Abstract
Abnormal phenotypic switch, migration, and proliferation of vascular smooth muscle cells (VSMCs) are hallmarks for pathogenesis of thoracic aortic dissection (TAD). In the current study, we identified miR-134-5p as a critical regulator controlling human VSMC phenotypic switch and migration to investigate whether miR-134-5p affects human VSMC functions and development of TAD. Using miRNA microarray of aorta specimens from 12 TAD and 12 controls, we identified miR-134-5p, which was significantly downregulated in TAD tissues. With qPCR detection, we found that miR-134-5p was also evidently decreased in human AoSMCs. Ectopic expression of miR-134-5p obviously promoted VSMC differentiation and expression of contractile markers, such as α-SMA, SM22α, and MYH11. miR-134-5p potently inhibited PDGF-BB-induced VSMC phenotypic switch and migration. We further identified STAT5B and ITGB1 as downstream targets of miR-134-5p in human VSMCs and proved them to be mediators in VSMC phenotypic switch and progression of TAD. Finally, Ad-miR-134-5p obviously suppressed the aorta dilatation and vascular media degeneration by 39% in TAD mice after vascular injury induced by Ang II. Our findings revealed that miR-134-5p was a novel regulator in vascular remodeling and pathological progress of TAD via targeting STAT5B/ITGB1 expression. Targeting miR-134-5p or its downstream molecules in VSMCs might develop new avenues in clinical treatment of TAD.
Keywords: miR-134-5p, media degeneration, VSMC, phenotypic switch, thoracic aortic dissection
Introduction
Vascular remodeling is mainly characterized as intima-media thickening and plays a central role in the pathological progression for aortopathy, such as aortic aneurysm.1 Thoracic aortic dissection (TAD) is a life-threatening macro-vascular disease by virtue of its predisposition of rupture. About 40% patients with aortic dissection have no time to reach the hospital and die immediately. In the pathogenesis of TAD, vascular smooth muscle cells (VSMCs) are essential in contraction and synthesis of aortic wall in response to various cellular stimuli, which facilitates vascular remodeling.2, 3, 4, 5, 6, 7, 8 With phenotype switching, VSMCs alternate from contractile (differentiated) phenotype to synthetic (dedifferentiated) phenotype. Dedifferentiated VSMCs presented elevated viability in proliferation, migration, and synthesis, as well as reduced expression of differentiation markers α-SMA and SM22α.2, 9 Pathological phenotype switching is a pivotal factor contributing to the development of aortic aneurysm and dissection.5, 6 Phenotype of VSMCs can be changed by numerous factors, including growth factor, chemotactic factors, cell adhesion molecules, extracellular matrix enzymes, and injury stimuli signal.10 Especially, PDGF promotes the phenotypic switch to synthetic VSMCs and enhances cell proliferation and migration into the neointima after artery injury.11 However, the molecular mechanisms underlying the VSMC phenotypic switch and neointima formation still remains unclear.
MicroRNA (miRNA), the endogenous 18–24 nucleotide non-coding RNA, is a novel regulator in vascular disease via affecting VSMC functions, including proliferation, apoptosis, differentiation, synthesis, and secretion.12 As an example, miR-143/145 is involved in vascular remodeling via affecting VSMC phenotypic switch,13, 14 plasticity,15 and matrix synthesis.16 The polymorphism on miR-34b/c significantly decreased the risk of aneurysm.17 In the present study, we identified miR-134-5p as a key molecule in inhibiting aortic dissection development via microarray screening. We further explored the impact of miR-134-5p on VSMC phenotypic switch and migration. Moreover, the potential molecular mechanism was investigated. Our study provided significant novel insights into the importance and functional complexities of miRNA for regulation of VSMC functions, vascular remodeling, as well as the development of macro-vascular diseases.
