE, Marechal A, Flynn RL. SMARCAL1 Resolves Replication Stress at ALT Telomeres. Cell Rep 2016;14:1032–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole LA, Zhao R, Glick GG, Lovejoy CA, Eischen CM, Cortez D. SMARCAL1 maintains telomere integrity during DNA replication. Proc Natl Acad Sci U S A 2015;112:14864–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985;318:533–8 [DOI] [PubMed] [Google Scholar]
- 25.Adams CM, Hiebert SW, Eischen CM. Myc Induces miRNA-Mediated Apoptosis in Response to HDAC Inhibition in Hematologic Malignancies. Cancer Res 2016;76:736–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alt JR, Greiner TC, Cleveland JL, Eischen CM. Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. EMBO J 2003;22:1442–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arrate MP, Vincent T, Odvody J, Kar R, Jones SN, Eischen CM. MicroRNA biogenesis is required for Myc-induced B-cell lymphoma development and survival. Cancer Res 2010;70:6083–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams CM, Eischen CM. Inactivation of p53 is insufficient to allow B cells and B-cell lymphomas to survive without Dicer. Cancer Res 2014;74:3923–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grieb BC, Gramling MW, Arrate MP, Chen X, Beauparlant SL, Haines DS, et al. Oncogenic protein MTBP interacts with MYC to promote tumorigenesis. Cancer Res 2014;74:3591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feeley KP, Adams CM, Mitra R, Eischen CM. Mdm2 Is Required for Survival and Growth of p53-Deficient Cancer Cells. Cancer Res 2017;77:3823–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 1998;12:2424–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puccetti MV, Fischer MA, Arrate MP, Boyd KL, Duszynski RJ, Betous R, et al. Defective replication stress response inhibits lymphomagenesis and impairs lymphocyte reconstitution. Oncogene 2017;36:2553–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alt JR, Bouska A, Fernandez MR, Cerny RL, Xiao H, Eischen CM. Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J Biol Chem 2005;280:18771–81 [DOI] [PubMed] [Google Scholar]
- 34.Bouska A, Lushnikova T, Plaza S, Eischen CM. Mdm2 promotes genetic instability and transformation independent of p53. Mol Cell Biol 2008;28:4862–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maya-Mendoza A, Ostrakova J, Kosar M, Hall A, Duskova P, Mistrik M, et al. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol Oncol 2015;9:601–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohban S, Campaner S. Myc induced replicative stress response: How to cope with it and exploit it. Biochim Biophys Acta 2015;1849:517–24 [DOI] [PubMed] [Google Scholar]
- 37.Srinivasan SV, Dominguez-Sola D, Wang LC, Hyrien O, Gautier J. Cdc45 is a critical effector of myc-dependent DNA replication stress. Cell Rep 2013;3:1629–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell 2002;9:1031–44 [DOI] [PubMed] [Google Scholar]
- 39.Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med 1988;167:353–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res 1995;23:1686–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMahon SB. MYC and the control of apoptosis. Cold Spring Harb Perspect Med 2014;4:a014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol 2015;16:207–20 [DOI] [PubMed] [Google Scholar]
- 43.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 2002;297:599–602 [DOI] [PubMed] [Google Scholar]
- 44.Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, et al. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol 2015;208:563–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell 2007;28:167–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaplan HS. The Role of Radiation on Experimental Leukemogenesis. Natl Cancer Inst Monogr 1964;14:207–20 [PubMed] [Google Scholar]
- 47.Kaplan HS, Brown MB. A quantitative dose-response study of lymphoid-tumor development in irradiated C 57 black mice. J Natl Cancer Inst 1952;13:185–208 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.