Biofilms are multicellular bacterial communities encased in a self-secreted extracellular matrix comprised of polysaccharides, proteinaceous fibers, and DNA. Organization of these components lends spatial organization in the biofilm community. Here we demonstrate that oxygen gradients in uropathogenic Escherichia coli (UPEC) biofilms lead to spatially distinct expression programs for quinol oxidases—components of the terminal electron transport chain. Our studies reveal that the cytochrome bd-expressing subpopulation is critical for biofilm development and matrix production. In addition, we show that quinol oxidases are heterogeneously expressed in planktonic populations and that this respiratory heterogeneity provides a fitness advantage during infection. These studies define the contributions of quinol oxidases to biofilm physiology and suggest the presence of respiratory bet-hedging behavior in UPEC.
KEYWORDS: Escherichia coli, biofilms, heterogeneity, oxygen gradients, respiration, urinary tract infection
ABSTRACT
Biofilms are multicellular bacterial communities encased in a self-secreted extracellular matrix comprised of polysaccharides, proteinaceous fibers, and DNA. Organization of these components lends spatial organization to the biofilm community such that biofilm residents can benefit from the production of common goods while being protected from exogenous insults. Spatial organization is driven by the presence of chemical gradients, such as oxygen. Here we show that two quinol oxidases found in Escherichia coli and other bacteria organize along the biofilm oxygen gradient and that this spatially coordinated expression controls architectural integrity. Cytochrome bd, a high-affinity quinol oxidase required for aerobic respiration under hypoxic conditions, is the most abundantly expressed respiratory complex in the biofilm community. Depletion of the cytochrome bd-expressing subpopulation compromises biofilm complexity by reducing the abundance of secreted extracellular matrix as well as increasing cellular sensitivity to exogenous stresses. Interrogation of the distribution of quinol oxidases in the planktonic state revealed that ∼15% of the population expresses cytochrome bd at atmospheric oxygen concentration, and this population dominates during acute urinary tract infection. These data point toward a bet-hedging mechanism in which heterogeneous expression of respiratory complexes ensures respiratory plasticity of E. coli across diverse host niches.
INTRODUCTION
Rather than existing as phenotypically uniform populations, bacterial colonies, biofilms, and cultures are characterized by subpopulations that are often phenotypically distinct. This intrastrain heterogeneity can be irreversible, through the acquisition of mutations, or transient and reversible if it is brought about by stochastic differences in the abundance and activity of regulators in each individual cell or by metabolic adaptation to local environmental conditions. Heterogeneity often confers a survival advantage to the population by allowing at least portion of the population to survive in a given niche.
In biofilms, bacteria assemble in an organized fashion in three-dimensional space. One of the most critical features of biofilms is a self-secreted extracellular matrix (ECM) that comprises a variety of exopolysaccharides, proteinaceous fibers, and extracellular DNA (1). The ECM protects the biofilm residents from predation, desiccation, assault by antimicrobial agents, and—when biofilms form in the host—the immune system. In addition to providing a physical barrier against external threats, the ECM also serves as a barrier to diffusion. Restricted diffusion, in conjunction with the metabolic activity of resident bacteria, leads to the establishment of various chemical gradients throughout the biofilm community (2, 3). Bacteria at different locales along the gradient respond to the microenvironment differently and as a result differentiate into distinct and often metabolically cooperative subpopulations (2–6). Previous studies in Pseudomonas aeruginosa and Escherichia coli indicated that oxygen gradients play a key role in regulating the differential expression of genes involved in biofilm formation and metabolic specialization (7–10). The presence of an oxygen gradient suggests the emergence of subpopulations that utilize different respiratory components as a function of the oxygen abundance to which they are exposed. This leads to the hypothesis that the metabolic programs of differentially respiring subpopulations are distinct from one another and contribute to differential production of biofilm goods that in turn enhance biofilm resilience.
Escherichia coli is a facultative anaerobe capable of utilizing multiple metabolic pathways to fulfill its energy requirements. In aerobically respiring E. coli, quinol oxidases comprise essential components of the terminal electron transport chain that couple the flow of electrons to the reduction of molecular oxygen into water (11, 12). E. coli encodes two classes of quinol oxidases with differing oxygen affinities: one heme copper oxidase, cytochrome bo (encoded by the cyoABCD gene cluster), and two bd-type oxidases, cytochromes bd (cydABX) and bd2 (appBC) (12, 13). Studies in K-12 E. coli indicated that cytochrome bo is induced at high (atmospheric, 21%) oxygen tensions, whereas the bd-type oxidases are induced at low (hypoxic, 2 to 15%) oxygen tensions (12, 14). Cytochromes bd and bd2 have approximately 60% amino acid identity, similar spectral properties, and indistinguishable reaction mechanisms (13, 15). Based on the in vitro expression patterns of these quinol oxidases, we hypothesized that cytochrome bo would be enriched on the air-exposed biofilm surface, whereas cytochromes bd and bd2 would be enriched in the hypoxic interior.
Here we report that the spatial distribution of quinol oxidases in biofilms formed by uropathogenic Escherichia coli (UPEC) is a fundamental driver of biofilm architecture. Peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) analyses assigned locations to each quinol oxidase-producing subpopulation, elucidating for the first time spatially distinct expression programs for respiratory oxidases in E. coli. Depletion of the cytochrome bd-expressing subpopulation from the biofilm significantly impaired diffusion resistance by altering the abundance and organization of the ECM. Assessment of deletion mutants in a well-established urinary tract infection murine model revealed that only the cytochrome bd mutant was significantly attenuated for virulence, although the infecting pool of bacteria in the parent strain exhibited heterogeneous expression of all three respiratory oxidases. In situ analysis of urine-associated bacteria demonstrated a shift of the population to cytochrome bd expression, suggesting that the bladder favors cytochrome bd-expressing bacteria and that heterogeneity in the input pool provides a fitness advantage to uropathogenic strains. Our studies, performed on one of the most commonly acquired human pathogens and a prolific biofilm producer in vivo, unveil a potential avenue for targeting heterogeneity and homogenizing bacterial programming as a therapeutic approach.
RESULTS
Cytochrome bd is the most abundant respiratory transcript in mature UPEC biofilms.
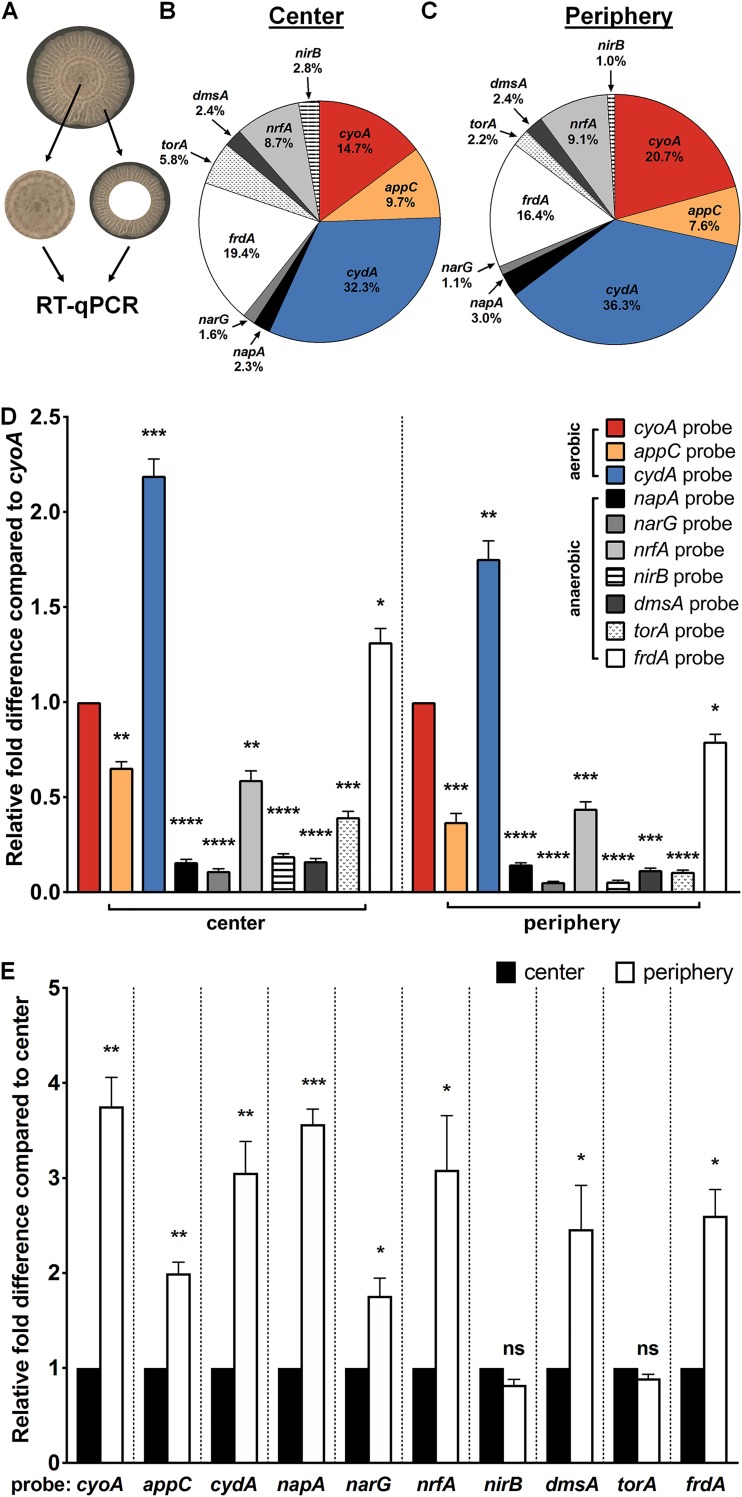
