Abstract
A current challenge in cancer treatment is drug resistance. Even the most effective therapies often fail to produce complete and durable tumor response and ultimately give rise to therapy resistance and tumor relapse. How resistance arises in cancer remains incompletely understood. While drug resistance in cancer is thought to be driven by irreversible genetic mutations, emerging evidence also implicates reversible proteomic and epigenetic mechanisms in the development of drug resistance. Tumor microenvironment-mediated mechanisms and tumor heterogeneity can significantly contribute to cancer treatment resistance. Here, we discuss the diverse and dynamic strategies that cancers use to evade drug response, the promise of upfront combination and intermittent therapies and therapy switching in forestalling resistance, and epigenetic reprogramming to combat resistance.
Keywords: targeted therapy, polytherapy, drug resistance, molecular targets, cancer evolution, genetics, epigenetics
Cancer therapy resistance: multi-factorial, heterogeneous and rapidly evolving.
Drug resistance is a barrier to long-term patient survival [1]. Cancers use different routes to escape therapy-induced cell killing and acquire drug resistance, and many of these routes remain unpredictable and incompletely characterized [2, 3]. Improved understanding of the molecular changes that drive tumor progression and drug resistance coupled with the identification of strategies that different cancers use to evade treatment response will aid in designing both cancer subtype-specific treatments and broadly-applicable therapies to combat resistance.
The phenomenon of drug resistance is defined as the inherited ability of cells to survive at clinically-relevant drug concentrations. In the current studies to understand how cancer cells evade drug response two related phenomena, ‘tolerance’ and ‘persistence’, are considered part of a spectrum of therapeutic sensitivity and resistance states [4–6]. While ‘tolerance’ is the ability of cells to survive transient exposure to clinically-relevant concentrations of a drug, ‘persistence’ is the ability of a sub-population of a clonal population of cells to survive such treatment. Although these phenomena are well-documented in the response to anti-microbials [7], their distinction and relationship to the evolution of the fully drug-resistant tumor state remain unclear [8].
Drug resistance in cancer can be classified as intrinsic and acquired (including adaptive) [9], canonical examples of which are recently reviewed elsewhere [10, 11]. Intrinsic resistance arises before therapy and refers to the ability of a population of cells within a treatment-naïve tumor to survive initial therapy due to a pre-existing genetic alteration or cell-state [12–15]. Acquired resistance develops during treatment by therapy-induced selection of pre-existing genetic alterations in the original tumor and/or by acquisition of new mutations or adaptations in the drug target itself, recruitment of another survival factor such as a parallel or downstream pathway protein, metabolic adaptations, and epigenetic changes [16–30]. The tumor microenvironment can also contribute to acquired resistance: growth factors secreted by tumor-cells or tumor-resident stromal cells can enable tumor cell survival during initial treatment [31–33].
Tumor heterogeneity is another challenge that promotes all modes of cancer drug resistance [34–39]. A high degree of biological heterogeneity exists within a tumor (intra-tumor heterogeneity) in individual tumor cells and in cells comprising the tumor microenvironment or within different tumors (inter-tumor heterogeneity) in an individual patient or between patients [40–46]. Large scale studies such as The Cancer Genome Atlas Project (TCGA) provide critical insight into the magnitude of biological heterogeneity present within individual cancer types and across all cancers [47–49]. Such heterogeneity can promote ‘varied’ or ‘no’ response to therapy [50–52].
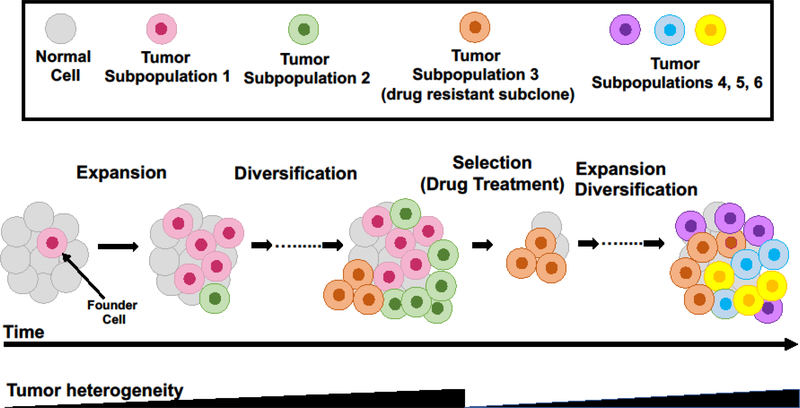
Tumor heterogeneity evolves temporally over time with tumor progression or with changes in therapy and spatially from primary to metastatic tumors [53–55]. Tumor evolution, which describes the emergence of multiple distinct sub-populations of cancer cells within the same tumor or patient, is a key feature of cancer progression and relapse and plays a key role in drug resistance [56]. Although, cancers usually arise by clonal outgrowth from a single founder cell [57], the expansion of the daughter cells of the founder cell after malignant transformation coupled with the continuous acquisition of alterations/mutations promotes the emergence of divergent cancer cell sub-populations and an increase in heterogeneity (as reviewed elsewhere [58], Figure 1). Alterations that occur early in tumor evolution are clonal whereas those acquired later are subclonal [54], Drug treatment leads to elimination of drug-sensitive sub-populations, tumor microenvironment alteration and positive selection of a drug-resistant sub-population of cells, which can result in a decrease in heterogeneity at least temporarily ([56, 58], Figure 1). Expansion of the resistant sub-population rapidly re-establishes heterogeneity through the acquisition of new mutations that provide survival benefits to the daughter cells of the resistant subclone ([58], Figure 1).
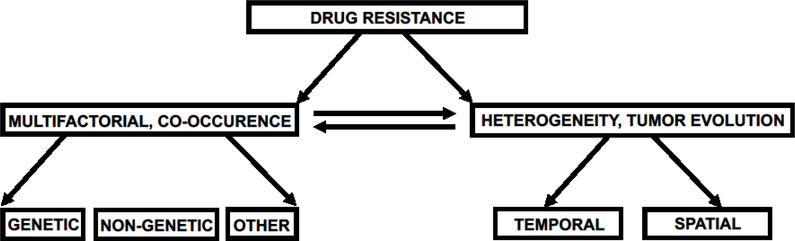
Figure 1. Different mechanisms of drug resistance in cancer.
Drug resistance is multifactorial and heterogenous and poses serious challenges to cancer treatment. The underlying mechanisms of drug resistance are diverse and complex, and often not mutually exclusive. Drug resistance is driven by both genetic and epigenetic mechanisms as well as by several other factors, including drug transporters and adaptive signaling events in tumor cells as well as tumor microenvironment features.
