Abstract
MicroRNAs (miRNAs), a class of endogenous, tiny, non-coding RNAs, are master regulators of gene expression among most eukaryotes. Intracellular miRNA abundance is regulated under multiple levels of control including transcription, processing, RNA modification, RNA-induced silencing complex (RISC) assembly, miRNA-target interaction, and turnover. In this review, we summarize our current understanding of the molecular components and mechanisms that influence miRNA biogenesis, homeostasis, and degradation in plants. We also make comparisons with findings from other organisms where necessary.
Keywords: miRNA biogenesis, DCL1, HEN1, Argonaute, target mimic, uridylation
Introduction
RNA silencing, which is mediated by small non-coding RNAs of 20–35 nucleotides in length, is an important and indispensable form of gene regulation among most eukaryotes. According to their origin, processing mode and effector protein association, small RNAs can be divided into four major categories: microRNA (miRNA), small interfering RNA (siRNA), PIWI-interacting RNA (piRNA, animals only), and transfer RNA-derived small RNAs (tsRNAs) (Borges and Martienssen, 2015; Czech et al., 2018; Zhu et al., 2018; Treiber et al., 2019). siRNAs can be further separated into different sub-categories such as heterochromatic siRNAs (hc-siRNAs), phased secondary siRNAs (phasiRNAs) and epigenetically activated siRNAs (easiRNAs) (Borges and Martienssen, 2015; Zhang et al., 2016). Unlike siRNAs and piRNAs, which can mediate gene silencing at either the transcriptional (TGS) or post-transcriptional (PTGS) level, miRNAs predominantly repress target genes post-transcriptionally (Borges and Martienssen, 2015; Bartel, 2018). Plant miRNAs are produced from specific stem regions of single-stranded hairpin precursors, which possess distinct features from other types of small RNAs. Detailed criteria for plant miRNA annotation were described recently (Axtell and Meyers, 2018). If not otherwise specified, we hereinafter refer to plant miRNAs as miRNAs. miRNAs play crucial roles in almost all aspects of normal plant growth and development, but also in response to environmental fluctuations such as light, nutrition, and various abiotic and biotic stresses (Budak et al., 2015; Shriram et al., 2016; Li S. J. et al., 2017; Brant and Budak, 2018). As such, the temporal-spatial expression of intracellular miRNAs is under multi-level control to ensure fine regulation of target genes. Here we review our current understanding of the biogenesis, homeostasis, and turnover of miRNAs, with a focus on the regulation of each step that affects the production or degradation of miRNAs.
Overview of miRNA Biogenesis, RISC Assembly, and Action
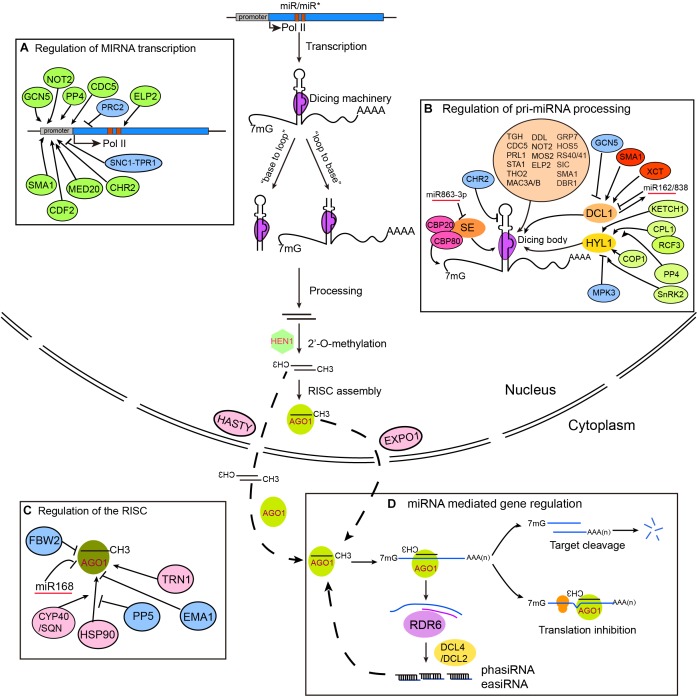
Plant genomes typically encode a hundred to several hundreds of MIRNA (MIR) genes, with many of them existing as families (Nozawa et al., 2012; Budak and Akpinar, 2015). According to their location in the genome, miRNAs are classified as either “intergenic” or “intronic.” Intergenic miRNAs are located between two protein-coding genes and are transcribed as independent units by DNA-dependent RNA Polymerase II (Pol II), while intronic miRNAs are processed from introns of their host transcripts (Millar and Waterhouse, 2005; Budak and Akpinar, 2015). As canonical Pol II products, primary transcripts of MIRs (termed pri-miRNAs) are 5′ capped, 3′ polyadenylated, and/or spliced (Xie et al., 2005; Rogers and Chen, 2013). pri-miRNAs are folded into hairpin-like structures consisting of a terminal loop, an upper stem, the miRNA/miRNA∗ region, a lower stem, and two arms, which can be recognized and processed by Dicer-like RNase III endonucleases (DCLs). Different plant species have different numbers of DCL proteins. In Arabidopsis thaliana, there are four DCL proteins. DCL1 catalyzes the production of most miRNAs with the assistance of accessory proteins including the double-stranded RNA-binding protein Hyponastic Leaves 1 (HYL1) and the zinc-finger protein Serrate (SE) (Fang and Spector, 2007; Dong et al., 2008). Other DCLs may also be involved in miRNA production. For instance, AtDCL4 is responsible for miR822 and miR839 production while OsDCL3a in rice generates a class of 24-nt miRNAs that direct DNA methylation like hc-siRNAs (Rajagopalan et al., 2006; Wu et al., 2010). The stem-loops of pri-miRNAs are much more variable in length (from 60 nt to over 500 nt) and bear more complex structures than their ∼70 nt animal counterparts (Xie et al., 2005; Bologna and Voinnet, 2014). As such, pri-miRNAs in plants can be processed from either the loop-distal site to the loop-proximal site or vice versa (Addo-Quaye et al., 2009; Bologna et al., 2009, 2013; Mateos et al., 2010; Song et al., 2010; Werner et al., 2010). The nascent miRNA/miRNA∗ duplex generated by DCL-mediated processing exhibits 2-nt 3′ overhangs at both strands and each strand possesses a 5′ end phosphate and two 3′ end hydroxyl groups (2′ OH and 3′ OH). While both hydroxyl groups are essential, only the 2′-OH position is methylated by the small RNA methyltransferase HUA Enhancer 1 (HEN1) (Yu et al., 2005; Yang Z. Y. et al., 2006).
Methylated miRNA/miRNA∗ duplexes are thought to be exported by the animal Exportin 5 (EXPO5) homologous protein Hasty (HST) (Park et al., 2005). For a long time, it was not known where the RISC assembled. Recently, Bologna et al. (2018) showed that RISC is mainly assembled in the nucleus and is then exported to the cytosol by EXPO1. However, current data do not exclude the possibility that some miRNAs are exported in their duplex forms and are assembled in the cytosol (Figure 1). One strand of the miRNA/miRNA∗ duplex (the guide strand, miRNA) is selectively assembled into the Argonaute (AGO) protein, and the other strand (the passenger strand, miRNA∗) is ejected and degraded. Arabidopsis has 10 AGO proteins, with AGO1 being the major effector protein for miRNAs (Zhang H. et al., 2015).
FIGURE 1.
Regulation of miRNA biogenesis, RISC loading, and action in plants. MIRNA genes are transcribed by RNA Polymerase II (Pol II) and fold into stem-loop structures called pri-miRNAs. pri-miRNAs are mainly processed by DCL1 from either “base-to-loop” or “loop-to-base” direction. Nascent miRNA/miRNA∗ duplexes are methylated by the small RNA methyltransferase HEN1. RISC loading occurs in the nucleus, but may also occur in the cytoplasm. miRNAs mediate gene silencing through either target cleavage or translation inhibition. Some miRNAs can trigger the production of secondary siRNAs through sequential actions of RDR6 and DCL2/4, generating 21–22 nt secondary siRNAs called phasiRNAs and easiRNAs, which in turn repress gene expression via PTGS (for phasiRNAs) or TGS (for easiRNAs) (D). It is important to note that although the steps in the model are separate, they could be closely coupled. Factors involved in the regulation of MIR transcription, pri-miRNA processing, and RISC assembly are shown in parts (A–C), respectively.
miRNAs guide the RISC to target genes via base pairing and predominantly mediate gene silencing through target cleavage and/or translation inhibition. Nevertheless, recent studies also suggest a role of RISC/AGO1 in transcriptional regulation (Dolata et al., 2016; Liu et al., 2018; Yang et al., 2019). Some miRNAs including miR390, miR173, and miR845 are capable of initiating the production of secondary siRNAs called phasiRNAs and/or easiRNAs (Figure 1D) (Fei et al., 2013; Creasey et al., 2014; Deng et al., 2018). In animals, a short base-pairing to the seed region of miRNAs (positions 2–8) is sufficient for target recognition, although non-canonical targeting has also been observed (Helwak et al., 2013; Agarwal et al., 2015). In contrast, a more stringent base-pairing rule is employed by plants, with near perfect pairing in the 5′ region (no more than 1 mismatch) and relaxed, but ample pairing in the 3′ region (no more than 4 mismatches and only small bulges allowed) (Schwab et al., 2005; Axtell and Meyers, 2018). Theoretically, plants have at least two orders of magnitude fewer target genes than animals. Although translation inhibition seems prevalent, target cleavage is more important as it is essential for post-germination plant development (Carbonell et al., 2012).