In previous studies we reported spatial organization of proteins as a function of oxygen gradients in surface-associated UPEC biofilms formed at the air-liquid interface of yeast extract-Casamino Acids medium (YESCA) (9), which induces expression of key matrix components (curli amyloid fibers, type 1 pili, secreted proteins Hu-α/β, and cellulose) that are also critical for fitness in the urinary tract (9, 16–18). We then went on to show that biofilm formation is greatly diminished under anaerobic conditions, despite the addition of alternative terminal electron acceptors used by E. coli for anaerobic respiration and irrespective of growth medium (19). Given that aerobic respiration is critical for UPEC colonization and the establishment of intracellular biofilms during bladder infection (20–22), we sought to determine the relative expression of aerobic and anaerobic respiratory machineries in mature colony biofilms formed under aerobic conditions on YESCA agar without supplementation of alternative terminal electron acceptors. Under these growth conditions, UPEC forms elaborate rugose-colony biofilms (Fig. 1A) that quickly establish an oxygen gradient from the surface to the interior of the biofilm (8). Previous studies demonstrated the presence of a matrix-rich region and a matrix-devoid region in colony biofilms imaged at 48 h and 6 days post-inoculation (8, 23–25) and additionally revealed distinct spatial expression of regulators at the growing edge versus the center of the biofilm (23). To investigate the role of quinol oxidases in biofilms, we first used RT-qPCR to measure the steady-state transcript at the growing edge (periphery) and the center of the colony (Fig. 1; see also Fig. S1 in the supplemental material). Although overall transcript abundance was significantly increased in the periphery relative to the center (Fig. 1E), consistent with the notion that cells at the periphery are more metabolically active, we observed a similar distribution of transcript at the center of the biofilm and the growing edge (Fig. 1B to D). Consistent with previous studies demonstrating the importance of aerobic respiration in UPEC biofilms, the majority of detected transcript corresponded to aerobic respiratory components (Fig. 1B and C). The most abundant transcript was that of cydA (Fig. 1B to D and Fig. S1), corresponding to cytochrome bd complex (cydABX) expression. The cydA transcript levels were approximately 2-fold higher than those corresponding to cyoABCD (Fig. 1B to D), which was the second most highly abundant oxidase under the conditions tested. Although most anaerobic respiratory operons exhibited baseline expression levels, we detected high levels of transcript corresponding to fumarate reductase (frdA) and periplasmic nitrite reductase (nrfA) (Fig. 1B to D and Fig. S1). These results reveal the presence of marked respiratory heterogeneity within UPEC biofilm communities and suggest that respiration via cytochrome bd may be preferred.
FIG 1.
Lateral expression of respiratory complexes in Escherichia coli biofilms. (A) Image of a mature colony biofilm formed by UPEC strain UTI89 on YESCA agar without supplementation of alternative terminal electron acceptors. The center and periphery of colony biofilms, including both the surface and interior of each region, were harvested and subjected to RNA extraction and RT-qPCR using probes targeting each respiratory operon present in UPEC. (B and C) Pie charts indicating the relative abundance of detected respiratory transcripts in the biofilm center (B) and periphery (C). Aerobic respiratory operons are presented in color, whereas anaerobic respiratory operons are presented in grayscale. (D) Graph depicting relative fold differences in respiratory transcript abundance in the biofilm center and periphery compared to cyoA abundance in the same region. (E) Graph depicting relative fold difference in abundance of each transcript in the biofilm periphery compared to abundance of the same transcript in the biofilm center. The graphs and pie charts depict the average from four biological replicates. Statistical analysis was performed in GraphPad Prism using a two-tailed paired t test. Data are presented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Expression of respiratory complexes compared to gyrB. (A) Graph depicting relative abundance of each respiratory transcript in the center and periphery of day 11 colony biofilms compared to gyrB. (B) Average CT values for each transcript in the center and periphery of colony biofilms. Data are presented as mean ± SEM. All data are representative of four biological replicates. Download FIG S1, TIF file, 0.9 MB (995.9KB, tif) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Spatial organization of quinol oxidase expression along the oxygen gradient.
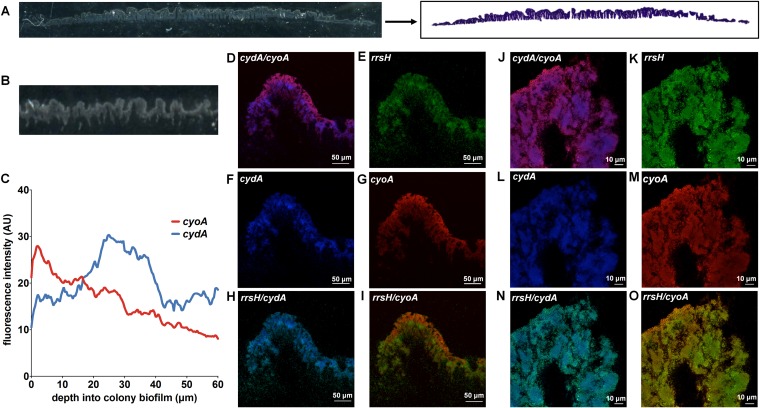
To define the spatial distribution of quinol oxidase-expressing subpopulations, we performed PNA-FISH on biofilm cryosections of mature colony biofilms using probes targeting each quinol oxidase operon (cyoA, appC, and cydA) as well as rrsH as an endogenous control (Fig. 2). Because cryosectioning captures both macroscopic and microscopic architecture of biofilms with minimal disruption to the overall structure or organization of the resident bacteria, this approach allows us to define the in situ distribution of transcripts in unperturbed communities (Fig. 2A and B). Each PNA-FISH probe was designed using the validated probe sequences used for qPCR (Table S1) to ensure comparable hybridization efficiencies for each probe. Specificity of each probe was confirmed using RT-qPCR (see Fig. 4H) and through staining of planktonic cells (Fig. S2). SYTO 9 staining of sections was used as an additional control to localize the entire biofilm community and account for possible hybridization inconsistencies with the rrsH control probe (Fig. 2E and K and Fig. S3). To account for possible mislocalization of signal due to biofilm breakage during the cryosectioning and staining procedure, we focused our analysis on regions devoid of significant breaks in the cryosection. Consistent with previous observations demonstrating that the highest oxygen abundance is at the air-exposed surface of the biomass (7, 8), we observed that cyoABCD transcript was most abundant in bacteria lining air-exposed surfaces of the biofilm (Fig. 2C, D, G, I, J, M, and O). In contrast, the highest abundance of cydABX transcript was found in densely packed clusters of bacteria in the interior of the biofilm (Fig. 2C, D, F, H, J, L, and N). Although cytochromes bd and bd2 are both induced under oxygen-limited conditions (13, 26), we observe different transcript distribution for these two gene clusters. Rather than organizing along the oxygen gradient, appBC transcript was observed to be evenly distributed throughout the community (Fig. S3). Interestingly, we also observe basal expression of cytochrome bo across the community with enrichment of cytochrome bd in pockets of cells in the interior (Fig. 2J to O), suggesting that individual cells may express multiple quinol oxidases simultaneously within biofilms. Additionally, while many biofilm wrinkles are empty or sparsely populated with rrsH-staining cells, we observe other wrinkles that are densely populated (Fig. S4). We observe reduced intensity of quinol oxidase staining in the interior of those populated wrinkles (Fig. S4), suggesting that—consistent with previous reports (7, 27)—respiration in the deeper layers of the biofilm respiration occurs anaerobically. Based on the qPCR results (Fig. 1B to D), respiration in the populated wrinkles may be occurring via fumarate reductase or periplasmic nitrite reductase. Here we focused our studies on understanding the contribution of cytochrome bd to biofilm architecture.
FIG 2.
Expression of quinol oxidases as a function of the oxygen gradient. (A) Representative images depicting a biofilm cryosection before (left) and after (right) fixation and crystal violet staining. (B) Magnified image of a cryosection allows visualization of architectural features of the biofilm. (C) Fluorescence intensity of cyoA and cydA PNA-FISH probes was quantified on ImageJ. Data are presented as the average fluorescence intensity as a function of depth obtained from four images, each with five measurements per image. (D to O) Representative images of PNA-FISH-stained biofilm cryosections at ×20 magnification (D to I) and ×63 magnification (J to O). Cryosections were stained with PNA-FISH probes targeting cyoA and cydA with rrsH (16S rRNA) as an endogenous control. Images are representative of three biological replicates.