Results
Differentiated Expression, Target Prediction, and Functional Enrichment for miRNA Signature in TAD Patients
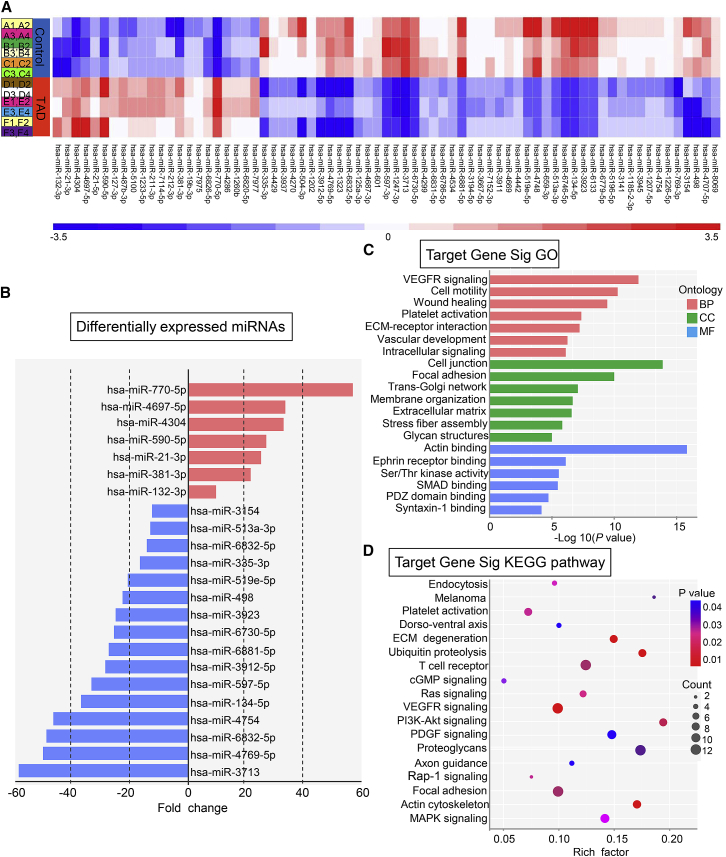
In miRNA microarray profiles, the hierarchical cluster of miRNA microarrays revealed that dissection aorta tissues had a significantly different miRNA expression signature from that of normal aorta tissues. Overall, 22 miRNAs, including miR-770-5p, miR-19b-3p, miR-212-3p, miR-127-3p, and miR-21-3p showed marked upregulation, and 49 miRNAs (such as miR-4769-5p, miR-6832-5p, miR-4754, miR-134-5p, and miR-597) exhibited evident downregulation in TAD tissues. In miRNA functional studies, identification of their target gene is pivotal. Thus, we performed target prediction applying three different algorithms—TargetScan, miRNA.org, PITA. Using the intersection set of target genes from the three databases, the Gene Ontology (GO) analysis were conducted (GO data are listed in Table S2). Enrichment was related to the significance of the specific function. For the TAD group, intersection target genes of candidate miRNAs showed a broad range of gene ontology. These genes were strikingly enriched in three categories: (1) biological process (BP): VEGFR signaling (GO: 0048010), cell motility (GO: 0048870), wound healing (GO: 0042060), platelet activation (GO: 0030168), extracellular matrix (ECM)-receptor interaction (GO: 0007160); (2) cellular component (CC): cell junction (GO: 0030054), focal adhesion (GO: 0005925), trans-Golgi network (GO: 0005802), membrane organization (GO: 0045121), extracellular matrix (GO: 0030314); (3) molecular function (MF): actin binding (GO: 0003779), ephrin receptor binding (GO: 0046875), Ser/Thr kinase activity (GO: 0043539), SMAD binding (GO: 0046332), PDZ domain binding (GO: 0030165) (Figure 1C). We further performed the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of target genes (PATHWAY data are listed in Table S3). The enriched pathways were endocytosis (path:hsa04144), melanoma (path:hsa05218), platelet activation (path:hsa04611), dorso-ventral axis (path:hsa04320), and ECM degeneration (path:hsa04512).
Figure 1.
Differentially Expressed miRNAs, GO, and KEGG Pathway Analysis Concerning miRNA Target Genes
(A) The hierarchical cluster profiles of miRNA microarrays differentiate human thoracic aortic dissection from normal thoracic aortas (n = 12/group). Downregulated and upregulated miRNAs are shown in blue and red, respectively. (B) Differentially expressed miRNAs and the corresponding fold changes. (C) Gene Ontology (GO) analysis of miRNA target genes. The y axis is GO categories. The x axis refers to GO enrichment. (D) KEGG pathways of miRNA target genes. The y axis is pathway categories. The x axis refers to pathway enrichment.