Tumor heterogeneity arises due to both genetic and epigenetic changes in tumor cells and in cells comprising the tumor microenvironment [54, 59]. Genetic changes due to chromosomal instability or genome duplication events as well as a variety of transcriptional states generated by epigenetic changes through altered DNA methylation and histone modifications contribute to tumor heterogeneity [54, 59, 60]. The presence of different lineage-specific gene expression programs within tumor cell sub-populations can also contribute to tumor heterogeneity [11]. Differences in the tumor microenvironment, such as, heterogeneity in the tumor-resident stromal cells from one tissue type to another (like, lung versus liver) is another form of tumor heterogeneity that can also contribute to residual disease and drug resistance [34, 42, 61].
Tumor heterogeneity is well-described in acute leukemias: clonal diseases with high inter-individual genetic variability [62, 63]. Recent whole-genome sequencing and single-cell genotyping studies in acute myeloid leukemia revealed a complex clonal diversity [64, 65]. These studies showed that the leukemic cell population is composed of different clones with high genetic heterogeneity. Further, the size and number of clones follow a complex evolution over the course of the disease [66–69]. Subclonal driver mutations have been shown to contribute to acquisition of drug resistance in chronic myeloid leukemia (CML) [58]. The BCR-ABL translocation is an established driver mutation in CML. The emergence of a subclone of leukemic cells carrying an imatinib resistance mutation in the BCR-ABL kinase domain lead to disease relapse after treatment with the BCR-ABL kinase inhibitor imatinib [70]. By whole exome sequencing and copy number analysis a recent study in chronic lymphocytic leukemia (CLL) identified clonal (MYD88, trisomy 12, and del(13q) or subclonal (Tp53 and SF3BP1) driver mutations corresponding to early or late events in clonal evolution and linked the subclonal driver mutations with rapid disease progression [71]. By genome-scale bisulfite sequencing in CLL another recent study described the contribution of DNA methylation to tumor heterogeneity [72]. Compared to normal B cells, CLL cells exhibited altered DNA methylation patterns. By bulk and single cell transcriptome analysis the DNA methylation disordered state was associated with transcriptional variation in CLL. The disordered DNA methylation state was further associated with adverse clinical outcome [72]. To more accurately identify subclonal driver mutations in CLL, another study developed methods for targeted mutation detection in DNA and RNA samples from single cells and suggested mutated LCP1 and WNK1 as candidate CLL drivers [73].
Tumor heterogeneity is also well-described in lung cancer and contributes to the emergence of EGFR TKI resistance [74, 75]. By whole-exome sequencing in a recent study, more than 75% of early-stage NSCLC (non-small cell lung cancer) tumors were found to harbor a subclonal driver alteration in PIK3CA, NF1, KRAS, TP53, and NOTCH family genes [74]. These subclonal driver mutations appeared clonal in one region but were either absent or subclonal in other regions of a single tumor [74]. EGFR T790M subclones can also co-exist with T790M-negative sub-clones not only after acquisition of resistance to first- and second-generation EGFR inhibitors but even in treatment-naïve lung cancer, although at a lower frequency [75]. A recent study in NSCLC shed new light on the genetic heterogeneity in lung cancer and showed that multiple driver mutations, such as, WNT/β-catenin and cell-cycle gene alterations/ mutations, co-occur and influence clinical outcomes in many EGFR-mutant advanced-stage lung cancers [76].
Mechanisms of drug resistance: multiple and diverse but not mutually exclusive.
The mechanisms of cancer drug resistance are multiple, diverse and complex (Figure 2). Since cancer is generally thought of as a genetic disease that evolves by clonal expansion and genetic diversification and selection, drug resistance is often considered to arise by genetic alterations [40, 77, 78]. Drug resistance, whether intrinsic or acquired, can be a deterministic and an irreversible phenomenon. Underscoring the genetic basis of drug resistance in cancer, a recent study in BRAF V600E-mutant cancers (melanoma as well as NSCLC) found selection and propagation of distinct BRAF-amplified (BRAFamp) subclones within the same tumor through parallel evolution shortly after continuous treatment with ERK inhibitor alone that enabled the tumor cells to adapt to their environment while maintaining their intra-tumoral heterogeneity [79]. By contrast, intermittent treatment with a vertical combination of RAF, MEK and ERK inhibitors potently suppressed tumor growth [79]. This effect is presumably due to the inability of tumor cells to adapt well to the changing fitness threshold (or barriers that sub-populations of tumor cells need to overcome to regain fitness in the presence of therapy) imposed by the varying selective pressures exerted by different ERK pathway inhibitors in the intermittent polytherapy.
Figure 2. Changes in tumor heterogeneity during tumor progression and treatment.
Tumor heterogeneity arises due to mutations acquired by daughter cells upon clonal outgrowth of a single founder cell (red) and increases sharply with further development into subclones (different colors reflect different subclones/subpopulations). Some new mutations lead to accelerated growth (for example green and orange subclones). Drug treatment leads to selective survival of a drug resistant clone (orange subclone) and elimination of drug-sensitive subclones (for example green and red subclones) that reduces genetic heterogeneity transiently. Heterogeneity is re-established rapidly through acquisition of mutations by daughter cells of the resistant clone.
Cancer drug resistance is multi-factorial and not driven solely by genetic mechanisms [80, 81]. Non-genetic mechanisms, such as lineage plasticity/switching (a change in cell identity) [82] or epigenetic factors that promote gene expression changes and phenotypic plasticity [83] are increasingly recognized in cancer drug resistance. Lineage plasticity occurs in EGFR-mutant NSCLC treated with EGFR inhibitors and in hormone therapy-resistant prostate cancer with RB or PTEN mutation combined with p53 inactivation [84, 85]. In EGFR-mutant NSCLC, relapse under EGFR inhibition is associated with a transition from NSCLC to a small cell lung cancer-like (SCLC-like) cell state and with sensitivity to the standard chemotherapy used in SCLC [85]. The original EGFR driver mutation is often preserved in the relapsing SCLC-like tumor [85]. The observed lineage plasticity in EGFR-mutant NSCLC is either due to clonal outgrowth of a SCLC-like sub-population pre-existing in the initial EGFR-mutant NSCLC tumor or due to de-novo, adaptive cell-state shift in the NSCLC tumor cells that survive drug exposure.