Regulation of Mirna Transcription
Similar properties of transcription, co-transcriptional capping, polyadenylation, and splicing of MIR genes to coding genes suggest that essentially all known regulatory mechanisms for mRNA transcription may be applied to MIR gene transcription (Figure 1A). For instance, changes in the phosphorylation of the Pol II C-terminal domain (CTD) by Cyclin-Dependent Kinase Ds (CDKDs) and CDKF;1 have been reported to modulate MIR transcription and co-transcriptional capping, polyadenylation, and splicing (Hajheidari et al., 2012). The transcription co-activator complex mediator plays a general role in recruiting Pol II to MIR promoters during transcription initiation (Kim et al., 2011). MIR transcription is not only regulated by locus-specific transcription factors and regulators, but is also globally modulated by the CCR4-NOT (for Carbon Catabolite Repression 4-Negative on TATA-less) complex subunit NOT2, the Elongator complex subunits ELP2 and ELP5, the MYB-R2R3 type transcription factor Cell Division Cycle 5 (CDC5), the DOF (for DNA binding with One Finger) transcription factor Cycling DOF Factor 2 (CDF2), the Protein Phosphatase 4 complex, the disease resistance R protein SNC1 (for Suppressor of npr1-1, Constitutive 1) and its transcriptional corepressor Topless-Related 1 (TPR1) (Wang et al., 2013, 2019; Zhang et al., 2013; Fang et al., 2015a; Sun et al., 2015; Cai et al., 2018). A general effect of these proteins on MIR transcription may be related to their interactions with members of the miRNA processing machinery such as DCL1. The expression levels of MIRs are also dynamically regulated by histone modifications (Luo et al., 2013; Zhao et al., 2015). GCN5 (for General Control Non-repressed Protein 5)-mediated H3K14 acetylation promotes the expression of a subset of MIR genes, whereas deposition of H3K27me3 by PRC2 (for Polycomb Repressive Complex 2) at MIR156A and MIR156C genes downregulates their expression and drives the juvenile to adult transition (Kim et al., 2009; Xu et al., 2016).
Regulation of the Core Processing Machinery
pri-miRNA processing takes place at subnuclear foci called Dicing-bodies (D-bodies) or SmD3/SmB bodies (Fang and Spector, 2007; Fujioka et al., 2007). Among many accessory proteins identified to date, HYL1 and SE are two core cofactors; defects in either HYL1 or SE result in global abolishment of miRNAs and dramatic accumulation of pri-miRNAs (Han et al., 2004; Vazquez et al., 2004; Lobbes et al., 2006; Yang L. et al., 2006). HYL1 and SE may form a complex with DCL1 in D-bodies (i.e., the core processing machinery) to ensure both the precise and efficient cleavage of pri-miRNAs (Fang and Spector, 2007; Dong et al., 2008; Liu et al., 2012; Zhu et al., 2013). The core processing machinery is monitored at multiple levels, including transcriptional, post-transcriptional, and post-translational regulation (Figure 1B).
Transcription of DCL1 is negatively regulated by the histone acetyltransferase GCN5, and positively regulated by XCT (for XAP5 Circadian Timekeeper) and the pre-mRNA processing factor 6 homolog Stabilized 1 (STA1) (Kim et al., 2009; Ben Chaabane et al., 2013; Fang et al., 2015b). Another splicing factor, SMALL 1 (SMA1), is required for the correct splicing of the ninth intron of DCL1 (Li et al., 2018). In addition, the abundance of DCL1 is fine-tuned by two negative feedback mechanisms. First, miR162, a miRNA generated by DCL1, can in turn target DCL1 mRNA for cleavage. Second, DCL1-mediated processing of MIR838, an intronic MIRNA gene that resides in the 14th intron of DCL1 pre-mRNAs, leads to abortive DCL1 transcription (Xie et al., 2003; Rajagopalan et al., 2006). Indeed, in hyl1 mutants where such feedback mechanisms are impaired, the abundance of DCL1 is significantly increased (Liu et al., 2012).
HYL1 functions as a dimer and binds to the stem region of pri-miRNAs (Yang et al., 2010; Yang et al., 2014). The N-terminal double-strand RNA binding domains (dsRBDs) of HYL1 are sufficient for pri-miRNA processing, while the C-terminal part appears dispensable (Wu et al., 2007). Interestingly, expression of the dsRBDs of DCL1 can fully rescue the phenotype of hyl1 (Liu et al., 2013). Moreover, a suppressor screen identified multiple dominant DCL1 alleles that rescue both the hyl1 mutant phenotype and miRNA processing defects (Tagami et al., 2009; Liu et al., 2012). These observations indicate an auxiliary, rather than indispensable, role for HYL1 in the recruitment and positioning of pri-miRNAs into DCL1 and facilitating processing. Overexpression of hairpin-like SINE (for Short Interspersed Elements) transposon RNAs can sequester HYL1 from miRNA precursors, leading to reductions in miRNA expression (Pouch-Pelissier et al., 2008). The accumulation of HYL1 is sensitive to light-dark transitions. HYL1 is degraded in the dark by a yet uncharacterized protease in the cytoplasm. While in the light, Constitutive Photomorphogenic 1 (COP1), a RING-finger E3 ligase, can move from the nucleus to the cytoplasm and prevent HYL1 degradation by inhibiting the protease activity (Cho et al., 2014). KETCH1, a member of the importin-β family, is responsible for the translocation of cytoplasmic HYL1 into the nucleus (Zhang et al., 2017b). Post-translational phosphorylation plays a crucial role in regulating both the activity and stability of HYL1. Phosphorylated HYL1 appears non-functional, but can be retained in the nucleus, protecting it from degradation in the dark until it can be reactivated through light-mediated de-phosphorylation (Achkar et al., 2018). Multiple kinases and phosphatases have been characterized in the last few years. Both Mitogen-Activated Protein Kinase 3 (MPK3) and SNF1-related Protein Kinase 2 (SnRK2) were shown to interact with HYL1 in vivo and to phosphorylate HYL1 in vitro (Raghuram et al., 2015; Yan et al., 2017). On the other hand, C-Terminal Domain Phosphatase-like 1 (CPL1) and the PP4/ Suppressor of MEK 1 (SMEK1) complex were demonstrated to dephosphorylate HYL1 (Manavella et al., 2012; Su et al., 2017). The K homology (KH) domain protein Regulator of CBF Gene Expression 3 (RCF3) promotes HYL1 dephosphorylation through interaction with CPL1 and CPL2 in apex tissues (Karlsson et al., 2015). However, inconsistent results have been observed and await future clarification. For instance, loss of MPK3 results in over-accumulation of HYL1 and increased miRNA levels, whereas the snrk2 mutant has reduced HYL1 protein levels and decreased miRNA accumulation. Moreover, phosphorylated HYL1 proteins in the smek1 mutant are unstable and degraded, whereas in another report, phosphorylation protects HYL1 from degradation in the dark by nuclear retention (Raghuram et al., 2015; Su et al., 2017; Yan et al., 2017; Achkar et al., 2018). It is possible that different phosphorylation sites, subcellular localizations, and/or experimental conditions could explain the discrepancy.
Similar to the DCL1-miR162 feedback loop, SE is targeted by miR863-3p. Such regulation occurs during later stages of bacterial infection (Niu et al., 2016). SE is phosphorylated by SnRK2 in vitro, although whether this is also the case in vivo and what the biological consequence of this modification is remain to be explored (Yan et al., 2017). Interestingly, SE may have dual functions in coordinating miRNA production. First, as a core cofactor of the miRNA processing machinery, SE not only serves as a scaffold protein during microprocessor complex assembly, but may also promote miRNA processing in vivo, although a direct role of SE in pri-miRNA processing in vitro remains debatable (Dong et al., 2008; Zhu et al., 2013). Second, SE is known to interact with Chromatin Remodeling 2 (CHR2), which remodels pri-miRNAs to inhibit their processing (Wang Z. Y. et al., 2018). It is also worth noting that SE has broader functions than miRNA processing. SE interacts with the cap-binding complex and is involved in pre-mRNA splicing (Laubinger et al., 2008). In a recent study, SE was shown to promote intronless gene expression via direct chromatin binding and facilitating Pol II association (Speth et al., 2018). Moreover, SE is involved in fine-tuning transposon expression via promoting H3K27me1 by the Arabidopsis Trithorax Related Protein 5 and 6 (ATXR5/6) and suppressing RNA Dependent RNA Polymerase 6 (RDR6)-mediated RNA silencing (Ma et al., 2018).
Other Regulatory Proteins Influencing miRNA Biogenesis
A battery of additional regulatory proteins influencing the folding, stability, and/or processing of pri-miRNAs have been identified during the last decade or so, which will not be discussed here (Figure 1B) (for recent reviews, see (Zhang S. X. et al., 2015; Yu et al., 2017b). One of the most important features revealed through these studies is that pri-miRNAs are modified, folded, and processed co-transcriptionally. This is supported by several lines of evidence. First, DCL1 is associated with chromatin regions of MIR genes (Fang et al., 2015a). Second, several regulatory proteins affecting miRNA transcription (e.g., CDC5, NOT2, and ELP2) have been shown to interact with processing machinery proteins (Wang et al., 2013; Zhang et al., 2013; Fang et al., 2015a). Notably, a recent study showed that mRNA adenosine methylase (MTA), a homologous protein of animal METTL3, deposits m6A onto pri-miRNAs and may impact miRNA biogenesis via its dual interaction with Pol II and Tough (TGH), a known miRNA processing regulator (Ren et al., 2012b; Bhat et al., 2019). Another intriguing finding is that many proteins affecting MIRNA gene transcription, pri-miRNA stability, and/or processing have reported or proposed functions in RNA splicing (Yu et al., 2017b).