Validation of Selected miRNAs, Network Analysis of Target miRNA-mRNA in TAD Patients
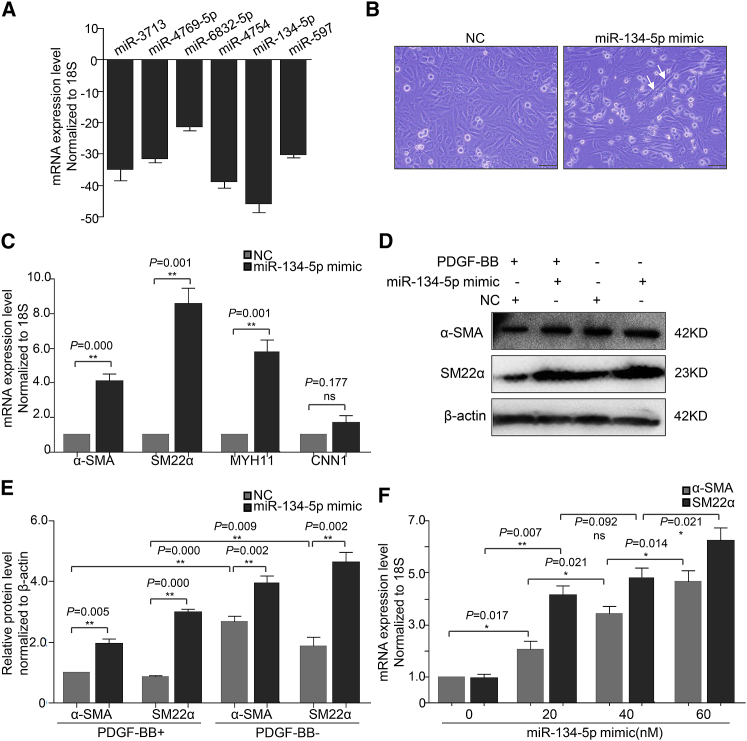
Differential miRNA expression was validated using quantitative real-time PCR. Overall, the top six significantly downregulated miRNAs in TAD specimens (Figure 1B) were selected for validation. The expression of tested miRNAs in new specimens consisting of normal aortas and TAD tissues (n = 8/group) showed an agreement with microarray data. miR-3713, miR-4769-5p, miR-6832-5p, miR-4754, miR-134-5p, and miR-597 were strikingly lower in TAD group compared to control group. Among them, miR-134-5p was lowest expressed in TAD specimens with quantitative real-time PCR examination (Figure 3A). Then we chose miR-134-5p for further functional analysis.
Figure 3.
miR-134-5p Promoted AoSMC Phenotypic Switch and Enhanced the Expression of AoSMC Contractile Genes
(A) The quantitative real-time PCR verification of microRNA expression for microarray data in human aortas and TAD specimens. (B) Morphology changes of AoSMCs transfected with miR-134-5p. Scale bar, 100 μm. (C) mRNA level of contractile genes in AoSMCs transfected with NC or miR-134-5p. (D) Western blot analysis of contractile genes in AoSMCs transfected with NC or miR-134-5p. (E) Densitometric analysis for (D). (F) Differentiated VSMC marker genes expression in AoSMCs treated with miR-134-5p in different doses. Each experiment was repeated at least three times. *p < 0.05; **p < 0.01; ns, not significant.