In anti-androgen therapy-resistant prostate cancer, mutation of RB and p53 leads to overexpression of a pluripotency transcription factor SOX2, which triggers a switch from an anti-androgen therapy responsive luminal prostate cell state to a multi-lineage cell state that does not rely on androgen receptor signaling [86]. Other studies offer evidence for a distinct WNT5A-driven cell state during acquisition of androgen independence [87]. The extent to which these cell-state transitions in EGFR-mutant NSCLC and prostate cancer are mediated by epigenetic alterations remains uncertain but studies in other tumor types are suggestive of this possibility. In T-cell acute lymphoblastic leukemia resistant to NOTCH1 targeting, transition to an epigenetic state upon gamma-secretase inhibition that prevents NOTCH1 activation is associated with a sensitivity to epigenetic therapy with bromodomain and extra-terminal domain (BET) inhibitors [88]. A recent study in BRAF V600E-mutant melanoma found no clear genetic cause for resistance emergence upon continuous RAF inhibitor therapy and raised the possibility of non-genetic mechanisms driving resistance [89]. No genetic mutations were detected in response to drug treatment, but a small population of melanoma cells that displayed profound transcriptional variability at the single-cell level was found and likely to be epigenetically controlled. RAF inhibitor treatment induced epigenetic reprogramming in this cell sub-population, converting the transient transcriptional state to a stable resistant state [89].
A growing body of evidence indicates that the genetic and non-genetic/epigenetic resistance-conferring mechanisms are not mutually exclusive but instead co-exist within a given cancer to drive resistance development and therapy failure [79, 89, 90]. A genetic/epigenetic duality model of drug resistance (as described in a recent review, see [8]) may reconcile this complexity. According to this model, pre-existing genetic alterations or transient transcriptional or proteomic variations may allow a minor sub-population of tumor cells to overcome the fitness threshold and survive drug exposure (drug tolerance or persistence) until some cells acquire epigenetic changes and/or secondary genetic mutations that ultimately drive the emergence of drug resistance and tumor progression during therapy.
Other mechanisms like bi-directional interactions between the tumor cells and the tumor microenvironment can also influence drug exposure/response and contribute to drug resistance and are reviewed elsewhere [11, 31–33]. Pharmacokinetic variabilities associated with drug solubility, abnormal expression of drug-uptake and drug-efflux transporters in the tumor cells as well as stromal and physical barriers that restrict drug delivery can reduce drug uptake, intracellular drug concentration and distribution and thereby can lead to incomplete anti-tumor effects, residual disease and drug resistance [91–96]. Several recent studies have linked dysfunctional metabolism of lipid second messengers, such as, ceramide to cancer drug resistance [97, 98]. Ceramide, the basic structural unit of sphingolipids, activates cell death signals induced by cytokines, chemotherapy and radiation therapy. Several anti-cancer agents such as N-(4-hydroxyphenyl) retinamide (4-HPR) have been shown to act, partly, by increasing ceramide synthesis in tumor cells [98, 99]. Impaired ceramide synthesis decreases the cytotoxic effects of chemotherapy and contributes to chemotherapy resistance [97].
Tumor containment versus aggressive therapy to delay resistance emergence.
Tumor cell states are traditionally perceived as dichotomous: drug-sensitive or drug-resistant. Current standard practice based on this binary view of drug sensitivity and resistance is to apply maximum dose-density therapy through multiple cycles to eradicate the tumor as quickly as possible so that resistance does not arise [100, 101]. Such continuous and aggressive treatment regimens eliminate the bulk of the tumor cells which are drug-sensitive but also select for the minor, drug resistant sub-population already present within the tumor which ultimately becomes the primary tumor clone and promotes therapy failure [101–103] (Figure 3). Drug-sensitive and drug-resistant sub-populations are not mutually exclusive and may co-exist within the same tumor, competing against each other for survival. In fact, competition (direct or indirect) for nutrients or other critical resources between drug-resistant and drug-sensitive sub-populations of tumor cells may be the only natural force suppressing resistance emergence [104].
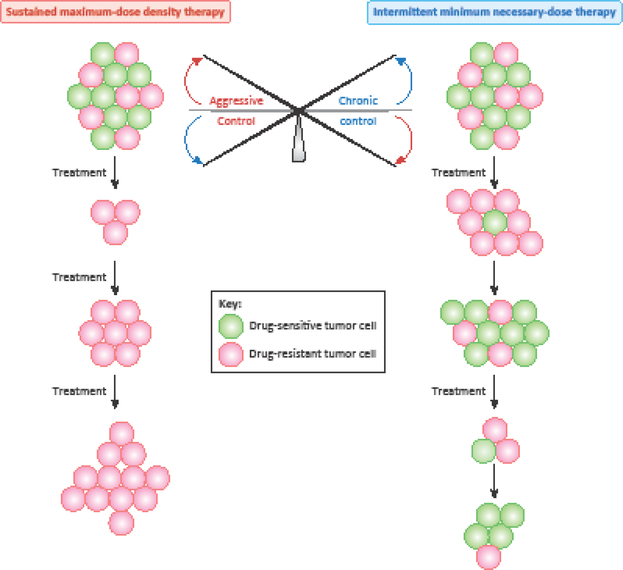
Figure 3. Aggressive therapy versus chronic control of tumors.
There is a fine balance between sustained maximum-dose density therapy and intermittent minimum necessary-dose therapy. Continuous maximum-dose density therapy is based on the principle of eradicating the tumor as swiftly as possible so that drug-resistance does not emerge. However, such rapid and aggressive treatment regimens eliminate the bulk of the tumor cells which are drug sensitive but also select for the minor, drug resistant sub-population already present within the tumor which ultimately takes over the entire tumor and promotes therapy failure. By contrast, in intermittent minimum necessary-dose therapy the idea is to contain the tumors at a fixed tolerable level to allow for expansion of drug sensitive cells at the expense of resistant ones. Although the tumor will increase in size between treatments, it will continue to remain sensitive to therapy overall. Adapted from [8, 102].
Intermittent and minimum necessary-dose treatment regimens to contain the tumors at a fixed tolerable level could allow expansion of drug-sensitive cells at the expense of resistant ones and may be more effective in delaying the emergence of resistance [8] (Figure 3). However, this treatment strategy should be practiced with caution as it could potentially be detrimental for the patient, for instance when the drug-sensitive cells acquire de-novo alterations and become drug-resistant [8, 104]. Mathematical models proposed in recent studies analyze the fitness thresholds to guide treatment decisions, such as aggressive therapy versus chronic control of tumors [104]. For example, when the patient cannot tolerate any tumor burden it is advisable to use a swift and attack-all aggressive therapy [104]. When tumors are rapidly mutating, it is advantageous to enforce a stable tumor burden by allowing the drug-sensitive, less aggressive cells to survive, which in turn will competitively suppress the aggressive drug-resistant cells to achieve chronic control of tumor evolution [8, 102–104]. Drug scheduling strategies based on these principles may be useful in halting and/or delaying resistance emergence by harnessing tumor heterogeneity and improving clinical outcomes and should be increasingly incorporated into clinical trials.
Polytherapy versus monotherapy in forestalling resistance development.
Drug resistance emergence in cancer may be suppressed by combination therapies that target multiple cancer dependencies [80]. In comparison to monotherapy, rational polytherapies that simultaneously target distinct mechanisms of resistance are less likely to fail, particularly when non-cross resistant treatments are applied together. Despite their promise, several challenges face the use of polytherapy such as limited drug options (which can be overcome partially with drug repurposing, as reviewed elsewhere [105, 106]) or drug access and treatment-related toxicity.