Regulation of RISC Assembly and Ago1 Stability
AGO proteins harbor conserved PAZ, MID, and PIWI domains. The MID and PAZ domains bind to the 5′ phosphate and 3′ end of small RNAs, respectively, while the PIWI domain cuts target RNAs through its endonuclease activity (Swarts et al., 2014). The 5′ terminal nucleotide and/or thermodynamic properties of the miRNA/miRNA∗ duplex are crucial for AGO sorting and strand selection (Khvorova et al., 2003; Schwarz et al., 2003; Mi et al., 2008; Takeda et al., 2008). AGO1 is the major effector protein and prefers miRNA cargos with a 5′ uracil (Mi et al., 2008). In contrast, AGO7 and AGO10 are predominantly associated with miR390 and miR165/166, respectively (Montgomery et al., 2008; Zhu et al., 2011). The miR390-AGO7 complex is specific for TAS3 ta-siRNA generation. AGO10 antagonizes AGO1 for miR165/166 loading, which ensures proper development of both shoot and floral apical meristems (Zhu et al., 2011). Interestingly, some miRNA∗s can preferentially and stably associate with AGO proteins and function like miRNAs (Mi et al., 2008). For example, miR393b∗-AGO2 targets MEMB12 for translation inhibition during Pseudomonas syringae pv. tomato infection (Zhang et al., 2011). Cumulative evidence shows that miRNA∗s can be as functional as miRNAs, and in some cases, both strands are abundantly expressed. In fact, the miR-5p/miR-3p name format is now adopted by the miRBase database to replace miRNA/miRNA∗ (Kozomara and Griffiths-Jones, 2014). Nevertheless, this change brings inconveniences and confusions, because it is almost impossible to know whether a given miR-X-3p refers to miR-X or miR-X∗ without checking the annotation.
The assembly of RISC requires the molecular chaperone HSP90, and this process is facilitated by Cyclophilin 40/Squint (CYP40/SQN) and inhibited by Protein Phosphatase 5 (PP5) (Figure 1C) (Iki et al., 2010, 2012; Iwasaki et al., 2010). Loss of CYP40/SQN phenotypically resembles hypomorphic alleles of ago1, likely due to reduced miRNA loading efficiency (Smith et al., 2009). CYP40 contains an N-terminal peptidyl-proline isomerase (PPIase) domain and a C-terminal tetratricopeptide repeat (TPR) domain. The TPR domain of CYP40 directly interacts with HSP90 variants containing a C-terminal MEEVD sequence, which is essential for CYP40 function in vivo (Earley and Poethig, 2011; Iki et al., 2012). In addition, the RISC loading process is negatively and positively regulated by two importin-β family proteins called Enhanced MiRNA Activity 1 (EMA1) and Transportin 1 (TRN1), respectively (Wang et al., 2011; Cui et al., 2016).
Expression of AGO1 is also under tight regulation. At the post-transcriptional level, AGO1 transcripts are targeted by miR168. On the other hand, AGO1 loading is important for the stabilization of miRNAs including miR168. Such feedback regulation is crucial for AGO1 homeostasis (Vaucheret et al., 2006). At the post-translational level, the endogenous F-Box protein FBW2 (for F-Box with WD-40 2) targets AGO1 for degradation (Earley et al., 2010). FBW2 was identified and characterized from a forward genetic screen for sqn suppressors. FBW2 mutations also suppress weak ago1 alleles, but not the null ago1-36 mutant. FBW2 may trigger AGO1 degradation through autophagy rather than the proteasome pathway, because treatment of MG132, a proteasome-specific inhibitor, does not affect AGO1 degradation (Earley et al., 2010). In addition to its roles in miRNA-related RNA degradation, AGO1 is also the major effector in siRNA-mediated post-transcriptional gene silencing and antiviral immunity (Carbonell and Carrington, 2015; Fang and Qi, 2016). AGO1 is one of the major targets of viral silencing suppressors. These include the F-Box Protein P0 of polerovirus and the RNA-binding protein P25 of potato virus X, which target AGO1 for degradation through autophagy and the proteasome pathways, respectively (Baumberger et al., 2007; Bortolamiol et al., 2007; Chiu et al., 2010; Csorba et al., 2010; Derrien et al., 2012).
miRNA Stability Control
Like all other RNAs, miRNAs have their own half-lives. Turnover and degradation of miRNAs are not only important for maintaining intracellular miRNA homeostasis, but also provide means for their clearance in response to developmental transitions and environmental changes. Factors affecting miRNA stability include 3′ end modification, AGO association, and miRNA-target RNA interaction (Figure 2). In addition, a growing number of exoribonulceases responsible for miRNA degradation at different stages have been characterized.
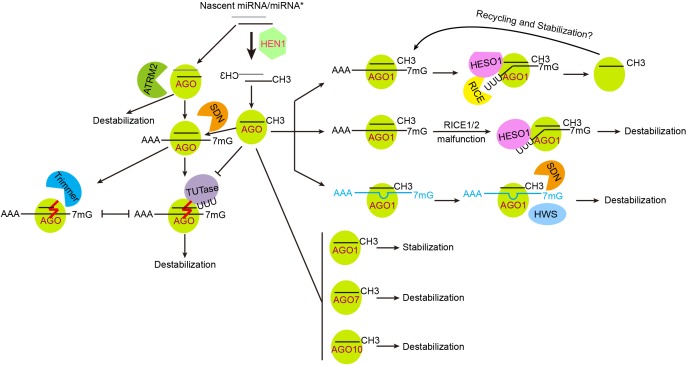
FIGURE 2.
Regulation of miRNA stability and turnover. Plant miRNAs are heavily methylated, which is crucial for their stabilization. ATRM2 is involved in the degradation of unmethylated miRNA/miRNA∗s, likely during the initiation of RISC loading. RISC-associated unmethylated miRNAs are destabilized via 3′ end tailing and 3′-to-5′ trimming. TUTase, terminal uridylyl transferase such as HESO1 and URT1 in Arabidopsis and MUT68 in Chlamydomonas reinhardtii. Trimmer, a yet uncharacterized enzyme catalyzing 3′-to-5′ trimming of unmethylated miRNAs. The effects of AGO proteins on miRNA stability are judged only by the results of ago mutants and/or over-expression transgenic plants, and are not necessarily absolute. Defects in the degradation of cleavage products (e.g., in the RICE1/2 malfunction backgrounds) cause miRNA reduction, suggesting that successful release of cleavage products may facilitate RISC recycling and stabilization. TM (in blue color) induced miRNA destabilization involves the actions of SDNs and an F-box protein HWS.
Regulation of miRNA Stability by 3′ End Modification
As a self-protection mechanism, mRNAs bear a 5′ cap and a 3′ poly(A) tail after transcription, which ensure their nuclear exportation and promote translation initiation (Garneau et al., 2007). Mature miRNAs lack such structures, but have 2′-O-methylation modification at their 3′ end. This modification, as catalyzed by HEN1, represents a critical step in miRNA stabilization in plants (Yu et al., 2005). Little is known about the regulation of HEN1 expression and activity. There is only one report showing that the expression of HEN1 is relatively low in the dark and is elevated upon light irradiation, which regulates photomorphogenesis by fine-tuning the expression of miR157d and miR319 (Tsai et al., 2014). Loss of HEN1 not only results in disrupted photomorphogenesis and skotomorphogenesis, but also other developmental defects including dwarfism, late flowering, and reduced fertility. At the molecular level, hen1 mutations cause miRNA destabilization and 3′ end heterogeneity due to combined actions of tailing and 3′-to-5′ truncation. Sequencing analysis of 3′ end tails reveals a dominant addition of uracils, a process that is termed uridylation (Li et al., 2005; Zhao et al., 2012; Wang et al., 2015). In Arabidopsis, HEN1 Suppressor 1 (HESO1) is the major terminal uridylyl transferase that catalyzes the addition of uracils to the 3′ end of unmethylated miRNAs (Ren et al., 2012a; Zhao et al., 2012). Loss of HESO1 greatly suppresses the hen1 phenotype while overexpression of HESO1 in hen1 results in further reduced miRNA levels and more drastic developmental defects, demonstrating that uridylation triggers miRNA degradation (Ren et al., 2012a). Another terminal uridylyl transferase, UTP:RNA uridylyltransferase 1 (URT1), only slightly affects the uridylation of a few miRNAs (e.g., miR158) when HESO1 is competent, but can significantly compensate for the loss of HESO1 (Tu et al., 2015; Wang et al., 2015). Different from HEN1, HESO1 and URT1 do not have apparent dsRNA binding domains, arguing against their actions on miRNA/miRNA∗ duplexes. Instead, both HESO1 and URT1 colocalize and interact with AGO1, and can uridylate AGO1-bound miRNAs (Ren et al., 2014b; Tu et al., 2015; Wang et al., 2015). Furthermore, miRNA tailing is largely diminished in two hypomorphic ago1 alleles (Zhai et al., 2013; Ren et al., 2014b). U-tails of unmethylated miRNAs were also abolished in the heso1-2 urt1-3 mutant, suggesting similar actions of HESO1 and URT1 on miRNAs at molecular level when HEN1 is fully functional (Wang et al., 2015). However, it is not known whether the plant genome encodes any miRNA “demethylases,” because it is difficult to distinguish whether unmetyhlated miRNAs in wild-type plants are demethylated products or those escaped from HEN1 targeting. Small RNA Degrading Nucleases (SDNs, see below for details), which harbors “demethylase”-like function by removing methylated 3′-end nucleotides, cooperate with HESO1/URT1 and contribute to the degradation of some methylated miRNAs (Ramachandran and Chen, 2008; Yu et al., 2017a).