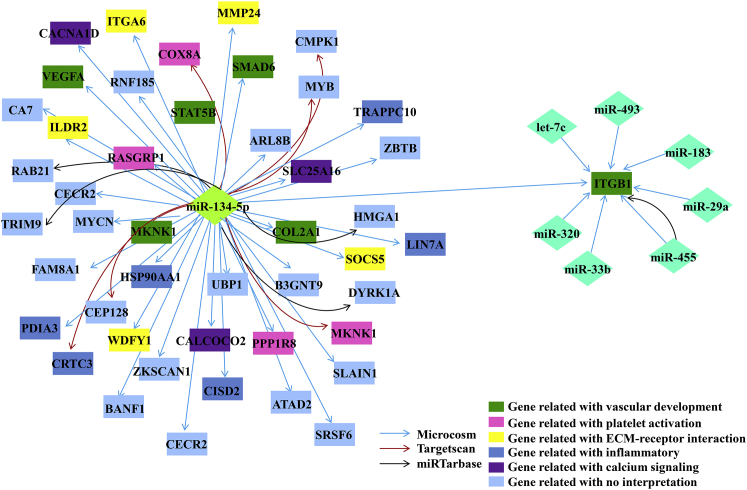
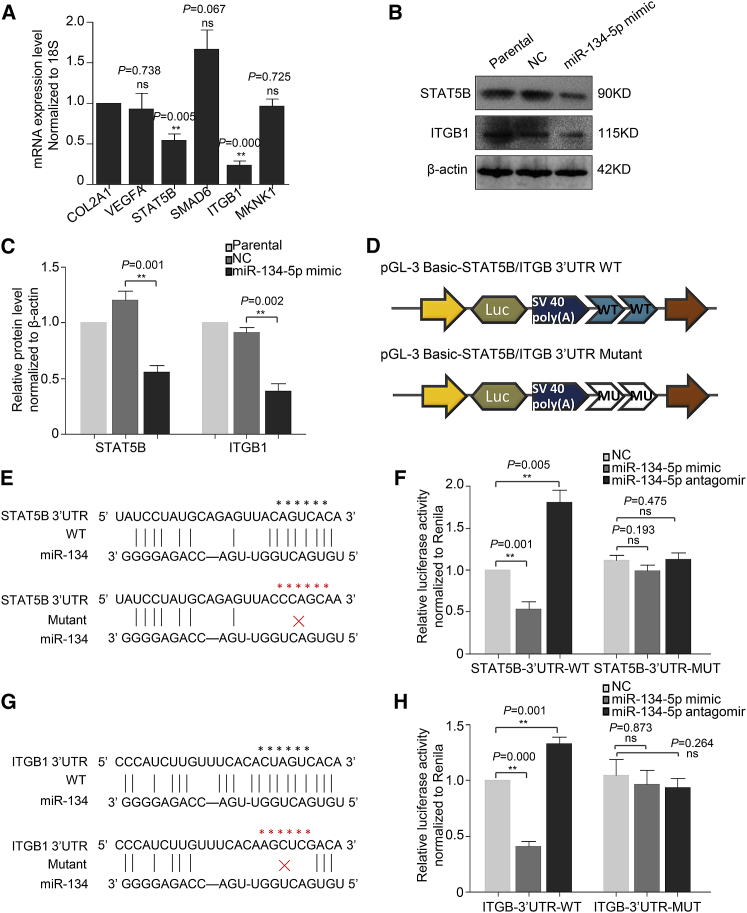
To speculate the potential functional modules of miR-134-5p, we constructed miRNA-mRNA gene network based on datasets including miRNA-target gene binding information and miRNA-mRNA expression files using bioinformatical tools-Cytoscape 3.6.1 software package (Figure 2). This network analysis indicated that miR-134-5p had a high degree of regulation in the miRNA gene network; 132 target genes of miR-134-5p were upregulated and highly enriched in vascular development, platelet activation, ECM-receptor interaction, inflammatory, and calcium-signaling modules. Specifically, COL2A1, VEGFA, STAT5B, SMAD6, ITGB1, and MKNK1 genes were notably associated with vascular development.
Figure 2.
The miRNA-Target mRNA Network
The diamond nodes represent miRNAs, and box nodes represent target mRNA. Arrows indicate the inhibitive effect of miRNAs on target mRNA. Dark green nodes describe genes related with vascular development. Pink nodes show platelet activation genes. Yellow nodes illustrate ECM-receptor interaction genes. Dark blue nodes mean inflammatory genes. Purple nodes mark calcium signaling genes. Light blue denotes genes without interpretation.
miR-134-5p Inhibits the Expression of Phenotype Marker and Phenotypic Switch of AoSMCs In Vitro
According to the miRNA-mRNA network analysis, the target genes of TAD-related miRNA-miR-134-5p were enriched in vascular development and platelet activation. These bioprocesses were consistent with the pathogenesis of TAD. Then, we investigated the impact of miR-134-5p on aortic smooth muscle cells (AoSMCs), the most pivotal cells in vascular pathophysiological responses of aortic wall. With transfection of miR-134-5p mimic (50 nM), we found a morphology change of AoSMCs. Cells were short, small, and round before (normal synthetic VSMC morphous), while long, thin, and polypodous after (contractile VSMC morphous) (Figure 3B). Next, we detected the expression of contractile markers in AoSMCs transfected with negative control (NC) or miR-134-5p mimic. miR-134-5p overexpression significantly increased α-SMA, SM22α, and MYH11 expression, while CNN1 was not significant (Figure 3C). PDGF is involved in VSMC differentiation, vascular remodeling, and aortic aneurysm model construction.11, 18 Accordingly, PDGF-BB was used in pathological VSMC models. Whether miR-134-5p repressed PDGF-BB-induced downregulation of contractile markers was measured by western blot assay. As shown in Figures 3D and 3E, miR-134-5p overexpression markedly increased SM22α and α-SMA protein expression at both quiescent and PDGF-BB-stimulated conditions. Furthermore, the increase exhibited a dose-dependent manner (Figure 3F). These findings supported the viewpoint that miR-134-5p was a novel regulator for phenotypic switch of AoSMCs.