Several resistance mechanisms are ‘pathway-based’ mechanisms and frequently driven by pathway reactivation of a drug-inhibited oncoprotein [3, 80]. One way to achieve downstream pathway reactivation is by rendering a protein drug-target in-sensitive to therapy through acquisition of second-site or third-site mutations (i.e. by drug-target alterations) with the original oncogenic driver mutation. A rational polytherapy strategy to suppress drug-target alteration and pathway reactivation is to inhibit the target with two non-overlapping drugs simultaneously that bind to the same target but at different sites or with different modes of action. Such a dual-blockade approach has been applied clinically in HER2-amplified breast cancer, where HER2 function is blocked by both pertuzumab and trastuzumab [107, 108] and by lapatinib and trastuzumab [109] combinations. In chronic myeloid leukemia (CML) driven by BCR-ABL1, combining ATP-competitive/catalytic and allosteric inhibitors of the ABL kinase helped to overcome acquired resistance in pre-clinical studies [110].
Alternatively, primary oncogenic pathway reactivation can be achieved by activating effector proteins acting either upstream or parallel or downstream of the primary drug target. In these cases, multiple nodes in the same pathway should be targeted to block pathway reactivation and prevent resistance. For example, in BRAF-mutant melanoma, vertical combination of RAF and MEK inhibitors is clinically superior to RAF-directed monotherapy in preventing MAPK pathway reactivation and combating resistance [111, 112]. In prostate cancer, combination of ADT (androgen deprivation therapy) and the second-generation anti-androgen drug abiraterone suppresses AR signaling more potently than ADT alone [113, 114]. Despite their initial promise to improve clinical outcomes in many settings, these approaches are often not durable and resistance eventually emerges due to the limitations of single-pathway inhibition in constraining cancer evolution. On the other hand, targeting parallel pathways/dependencies with horizontal cancer drug combinations holds promise. But these regimens are often toxic, because the pathways targeted are often essential for the survival of normal cells, and hence are not always clinically-feasible [115, 116]. Alternatively, other convergent molecular targets that can compensate for single-pathway inhibition, for instance activation of the Hippo pathway effector YAP1, or its key gene targets, in mutant BRAF and RAS-driven cancers may need to be blocked in a non-cross resistant manner to achieve durable clinical remissions [117–123].
Along these lines, many oncogenic signal transduction cascades alter the function of downstream transcriptional programs to implement gene expression changes that drive cell transformation. Targeting an overactive, cancer specific transcription factor/regulator (e.g. YAP1) may combat several upstream oncogenes more effectively. If an oncogenic transcription factor is a broad integrator of expression of many different genes, like YAP1 or the master regulatory transcription factor MYC, its function may not be easily replaced by any other pathway or mechanism, decreasing the risk of resistance development.
Targeting transcriptional dependencies in cancer is challenging, as transcription factors often lack structural features that can be readily and selectively targeted with small molecule inhibitors. This limitation can be overcome indirectly by targeting the chromatin regulatory factors that influence transcription factor expression, activity, and stability and this approach is being tested in the clinic using small molecule inhibitors. For example, histone deacetylase (HDAC) inhibitors, which indirectly inhibit certain oncoproteins such as the melanocyte lineage-specific transcription factor MITF, are combined with MAPK pathway inhibitors to overcome MITF-driven resistance in melanoma [124]. The bromodomain inhibitors, targeting the BET family chromatin regulators and transcriptional co-activators (BRD4, 3, 2 and T), when administered as monotherapy produce less durable response in several cancers and are being combined with other agents, such as kinase inhibitors (e.g., RAF [125], MEK [126] and PI3K [127] inhibitors), cell-cycle inhibitors (e.g., CDK9 inhibitor [128]), DNA-damage repair inhibitors (e.g., ATR inhibitor, AZ20 [129]), immune-checkpoint inhibitors (e.g., PD-1 and PD-L1 inhibitors) [130], cytotoxic chemotherapeutic agents and other epigenetic drugs (e.g., HDAC [131] inhibitors) to enhance therapy response and forestall resistance. These combinations either induce a greater cytotoxic effect where merely a cytostatic effect was observed with either agent alone or prevent/delay tumor progression in-vivo suggesting that they may be able to overcome resistance to a single agent. Such combinations may be effective with reduced doses of each drug, potentially limiting toxicity issues. Yet, in some cases the combined toxicity of the combination regimens may be significant, and remains a major focus when designing combinatorial therapies. One potential solution is to consider intermittent or alternating drug administration schedules in the clinic.
Reversion to drug sensitivity by epigenetic reprogramming.
Another pertinent question in cancer treatment is how to tackle drug resistance after it emerges. Following emergence of drug resistance, drug discontinuation can lead to drug-sensitive populations outcompeting drug-resistant populations, restoration of drug-sensitivity, and clinical response upon drug re-exposure. For example, after a drug-free interval, drug-resistant lung cancers can re-respond to EGFR TKIs [132, 133]. In lung cancer patients whose tumors were assessed at multiple points along their treatment course, it was observed that genetic resistance mechanisms were lost without continued TKI treatment, providing a mechanistic basis for the re-treatment clinical responses [36]. Drug scheduling strategies based on these principles may be helpful in achieving reacquisition of drug sensitivity and response to therapy after a drug holiday to achieve improved chronic cancer control.
Cancer cells can use epigenetic mechanisms to control cell state. Emerging evidence shows the potential to reverse cancer-associated epigenetic abnormalities to reprogram neoplastic cells. While genetic alterations are deterministic and irreversible, epigenetic modifications are not caused by changes in the DNA sequence and are inherited at cell division. Epigenetic modifications are frequently reversible. Because of this inherent plasticity, there is an opportunity to revert back to a heritable drug-sensitive state by epigenetic reprogramming after resistance emerges. In a recent study in EGFR-mutant NSCLC PC9 cells, exposure to high concentrations of the EGFR inhibitor erlotinib killed nearly all parental PC9 cells but spared a small population of non-dividing cells [6]. These drug-tolerant persister cells re-initiated growth in the presence of erlotinib and displayed global chromatin alterations rather than genetic changes (such as EGFR-T790M or MET amplification [26]) known to confer erlotinib resistance. The drug-tolerant state could be reversed, converting the persister cells to drug-sensitive, either by continuous culture of these cells in the absence of erlotinib or by concomitantly treating them with an epigenetic drug (HDAC inhibitor) [6]. A recent study in BRAF-mutant melanoma showed that epigenetic therapy with BET bromodomain inhibitors could re-sensitize cells that acquired resistance to the RAF inhibitor vemurafenib in a YAP/TAZ-dependent manner after chronic exposure to vemurafenib [117]. In prostate cancer where regulation of GR signaling via a BET-dependent tissue-specific enhancer drives enzalutamide resistance, BET bromodomain inhibition re-sensitized drug-resistant tumors to enzalutamide by selectively impairing the GR signaling axis via this enhancer [134]. These studies show that cancer cells can become recalcitrant to therapy by acquiring a reversible drug-tolerant state during treatment that is epigenetically programmed and can be reversed by epigenetic re-programming, providing a rationale for innovative clinical trials.