In the unicellular green algae Chlamydomonas reinhardtii, the HESO1 homologous protein Mutator 68 (MUT68) uridylates both miRNAs and siRNAs for degradation, implying that uridylation triggered small RNA destabilization is highly conserved (Ibrahim et al., 2010). Interestingly, animal miRNAs do not require protection by methylation, probably due to different complementarity degrees required for target recognition (Ren et al., 2014a,b; Moran et al., 2017). Indeed, incorporation of perfect or near perfect complementary targets results in tailing and trimming of corresponding miRNA in Drosophila (Ameres et al., 2010). In addition to uridylation, miRNAs are also subject to oligo-adenylation modification. In Populus trichocarpa (black cottonwood), a substantial proportion of miRNAs contain one or more non-template adenines, which play a protective role (Lu et al., 2009). Other types of non-templated nucleotide addition are observed, and become more evident in the hen1 heso1 urt1 triple mutant, suggesting both hierarchical actions and compensation effects among different terminal nucleotidyl transferases (Wang et al., 2015).
Exoribonucleases Involved in miRNA Surveillance and Degradation
The requirement of 3′ end methylation for stabilization and 3′-to-5′ truncation of miRNAs in the hen1 mutant indicates that miRNAs are more likely degraded from 3′ to 5′, although theoretically they could be degraded from both directions by exoribonucleases, or cleaved by endoribonucleases. In Caenorhabditis elegans, the 5′-to-3′ exoribonuclease XRN-2 has been implicated in miRNA decay (Chatterjee and Grosshans, 2009). In human HEK293 cells, a subset of miRNAs, including miR382, is degraded by the 3′-to-5′ exosome exoribonucleases RRP41 and XRN-1 (Bail et al., 2010). More recently, the human endonuclease Tudor-SN (TSN) was shown to degrade both AGO2-bound and effector-free miRNAs bearing CA and/or UA dinucleotides (Elbarbary et al., 2017). In addition, a number of 3′-to-5′ exoribonucleases have been reported to target small RNAs in a variety of organisms, such as MUT-7 and Poly(A)-specific Ribonuclease (PARN-1) in C. elegans, QDE-2-interacting protein (QIP) in Neurospora crassa, Nibbler in Drosophila, and PARN-Like Domain Containing 1 (PNLDC1) in silkworms and mice (Tops et al., 2005; Maiti et al., 2007; Han et al., 2011; Liu et al., 2011; Izumi et al., 2016; Tang et al., 2016; Zhang et al., 2017a). The 3′-to-5′ exoribonucleases consist of five superfamilies, DEDD, RRP4, PDX, RBN, and RNR, with each having multiple members in plants (Wang X. Y. et al., 2018). Searching for 3′-to-5′ exoribonucleases involved in miRNA degradation led to the identification of SDNs, which belong to the DEDDh subclass of exoribonucleases (Ramachandran and Chen, 2008). SDN1 is capable of degrading single-stranded small RNAs of 17-27 nt in length in vitro, but is inactive on miRNA/miRNA∗ duplexes and long single-stranded RNAs. Importantly, methylation only slightly impedes SDN1 activity, making it an excellent candidate for degrading methylated miRNAs (Ramachandran and Chen, 2008). Deep sequencing analysis of AGO1- and AGO10-bound miRNAs after SDN1 treatment suggests that SDN1 is responsible for the truncation of both methylated and unmethylated miRNAs, which are further degraded via uridylation-dependent or -independent pathways (Yu et al., 2017a; Chen et al., 2018). Structural and biochemical analysis of SDN1 further reveals a dynamic interaction among SDN1, AGO1 and target RNA, which provides detailed insights into the action of SDN1 on AGO-bound miRNAs (Chen et al., 2018). Since SDN1 is unresponsive to miRNAs with U-tails in vitro, the nuclease that degrades uridylated miRNAs remains to be identified. In animals, the RNase II family exoribonuclease DIS3L2 specifically binds and degrades uridylated RNA substrates including uridylated pre-let-7, 7SL, and snRNAs (Chang et al., 2013; Labno et al., 2016; Pirouz et al., 2016). DIS3L2 belongs to the RNR superfamily and the closest homologous gene to DIS3L2 in Arabidopsis is SOV. However, SOV is inactive in the Col-0 background, reducing its likelihood of being the enzyme that degrades uridylated miRNAs (Zhang et al., 2010). In Chlamydomonas, the exosome auxiliary nuclease RRP6 acts cooperatively with MUT68 in the degradation of unmethylated miRNAs (Ibrahim et al., 2010). There are three RRP6 homologous proteins with different subcellular locations in Arabidopsis. However, the quadruple mutant hen1 rrp6l1 rrp6l2 rrp6l3 is morphologically indistinguishable from the hen1 single mutant, suggesting that RRP6-like proteins are unlikely involved in degrading uridylated miRNAs (Wang X. Y. et al., 2018). Interestingly, knock-out of ATRM2, a DEDDy type exoribonucleases, significantly rescues the hen1 phenotype. ATRM2 acts downstream of HEN1 and may be involved in the degradation of unmethylated miRNA/miRNA∗ duplexes during RISC assembly (Figure 2) (Wang X. Y. et al., 2018).
Ago Proteins Affecting miRNA Stability
AGO proteins not only serve as the effector protein, but also influence the stability of miRNAs. Arabidopsis AGO1 is suggested to stabilize miRNAs in general since miRNA abundances are decreased in the ago1 knock-out mutant (Vaucheret et al., 2004). Similar observations are obtained in human cells, where overexpression of AGO2 promotes miRNA accumulation and increases the half-lives of some miRNAs, while knocking out AGO2 reduces miRNA levels (Diederichs and Haber, 2007). The effect of AGO2 on miRNA stability appears independent of its endonuclease activity as miRNA abundances are not affected in an ago2 mutant specifically defective in cleavage (Diederichs and Haber, 2007). However, different AGO proteins may have different effects on the stability of their bound miRNAs (Figure 2). For instance, Arabidopsis ago10 mutants exhibit over-accumulation of miR165/166, whereas overexpression of AGO10 results in miR165/166 reduction (Yu et al., 2017a). Similarly, in both tomato and Arabidopsis ago7 mutants, increases in miR390 abundance are observed, although the underlying mechanism is unknown (Yifhar et al., 2012; Li J. et al., 2017). As multiple degradative enzymes, such as SDN1, HESO1, and URT1, act on AGO-associated miRNAs, it is not surprising that AGO proteins may simultaneously have two opposing roles on miRNA stability (i.e., protect miRNAs from exposure to various intracellular RNases on the one hand, while recruiting degradative factors for active turnover on the other hand).
Target Rnas Affecting miRNA Stability
As mentioned above, introduction of highly complementary target RNAs initiates miRNA destabilization in animals via tailing and truncation at their 3′ ends, mimicking results observed in plant hen1 mutants (Li et al., 2005; Ameres et al., 2010). The phenomenon is termed target RNA-directed miRNA degradation (TDMD) in animals (de la Mata et al., 2015). This mutual degradation mechanism (i.e., between a miRNA and its target genes), together with a miRNA gradient generated by cell-to-cell diffusion, sharpens spatial expression of target genes, which plays a critical role in morphogenesis such as root vascular patterning and leaf polarity establishment (Hamelryck et al., 2006; Muraro et al., 2014; Ramachandran et al., 2017). In addition to highly complementary target RNAs, mimic targets can also trigger destabilization of miRNAs (Figure 2). Target mimicry was initially reported in plants (Franco-Zorrilla et al., 2007). A non-coding gene, IPS1, harbors a non-canonical miR399 targeting site by the presence of a 3-nt bulge at the miR399 cleavage site, which inhibits its cleavage by miR399. IPS1 serves as a target mimic (TM) to sequester miR399 from its endogenous true target genes (Franco-Zorrilla et al., 2007). Target mimicry may be widespread since many potential endogenous target mimics (eTM) were found in the Arabidopsis, rice and wild emmer wheat genomes based on bioinformatic predictions (Wu et al., 2013; Akpinar et al., 2018). MIM and Short Tandem Target Mimic (STTM) are artificially designed TM technologies. Both TM strategies effectively reduce miRNA abundance (Todesco et al., 2010; Yan et al., 2012). SDN1/2 is involved in this process, as the sdn1 sdn2 double knock-out mutant significantly suppresses developmental defects caused by STTM (Yan et al., 2012). It is probable that TM may lead to conformational and/or post-translational modification changes in AGO1 (Golden et al., 2017; Huberdeau et al., 2017; Kobayashi et al., 2019), which releases the 3′ end of miRNAs from the PAZ domain of AGO1 and increases their susceptibility to SDN1/2 (Chen et al., 2018). Moreover, loss of function in an F-box family protein HWS (Hawaiian Skirt) suppresses both MIM and STTM-induced developmental defects (Lang et al., 2018; Mei et al., 2018). Intriguingly, miRNA and its corresponding mimicry targets are stably coexisted in the AGO1 immunoprecipitates when HWS is compromised, suggesting that HWS may specifically trigger degradation of non-optimal RISCs (RISCs associated with mimicry target RNAs) (Mei et al., 2018) (Figure 2). The relationship between SDN1/2 and HWS in TM-induced miRNA decay awaits future investigation. Although targets with high complementary cause miRNA destabilization, mRNA targeting could be beneficial for miRNA stabilization (Figure 2). In C. elegans, efficient targeting protects let-7 miRNA from XRN2-mediated 5′-to-3′ clearance (Chatterjee and Grosshans, 2009). In Arabidopsis, DnaQ-like exonucleases RICE1 and 2 (for RISC-interacting Clearing 3′-to-5′ exoribonucleases) interact with AGO1 and AGO10, and degrade uridylated miRNA cleavage products for their clearance (Ren et al., 2014b; Zhang et al., 2017c; Zuber et al., 2018). Disruptions of RICE function by either simultaneously knocking down RICE1 and 2 or overexpressing catalytically inactive RICE1 leads to reduced miRNA levels. These data suggest that over-accumulation of cleaved targets could also affect the stability of RISC-associated miRNAs (Zhang et al., 2017c).