PNA-FISH on quinol oxidase deletion mutants. Representative images of ΔcydAB (A to D) and ΔcyoAB (E to H) cells stained with PNA-FISH probes to assess specificity. Download FIG S2, TIF file, 2.0 MB (2.1MB, tif) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Localization of appC transcript in biofilm cryosections. (A) Representative image of SYTO 9-stained biofilm cryosection at ×20 magnification. (B and C) Representative images of biofilm cryosections stained with an appC PNA-FISH probe at ×20 (B) and ×63 (C) magnification. Download FIG S3, JPG file, 0.4 MB (384.7KB, jpg) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Localization of quinol oxidase transcripts in biofilm wrinkles. Representative image of a PNA-FISH-stained cryosection depicting a biofilm wrinkle with a central region filled with rrsH-stained cells at ×20 magnification. Download FIG S4, JPG file, 2.4 MB (2.5MB, jpg) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers and probes. Download Table S1, DOCX file, 0.1 MB (104.4KB, docx) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Loss of cytochrome bd alters biofilm architecture, development, and ECM abundance.
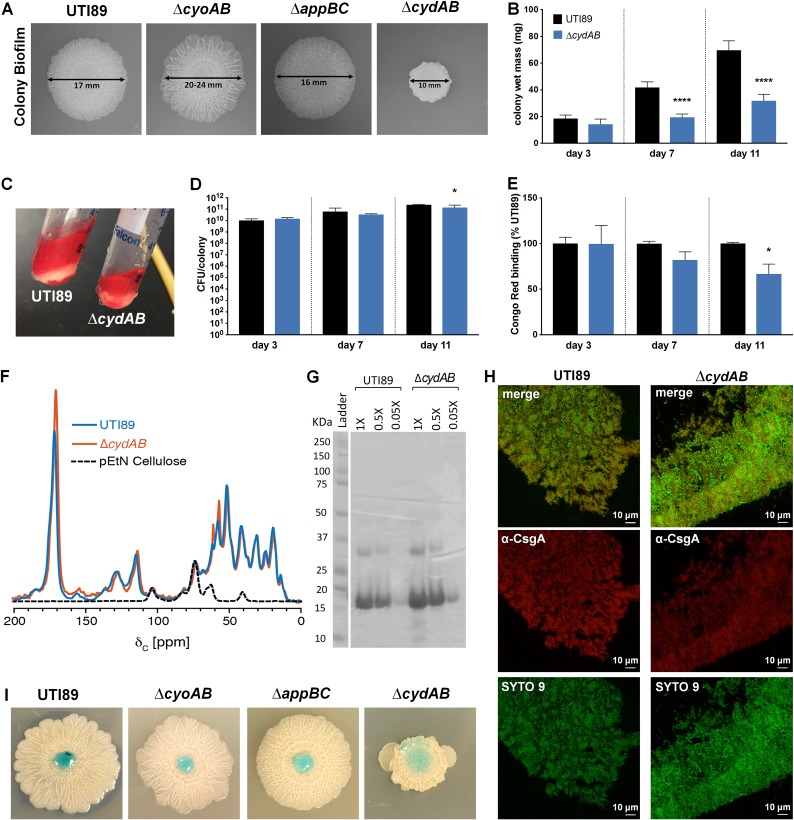
The highly ordered spatial organization of cytochrome bo and cytochrome bd in the biofilm raised the hypothesis that each of these quinol oxidase-expressing subpopulations uniquely contributes to overall biofilm architecture. To test this hypothesis, we created isogenic deletion mutants lacking the cyoAB, appBC, or cydAB genes and compared the biofilms formed by the resulting strains (Fig. 3A). Colony biofilms formed by the parental strain expand to an average diameter of 16.8 mm over an 11-day incubation period and exhibit elaborate rugose architecture with distinct central and peripheral regions (Fig. 3A and Fig. S5). Strains deleted for cyoAB and appBC exhibited inverse phenotypes to each other, with the ΔcyoAB colony biofilms expanding more than the parental strain (average diameter, 19.9 mm) and the ΔappBC colony biofilms appearing more compact and with apparently higher rugosity (Fig. 3A and Fig. S5). Strikingly, while ΔcyoAB and ΔappBC colony biofilms displayed only minor architectural changes, ΔcydAB colony biofilms exhibited pronounced defects in both development and architecture (Fig. 3A and Fig. S5). Colony biofilms from all strains grew at similar rates for the first 72 h (Fig. S5). However, ΔcydAB colony growth was significantly stunted between days 3 and 11, with radial expansion remaining at an average diameter of 10.3 mm and colonies exhibiting a wet mass approximately 50% of the parental strain after 7 days of growth, even though the CFU produced by the two strains were comparable (Fig. 3B and D and Fig. S5). Complementation of the ΔcydAB strain with an extrachromosomal construct expressing cydABX under its native promoter rescued the deletion phenotype, indicating that the defects observed in the ΔcydAB mutant stem solely from the removal of the cydABX cluster (Fig. S7). Furthermore, deletion of both cyoAB and appBC from the same strain led to an early onset of rugose phenotype (Fig. S6). Together, these results demonstrate that cytochrome bd is a key contributor to biofilm development and suggest that loss of cydAB alters the synthesis and organization of the ECM.
FIG 3.
Cytochrome bd organizes biofilm architecture and ECM production. (A) Colony biofilms of UTI89 and quinol oxidase mutants grown on YESCA agar for 11 days. Images are representative of at least 30 biological replicates. (B) Graph depicting wet mass of individual colony biofilms at days 3, 7, and 11 of growth. Data are the average from five biological replicates per day. Data are presented as mean ± SD. (C) Image depicting gross changes to extracellular matrix (ECM) abundance between UTI89 and ΔcydAB colony biofilms. ECM is stained red by the presence of Congo red in the growth medium. (D) CFU per colony biofilm was measured at days 3, 7, and 11 of growth. Data are presented as mean ± SD. Data are representative of five biological replicates. (E) Congo red binding as a percentage of binding in UTI89. Data are presented as mean ± SEM. (F) Solid-state NMR spectra of the ECM of UTI89 (blue), ΔcydAB strain (orange), and isolated pEtN cellulose (black). (G) SDS-PAGE gel of UTI89 and ΔcydAB ECM. ECM was treated with 98% formic acid and vacuum centrifuged prior to analysis to dissociate curli amyloid fibers. (H) Immunofluorescence images of curli (α-CsgA, red) localization in UTI89 and ΔcydAB colony biofilm cryosections. (I) Colored water droplets were added to the top of day 11 colony biofilms to probe biofilm barrier function. All statistical analysis was performed in GraphPad Prism using a two-tailed unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Temporal development of colony biofilms by UTI89 and quinol oxidase mutants. (A) Representative images of UTI89 and quinol oxidase mutant colony biofilms grown on YESCA agar taken on days 3, 7, and 11 of growth. (B) Graph depicting colony biofilm diameter at days 3, 7, and 11 of growth. Each triangle represents an individual colony biofilm. Data are representative of at least 30 biological replicates. Statistical analysis was performed in GraphPad Prism using Welch’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Download FIG S5, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of ΔappBC ΔcyoAB::kanR colony biofilms. Comparison of colony biofilms formed by UTI89, ΔcyoAB, ΔappBC, ΔappBC ΔcyoAB::kanR, and ΔcydAB strains after 6 days of growth on YESCA agar. Images are representative of five biological replicates. Download FIG S6, TIF file, 2.3 MB (2.3MB, tif) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Extrachromosomal complementation of ΔcydAB rescues biofilm defects. Representative images on UTI89_pTRC99a, ΔcydAB_pTRC99a, and complemented ΔcydAB_pCydABX strains under the control of a native promoter. Images were taken of colony biofilms grown on YESCA agar at days 3, 7, and 11 of growth. Images are representative of at least five biological replicates. Download FIG S7, JPG file, 0.2 MB (251.4KB, jpg) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Under the conditions used, the ECM of E. coli comprises primarily cellulose and curli amyloid fibers (28). Previous solid-state nuclear magnetic resonance (NMR) spectroscopy analyses on intact ECM material defined the contributions of cellulose and curli to the E. coli biofilm ECM and determined that curli and cellulose are present in a 6-to-1 ratio (28). More recently, the ECM cellulose was determined to be a chemically modified form of cellulose, specifically phosphoethanolamine (pEtN) cellulose (29). To interrogate the effects of cydAB deletion on curli and exopolysaccharide production, we extracted ECM and performed solid-state NMR analysis to evaluate the abundance of curli and cellulose components (Fig. 3F and G). The NMR spectra obtained for the parent and ΔcydAB ECM are very similar overall, indicating a comparable protein-to-polysaccharide ratio between the samples (Fig. 3F). Consistent with this analysis, we observe no change in protein composition between the parent and ΔcydAB ECM samples when analyzed on SDS-PAGE gels (Fig. 3G). We additionally do not observe any overt alterations to curli abundance or localization between UTI89 and ΔcydAB biofilm cryosections using immunofluorescence (Fig. 3H). Despite the similar composition, the total amount of ECM recovered was reduced in the ΔcydAB biofilm, indicative of a decrease in ECM production. When quantified by Congo red depletion assays, ΔcydAB colony biofilms exhibited a trend toward reduced total ECM abundance at 7 days (82.1% of parental value) and significantly reduced abundance at 11 days (66.6% of parental value) (Fig. 3E), which could be the result of reduced CFU at the 11-day time point. Because the protein-to-polysaccharide ratio and curli abundance are unchanged between the parent and ΔcydAB biofilms, these data are suggestive of a change to the overall mixture of matrix components in ΔcydAB biofilm, with particular reductions in the abundance of non-curli and non-pEtN cellulose ECM components.