Concluding Remarks
Tumor heterogeneity and cancer evolution underlie the emergence of therapeutic resistance and eventually disease relapse. Therefore, understanding how tumor heterogeneity evolves during the course of the disease and how to measure tumor evolution are essential to design more effective and durable rational polytherapies as well as to determine the timing of specific therapy deployment, therapy interruption, dose alteration or therapy switch in order to tackle the multifaceted problem of drug resistance in cancer (see Outstanding Questions). The analysis of the tumor composition (clones, subclones and the tumor microenvironment) in so-called “super-responder” patients who exhibit a dramatic anti-tumor response to a molecular therapeutic may help yield insight into the factors influencing exceptional response (or resistance) [135].
Current treatment protocols apply continuous maximum dose-density therapy to achieve maximum cell kill and eradicate the tumor as quickly as possible so that resistance does not arise. In the context of tumor evolution, however, it will be beneficial to employ intermittent or adaptive therapy [56, 136] to maintain a fine balance between drug-sensitive and drug-resistant tumor cell sub-populations (Figure 3), while avoiding excessive toxicity. Consistent with this prediction, in a recent study in breast cancer by employing adaptive therapy tumor progression was acutely controlled with an intensive chemotherapy followed by tumor maintenance with progressively smaller drug doses, in contrast, to rapid re-growth of tumor after the completion of maximum dose-density therapy [46, 56]. Similarly, in targeted therapy-based BRAF-V600E mutant melanoma and KRAS-mutant colorectal cancer models the onset of resistance was also stalled by intermittent BRAF inhibition or EGFR inhibition. In the chemotherapy-based breast cancer model, however, intermittent dosing strategy was less successful than variable dosing schedules [46, 56]. Observations from these studies suggest that intermittent and adaptive therapy and chronic control of tumors hold promise for combating resistance and should be tested more often in clinical trials in the future. These trials should be guided by future research that capitalizes on emerging preclinical models such as patient-derived organoids (PDOs) and patient-derived xenografts (PDXs), which can offer more clinically-relevant systems to facilitate high fidelity clinical translation [137–139].
Further, repeated molecular assessment will be critical to measure changes in tumor heterogeneity and cancer evolution during therapy to determine or adjust drug combinations or scheduling. Tumor profiling at multiple metastatic sites or at multiple regions within a single tumor can provide a comprehensive but only a static overview of genetic alterations. Single-site or even multi-site single biopsies are sub-optimal and will not detect all the molecular alterations within a tumor. Monitoring clonal dynamics and capturing changes in subclonal alterations as the tumor evolves (either temporally or spatially or both) through the analysis of circulating tumor DNA in the blood is becoming an increasingly useful tool. Techniques to perform tumor-sampling (such as, serial biopsies) and blood sampling (such as, analysis of circulating tumor DNA) should be further developed to allow for a more comprehensive analysis of the tumor heterogeneity and clonal evolution. The adjustment of the treatment strategy by incorporating the genetic heterogeneity factors, which is currently lacking in the standard protocols of cancer therapy, will be critical and should help prevent or delay the emergence of drug resistance to transform more cancers into chronic diseases.
Highlights.
The emergence of drug resistance is a barrier to effective cancer treatment. Resistance develops during chemotherapy, radiotherapy, molecularly-targeted therapy, and immunotherapy in most cancer patients and prevents their long-term survival.
Multiple mechanisms can promote the emergence of drug resistance and therapy failure. While gene mutations pre-existing in the tumor or acquired during therapy can drive drug resistance, cancers can also evade drug response through non-genetic and epigenetic mechanisms. Other mechanisms contributing to therapy resistance include tumor microenvironment influences, which can modify both drug exposure and response, and tumor heterogeneity within or between patients that underlies varied or minimal therapy response.
Cancer drug resistance is multi-factorial and evolves dynamically. Tumors evolve rapidly and studying how tumors adapt or change during therapy could be the key to tackling drug resistance. There is a need to recognize that drug sensitivity and resistance in cancer, and the various resistance mechanisms are not mutually exclusive but can operate together within the same tumor, or across different metastatic tumors in an individual patient. This reality must be considered in designing therapies and determining therapy duration, drug-dosing and timing for deployment of monotherapy or combination therapy. An up-front combination therapy that prevents or delays the evolution of tumors and/or dynamic switching of polytherapies during initial tumor response before resistance fully emerges holds promise for more effectively attenuating drug resistance to increase patient survival.
Outstanding questions.
How does tumor heterogeneity impact drug response and resistance?
How do genetic and epigenetic factors cooperate to promote drug resistance?
What are the most appropriate treatment strategies to forestall the evolution of drug resistance?
Can transcriptional nodes be effectively targeted to blunt resistance-associated cell state programs?
What are the most appropriate polytherapy strategies for clinical use to achieve chronic cancer control: co-administration therapy, alternating therapy, and/or intermittent and therapy switching regimens?
Acknowledgements.
The authors acknowledge funding support from NIHT32CA108462-14 (to N.C.), NIH / NCI U01CA217882, NIH / NCI U54CA224081, NIH / NCI R01CA204302, NIH / NCI R01CA211052, NIH / NCI R01CA169338, and the Pew-Stewart Foundations (to T.G.B.)
T.G.B is a consultant/advisor to Novartis, AstraZeneca, Revolution Medicines, Takeda, and Array Biopharma and receives research funding from Novartis and Revolution Medicines.
Glossary
- Drug resistance
The phenomenon of drug resistance is defined as the inherited ability of cells to survive at high drug concentrations.
- Tolerance and persistence
Two other related phenomena, ‘tolerance’ and ‘persistence’, are considered part of a spectrum of therapeutic sensitivity and resistance states [4–6]. While tolerance is the ability of cells to survive transient exposure to clinically-relevant concentrations of a drug, persistence is the ability of a sub-population of a clonal population of cells to survive such treatment. Drug resistance, tolerance and persistence are well documented in the response to anti-microbials in microbiology [7].
- Intrinsic and acquired resistance
Drug resistance in cancer can be classified as intrinsic and acquired (including adaptive) [9], for canonical examples refer to [10, 11]. Intrinsic resistance arises before therapy and refers to the ability of a population of cells within a treatment-naïve tumor to survive initial therapy due to a pre-existing genetic alteration or cell-state [12–15]. Acquired resistance develops during treatment by therapy-induced selection of pre-existing genetic alterations in the original tumor and/or by acquisition of new mutations or adaptations in the drug target itself, recruitment of another survival factor such as a parallel or downstream pathway protein, metabolic adaptations, and epigenetic changes [16–30].