Future Perspectives
The identification of many players in miRNA transcription and processing has shed light on co-transcriptional splicing, modification, and processing of pri-miRNAs. However, key challenges remain with respect to the relationships and precise biochemical contributions of these players. At the subcellular level, it is of particular interest to investigate how the dicing body is formed, its composition, and its role in co-transcriptional pri-miRNA processing. Implementation of novel techniques such as single-cell biology and in vitro re-constitution of the dicing machinery will be crucial to tackle these problems. Although many factors, such as AGOs, targets, and 3′ modifications are known to affect miRNA stability, not much is known about the underlying mechanisms. Future studies are needed to identify and characterize additional enzymes and modulators involved in these processes. It will also be important to understand the biological significance of either global or sequence-specific miRNA degradation during developmental transitions and in response to environmental stimuli.
Author Contributions
JW and JM drafted the manuscript and the figures. GR conceived the idea and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. Work in the Ren laboratory is supported by grants from the National Natural Science Foundation of China (91740101 and 31622009), the National Key R&D Program of China (2016YFA0503200), and the Science and Technology Commission of Shanghai Municipality (15JC1400800).
References
- Achkar N. P., Cho S. K., Poulsen C., Arce A. L., Re D. A., Giudicatti A. J., et al. (2018). A Quick HYL1-dependent reactivation of MicroRNA production is required for a proper developmental response after extended periods of light deprivation. Dev Cell 46 236–247. 10.1016/j.devcel.2018.06.014 [DOI] [PubMed] [Google Scholar]
- Addo-Quaye C., Snyder J. A., Park Y. B., Li Y. F., Sunkar R., Axtell M. J. (2009). Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 15 2112–2121. 10.1261/rna.1774909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V., Bell G. W., Nam J. W., Bartel D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:e05005. 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akpinar B. A., Biyiklioglu S., Alptekin B., Havrankova M., Vrana J., Dolezel J., et al. (2018). Chromosome-based survey sequencing reveals the genome organization of wild wheat progenitor Triticum dicoccoides. Plant Biotechnol. J. 16 2077–2087. 10.1111/pbi.12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres S. L., Horwich M. D., Hung J. H., Xu J., Ghildiyal M., Weng Z. P., et al. (2010). Target RNA-directed trimming and tailing of small silencing RNAs. Science 328 1534–1539. 10.1126/science.1187058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M. J., Meyers B. C. (2018). Revisiting criteria for plant MicroRNA annotation in the era of big data. Plant Cell 30 272–284. 10.1105/tpc.17.00851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bail S., Swerdel M., Liu H. D., Jiao X. F., Goff L. A., Hart R. P., et al. (2010). Differential regulation of microRNA stability. RNA 16 1032–1039. 10.1261/rna.1851510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2018). Metazoan MicroRNAs. Cell 173 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Tsai C. H., Lie M., Havecker E., Baulcombe D. C. (2007). The polerovirus silencing suppressor PO targets ARGONAUTE proteins for degradation. Curr. Biol. 17 1609–1614. 10.1016/j.cub.2007.08.039 [DOI] [PubMed] [Google Scholar]
- Ben Chaabane S., Liu R. Y., Chinnusamy V., Kwon Y., Park J. H., Kim S. Y., et al. (2013). STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res. 41 1984–1997. 10.1093/nar/gks1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S. S., Bielewicz D., Grzelak N., Gulanicz T., Bodi Z., Szewc L., et al. (2019). mRNA adenosine methylase (MTA) deposits m6A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana. bioRxiv. 10.1101/557900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna N. G., Iselin R., Abriata L. A., Sarazin A., Pumplin N., Jay F., et al. (2018). Nucleo-cytosolic Shuttling of ARGONAUTE1 prompts a revised model of the plant MicroRNA Pathway. Mol. Cell 69 709–719. 10.1016/j.molcel.2018.01.007 [DOI] [PubMed] [Google Scholar]
- Bologna N. G., Mateos J. L., Bresso E. G., Palatnik J. F. (2009). A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. Embo J. 28 3646–3656. 10.1038/emboj.2009.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna N. G., Schapire A. L., Zhai J. X., Chorostecki U., Boisbouvier J., Meyers B. C., et al. (2013). Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 23 1675–1689. 10.1101/gr.153387.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna N. G., Voinnet O. (2014). The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65 473–503. 10.1146/annurev-arplant-050213-035728 [DOI] [PubMed] [Google Scholar]
- Borges F., Martienssen R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16 727–741. 10.1038/nrm4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolamiol D., Pazhouhandeh M., Marrocco K., Genschik P., Ziegler-Graff V. (2007). The polerovirus F box protein PO targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 17 1615–1621. 10.1016/j.cub.2007.07.061 [DOI] [PubMed] [Google Scholar]
- Brant E. J., Budak H. (2018). Plant small non-coding RNAs and their roles in biotic stresses. Front. Plant Sci. 9:1038. 10.3389/fpls.2018.01038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budak H., Akpinar B. A. (2015). Plant miRNAs: biogenesis, organization and origins. Funct. Integr. Genomic 15 523–531. 10.1007/s10142-015-0451-2 [DOI] [PubMed] [Google Scholar]
- Budak H., Kantar M., Bulut R., Akpinar B. A. (2015). Stress responsive miRNAs and isomiRs in cereals. Plant Sci. 235 1–13. 10.1016/j.plantsci.2015.02.008 [DOI] [PubMed] [Google Scholar]
- Cai Q., Liang C., Wang S. K., Hou Y. N., Gao L., Liu L., et al. (2018). The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat. Commun. 9:5080. 10.1038/s41467-018-07516-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A., Carrington J. C. (2015). Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 27 111–117. 10.1016/j.pbi.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A., Fahlgren N., Garcia-Ruiz H., Gilbert K. B., Montgomery T. A., Nguyen T., et al. (2012). Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24 3613–3629. 10.1105/tpc.112.099945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. M., Triboulet R., Thornton J. E., Gregory R. I. (2013). A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 497 244–248. 10.1038/nature12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Grosshans H. (2009). Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461 546–549. 10.1038/nature08349 [DOI] [PubMed] [Google Scholar]
- Chen J., Liu L., You C. J., Gu J. Q., Ruan W. J., Zhang L., et al. (2018). Structural and biochemical insights into small RNA 3′ end trimming by Arabidopsis SDN1. Nat. Commun. 9:3585. 10.1038/s41467-018-05942-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu M. H., Chen I. H., Baulcombe D. C., Tsai C. H. (2010). The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 11 641–649. 10.1111/J.1364-3703.2010.00634.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. K., Ben Chaabane S., Shah P., Poulsen C. P., Yang S. W. (2014). COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat. Commun. 5:5867. 10.1038/ncomms6867 [DOI] [PubMed] [Google Scholar]
- Creasey K. M., Zhai J. X., Borges F., Van Ex F., Regulski M., Meyers B. C., et al. (2014). miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508 411–415. 10.1038/nature13069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T., Lozsa R., Hutvagner G., Burgyan J. (2010). Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 62 463–472. 10.1111/j.1365-313X.2010.04163.x [DOI] [PubMed] [Google Scholar]
- Cui Y. W., Fang X. F., Qi Y. J. (2016). TRANSPORTIN1 Promotes the Association of MicroRNA with ARGONAUTE1 in Arabidopsis. Plant Cell 28 2576–2585. 10.1105/tpc.16.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B., Munafo M., Ciabrelli F., Eastwood E. L., Fabry M. H., Kneuss E., et al. (2018). piRNA-guided genome defense: from biogenesis to silencing. Annu. Rev. Genet. 52 131–157. 10.1146/annurev-genet-120417-031441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M., Gaidatzis D., Vitanescu M., Stadler M. B., Wentzel C., Scheiffele P., et al. (2015). Potent degradation of neuronal miRNAs induced by highly complementary targets. Embo Rep. 16 500–511. 10.15252/embr.201540078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P. C., Muhammad S., Cao M., Wu L. (2018). Biogenesis and regulatory hierarchy of phased small interfering RNAs in plants. Plant Biotechnol. J. 16 965–975. 10.1111/pbi.12882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien B., Baumberger N., Schepetilnikov M., Viotti C., De Cillia J., Ziegler-Graff V., et al. (2012). Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. U.S.A. 109 15942–15946. 10.1073/pnas.1209487109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S., Haber D. A. (2007). Dual role for argonautes in MicroRNA processing and Posttranscriptional regulation of MicroRNA expression. Cell 131 1097–1108. 10.1016/j.cell.2007.10.032 [DOI] [PubMed] [Google Scholar]