miR-134-5p Is a Novel Regulator of Matrix Metalloproteinase Excretion and AoSMC Migration
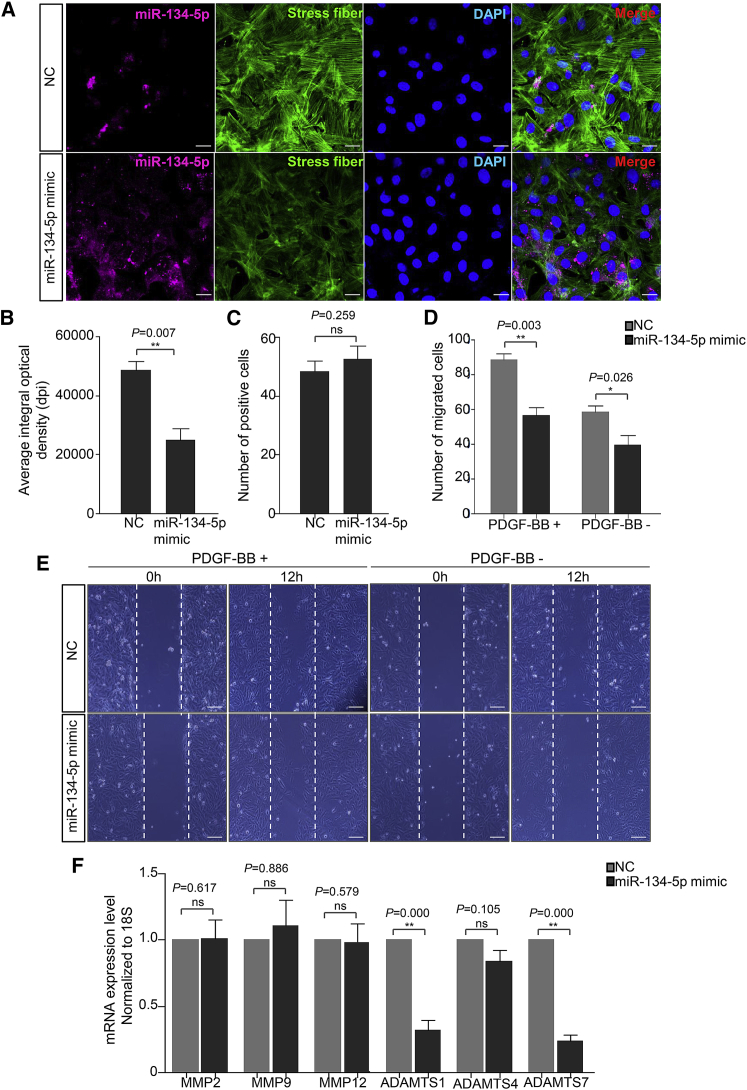
Concerning the pathological mechanism of TAD, VSMC dedifferentiation is accompanied with increased migration potential. To corroborate the role of miR-134-5p in VSMC migration, AoSMCs were transfected with NC and miR-134-5p mimic. In immunofluorescence analysis, miR-134-5p mimic dramatically promoted the formation of migration-related stress fiber (Figure 4A). The expression of F-actin was significantly augmented in miR-134-5p mimic group (NC versus miR-134-5p mimic, 24489 ± 3839 versus 48780 ± 2890 pixels), whereas there was no change in cell numbers (Figures 4B and 4C). We further assessed the migration potential of AoSMCs transfected with NC or miR-134-5p mimic by scratch-wound healing assay. As presented in Figures 4D and 4E, miR-134-5p mimic markedly inhibited the migration of AoSMCs in contrast to the NC group with or without PDGF-BB-induction. In addition, we demonstrated that matrix metalloproteinases ADAMTS-1 and ADAMTS-7 were significantly downregulated by miR-134-5p overexpression. However, no alteration was observed in MMP2, MMP9, MMP12, and ADAMTS-4 expression (Figure 4F). Of note, ADAMTS family members were newly found metalloproteinases that were implicated in the progression of thoracic aortic aneurysm and TAD.19 Collectively, these results revealed that miR-134-5p is a novel regulator for matrix metalloproteinase excretion AoSMC migration in pathogenesis of TAD.