The ECM plays a central role in biofilm physiology by providing physical protection against exogenous insults, serving as a structural scaffold, and helping to establish chemical gradients which lead to metabolic differentiation and subpopulation formation (2, 3, 16, 30). As such, disruptions to the matrix can have catastrophic consequences for the biofilm community. We hypothesized that the altered matrix abundance and architecture in the ΔcydAB mutant would render the biofilm more susceptible to exogenous insults. To investigate this possibility, we probed the barrier function of the quinol oxidase mutant biofilms by applying a drop of colored water to the surface of mature colony biofilms (Fig. 3I) (31). While the parent strain, ΔcyoAB, and ΔappBC biofilms repelled the drop, the solution readily penetrated ΔcydAB biofilms, demonstrating that the alterations to ΔcydAB biofilm architecture and matrix abundance increase penetrance of aqueous solutions.
Loss of cytochrome bd increases population sensitivity to nitrosative stress under ambient oxygen concentrations.
Together, our studies indicate that cytochrome bd is highly expressed in biofilms and that loss of the cytochrome bd-expressing subpopulation impairs barrier function and reduces the abundance of extracellular matrix. These data suggest that the cytochrome bd-expressing subpopulation plays a critical role in promoting ECM synthesis and providing structural integrity to the community. However, it is also possible that cytochrome bd is preferentially expressed in the biofilm because cytochrome bd provides protection against oxidative and nitrosative stress—by-products of biofilm metabolism (32, 33) and, in the case of infection, components of the innate immune response (34–36). In addition to functioning as a respiratory quinol:O2 oxidoreductase, previous studies demonstrated that cytochrome bd has catalase activity, is capable of oxidizing the respiratory inhibitor nitric oxide, and is insensitive to nitrosative stress due to its unusually high nitric oxide dissociation rate (34). These biochemical activities are thought to occur at unique locations on the protein; quinol oxidation occurs at the periplasmic Q loop, oxygen reduction and nitric oxide binding occur at heme d, and catalase activity is thought to occur through heme b595 (13, 37, 38). In contrast, cytochrome bo affords no protection against nitrosative stress and is irreversibly inhibited by nitric oxide (34).
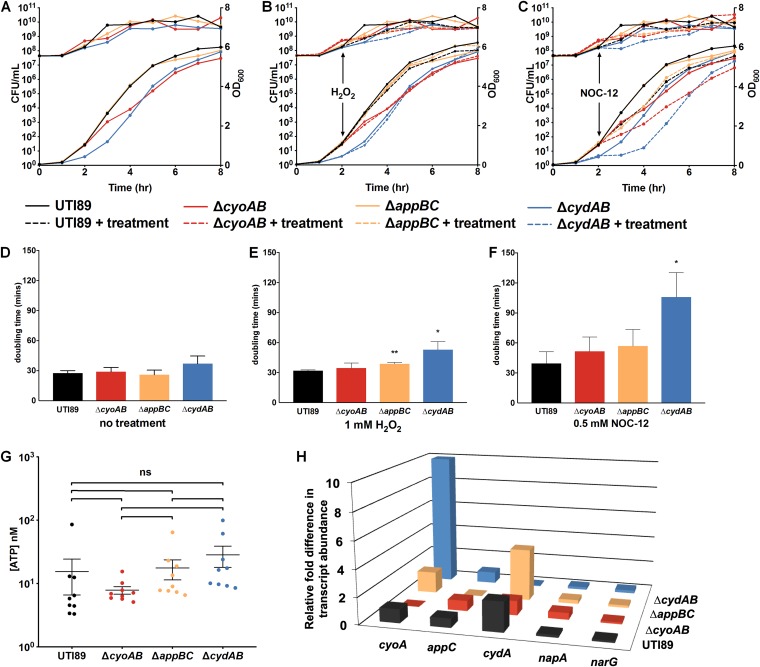
Given these additional functions of cytochrome bd, we performed growth curves at ambient oxygen concentration and evaluated the effects of nitrosative and oxidative stress on the fitness of cells lacking each quinol oxidase compared to the parental strain. Without the addition of stressors, both ΔcydAB and ΔcyoAB mutants exhibited a delay in growth, but growth of the ΔappBC strain closely mirrored the parental strain (Fig. 4A and D). Despite the delay, all strains reached similar maximal CFU/ml by the end of the experiment (Fig. 4A). ATP measurements of normalized samples taken from each strain during logarithmic phase revealed no significant overall differences in ATP concentrations (Fig. 4G). Next, to determine whether loss of cytochrome bd impairs resistance to oxidative and nitrosative stress, we measured growth with and without these stressors. Consistent with the reported catalase activity of cytochrome bd, significant increases in the doubling time of both ΔcydAB and ΔappBC strains were observed after treatment with 1 mM H2O2 (Fig. 4B and E). Although previous studies in K-12 E. coli demonstrated that treatment with 1 mM H2O2 reduced the growth rate of the ΔcyoAB strain by ∼70% relative to wild type (39), we did not observe significant reductions in growth rate of the ΔcyoAB strain after treatment (Fig. 4B and E). Addition of the nitric oxide donor NOC-12 to planktonic cultures induced an apparent growth delay in all strains but only significantly reduced the growth rate of the ΔcydAB mutant (Fig. 4C and F). Whereas treatment with NOC-12 increased the doubling time from 27 to 39 min in UTI89, in the ΔcydAB strain the doubling time increased from 37 to 106 min after treatment (Fig. 4F). Together, these data demonstrate that although cytochrome bd is dispensable for energy generation during planktonic growth, loss of cytochrome bd sensitizes bacteria to oxidative and nitrosative stress, consistent with previous studies on K-12 E. coli and the multidrug-resistant strain ST131 (35, 36).
FIG 4.
Cytochrome bd provides nitrosative stress resistance. (A to C) Growth curves for UTI89, ΔcyoAB, ΔappBC, and ΔcydAB strains as measured by CFU-per-ml (upper lines, left axis) and OD600 (lower lines, right axis) with no treatment (A), treatment with 1 mM hydrogen peroxide (B), or treatment with 0.5 mM nitric oxide donor NOC-12 (C). (D to F) Doubling time in minutes of each strain between hours 2 and 4 was calculated using CFU-per-ml data shown in panels A to C. (D) No treatment. (E) Treatment with 1 mM hydrogen peroxide. (F) Treatment with 0.5 mM NOC-12. (G) ATP levels measured from logarithmic cultures of each strain normalized to OD600 = 0.5. (H) RT-qPCR data depicting relative fold difference in respiratory transcript abundance in the center of day 11 colony biofilms in UTI89 and quinol oxidase mutant strains. In UTI89 (black), data are presented as relative fold difference in abundance of each transcript compared to cyoA abundance. In each mutant strain, data are presented as relative fold difference in transcript abundance compared to the abundance of the same transcript in UTI89. Statistical analysis was performed on GraphPad Prism using a two-tailed unpaired t test. All data are presented as mean ± SEM and are representative of at least three biological replicates.
While there was a trend toward increased doubling time in all strains after treatment with NOC-12, treatment of the ΔcydAB strain increased doubling time approximately 3-fold relative to its untreated control. This observation suggests that during aerobic growth cytochrome bd serves as an NO sink that reversibly sequesters NO and protects the more efficient cytochrome bo-mediated respiration. Accordingly, loss of cytochrome bd would decrease nitrosative stress resistance and render the dominant respiratory complex, cytochrome bo, susceptible to irreversible inhibition by NO. As such, treatment of the ΔcydAB strain with NO would poison all preformed cytochrome bo complexes in the membrane and force the bacteria to synthesize new oxidases prior to resuming growth. Consistent with this hypothesis, we observe a marked increase (∼10-fold relative to UTI89) of cyoABCD transcript in the interior of ΔcydAB colony biofilms, where NO is expected to be most abundant (Fig. 4H). These results contrast with previous studies in K-12 E. coli, in which loss of cytochrome bd induces a marked upregulation of appBC (26). These observations demonstrate that the regulation of quinol oxidases in UPEC is distinct from that previously defined in K-12 and suggest that cytochrome bd may serve as an NO sink in biofilms. In conjunction with the disrupted biofilm architecture and altered ECM abundance in ΔcydAB biofilms, these data suggest that cytochrome bd-expressing subpopulations are critical, not only for directing ECM biosynthesis but also for withstanding harmful metabolic by-products while in the biofilm state.
Heterogeneous expression of quinol oxidases at the population level.
Our data thus far indicate that in addition to heterogeneity in quinol oxidase expression in the biofilm state, heterogeneous expression of quinol oxidases must also be occurring in the planktonic population. Our planktonic studies revealed a lag in growth of the ΔcyoAB and the ΔcydAB mutants when these strains were grown under ambient oxygen concentrations, suggesting that in a given culture there are subpopulations—like in the biofilm—that stochastically or deterministically express different respiratory components. Such a bet-hedging approach could provide UPEC with the flexibility to quickly adapt to a given niche, be it different locales in the genitourinary tract or in the gastrointestinal tract during host colonization. In the context of urinary tract infection, E. coli traverses from the nearly anoxic gut to the perineum, where it encounters atmospheric oxygen concentrations, prior to ascending the urethra to enter the hypoxic bladder, where the dissolved urinary oxygen concentration is 4 to 6% (40). This microbial journey is performed by planktonic cells, which can then expand into multicellular communities on and within bladder epithelial cells, as well as on urinary catheters (1, 41). In previous studies, we and others demonstrated that UPEC respire aerobically during infection (20–22) and that biofilm formation is favored under conditions that mimic oxygen levels in the bladder (19).