- Lineage plasticity/switching
It is a change in cell identity as reviewed elsewhere [82]. Many cancers evade targeted therapies through this mechanism known as lineage plasticity, whereby tumor cells acquire phenotypic characteristics of a cell lineage whose survival no longer depends on the drug target.
- Phenotypic plasticity
It is the ability of an organism to alter its phenotype in response to environmental influences, without altering its genome as reviewed elsewhere [83].
- Maximum dose-density therapy
This therapeutic regimen is applied through multiple cycles to eradicate the tumor as quickly as possible so that resistance does not arise [100, 101].
- Intermittent and minimum necessary-dose treatment
regimens are employed to contain the tumors at a fixed tolerable level could allow expansion of drug-sensitive cells at the expense of resistant ones and may be more effective in delaying the emergence of resistance [8]
- Tumor heterogeneity
another challenge that promotes all modes of cancer drug resistance [34–39]. A high degree of molecular and genetic heterogeneity exists within a tumor (intra-tumor heterogeneity) in individual tumor cells and in cells comprising the tumor microenvironment or within different tumors (inter-tumor heterogeneity) in an individual patient or between patients [40–46].
Footnotes
Potential Conflicts of Interest.
N.C. declares no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nikolaou M et al. (2018) The challenge of drug resistance in cancer treatment: a current overview. Clin Exp Metastasis 35 (4), 309–318. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman MM et al. (2016) Toward a Better Understanding of the Complexity of Cancer Drug Resistance. Annu Rev Pharmacol Toxicol 56, 85–102. [DOI] [PubMed] [Google Scholar]
- 3.Garraway LA and Janne PA (2012) Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov 2 (3), 214–26. [DOI] [PubMed] [Google Scholar]
- 4.Blakely CM et al. (2015) NF-kappaB-activating complex engaged in response to EGFRoncogene inhibition drives tumor cell survival and residual disease in lung cancer. Cell Rep 11 (1), 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HJ et al. (2014) Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 26 (2), 207–21. [DOI] [PubMed] [Google Scholar]
- 6.Sharma SV et al. (2010) A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141 (1), 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brauner A et al. (2016) Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14 (5), 320–30. [DOI] [PubMed] [Google Scholar]
- 8.Salgia R and Kulkarni P (2018) The Genetic/Non-genetic Duality of Drug ‘Resistance’ in Cancer. Trends Cancer 4 (2), 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holohan C et al. (2013) Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 13 (10), 714–26. [DOI] [PubMed] [Google Scholar]
- 10.Bivona TG and Doebele RC (2016) A framework for understanding and targeting residual disease in oncogene-driven solid cancers. Nat Med 22 (5), 472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rotow J and Bivona TG (2017) Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer 17 (11), 637–658. [DOI] [PubMed] [Google Scholar]
- 12.Wu JY et al. (2008) Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res 14 (15), 4877–82. [DOI] [PubMed] [Google Scholar]
- 13.Bivona TG et al. (2011) FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature 471 (7339), 523–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng KP et al. (2012) A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med 18 (4), 521–8. [DOI] [PubMed] [Google Scholar]
- 15.Konieczkowski DJ et al. (2014) A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov 4 (7), 816–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi S et al. (2005) EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 352 (8), 786–92. [DOI] [PubMed] [Google Scholar]
- 17.Pao W et al. (2005) Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2 (3), e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun CH et al. (2008) The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 105 (6), 2070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cross DA et al. (2014) AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 4 (9), 1046–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu HA et al. (2015) Acquired Resistance of EGFR-Mutant Lung Cancer to a T790M-Specific EGFR Inhibitor: Emergence of a Third Mutation (C797S) in the EGFR Tyrosine Kinase Domain. JAMA Oncol 1 (7), 982–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thress KS et al. (2015) Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 21 (6), 560–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim TM et al. (2015) Mechanisms of Acquired Resistance to AZD9291: A Mutation-Selective, Irreversible EGFR Inhibitor. J Thorac Oncol 10 (12), 1736–44. [DOI] [PubMed] [Google Scholar]
- 23.Hata AN et al. (2016) Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 22 (3), 262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turke AB et al. (2010) Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 17 (1), 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bean J et al. (2007) MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 104 (52), 20932–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelman JA et al. (2007) MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316 (5827), 1039–43. [DOI] [PubMed] [Google Scholar]
- 27.Arora VK et al. (2013) Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 155 (6), 1309–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J et al. (2017) Aberrant corticosteroid metabolism in tumor cells enables GR takeover in enzalutamide resistant prostate cancer. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J et al. (2015) Epigenetic changes of EGFR have an important role in BRAF inhibitor-resistant cutaneous melanomas. J Invest Dermatol 135 (2), 532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flinders C et al. (2016) Epigenetic changes mediated by polycomb repressive complex 2 and E2a are associated with drug resistance in a mouse model of lymphoma. Genome Med 8 (1), 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Straussman R et al. (2012) Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487 (7408), 500–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson TR et al. (2012) Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487 (7408), 505–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obenauf AC et al. (2015) Therapy-induced tumour secretomes promote resistance and tumour progression. Nature 520 (7547), 368–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu D et al. (2017) Roles of tumor heterogeneity in the development of drug resistance: A call for precision therapy. Semin Cancer Biol 42, 13–19. [DOI] [PubMed] [Google Scholar]
- 35.Turner NC and Reis-Filho JS (2012) Genetic heterogeneity and cancer drug resistance. Lancet Oncol 13 (4), e178–85. [DOI] [PubMed] [Google Scholar]
- 36.Sequist LV et al. (2011) Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 3 (75), 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagle N et al. (2014) MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov 4 (1), 61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel AP et al. (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344 (6190), 1396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai W et al. (2015) Intratumoral Heterogeneity of ALK-Rearranged and ALK/EGFR Coaltered Lung Adenocarcinoma. J Clin Oncol 33 (32), 3701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogelstein B et al. (2013) Cancer genome landscapes. Science 339 (6127), 1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meric-Bernstam F and Mills GB (2012) Overcoming implementation challenges of personalized cancer therapy. Nat Rev Clin Oncol 9 (9), 542–8. [DOI] [PubMed] [Google Scholar]