Figure 4.
Role of miR-134-5p in AoSMC Migration
(A) Representative confocal microscopy images for stress fiber formation in AoSMCs transfected with NC or miR-134-5p mimic. Deep red, miR-134-5p FISH probe; green, stress fibers (F-actin); blue, DAPI. Scale bar, 50 μm. (B) The plot of average integral optical density for F-actin expression in different groups, n = 5/group. Data are means ± SD. (C) Quantitative analysis of positive-staining cell. (D) Quantification of migrated cells in different groups. (E) Representative pictures of scratch-wound assay in different groups with or without PDGF-BB treatment. Scale bar, 100 μm. (F) Matrix metalloproteinase expression in human AoSMCs transfected with NC or miR-134-5p mimic were detected using quantitative real-time PCR (n = 5). Stress fiber formation was detected by staining cells with Alexa Fluor 488 phalloidin. *p < 0.05; **p < 0.01; ns, not significant.
Identification of STAT5B and ITGB1 as Target Genes of miR-134-5p in Human AoSMCs
Based on the target gene prediction and miRNA-mRNA network analysis of miR-134-5p, we screened six target genes closely related to vascular disease for next step of corroboration. According to the quantitative real-time PCR data, STAT5B and ITGB1 exhibited a striking downregulation after miR-134-5p overexpression, while COL2A1, VEGFA, SMAD6, and MKNK1 had no discrimination (Figure 5A). Western blot determination further consolidated the regulation role of miR-134-5p on STAT5B and ITGB1 (Figures 5B and 5C). Bioinformatical prediction for binding site of target genes and miR-134-5p indicated the 6-mer conserved sites in the 3′ UTR of STAT5B and ITGB1. In order to detect the substantial binding and impact of miR-134-5p on targets, we constructed luciferase reporters on the basis of pGL-3 basic backbone. Two copies of ∼100-bp wild-type (WT) or mutant 3′ UTR sequence of STAT5B and ITGB1 were inserted into pGL-3 basic plasmid after the poly(A) tail of the luciferase gene (Figures 5D, 5E, and 5G). Dual luciferase reporter assay revealed that miR-134-5p mimic notably inhibited the relative luciferase activity of plasmid fused with 3′ UTR-WT of STAT5B and ITGB1 but was not significant for 3′ UTR mutant (Figures 5F and 5H). In contrast, miR-134-5p antagomir dramatically enhanced relative luciferase activity in the 3′ UTR-WT group of STAT5B and ITGB1. These data confirmed the direct binding of miR-134-5p on the 3′ UTR of STAT5B and ITGB1 and its modulation role of gene expression.
Figure 5.
Identification of STAT5B and ITGB1 as Target Genes of miR-134-5p in AoSMCs
(A) Quantitative real-time PCR analysis for target gene expression in AoSMCs transfected with miR-134-5p mimic (normalized to NC group). (B) Western blot assay for STAT5B and ITGB1 expression in AoSMCs transfected with NC or miR-134-5p mimic. (C) Densitometric analysis for (B). (D) The schematic of luciferase reporter plasmids construction. Two copies of STAT5B or ITGB1 3′ UTR (100 bp) around miR-134-5p binding site were inserted in pGL-3 basic plasmid. The theoretical miRNA-mRNA duplex between STAT5B (E) or ITGB1 (G) and miR-134-5p. The pivotal binding sites and mutant sites are highlighted with asterisks (*). Relative luciferase activities (normalized to Renilla luciferas activities) of plasmids carrying STAT5B (F) or ITGB1 (H) 3′ UTR/MUT were examined in AoSMCs with different treatment. There were five replicates in each group, and the experiments were repeated for three times. **p < 0.01; ns, non-significant.