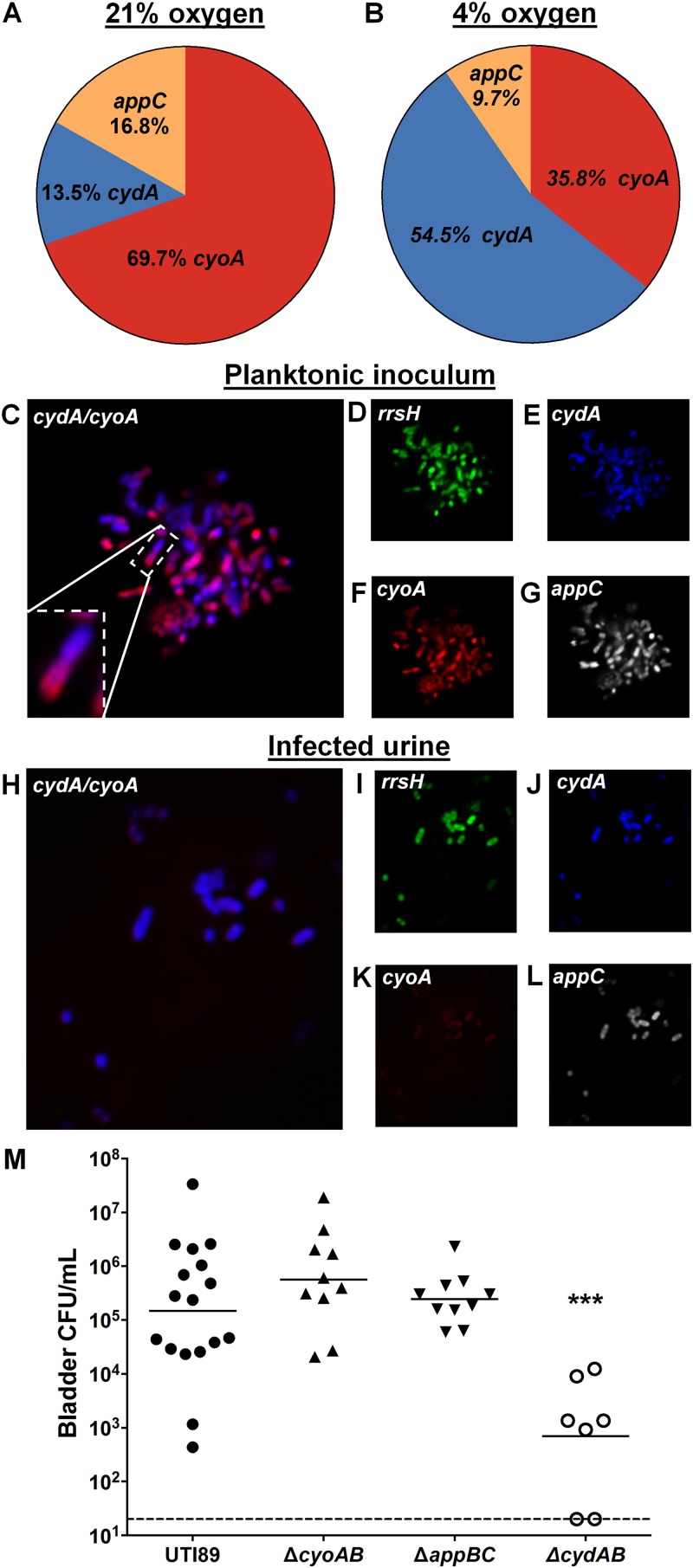
The high abundance of cydABX transcript in the hypoxic areas of the biofilm, in conjunction with the defects observed in aerobically grown ΔcydAB planktonic cultures, raised the hypothesis that a cytochrome bd-expressing subpopulation exists in the planktonic state under ambient oxygen conditions and that this cytochrome bd-expressing subpopulation exhibits the greatest fitness advantage during infection. To test this hypothesis, we first analyzed transcript abundance in aerobic cultures used for inoculation during murine infections with RT-qPCR and PNA-FISH (Fig. 5). Under these conditions, the majority of transcript corresponds to cyoABCD (69.7%), with cydABX and appBC transcripts each comprising approximately 15% of detected transcripts (Fig. 5A). Transcript abundance was altered by decreasing ambient oxygen concentrations, with the most abundant transcript corresponding to cydABX in 12%, 8%, and 4% oxygen, the last being the concentration of dissolved oxygen concentration in the urine (Fig. 5A and B and Fig. S8) (40). This shift in transcript abundance is largely due to a marked induction of cydABX expression under hypoxic conditions (Fig. S9). PNA-FISH analysis revealed the presence of bacteria which uniquely express cytochrome bo (Fig. 5F), bd (Fig. 5E), or bd2 (Fig. 5G), as well as some cells that have transcript of all three operons (Fig. 5C and E to G). Intriguingly, we observed dividing cells in which each daughter had distinct quinol oxidase transcript abundance (Fig. 5C, inset), suggesting that asymmetric distribution of respiratory transcripts during division may be a mechanism by which these subpopulations are generated. This hypothesis is supported by previous studies in E. coli demonstrating that quinol oxidases exhibit unusually noisy gene expression and that asymmetric cell division is a major generator of heterogeneity (42–44).
FIG 5.
Respiratory heterogeneity provides a fitness advantage during urinary tract infection. (A and B) Pie charts depicting relative abundance of cydA, cyoA, and appC transcripts detected using RT-qPCR in planktonic cultures grown at 21% oxygen in the manner used to prepare cultures to inoculate mice (A), as well as planktonic cultures grown at 4% oxygen (B). Data are representative of three biological replicates. (C to G) PNA-FISH was used to detect quinol oxidase transcripts from cultures used to inoculate mice. Data are representative of three biological replicates. (H to L) PNA-FISH was used to detected quinol oxidase transcripts in the urine of mice infected with UTI89. Urine was pooled from 20 mice. (M) Graph depicting bladder titers obtained from mice infected with UTI89 or quinol oxidase mutant strains at 24 h postinfection. Each point represents a mouse. UTI89 and ΔcydAB strains are representative of two independent experiments. ΔcyoAB and ΔappBC strains are representative of one experiment. Statistical analysis was performed in GraphPad Prism using a two-tailed Mann-Whitney test. Line represents geometric mean. ***, P < 0.001.
Ambient oxygen concentration influences quinol oxidase transcript abundance. Pie charts depicting relative abundance of cydA, cyoA, and appC transcripts detected using RT-qPCR in planktonic cultures grown at 12% (A) or 8% (B) oxygen. Data are representative of three biological replicates. Download FIG S8, TIF file, 2.0 MB (2MB, tif) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of quinol oxidases as a function of oxygen tension. (A) RT-qPCR data depicting relative fold difference in abundance of each quinol oxidase transcript of planktonic bacteria grown at 4% oxygen compared to 21% oxygen. (B) Relative abundance of each transcript at 21% and 4% oxygen compared to gyrB. (C) Raw CT values for each transcript at 21% and 4% oxygen. Data are presented as mean ± SEM. All data are representative of three biological replicates. Download FIG S9, TIF file, 0.9 MB (935.6KB, tif) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of cytochrome bd is dominant during acute urinary tract infection.
Previous studies reported that deletion of cytochrome bd impairs UPEC virulence in a UTI model (36). To gauge the contribution of each quinol oxidase during infection, we evaluated the fitness of ΔcyoAB, ΔappBC, and ΔcydAB mutants compared to the parent strain in a murine model of acute urinary tract infection. Consistent with the previous report (36), the ΔcydAB mutant exhibited an ∼2-log decrease in bladder colonization by 24 h relative to the parent strain, while the mutants deleted for cyoAB and appBC colonized mice at the same level as the parent strain (Fig. 5M). Subsequent PNA-FISH on pooled urine obtained from mice infected with the parent strain revealed a marked enrichment in cytochrome bd-expressing cells and a corresponding reduction in the number of cells expressing cytochrome bo (Fig. 5H to L). This suggests either that the bladder environment induces transcription of cydABX or that only subpopulations of bacteria expressing cydABX are capable of efficiently colonizing the bladder. Together these data reveal the presence of subpopulations of bacteria that differentially express quinol oxidases as a potential bet-hedging mechanism to promote bladder colonization.