- 42.Alizadeh AA et al. (2015) Toward understanding and exploiting tumor heterogeneity. Nat Med 21 (8), 846–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiley C et al. (2014) Deciphering intratumor heterogeneity and temporal acquisition of driver events to refine precision medicine. Genome Biol 15 (8), 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerlinger M et al. (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366 (10), 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGranahan N and Swanton C (2015) Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27 (1), 15–26. [DOI] [PubMed] [Google Scholar]
- 46.McGranahan N and Swanton C (2017) Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 168 (4), 613–628. [DOI] [PubMed] [Google Scholar]
- 47.Sanchez-Vega F et al. (2018) Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 173 (2), 321–337 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris LG et al. (2016) Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 7 (9), 10051–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raynaud F et al. (2018) Pan-cancer inference of intra-tumor heterogeneity reveals associations with different forms of genomic instability. PLoS Genet 14 (9), e1007669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlino MS et al. (2015) Preexisting MEK1P124 mutations diminish response to BRAF inhibitors in metastatic melanoma patients. Clin Cancer Res 21 (1), 98–105. [DOI] [PubMed] [Google Scholar]
- 51.Menzies AM et al. (2014) Inter- and intra-patient heterogeneity of response and progression to targeted therapy in metastatic melanoma. PLoS One 9 (1), e85004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russo M et al. (2016) Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov 6 (2), 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Bruin EC et al. (2014) Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346 (6206), 251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dagogo-Jack I and Shaw AT (2018) Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 15 (2), 81–94. [DOI] [PubMed] [Google Scholar]
- 55.Swanton C (2012) Intratumor heterogeneity: evolution through space and time. Cancer Res 72 (19), 4875–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turajlic S and Swanton C (2017) Implications of cancer evolution for drug development. Nat Rev Drug Discov 16 (7), 441–442. [DOI] [PubMed] [Google Scholar]
- 57.Fialkow PJ (1979) Clonal origin of human tumors. Annu Rev Med 30, 135–43. [DOI] [PubMed] [Google Scholar]
- 58.Gerlinger M and Swanton C (2010) How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer 103 (8), 1139–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilting RH and Dannenberg JH (2012) Epigenetic mechanisms in tumorigenesis, tumor cell heterogeneity and drug resistance. Drug Resist Updat 15 (1–2), 21–38. [DOI] [PubMed] [Google Scholar]
- 60.Negrini S et al. (2010) Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11 (3), 220–8. [DOI] [PubMed] [Google Scholar]
- 61.Junttila MR and de Sauvage FJ (2013) Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501 (7467), 346–54. [DOI] [PubMed] [Google Scholar]
- 62.Haegert DG et al. (1975) Acute lymphoblastic leukaemia: a heterogenous disease. Br Med J 1 (5953), 312–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stiehl T et al. (2016) Emergence of heterogeneity in acute leukemias. Biol Direct 11 (1), 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ley TJ et al. (2008) DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456 (7218), 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paguirigan AL et al. (2015) Single-cell genotyping demonstrates complex clonal diversity in acute myeloid leukemia. Sci Transl Med 7 (281), 281re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anderson K et al. (2011) Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469 (7330), 356–61. [DOI] [PubMed] [Google Scholar]
- 67.Ding L et al. (2012) Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481 (7382), 506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim T et al. (2016) Clonal dynamics in a single AML case tracked for 9 years reveals the complexity of leukemia progression. Leukemia 30 (2), 295–302. [DOI] [PubMed] [Google Scholar]
- 69.Welch JS et al. (2012) The origin and evolution of mutations in acute myeloid leukemia. Cell 150 (2), 264–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gorre ME et al. (2001) Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293 (5531), 876–80. [DOI] [PubMed] [Google Scholar]
- 71.Landau DA et al. (2013) Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152 (4), 714–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Landau DA et al. (2014) Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell 26 (6), 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang L et al. (2017) Integrated single-cell genetic and transcriptional analysis suggests novel drivers of chronic lymphocytic leukemia. Genome Res 27 (8), 1300–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jamal-Hanjani M et al. (2017) Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 376 (22), 2109–2121. [DOI] [PubMed] [Google Scholar]
- 75.Piotrowska Z et al. (2015) Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov 5 (7), 713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blakely CM et al. (2017) Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet 49 (12), 1693–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stratton MR et al. (2009) The cancer genome. Nature 458 (7239), 719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Greaves M and Maley CC (2012) Clonal evolution in cancer. Nature 481 (7381), 306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xue Y et al. (2017) An approach to suppress the evolution of resistance in BRAF (V600E)-mutant cancer. Nat Med 23 (8), 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Konieczkowski DJ et al. (2018) A Convergence-Based Framework for Cancer Drug Resistance. Cancer Cell 33 (5), 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garraway LA and Lander ES (2013) Lessons from the cancer genome. Cell 153 (1), 17–37. [DOI] [PubMed] [Google Scholar]
- 82.Le Magnen C et al. (2018) Lineage Plasticity in Cancer Progression and Treatment. Annu Rev Cancer Biol 2, 271–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Flavahan WA et al. (2017) Epigenetic plasticity and the hallmarks of cancer. Science 357 (6348). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ku SY et al. (2017) Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355 (6320), 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oser MG et al. (2015) Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol 16 (4), e165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mu P et al. (2017) SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355 (6320), 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miyamoto DT et al. (2015) RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 349 (6254), 1351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Knoechel B et al. (2014) An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet 46 (4), 364–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shaffer SM et al. (2017) Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546 (7658), 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Su Y et al. (2017) Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc Natl Acad Sci U S A 114 (52), 13679–13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Szmulewitz RZ and Ratain MJ (2014) Vemurafenib oral bioavailability: an insoluble problem. J Clin Pharmacol 54 (4), 375–7. [DOI] [PubMed] [Google Scholar]
- 92.Namisaki T et al. (2014) Differential expression of drug uptake and efflux transporters in Japanese patients with hepatocellular carcinoma. Drug Metab Dispos 42 (12), 2033–40. [DOI] [PubMed] [Google Scholar]
- 93.Elkind NB et al. (2005) Multidrug transporter ABCG2 prevents tumor cell death induced by the epidermal growth factor receptor inhibitor Iressa (ZD1839, Gefitinib). Cancer Res 65 (5), 1770–7. [DOI] [PubMed] [Google Scholar]