- Dolata J., Bajczyk M., Bielewicz D., Niedojadlo K., Niedojadlo J., Pietrykowska H., et al. (2016). Salt Stress Reveals a New Role for ARGONAUTE1 in miRNA biogenesis at the transcriptional and posttranscriptional levels. Plant Physiol. 172 297–312. 10.1104/pp.16.00830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Han M. H., Fedoroff N. (2008). The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. U.S.A. 105 9970–9975. 10.1073/pnas.0803356105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K., Smith M., Weber R., Gregory B., Poethig R. (2010). An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana. Silence 1:15. 10.1186/1758-907X-1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K. W., Poethig R. S. (2011). Binding of the Cyclophilin 40 Ortholog SQUINT to Hsp90 protein is required for SQUINT function in Arabidopsis. J. Biol. Chem. 286 38184–38189. 10.1074/jbc.M111.290130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary R. A., Miyoshi K., Myers J. R., Du P. C., Ashton J. M., Tian B., et al. (2017). Tudor-SN-mediated endonucleolytic decay of human cell microRNAs promotes G(1)/S phase transition. Science 356 859–862. 10.1126/science.aai9372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X. F., Cui Y. W., Li Y. X., Qi Y. J. (2015a). Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat. Plants 1:15075. 10.1038/Nplants.2015.75 [DOI] [PubMed] [Google Scholar]
- Fang X. F., Qi Y. J. (2016). RNAi in plants: an argonaute-centered view. Plant Cell 28 272–285. 10.1105/tpc.15.00920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X. F., Shi Y. P., Lu X. L., Chen Z. L., Qi Y. J. (2015b). CMA33/XCT regulates small RNA production through modulating the transcription of dicer-like genes in Arabidopsis. Mol. Plant 8 1227–1236. 10.1016/j.molp.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Fang Y. D., Spector D. L. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17 818–823. 10.1016/j.cub.2007.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q. L., Xia R., Meyers B. C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25 2400–2415. 10.1105/tpc.113.114652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Valli A., Todesco M., Mateos I., Puga M. I., Rubio-Somoza I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet 39 1033–1037. 10.1038/ng2079 [DOI] [PubMed] [Google Scholar]
- Fujioka Y., Utsumi M., Ohba Y., Watanabe Y. (2007). Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 48 1243–1253. 10.1093/pep/pcm099 [DOI] [PubMed] [Google Scholar]
- Garneau N. L., Wilusz J., Wilusz C. J. (2007). The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8 113–126. 10.1038/nrm2104 [DOI] [PubMed] [Google Scholar]
- Golden R. J., Chen B., Li T., Braun J., Manjunath H., Chen X., et al. (2017). An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 542 197–202. 10.1038/nature21025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajheidari M., Farrona S., Huettel B., Koncz Z., Koncz C. (2012). CDKF;1 and CDKD Protein Kinases Regulate Phosphorylation of Serine Residues in the C-Terminal Domain of Arabidopsis RNA Polymerase II. Plant Cell 24 1626–1642. 10.1105/tpc.112.096834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelryck T., Kent J. T., Krogh A. (2006). Sampling realistic protein conformations using local structural bias. PLoS Comput. Biol. 2:e131. 10.1371/journal.pcbi.0020131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B. W., Hung J. H., Weng Z. P., Zamore P. D., Ameres S. L. (2011). The 3′-to-5′ Exoribonuclease Nibbler Shapes the 3′ Ends of MicroRNAs Bound to Drosophila Argonaute1. Curr. Biol. 21 1878–1887. 10.1016/j.cub.2011.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M. H., Goud S., Song L., Fedoroff N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. U.S.A. 101 1093–1098. 10.1073/pnas.0307969100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A., Kudla G., Dudnakova T., Tollervey D. (2013). Mapping the Human miRNA Interactome by CLASH reveals frequent noncanonical binding. Cell 153 654–665. 10.1016/j.cell.2013.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberdeau M. Q., Zeitler D. M., Hauptmann J., Bruckmann A., Fressigne L., Danner J., et al. (2017). Phosphorylation of Argonaute proteins affects mRNA binding and is essential for microRNA-guided gene silencing in vivo. Embo J. 36 2088–2106. 10.15252/embj.201696386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim F., Rymarquis L. A., Kim E. J., Becker J., Balassa E., Green P. J., et al. (2010). Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 107 3906–3911. 10.1073/pnas.0912632107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Meshi T., Ishikawa M. (2012). Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. Embo J. 31 267–278. 10.1038/emboj.2011.395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Nishikiori M., Jaudal M. C., Matsumoto-Yokoyama E., Mitsuhara I., et al. (2010). In vitro assembly of Plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell 39 282–291. 10.1016/j.molcel.2010.05.014 [DOI] [PubMed] [Google Scholar]
- Iwasaki S., Kobayashi M., Yoda M., Sakaguchi Y., Katsuma S., Suzuki T., et al. (2010). Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 39 292–299. 10.1016/j.molcel.2010.05.015 [DOI] [PubMed] [Google Scholar]
- Izumi N., Shoji K., Sakaguchi Y., Honda S., Kirino Y., Suzuki T., et al. (2016). Identification and Functional Analysis of the Pre-piRNA 3′ Trimmer in Silkworms. Cell 164 962–973. 10.1016/j.cell.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson P., Christie M. D., Seymour D. K., Wang H., Wang X., Hagmann J., et al. (2015). KH domain protein RCF3 is a tissue-biased regulator of the plant miRNA biogenesis cofactor HYL1. Proc. Natl. Acad. Sci. U.S.A. 112 14096–14101. 10.1073/pnas.1512865112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S. D. (2003). Functional siRNAs and miRNAs exhibit strand bias. Cell 115 505–505. 10.1016/S0092-8674(03)00893-6 [DOI] [PubMed] [Google Scholar]
- Kim W., Benhamed M., Servet C., Latrasse D., Zhang W., Delarue M., et al. (2009). Histone acetyltransferase GCN5 interferes with the miRNA pathway in Arabidopsis. Cell Res. 19 899–909. 10.1038/cr.2009.59 [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Zheng B. L., Yu Y., Won S. Y., Mo B. X., Chen X. M. (2011). The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. Embo J. 30 814–822. 10.1038/emboj.2011.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H., Shoji K., Kiyokawa K., Negishi L., Tomari Y. (2019). Iruka Eliminates Dysfunctional Argonaute by Selective Ubiquitination of Its Empty State. Mol Cell 73 119–129.e5. 10.1016/j.molcel.2018.10.033 [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42 D68–D73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labno A., Warkocki Z., Kulinski T., Krawczyk P. S., Bijata K., Tomecki R., et al. (2016). Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Res. 44 10437–10453. 10.1093/nar/gkw649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang P. L. M., Christie M. D., Dogan E. S., Schwab R., Hagmann J., van de Weyer A. L., et al. (2018). A Role for the F-Box Protein HAWAIIAN SKIRT in Plant microRNA Function. Plant Physiol. 176 730–741. 10.1104/pp.17.01313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J. U., Rascht G., et al. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105 8795–8800. 10.1073/pnas.0802493105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang Z., Hu Y. G., Cao Y., Ma L. G. (2017). Polycomb group proteins RING1A and RING1B regulate the vegetative phase transition in Arabidopsis. Front. Plant Sci. 8:867. 10.3389/fpls.2017.00867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. J., Yang Z. Y., Yu B., Liu J., Chen X. M. (2005). Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabildopsis. Curr. Biol. 15 1501–1507. 10.1016/j.cub.2005.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Xu R., Li A., Liu K., Gu L., Li M., et al. (2018). SMA1, a homolog of the splicing factor Prp28, has a multifaceted role in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 46 9148–9159. 10.1093/nar/gky591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. J., Castillo-Gonzalez C., Yu B., Zhang X. R. (2017). The functions of plant small RNAs in development and in stress responses. Plant J. 90 654–670. 10.1111/tpj.13444 [DOI] [PubMed] [Google Scholar]
- Liu C., Xin Y., Xu L., Cai Z. K., Xue Y. C., Liu Y., et al. (2018). Arabidopsis ARGONAUTE 1 binds chromatin to promote gene transcription in response to hormones and stresses. Dev. Cell 44 348–361.e7. 10.1016/j.devcel.2017.12.002 [DOI] [PubMed] [Google Scholar]
- Liu C. G., Axtell M. J., Fedoroff N. V. (2012). The helicase and RNaseIIIa domains of Arabidopsis dicer-like1 modulate catalytic parameters during MicroRNA Biogenesis. Plant Physiol. 159 748–758. 10.1104/pp.112.193508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Abe M., Sabin L. R., Hendriks G. J., Naqvi A. S., Yu Z. M., et al. (2011). The Exoribonuclease Nibbler Controls 3′ End Processing of MicroRNAs in Drosophila. Curr. Biol. 21 1888–1893. 10.1016/j.cub.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yan Q. Q., Liu Y., Hong F., Sun Z. F., Shi L. L., et al. (2013). Complementation of hyponastic leaves1 by double-strand RNA-binding domains of dicer-like1 in nuclear dicing bodies. Plant Physiol. 163 108–117. 10.1104/pp.113.219071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbes D., Rallapalli G., Schmidt D. D., Martin C., Clarke J. (2006). SERRATE: a new player on the plant microRNA scene. Embo Rep. 7 1052–1058. 10.1038/sj.embor.7400806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. F., Sun Y. H., Chiang V. L. (2009). Adenylation of plant miRNAs. Nucleic Acids Res. 37 1878–1885. 10.1093/nar/gkp031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C. Y., Sidote D. J., Zhang Y., Kerstetter R. A., Michael T. P., Lam E. (2013). Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J. 73 77–90. 10.1111/tpj.12017 [DOI] [PubMed] [Google Scholar]