DISCUSSION
Cytochrome bd is a multifunctional protein that is central to respiration and can maintain activity in the face of nitrosative stress (34). As such, bacteria expressing cytochrome bd presumably exhibit a fitness advantage under growth conditions that are low in oxygen or high in metabolic by-products that increase nitric oxide concentration. The biofilm state, while protecting the bacterial residents from predation and desiccation, constitutes a high-density environment with several chemical gradients that result from the consumption and production of metabolites. Accordingly, expressing an enzyme that can facilitate tolerance to metabolic by-products, such as nitric oxide, would ensure that biofilm residents do not perish as a consequence of their own metabolic excretions. Our study elucidates the distribution of quinol oxidase expression in the biofilm state and indicates that the bulk of biofilm residents express cytochrome bd, particularly in the densely populated interior. The cytochrome bd-expressing bacteria are not necessarily using cytochrome bd for respiration, as many of them also have low levels of cytochrome bo and bd2 transcripts (Fig. 2 and Fig. S3 and S4). Rather, the production of cytochrome bd may be leveraged toward providing tolerance to nitrosative stress, which irreversibly inhibits cytochrome bo. Indeed, in ΔcydAB biofilms we observe a marked increase in cytochrome bo expression (Fig. 4H), suggesting that loss of cytochrome bd impairs nitric oxide tolerance and that increased production of cytochrome bo may be a compensatory mechanism that allows biofilm bacteria to respire in the presence of high levels of nitric oxide.
In addition to acting as a respiratory inhibitor, nitric oxide regulates cyclic di-GMP abundance and thereby governs the switch from motility to aggregation and biofilm expansion (45, 46). Consequently, if cytochrome bd decreases nitric oxide availability, it would indirectly influence ECM production. Consistent with this hypothesis, loss of the cytochrome bd-expressing subpopulation reduces the total abundance of matrix components and leads to gross alterations of biofilm architecture (Fig. 3). It is thus possible that the cytochrome bd-expressing subpopulation is critical for promoting the biosynthesis of the ECM by influencing the nitric oxide–cyclic di-GMP signaling axis. We are currently investigating this possibility.
Most importantly, this work revealed the presence of planktonic subpopulations that express distinct quinol oxidases during growth. In conjunction with the observation that only cytochrome bd expression is critical for fitness during infection, this finding suggests that basal expression of cytochrome bd under aerobic conditions serves as a bet-hedging mechanism that promotes the expansion of bacteria during the transition from the aerobic perineum to the hypoxic bladder. In addition to allowing for efficient respiration in the hypoxic bladder, expression of cytochrome bd provides resistance against nitrosative stress—a metabolic by-product and component of the innate immune response—and promotes the formation of resilient biofilm communities. Alternatively, cytochrome bd may serve as an oxygen scavenger, thereby reducing oxygen tension and allowing distinct UPEC subpopulations to utilize anaerobic respiratory pathways. Consistent with this hypothesis, the alternative terminal electron acceptors nitrate and trimethylamine oxide (TMAO) are known to be present in the urine, and the anaerobic reduction of nitrate to nitrite by Enterobacteriaceae is the basis of a commonly used clinical test used to diagnose urinary tract infection. Together our observations suggest the presence of respiratory bet-hedging behavior in UPEC and additionally suggest the possibility of targeting heterogeneity as a method for homogenizing bacterial populations and impeding their ability to colonize the urinary tract.
MATERIALS AND METHODS
Bacterial strains.
All studies were performed in Escherichia coli cystitis isolate UTI89 (47). All gene deletions (ΔcyoAB, ΔappBC, and ΔcydAB) were performed using the λ-red recombinase system (48). Complementation constructs were created in plasmid pTRC99a with cydABX under the control of its native promoter as previously described (49). Primers used for gene deletions and complementation plasmid construction are listed in Table S1 in the supplemental material.
Growth conditions.
For all analyses, strains were propagated overnight at 37°C with shaking in lysogeny broth (LB) (Fisher) at pH 7.4. To form colony biofilms, 10 µl of overnight culture was spotted onto 1.2× yeast extract-Casamino Acids (YESCA) agar (8) and allowed to grow at room temperature. Growth curves to assess tolerance to nitrosative or oxidative stress were performed in LB broth at 37°C with shaking, starting from an overnight culture normalized to optical density at 600 nm (OD600) of 0.05. At 2 h post-inoculation, cultures were split into equal volumes and treated with 0.5 mM NOC-12, 1 mM H2O2 or left unperturbed. OD600 and CFU-per-ml measurements were taken every hour for 8 h.
RT-qPCR.
RNA was extracted from day 11 colony biofilms or planktonic cultures using the RNeasy kit (Qiagen). RNA was DNase treated using Turbo DNase I (Invitrogen) and reverse transcribed using SuperScript III reverse transcriptase (Invitrogen). cDNA was amplified in an Applied Biosystems StepOne Plus Real-Time PCR machine using TaqMan MGB chemistry with the primers and probes listed in Table S1. All reactions were performed in triplicate with four different cDNA concentrations (100, 50, 25, or 12.5 ng per reaction). Relative fold difference in transcript abundance was determined using the ΔΔCT method (50) with target transcripts normalized to gyrB abundance from a total of 3 to 4 biological replicates.
Peptide nucleic acid fluorescence in situ hybridization (PNA-FISH).
Day 11 biofilms were flash frozen in Tissue-Tek O.C.T. compound (Electron Microscopy Sciences) and cryosectioned as described previously (4). The PNA-FISH hybridization protocol was adapted from the work of Almeida et al. (51). Biofilm cryosections were fixed in 4% paraformaldehyde (PFA) for 30 min at room temperature and then dehydrated for 10 min in 50% ethanol. After dehydration, 100 μl of hybridization solution (see below for details) was applied to the slides. All hybridizations were performed at 60°C for 30 min. Next, slides were submerged in prewarmed wash solution for 30 min, mounted using ProLong Diamond (ThermoFisher), and imaged using a Zeiss 710 confocal laser scanning microscope (CLSM). For planktonic cells, 1 ml of culture was sedimented, fixed in 4% PFA, resuspended in 50% ethanol, incubated at −20°C for 30 min, and resuspended in 100 μl hybridization solution. After hybridization, cells were pelleted, resuspended in 500 μl prewarmed wash solution, and incubated at 60°C for 30 min. Finally, cells were pelleted and resuspended in 100 μl sterile water before being applied to microscope slides for imaging. Wash solution contained 5 mM Tris-HCl (pH 7.4), 15 mM NaCl, and 1% Triton X-100. Hybridization solution contained 10% (wt/vol) dextran sulfate, 30% formamide, 50 mM Tris-HCl (pH 7.4), 10 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, and 200 nM (each) PNA-FISH probe. Probe sequences were based on the probes used for qPCR (efficiency of hybridization, rrsH, 81; cydA, 107; cyoA, 115; appC, 73) and were synthesized by PNA Bio (Newbury Park, CA).
ATP measurements.
ATP was quantified from mid-log (4 h after subculture) planktonic cultures using the Cell-Glo Titer kit (Promega). Cultures were normalized to an OD600 of 0.5, pelleted, and resuspended in PBS. Fifty microliters of bacterial suspension was mixed with an equal volume of Cell-Glo Titer reagent and incubated with shaking at room temperature for 15 min. After incubation, luminescence was measured on a SpectraMax i3 plate reader (Molecular Devices). Luminescence was converted to concentration of ATP using a standard curve on the same plate.
Extracellular matrix extraction.
Extracellular matrix was extracted using established methods (28). Briefly, biofilms were grown on YESCA agar containing 25 µg/ml Congo red. After 60 h, biofilms were homogenized in cold 10 mM Tris-HCl (pH 7.4) using an Omni Tissue Homogenizer (motor speed 9) five times for 1 min per cycle. Next, the homogenate was centrifuged three times for 10 min at 5,000 × g to remove cells. The supernatant was spiked with NaCl (final concentration, 170 mM) and centrifuged for 1 h at 13,000 × g to pellet the matrix. The ECM pellet was washed in 10 mM Tris-HCl, pH 7.4, with 4% SDS and incubated at room temperature with rocking overnight. Next, the suspended ECM was centrifuged at 13,000 × g for 1 h, resuspended in cold 10 mM Tris-HCl (pH 7.4), and centrifuged at 30,000 × g for 20 min. Pelleted ECM was resuspended in MQ water and flash frozen.
Congo red depletion assays.
ECM abundance was quantified using Congo red depletion assays adapted from established protocols (52). Colony biofilms grown on YESCA agar were harvested into PBS at specific time points and homogenized. Congo red (40 µg/ml, final concentration) was added to homogenized biofilms, which were then incubated at 37°C for 1 h. After incubation, ECM was pelleted by centrifugation, the supernatant was removed, and supernatant absorbance (490 nm) was measured using a SpectraMax i3 plate reader (Molecular Devices).
Solid-state NMR measurements.
All NMR experiments were performed in an 89-mm-bore 11.7T magnet using either an HCN Agilent probe with a DD2 console (Agilent Technologies) or a home-built four-frequency transmission line probe with a Varian console. Samples were spun at 7,143 Hz in either 36-µl-capacity 3.2-mm zirconia rotors or thin-walled 5-mm-outer-diameter zirconia rotors. The temperature was maintained at 5°C with an FTS chiller (FTS Thermal Products, SP Scientific, Warminster, PA) supplying nitrogen at −10°C. The field strength for 13C cross-polarization was 50 kHz with a 10% 1H linear ramp centered at 57 kHz. The cross-polarization magic angle spinning (CPMAS) recycle time was 2 s for all experiments. 1H decoupling was performed with continuous wave decoupling. 13C chemical shifts were referenced to tetramethylsilane as 0 ppm using a solid adamantine sample at 38.5 ppm. The 15.6-mg wild-type 13C CPMAS spectrum was the result of 32,768 scans, and the 8.3-mg mutant spectrum was the result of 100,000 scans. NMR spectra were processed with 80-Hz line broadening.