- 94.Neesse A et al. (2015) Stromal biology and therapy in pancreatic cancer: a changing paradigm. Gut 64 (9), 1476–84. [DOI] [PubMed] [Google Scholar]
- 95.Provenzano PP et al. (2012) Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell 21 (3), 418–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Undevia SD et al. (2005) Pharmacokinetic variability of anticancer agents. Nat Rev Cancer 5 (6), 447–58. [DOI] [PubMed] [Google Scholar]
- 97.Senchenkov A et al. (2001) Targeting ceramide metabolism--a strategy for overcoming drug resistance. J Natl Cancer Inst 93 (5), 347–57. [DOI] [PubMed] [Google Scholar]
- 98.Reynolds CP et al. (2004) Ceramide synthesis and metabolism as a target for cancer therapy. Cancer Lett 206 (2), 169–80. [DOI] [PubMed] [Google Scholar]
- 99.Wang H et al. (2003) Increasing intracellular ceramide: an approach that enhances the cytotoxic response in prostate cancer cells. Urology 61 (5), 1047–52. [DOI] [PubMed] [Google Scholar]
- 100.Read AF et al. (2011) The evolution of drug resistance and the curious orthodoxy of aggressive chemotherapy. Proc Natl Acad Sci U S A 108 Suppl 2, 10871–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gatenby RA (2009) A change of strategy in the war on cancer. Nature 459 (7246), 508–9. [DOI] [PubMed] [Google Scholar]
- 102.Enriquez-Navas PM et al. (2015) Application of Evolutionary Principles to Cancer Therapy. Cancer Res 75 (22), 4675–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Enriquez-Navas PM et al. (2016) Exploiting evolutionary principles to prolong tumor control in preclinical models of breast cancer. Sci Transl Med 8 (327), 327ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hansen E et al. (2017) How to Use a Chemotherapeutic Agent When Resistance to It Threatens the Patient. PLoS Biol 15 (2), e2001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pushpakom S et al. (2018) Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. [DOI] [PubMed] [Google Scholar]
- 106.Sleire L et al. (2017) Drug repurposing in cancer. Pharmacol Res 124, 74–91. [DOI] [PubMed] [Google Scholar]
- 107.Swain SM et al. (2015) Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med 372 (8), 724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.von Minckwitz G et al. (2017) Adjuvant Pertuzumab and Trastuzumab in Early HER2-Positive Breast Cancer. N Engl J Med 377 (2), 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Blackwell KL et al. (2012) Overall survival benefit with lapatinib in combination with trastuzumab for patients with human epidermal growth factor receptor 2-positive metastatic breast cancer: final results from the EGF104900 Study. J Clin Oncol 30 (21), 2585–92. [DOI] [PubMed] [Google Scholar]
- 110.Wylie AA et al. (2017) The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature 543 (7647), 733–737. [DOI] [PubMed] [Google Scholar]
- 111.Robert C et al. (2015) Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med 372 (1), 30–9. [DOI] [PubMed] [Google Scholar]
- 112.Johannessen CM et al. (2010) COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468 (7326), 968–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fizazi K et al. (2017) Abiraterone plus Prednisone in Metastatic, Castration-Sensitive Prostate Cancer. N Engl J Med 377 (4), 352–360. [DOI] [PubMed] [Google Scholar]
- 114.James ND et al. (2017) Abiraterone for Prostate Cancer Not Previously Treated with Hormone Therapy. N Engl J Med 377 (4), 338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Park SR et al. (2013) Safety and feasibility of targeted agent combinations in solid tumours. Nat Rev Clin Oncol 10 (3), 154–68. [DOI] [PubMed] [Google Scholar]
- 116.Yap TA et al. (2013) Development of therapeutic combinations targeting major cancer signaling pathways. J Clin Oncol 31 (12), 1592–605. [DOI] [PubMed] [Google Scholar]
- 117.Lin L et al. (2015) The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet 47 (3), 250–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Song R et al. (2018) Functional significance of Hippo/YAP signaling for drug resistance in colorectal cancer. Mol Carcinog 57 (11), 1608–1615. [DOI] [PubMed] [Google Scholar]
- 119.Song J et al. (2018) Role of YAP in lung cancer resistance to cisplatin. Oncol Lett 16 (3), 3949–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu BS et al. (2018) Inhibition of YAP reverses primary resistance to EGFR inhibitors in colorectal cancer cells. Oncol Rep 40 (4), 2171–2182. [DOI] [PubMed] [Google Scholar]
- 121.Shao DD et al. (2014) KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158 (1), 171–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zanconato F et al. (2016) YAP/TAZ at the Roots of Cancer. Cancer Cell 29 (6), 783–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hsu PC et al. (2016) YAP promotes erlotinib resistance in human non-small cell lung cancer cells. Oncotarget 7 (32), 51922–51933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Muller J et al. (2014) Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun 5, 5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Paoluzzi L et al. (2016) BET and BRAF inhibitors act synergistically against BRAF-mutant melanoma. Cancer Med 5 (6), 1183–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jing Y et al. (2016) Concomitant BET and MAPK blockade for effective treatment of ovarian cancer. Oncotarget 7 (3), 2545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stratikopoulos EE et al. (2015) Kinase and BET Inhibitors Together Clamp Inhibition of PI3K Signaling and Overcome Resistance to Therapy. Cancer Cell 27 (6), 837–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gerlach D et al. (2018) The novel BET bromodomain inhibitor BI 894999 represses super-enhancer-associated transcription and synergizes with CDK9 inhibition in AML. Oncogene 37 (20), 2687–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Muralidharan SV et al. (2016) BET bromodomain inhibitors synergize with ATR inhibitors to induce DNA damage, apoptosis, senescence-associated secretory pathway and ER stress in Myc-induced lymphoma cells. Oncogene 35 (36), 4689–97. [DOI] [PubMed] [Google Scholar]
- 130.Adeegbe DO et al. (2018) BET Bromodomain Inhibition Cooperates with PD-1 Blockade to Facilitate Antitumor Response in Kras-Mutant Non-Small Cell Lung Cancer. Cancer Immunol Res 6 (10), 1234–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heinemann A et al. (2015) Combining BET and HDAC inhibitors synergistically induces apoptosis of melanoma and suppresses AKT and YAP signaling. Oncotarget 6 (25), 21507–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yano S et al. (2005) Retreatment of lung adenocarcinoma patients with gefitinib who had experienced favorable results from their initial treatment with this selective epidermal growth factor receptor inhibitor: a report of three cases. Oncol Res 15 (2), 107–11. [PubMed] [Google Scholar]
- 133.Yoshimoto A et al. (2007) Remarkable effect of gefitinib retreatment in a patient with nonsmall cell lung cancer who had a complete response to initial gefitinib. Am J Med Sci 333 (4), 221–5. [DOI] [PubMed] [Google Scholar]
- 134.Shah N et al. (2017) Regulation of the glucocorticoid receptor via a BET-dependent enhancer drives antiandrogen resistance in prostate cancer. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nishikawa G et al. (2018) A comprehensive review of exceptional responders to anticancer drugs in the biomedical literature. Eur J Cancer 101, 143–151. [DOI] [PubMed] [Google Scholar]
- 136.Gatenby RA et al. (2009) Adaptive therapy. Cancer Res 69 (11), 4894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kuo CJ and Curtis C (2018) Organoids reveal cancer dynamics. Nature 556 (7702), 441–442. [DOI] [PubMed] [Google Scholar]
- 138.Neal JT et al. (2018) Organoid Modeling of the Tumor Immune Microenvironment. Cell 175 (7), 1972–1988 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tiriac H et al. (2018) Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov 8 (9), 1112–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]