- Ma Z. Y., Castillo-Gonzalez C., Wang Z. Y., Sun D., Hu X. M., Shen X. F., et al. (2018). Arabidopsis serrate coordinates histone methyltransferases ATXR5/6 and RNA processing factor RDR6 to regulate transposon expression. Dev. Cell 45 769–784. 10.1016/j.devcel.2018.05.023 [DOI] [PubMed] [Google Scholar]
- Maiti M., Lee H. C., Liu Y. (2007). QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Gene Dev. 21 590–600. 10.1101/gad.1497607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P. A., Hagmann J., Ott F., Laubinger S., Franz M., Macek B., et al. (2012). Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151 859–870. 10.1016/j.cell.2012.09.039 [DOI] [PubMed] [Google Scholar]
- Mateos J. L., Bologna N. G., Chorostecki U., Palatnik J. F. (2010). Identification of MicroRNA processing determinants by random mutagenesis of Arabidopsis MIR172a Precursor. Curr. Biol. 20 49–54. 10.1016/j.cub.2009.10.072 [DOI] [PubMed] [Google Scholar]
- Mei J., Jiang N., Ren G. (2018). The F-box protein HAWAIIAN SKIRT is required for mimicry target induced microRNA degradation in Arabidopsis. J. Integr. Plant Biol. 10.1111/jipb.12761 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Mi S. J., Cai T., Hu Y. G., Chen Y., Hodges E., Ni F. R., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133 116–127. 10.1016/j.cell.2008.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. A., Waterhouse P. M. (2005). Plant and animal microRNAs: similarities and differences. Funct. Integr. Genomic 5 129–135. 10.1007/s10142-005-0145-2 [DOI] [PubMed] [Google Scholar]
- Montgomery T. A., Howell M. D., Cuperus J. T., Li D. W., Hansen J. E., Alexander A. L., et al. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133 128–141. 10.1016/j.cell.2008.02.033 [DOI] [PubMed] [Google Scholar]
- Moran Y., Agron M., Praher D., Technau U. (2017). The evolutionary origin of plant and animal microRNAs. Nat. Ecol. Evol. 1:27. 10.1038/s41559-016-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro D., Mellor N., Pound M. P., Help H., Lucas M., Chopard J., et al. (2014). Integration of hormonal signaling networks and mobile microRNAs is required for vascular patterning in Arabidopsis roots. Proc. Natl. Acad. Sci. U.S.A. 111 857–862. 10.1073/pnas.1221766111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu D. D., Lii Y. E., Chellappan P., Lei L., Peralta K., Jiang C. H., et al. (2016). miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat. Commun. 7:11324. 10.1038/ncomms11324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M., Miura S., Nei M. (2012). Origins and Evolution of MicroRNA Genes in Plant Species. Genome Biol. Evol. 4 230–239. 10.1093/gbe/evs002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. Y., Wu G., Gonzalez-Sulser A., Vaucheret H., Poethig R. S. (2005). Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102 3691–3696. 10.1073/pnas.0405570102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirouz M., Du P., Munafo M., Gregory R. I. (2016). Dis3l2-mediated decay is a quality control pathway for noncoding RNAs. Cell Rep. 16 1861–1873. 10.1016/j.celrep.2016.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouch-Pelissier M. N., Pelissier T., Elmayan T., Vaucheret H., Boko D., Jantsch M. F., et al. (2008). SINE RNA induces severe developmental defects in Arabidopsis thaliana and interacts with HYL1 (DRB1), a Key Member of the DCL1 Complex. PLoS Genet. 4:e1000096. 10.1371/journal.pgen.1000096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram B., Sheikh A. H., Rustagi Y., Sinha A. K. (2015). MicroRNA biogenesis factor DRB1 is a phosphorylation target of mitogen activated protein kinase MPK3 in both rice and Arabidopsis. Febs J. 282 521–536. 10.1111/febs.13159 [DOI] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Gene Dev. 20 3407–3425. 10.1101/gad.1476406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P., Carlsbecker A., Etchells J. P. (2017). Class III HD- ZIPs govern vascular cell fate: an HD view on patterning and differentiation. J. Exp. Bot. 68 55–69. 10.1093/jxb/erw370 [DOI] [PubMed] [Google Scholar]
- Ramachandran V., Chen X. M. (2008). Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321 1490–1492. 10.1126/science.1163728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G. D., Chen X. M., Yu B. (2012a). Uridylation of miRNAs by HEN1 SUPPRESSOR1 in Arabidopsis. Curr. Biol. 22 695–700. 10.1016/j.cub.2012.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G. D., Chen X. M., Yu B. (2014a). Small RNAs meet their targets: When methylation defends miRNAs from uridylation. RNA Biol. 11 1099–1104. 10.4161/rna.36243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G. D., Xie M., Dou Y. C., Zhang S. X., Zhang C., Yu B. (2012b). Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109 12817–12821. 10.1073/pnas.1204915109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G. D., Xie M., Zhang S. X., Vinovskis C., Chen X. M., Yu B. (2014b). Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc. Natl. Acad. Sci. U.S.A. 111 6365–6370. 10.1073/pnas.1405083111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K., Chen X. M. (2013). Biogenesis, turnover, and mode of action of plant MicroRNAs. Plant Cell 25 2383–2399. 10.1105/tpc.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Palatnik J. F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of MicroRNAs on the plant transcriptome. Dev. Cell 8 517–527. 10.1016/j.devcel.2005.01.018 [DOI] [PubMed] [Google Scholar]
- Schwarz D. S., Hutvagner G., Du T., Xu Z. S., Aronin N., Zamore P. D. (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cell 115 199–208. 10.1016/S0092-8674(03)00759-1 [DOI] [PubMed] [Google Scholar]
- Shriram V., Kumar V., Devarumath R. M., Khare T. S., Wani S. H. (2016). MicroRNAs As potential targets for abiotic stress tolerance in plants. Front. Plant Sci. 7:817. 10.3389/fpls.2016.00817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. R., Willmann M. R., Wu G., Berardini T. Z., Moller B., Weijers D., et al. (2009). Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 5424–5429. 10.1073/pnas.0812729106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Axtell M. J., Fedoroff N. V. (2010). RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 20 37–41. 10.1016/j.cub.2009.10.076 [DOI] [PubMed] [Google Scholar]
- Speth C., Szabo E. X., Martinho C., Collani S., zur Oven-Krockhaus S., Richter S., et al. (2018). Arabidopsis RNA processing factor SERRATE regulates the transcription of intronless genes. Elife 7:e37078. 10.7554/eLife.37078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C. B., Li Z. W., Cheng J. P., Li L., Zhong S. X., Liu L., et al. (2017). The Protein Phosphatase 4 and SMEK1 Complex Dephosphorylates HYL1 to Promotemi RNA Biogenesis by Antagonizing the MAPK Cascade in Arabidopsis. Dev. Cell 41 527–539. 10.1016/j.devcel.2017.05.008 [DOI] [PubMed] [Google Scholar]
- Sun Z. F., Guo T. T., Liu Y., Liu Q., Fang Y. D. (2015). The Roles of Arabidopsis CDF2 in transcriptional and posttranscriptional regulation of primary MicroRNAs. PLoS Genet. 11:e1005598. 10.1371/journal.pgen.1005598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts D. C., Makarova K., Wang Y. L., Nakanishi K., Ketting R. F., Koonin E. V., et al. (2014). The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 21 743–753. 10.1038/nsmb.2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami Y., Motose H., Watanabe Y. (2009). A dominant mutation in DCL1 suppresses the hyl1 mutant phenotype by promoting the processing of miRNA. RNA 15 450–458. 10.1261/rna.1297109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A., Iwasaki S., Watanabe T., Utsumi M., Watanabe Y. (2008). The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant Cell Physiol. 49 493–500. 10.1093/pcp/pcn043 [DOI] [PubMed] [Google Scholar]
- Tang W., Tu S., Lee H. C., Weng Z. P., Mello C. C. (2016). The RNase PARN-1 Trims piRNA 3′ ends to promote transcriptome surveillance in C. elegans. Cell 164 974–984. 10.1016/j.cell.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. (2010). A collection of target mimics for comprehensive analysis of MicroRNA function in Arabidopsis thaliana. PLoS Genet. 6:e1001031. 10.1371/journal.pgen.1001031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tops B. B. J., Tabara H., Sijen T., Simmer F., Mello C. C., Plasterk R. H. A., et al. (2005). RDE-2 interacts with MUT-7 to mediate RNA interference in Caenorhabditis elegans. Nucleic Acids Res. 33 347–355. 10.1093/nar/gki183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T., Treiber N., Meister G. (2019). Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 20 5–20. 10.1038/s41580-018-0059-1 [DOI] [PubMed] [Google Scholar]
- Tsai H. L., Li Y. H., Hsieh W. P., Lin M. C., Ahn J. H., Wu S. H. (2014). HUA ENHANCER1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis. Plant Cell 26 2858–2872. 10.1105/tpc.114.126722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu B., Liu L., Xu C., Zhai J. X., Li S. B., Lopez M. A., et al. (2015). Distinct and Cooperative Activities of HESO1 and URT1 Nucleotidyl Transferases in MicroRNA Turnover in Arabidopsis. PLoS Genet. 11:e1005119. 10.1371/journal.pgen.1005119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Mallory A. C., Bartel D. P. (2006). AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 22 129–136. 10.1016/j.molcel.2006.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crete P., Bartel D. P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Gene Dev. 18 1187–1197. 10.1101/gad.1201404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Gasciolli V., Crete P., Vaucheret H. (2004). The nuclear dsRNA binding protein HYL1 is required for MicroRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14 346–351. 10.1016/j.cub.2004.01.035 [DOI] [PubMed] [Google Scholar]