SDS-PAGE gels.
A portion of the lyophilized ECM sample used for solid-state NMR analysis was resuspended in 98% formic acid and vacuum centrifuged. The samples were then reconstituted in SDS-PAGE sample buffer containing 8 M urea and 50 mM DTT and further diluted to desired concentrations. All samples were centrifuged briefly at 10,000 × g to remove any insoluble material and used for electrophoresis. The gels were stained with instant blue and destained in water.
Immunofluorescence.
An immunofluorescence assay targeting CsgA, the major curli subunit, was performed as previously described (9). Biofilm cryosections were fixed in 4% PFA for 30 min at room temperature and blocked overnight in 5% BSA at 4°C. Sections were washed in PBS, incubated with rabbit anti-CsgA antibodies (GenScript) (1:1,000) at room temperature for 1 h, washed in PBS, and incubated with Alexa Fluor 647 goat anti-rabbit IgG (ThermoFisher) (1:1,000) at room temperature for 1 h. Slides were counterstained with SYTO 9 and imaged using confocal laser scanning microscopy (CLSM).
Murine infections.
Murine infections were performed as described previously (53). In brief, UTI89 and each mutant strain were inoculated individually into 5 ml LB medium and grown with shaking at 37°C for 4 h. Next, this culture was diluted 1:1,000 into 10 ml fresh medium and grown statically at 37°C for 24 h. After 24 h, this culture was diluted 1:1,000 into 10 ml fresh medium and grown for another 24 h at 37°C statically. Next, 7- to 8-week-old C3H/HeN female mice were transurethrally inoculated with 50 µl PBS containing 107 CFU bacteria. Mice were sacrificed at 24 h postinfection, after which bladders were removed and homogenized for CFU enumeration. All animal studies were approved by the Vanderbilt University Medical Center Institutional Animal Care and Use Committee (IACUC) (protocol numbers M/12/191 and M1500017-01) and carried out in accordance with all recommendations in the Guide for the Care and Use of Laboratory Animals (54) of the National Institutes of Health and the IACUC.
Statistical analysis.
All statistical analyses were performed in GraphPad Prism using the most appropriate test. Details of test used, error bars, and statistical significance cutoffs are presented in the figure legends.
ACKNOWLEDGMENTS
We thank Jonathan Schmitz, Gerald Van Horn, Mariana X. Byndloss, and members of the Hadjifrangiskou laboratory for critical evaluation of the manuscript and helpful discussions.
This work was supported by the following NIH grants: R01 AI107052 (M.H.), DiaComp DK076169 (M.H.), T32 GM007347 (C.J.B.), K08 DK106472 (D.B.C.), and the Vanderbilt University Medical Center Pediatric Urology Research Fund. L.C. acknowledges support from the National Science Foundation Career Award 1453247. S.H.W. is a recipient of an NSF Predoctoral Fellowship.
C.J.B., A.R.E., and M.H. conceived the study, performed the experiments, analyzed the data, and composed the manuscript. S.A.R. performed the nitrosative and oxidative stress assays. M.J.F. and M.M.H. constructed deletion mutants and complementation constructs under the supervision of C.J.B. and A.R.E. J.R.B. assisted with imaging data acquisition. S.D. and D.B.C. performed the animal experiments along with C.J.B. and A.R.E., assisted in data analysis, and edited the manuscript. S.H.W. and L.C. performed the extracellular matrix component measurements and edited the manuscript.
We declare no conflicts of interest at the time of submission of the manuscript.
Footnotes
Citation Beebout CJ, Eberly AR, Werby SH, Reasoner SA, Brannon JR, De S, Fitzgerald MJ, Huggins MM, Clayton DB, Cegelski L, Hadjifrangiskou M. 2019. Respiratory heterogeneity shapes biofilm formation and host colonization in uropathogenic Escherichia coli. mBio 10:e02400-18. https://doi.org/10.1128/mBio.02400-18.
Contributor Information
Lars E. P. Dietrich, Columbia University.
Dianne K. Newman, California Institute of Technology.
REFERENCES
- 1.Kostakioti M, Hadjifrangiskou M, Hultgren SJ. 2013. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb Perspect Med 3:a010306. doi: 10.1101/cshperspect.a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 3.Nadell CD, Drescher K, Foster KR. 2016. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol 14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 4.Vlamakis H, Aguilar C, Losick R, Kolter R. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Gestel J, Vlamakis H, Kolter R. 2015. Division of labor in biofilms: the ecology of cell differentiation. Microbiol Spectr 3:MB-0002-2014. doi: 10.1128/microbiolspec.MB-0002-2014. [DOI] [PubMed] [Google Scholar]
- 6.Dragoš A, Kiesewalter H, Martin M, Hsu CY, Hartmann R, Wechsler T, Eriksen C, Brix S, Drescher K, Stanley-Wall N, Kümmerli R, Kovács Á. 2018. Division of labor during biofilm matrix production. Curr Biol 28:1903–1913.e5. doi: 10.1016/j.cub.2018.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietrich LE, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. 2013. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol 195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePas WH, Hufnagel DA, Lee JS, Blanco LP, Bernstein HC, Fisher ST, James GA, Stewart PS, Chapman MR. 2013. Iron induces bimodal population development by Escherichia coli. Proc Natl Acad Sci U S A 110:2629–2634. doi: 10.1073/pnas.1218703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Floyd KA, Moore JL, Eberly AR, Good JA, Shaffer CL, Zaver H, Almqvist F, Skaar EP, Caprioli RM, Hadjifrangiskou M. 2015. Adhesive fiber stratification in uropathogenic Escherichia coli biofilms unveils oxygen-mediated control of type 1 pili. PLoS Pathog 11:e1004697. doi: 10.1371/journal.ppat.1004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jo J, Cortez KL, Cornell WC, Price-Whelan A, Dietrich LE. 2017. An orphan cbb3-type cytochrome oxidase subunit supports Pseudomonas aeruginosa biofilm growth and virulence. Elife 6:30205. doi: 10.7554/eLife.30205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unden G, Bongaerts J. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta 1320:217–234. doi: 10.1016/S0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 12.Borisov VB, Verkhovsky MI. 2015. Oxygen as acceptor. EcoSal Plus 6(2). doi: 10.1128/ecosalplus.ESP-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. 2011. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta 1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris RL, Schmidt TM. 2013. Shallow breathing: bacterial life at low O(2). Nat Rev Microbiol 11:205–212. doi: 10.1038/nrmicro2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borisov VB, Murali R, Verkhovskaya ML, Bloch DA, Han H, Gennis RB, Verkhovsky MI. 2011. Aerobic respiratory chain of Escherichia coli is not allowed to work in fully uncoupled mode. Proc Natl Acad Sci U S A 108:17320–17324. doi: 10.1073/pnas.1108217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, Chapman MR, Hultgren SJ. 2009. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol 5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurcisek JA, Brockman KL, Novotny LA, Goodman SD, Bakaletz LO. 2017. Nontypeable Haemophilus influenzae releases DNA and DNABII proteins via a T4SS-like complex and ComE of the type IV pilus machinery. Proc Natl Acad Sci U S A 114:E6632–E6641. doi: 10.1073/pnas.1705508114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rapsinski GJ, Wynosky-Dolfi MA, Oppong GO, Tursi SA, Wilson RP, Brodsky IE, Tükel Ç. 2015. Toll-like receptor 2 and NLRP3 cooperate to recognize a functional bacterial amyloid, curli. Infect Immun 83:693–701. doi: 10.1128/IAI.02370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberly AR, Floyd KA, Beebout CJ, Colling SJ, Fitzgerald MJ, Stratton CW, Schmitz JE, Hadjifrangiskou M. 2017. Biofilm formation by uropathogenic Escherichia coli is favored under oxygen conditions that mimic the bladder environment. Int J Mol Sci 18:E2077. doi: 10.3390/ijms18102077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Floyd KA, Mitchell CA, Eberly AR, Colling SJ, Zhang EW, DePas W, Chapman MR, Conover M, Rogers BR, Hultgren SJ, Hadjifrangiskou M. 2016. The UbiI (VisC) aerobic ubiquinone synthase is required for expression of type 1 pili, biofilm formation, and pathogenesis in uropathogenic Escherichia coli. J Bacteriol 198:2662–2672. doi: 10.1128/JB.00030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alteri CJ, Smith SN, Mobley HL. 2009. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog 5:e1000448. doi: 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadjifrangiskou M, Kostakioti M, Chen SL, Henderson JP, Greene SE, Hultgren SJ. 2011. A central metabolic circuit controlled by QseC in pathogenic Escherichia coli. Mol Microbiol 80:1516–1529. doi: 10.1111/j.1365-2958.2011.07660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klauck G, Serra DO, Possling A, Hengge R. 2018. Spatial organization of different sigma factor activities and c-di-GMP signalling within the three-dimensional landscape of a bacterial biofilm. Open Biol 8:180066. doi: 10.1098/rsob.180066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serra DO, Klauck G, Hengge R. 2015. Vertical stratification of matrix production is essential for physical integrity and architecture of macrocolony biofilms of Escherichia coli. Environ Microbiol 17:5073–5088. doi: 10.1111/1462-2920.12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serra DO, Richter AM, Hengge R. 2013. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol 195:5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shepherd M, Sanguinetti G, Cook GM, Poole RK. 2010. Compensations for diminished terminal oxidase activity in Escherichia coli: cytochrome bd-II-mediated respiration and glutamate metabolism. J Biol Chem 285:18464–18472. doi: 10.1074/jbc.M110.118448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin YC, Sekedat MD, Cornell WC, Silva GM, Okegbe C, Price-Whelan A, Vogel C, Dietrich LEP. 2018. Phenazines regulate Nap-dependent denitrification in Pseudomonas aeruginosa biofilms. J Bacteriol 200:e00031-18. doi: 10.1128/JB.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCrate OA, Zhou X, Reichhardt C, Cegelski L. 2013. Sum of the parts: composition and architecture of the bacterial extracellular matrix. J Mol Biol 425:4286–4294. doi: 10.1016/j.jmb.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thongsomboon W, Serra DO, Possling A, Hadjineophytou C, Hengge R, Cegelski L. 2018. Phosphoethanolamine cellulose: a naturally produced chemically modified cellulose. Science 359:334–338. doi: 10.1126/science.aao4096. [DOI] [PubMed] [Google Scholar]
- 30.Hufnagel DA, Depas WH, Chapman MR. 2015. The biology of the Escherichia coli extracellular matrix. Microbiol Spectr 3:MB-0014-2014. doi: 10.1128/microbiolspec.MB-0014-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hobley L, Ostrowski A, Rao FV, Bromley KM, Porter M, Prescott AR, MacPhee CE, van Aalten DM, Stanley-Wall NR. 2013. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci U S A 110:13600–13605. doi: 10.1073/pnas.1306390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asally M, Kittisopikul M, Rue P, Du Y, Hu Z, Cagatay T, Robinson AB, Lu H, Garcia-Ojalvo J, Suel GM. 2012. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc Natl Acad Sci U S A 109:18891–18896. doi: 10.1073/pnas.1212429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cutruzzolà F, Frankenberg-Dinkel N. 2016. Origin and impact of nitric oxide in Pseudomonas aeruginosa biofilms. J Bacteriol 198:55–65. doi: 10.1128/JB.00371-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giuffrè A, Borisov VB, Arese M, Sarti P, Forte E. 2014. Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim Biophys Acta 1837:1178–1187. doi: 10.1016/j.bbabio.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Mason MG, Shepherd M, Nicholls P, Dobbin PS, Dodsworth KS, Poole RK, Cooper CE. 2009. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat Chem Biol 5:94–96. doi: 10.1038/nchembio.135. [DOI] [PubMed] [Google Scholar]
- 36.Shepherd M, Achard ME, Idris A, Totsika M, Phan MD, Peters KM, Sarkar S, Ribeiro CA, Holyoake LV, Ladakis D, Ulett GC, Sweet MJ, Poole RK, McEwan AG, Schembri MA. 2016. The cytochrome bd-I respiratory oxidase augments survival of multidrug-resistant Escherichia coli during infection. Sci Rep 6:35285. doi: 10.1038/srep35285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borisov VB, Forte E, Davletshin A, Mastronicola D, Sarti P, Giuffrè A. 2013. Cytochrome bd oxidase from Escherichia coli displays high catalase activity: an additional defense against oxidative stress. FEBS Lett 587:2214–2218. doi: 10.1016/j.febslet.2013.05.047. [DOI] [PubMed] [Google Scholar]
- 38.Safarian S, Rajendran C, Müller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. 2016. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science 352:583–586. doi: 10.1126/science.aaf2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindqvist A, Membrillo-Hernańdez J, Poole RK, Cook GM. 2000. Roles of respiratory oxidases in protecting Escherichia coli K12 from oxidative stress. Antonie Van Leeuwenhoek 78:23–31. doi: 10.1023/A:1002779201379. [DOI] [PubMed] [Google Scholar]
- 40.Wang ZJ, Joe BN, Coakley FV, Zaharchuk G, Busse R, Yeh BM. 2008. Urinary oxygen tension measurement in humans using magnetic resonance imaging. Acad Radiol 15:1467–1473. doi: 10.1016/j.acra.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. 2010. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silander OK, Nikolic N, Zaslaver A, Bren A, Kikoin I, Alon U, Ackermann M. 2012. A genome-wide analysis of promoter-mediated phenotypic noise in Escherichia coli. PLoS Genet 8:e1002443. doi: 10.1371/journal.pgen.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huh D, Paulsson J. 2011. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat Genet 43:95–100. doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol 191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 47.Mulvey MA, Schilling JD, Hultgren SJ. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun 69:4572–4579. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy KC, Campellone KG. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol 4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaffer CL, Zhang EW, Dudley AG, Dixon BR, Guckes KR, Breland EJ, Floyd KA, Casella DP, Algood HM, Clayton DB, Hadjifrangiskou M. 2017. Purine biosynthesis metabolically constrains intracellular survival of uropathogenic Escherichia coli. Infect Immun 85:e00471-16. doi: 10.1128/IAI.00471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almeida C, Azevedo NF, Santos S, Keevil CW, Vieira MJ. 2011. Discriminating multi-species populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (PNA FISH). PLoS One 6:e14786. doi: 10.1371/journal.pone.0014786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okegbe C, Fields BL, Cole SJ, Beierschmitt C, Morgan CJ, Price-Whelan A, Stewart RC, Lee VT, Dietrich LEP. 2017. Electron-shuttling antibiotics structure bacterial communities by modulating cellular levels of c-di-GMP. Proc Natl Acad Sci U S A 114:E5236–E5245. doi: 10.1073/pnas.1700264114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hung CS, Dodson KW, Hultgren SJ. 2009. A murine model of urinary tract infection. Nat Protoc 4:1230–1243. doi: 10.1038/nprot.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of respiratory complexes compared to gyrB. (A) Graph depicting relative abundance of each respiratory transcript in the center and periphery of day 11 colony biofilms compared to gyrB. (B) Average CT values for each transcript in the center and periphery of colony biofilms. Data are presented as mean ± SEM. All data are representative of four biological replicates. Download FIG S1, TIF file, 0.9 MB (995.9KB, tif) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PNA-FISH on quinol oxidase deletion mutants. Representative images of ΔcydAB (A to D) and ΔcyoAB (E to H) cells stained with PNA-FISH probes to assess specificity. Download FIG S2, TIF file, 2.0 MB (2.1MB, tif) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Localization of appC transcript in biofilm cryosections. (A) Representative image of SYTO 9-stained biofilm cryosection at ×20 magnification. (B and C) Representative images of biofilm cryosections stained with an appC PNA-FISH probe at ×20 (B) and ×63 (C) magnification. Download FIG S3, JPG file, 0.4 MB (384.7KB, jpg) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Localization of quinol oxidase transcripts in biofilm wrinkles. Representative image of a PNA-FISH-stained cryosection depicting a biofilm wrinkle with a central region filled with rrsH-stained cells at ×20 magnification. Download FIG S4, JPG file, 2.4 MB (2.5MB, jpg) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers and probes. Download Table S1, DOCX file, 0.1 MB (104.4KB, docx) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Temporal development of colony biofilms by UTI89 and quinol oxidase mutants. (A) Representative images of UTI89 and quinol oxidase mutant colony biofilms grown on YESCA agar taken on days 3, 7, and 11 of growth. (B) Graph depicting colony biofilm diameter at days 3, 7, and 11 of growth. Each triangle represents an individual colony biofilm. Data are representative of at least 30 biological replicates. Statistical analysis was performed in GraphPad Prism using Welch’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Download FIG S5, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of ΔappBC ΔcyoAB::kanR colony biofilms. Comparison of colony biofilms formed by UTI89, ΔcyoAB, ΔappBC, ΔappBC ΔcyoAB::kanR, and ΔcydAB strains after 6 days of growth on YESCA agar. Images are representative of five biological replicates. Download FIG S6, TIF file, 2.3 MB (2.3MB, tif) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Extrachromosomal complementation of ΔcydAB rescues biofilm defects. Representative images on UTI89_pTRC99a, ΔcydAB_pTRC99a, and complemented ΔcydAB_pCydABX strains under the control of a native promoter. Images were taken of colony biofilms grown on YESCA agar at days 3, 7, and 11 of growth. Images are representative of at least five biological replicates. Download FIG S7, JPG file, 0.2 MB (251.4KB, jpg) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ambient oxygen concentration influences quinol oxidase transcript abundance. Pie charts depicting relative abundance of cydA, cyoA, and appC transcripts detected using RT-qPCR in planktonic cultures grown at 12% (A) or 8% (B) oxygen. Data are representative of three biological replicates. Download FIG S8, TIF file, 2.0 MB (2MB, tif) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of quinol oxidases as a function of oxygen tension. (A) RT-qPCR data depicting relative fold difference in abundance of each quinol oxidase transcript of planktonic bacteria grown at 4% oxygen compared to 21% oxygen. (B) Relative abundance of each transcript at 21% and 4% oxygen compared to gyrB. (C) Raw CT values for each transcript at 21% and 4% oxygen. Data are presented as mean ± SEM. All data are representative of three biological replicates. Download FIG S9, TIF file, 0.9 MB (935.6KB, tif) .
Copyright © 2019 Beebout et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.