- Wang L. L., Song X. W., Gu L. F., Li X., Cao S. Y., Chu C. C., et al. (2013). NOT2 Proteins Promote Polymerase II-Dependent Transcription and Interact with Multiple MicroRNA Biogenesis Factors in Arabidopsis. Plant Cell 25 715–727. 10.1105/tpc.112.105882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Quan L., Li S., You C., Zhang Y., Gao L., et al. (2019). The protein phosphatase 4 complex promotes transcription and processing of primary microRNAs in Arabidopsis. Plant Cell 10.1105/tpc.18.00556 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ye R. Q., Xin Y., Fang X. F., Li C. L., Shi H. Q., et al. (2011). An importin beta protein negatively regulates MicroRNA activity in Arabidopsis. Plant Cell 23 3565–3576. 10.1105/tpc.111.091058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Y., Wang Y., Dou Y. C., Chen L., Wang J. L., Jiang N., et al. (2018). Degradation of unmethylated miRNA/miRNA∗s by a DEDDy-type 3′ to 5′ exoribonuclease Atrimmer 2 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115 E6659–E6667. 10.1073/pnas.1721917115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Y., Ma Z. Y., Castillo-Gonzalez C., Sun D., Li Y. J., Yu B., et al. (2018). SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature 557 516–521. 10.1038/s41586-018-0135-x [DOI] [PubMed] [Google Scholar]
- Wang X. Y., Zhang S. X., Dou Y. C., Zhang C., Chen X. M., Yu B., et al. (2015). Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3′ Tailing of Small RNAs in Arabidopsis. PLoS Genet. 11:e1005091. 10.1371/journal.pgen.1005091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S., Wollmann H., Schneeberger K., Weigel D. (2010). Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr. Biol. 20 42–48. 10.1016/j.cub.2009.10.073 [DOI] [PubMed] [Google Scholar]
- Wu F. J., Yu L., Cao W. G., Mao Y. F., Liu Z. Y., He Y. K. (2007). The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing. Plant Cell 19 914–925. 10.1105/tpc.106.048637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. J., Wang Z. M., Wang M., Wang X. J. (2013). Widespread long noncoding RNAs as endogenous target mimics for MicroRNAs in Plants. Plant Physiol. 161 1875–1884. 10.1104/pp.113.215962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Zhou H. Y., Zhang Q. Q., Zhang J. G., Ni F. R., Liu C., et al. (2010). DNA Methylation Mediated by a MicroRNA Pathway. Mol. Cell 38 465–475. 10.1016/j.molcel.2010.03.008 [DOI] [PubMed] [Google Scholar]
- Xie Z. X., Allen E., Fahlgren N., Calamar A., Givan S. A., Carrington J. C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138 2145–2154. 10.1104/pp.105.062943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. X., Kasschau K. D., Carrington J. C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13 784–789. 10.1016/S0960-9822(03)00281-1 [DOI] [PubMed] [Google Scholar]
- Xu M. L., Hu T. Q., Smith M. R., Poethig R. S. (2016). Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell 28 28–41. 10.1105/tpc.15.00854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Gu Y. Y., Jia X. Y., Kang W. J., Pan S. J., Tang X. Q., et al. (2012). Effective Small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24 415–427. 10.1105/tpc.111.094144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Wang P. C., Wang B. S., Hsu C. C., Tang K., Zhang H. R., et al. (2017). The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLoS Genet. 13:e1006753. 10.1371/journal.pgen.1006753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Li Y., Wu B., Zhang K., Gao L., Zheng C. (2019). MicroRNAs transcriptionally regulate promoter activity in Arabidopsis thaliana. J. Integr. Plant Biol. 10.1111/jipb.12775 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Yang L., Liu Z. Q., Lu F., Dong A. W., Huang H. (2006). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47 841–850. 10.1111/j.1365-313X.2006.02835.x [DOI] [PubMed] [Google Scholar]
- Yang S. W., Chen H. Y., Yang J., Machida S., Chua N. H., Yuan Y. A. (2010). Structure of Arabidopsis HYPONASTIC LEAVES1 and Its Molecular Implications for miRNA Processing. Structure 18 594–605. 10.1016/j.str.2010.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Ren W. Q., Zhao Q. X., Zhang P., Wu F. J., He Y. K. (2014). Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic Acids Res. 42 12224–12236. 10.1093/nar/gku907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Y., Ebright Y. W., Yu B., Chen X. M. (2006). HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 34 667–675. 10.1093/nar/gkj474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yifhar T., Pekker I., Peled D., Friedlander G., Pistunov A., Sabban M., et al. (2012). Failure of the Tomato Trans-Acting Short Interfering RNA Program to Regulate AUXIN RESPONSE FACTOR3 and ARF4 Underlies the Wiry Leaf Syndrome. Plant Cell 24 3575–3589. 10.1105/tpc.112.100222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Yang Z. Y., Li J. J., Minakhina S., Yang M. C., Padgett R. W., et al. (2005). Methylation as a crucial step in plant microRNA biogenesis. Science 307 932–935. 10.1126/science.1107130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Ji L. J., Le B. H., Zhai J. X., Chen J. Y., Luscher E., et al. (2017a). ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 15:e2001272. 10.1371/journal.pbio.2001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Jia T. R., Chen X. M. (2017b). The "how’ and "where’ of plant microRNAs. New Phytol. 216 1002–1017. 10.1111/nph.14834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J. X., Zhao Y. Y., Simon S. A., Huang S., Petsch K., Arikit S., et al. (2013). Plant MicroRNAs display differential 3′ truncation and tailing modifications that Are ARGONAUTE1 dependent and conserved across species. Plant Cell 25 2417–2428. 10.1105/tpc.113.114603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Xia R., Meyers B. C., Walbot V. (2015). Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr. Opin. Plant Biol. 27 84–90. 10.1016/j.pbi.2015.06.011 [DOI] [PubMed] [Google Scholar]
- Zhang S. X., Liu Y. H., Yu B. (2015). New insights into pri-miRNA processing and accumulation in plants. Wires RNA 6 533–545. 10.1002/wrna.1292 [DOI] [PubMed] [Google Scholar]
- Zhang S. X., Xie M., Ren G. D., Yu B. (2013). CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc. Natl. Acad. Sci. U.S.A. 110 17588–17593. 10.1073/pnas.1310644110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. P., Murphy C., Sieburth L. E. (2010). Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc. Natl. Acad. Sci. U.S.A. 107 15981–15985. 10.1073/pnas.1007060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. M., Zhao H. W., Gao S., Wang W. C., Katiyar-Agarwal S., Huang H. D., et al. (2011). Arabidopsis Argonaute 2 Regulates Innate Immunity via miRNA393∗-Mediated Silencing of a Golgi-Localized SNARE Gene, MEMB12. Mol. Cell 42 356–366. 10.1016/j.molcel.2011.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Zhu Y., Wu H. H., Guo H. W. (2016). Post-transcriptional gene silencing in plants: a double-edged sword. Sci. China Life Sci. 59 271–276. 10.1007/s11427-015-4972-7 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Guo R., Cui Y. Q., Zhu Z. P., Zhang Y. W., Wu H., et al. (2017a). An essential role for PNLDC1 in piRNA 3′ end trimming and male fertility in mice. Cell Res. 27 1392–1396. 10.1038/cr.2017.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. H., Guo X. W., Ge C. X., Ma Z. Y., Jiang M. Q., Li T. H., et al. (2017b). KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114 4011–4016. 10.1073/pnas.1619755114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. H., Hu F. Q., Sung M. W., Shu C., Castillo-Gonzalez C., Koiwa H., et al. (2017c). RISC-interacting clearing 3′-5′ exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. Elife 6:e24466. 10.7554/eLife.24466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. M., Wang F., Juan L. R. (2015). MicroRNA promoter identification in Arabidopsis using multiple histone markers. Biomed. Res. Int. 2015:861402. 10.1155/2015/861402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Y., Yu Y., Zhai J. X., Ramachandran V., Dinh T. T., Meyers B. C., et al. (2012). The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 22 689–694. 10.1016/j.cub.2012.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. L., Hu F. Q., Wang R. H., Zhou X., Sze S. H., Liou L. W., et al. (2011). Arabidopsis Argonaute10 Specifically Sequesters miR166/165 to regulate shoot apical meristem development. Cell 145 242–256. 10.1016/j.cell.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. L., Zhou Y. Y., Castillo-Gonzalez C., Lu A., Ge C. X., Zhao Y. T., et al. (2013). Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat. Struct. Mol. Biol. 20 1106–1115. 10.1038/nsmb.2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Ow D. W., Dong Z. C. (2018). Transfer RNA-derived small RNAs in plants. Sci. China Life Sci. 61 155–161. 10.1007/s11427-017-9167-5 [DOI] [PubMed] [Google Scholar]
- Zuber H., Scheer H., Joly A. C., Gagliardi D. (2018). Respective Contributions of URT1 and HESO1 to the Uridylation of 5′ Fragments Produced From RISC-Cleaved mRNAs. Front. Plant Sci. 9:1438. 10.3389/fpls.2018.01438 [DOI] [PMC free article] [PubMed] [Google Scholar]