Role of STAT5B and ITGB1 in AoSMC Proliferation, Differentiation, and Migration
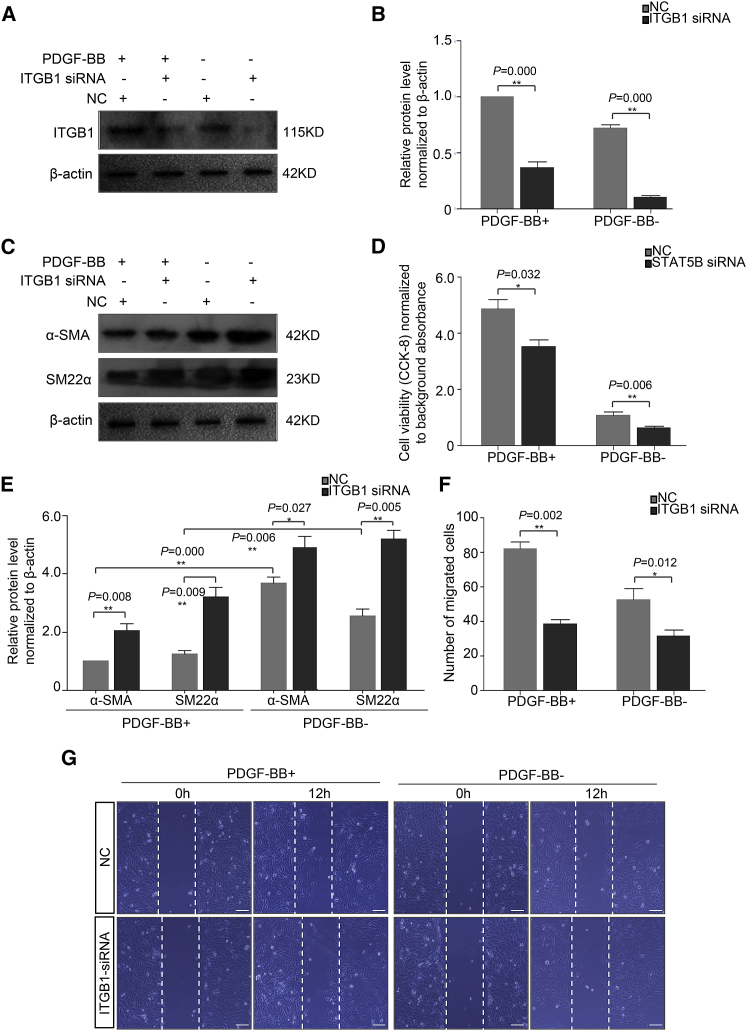
Signal transducer and activator of transcription 5B (STAT5B) is a member of STAT family of transcription factors. It is implicated in response to growth factors and cell proliferation.20 ITGB1 is also reported as a crucial mediator in cell motility, metastasis, and invasion.21 To further interpret the essential impact of STAT5B and ITGB1 in AoSMC function, we conducted the loss-of-function examination with small interfering RNAs (siRNAs). As displayed in Figures 6A and 6B, ITGB1 siRNA significantly inhibited ITGB1 expression with or without PDGF-BB stimuli, while PDGF-BB treatment increased ITGB1 expression. Western blot data suggested that ITGB1 knockdown obviously facilitated the VSMC contractile gene expression, e.g., α-SMA and SM22α (Figures 6C and 6E). STAT5B knockdown significantly repressed the proliferation of AoSMCs both in basal and PDGF-BB-induced proliferative status (Figure 6D). In addition, PDGF-BB stimulated AoSMC migration was notably attenuated by ITGB1 knockdown (Figures 6F and 6G). Taken together, these findings confirmed that ITGB1 and STAT5B were pivotal mediators in AoSMC phenotypic switch, proliferation, and migration. They were implicated in miR-134-5p regulation on AoSMC function.
Figure 6.
STAT5B and ITGB1 Are Involved in AoSMC Proliferation, Phenotype Switch, and Migration
(A) Western blot assay for ITGB1 expression in AoSMCs from different groups with or without PDGF-BB treatment. (B) Densitometric analysis for (A). (C) Western blot assay for contractile gene expression in AoSMCs from different groups with or without PDGF-BB treatment. (D) The proliferation potential of AoSMCs in NC or STAT5B siRNA group with or without PDGF-BB treatment. (E) Densitometric analysis for (C). (F) Quantification of migrated cells in different groups. (G) Representative images of scratch-wound assay in different groups with or without PDGF-BB treatment. Scale bar, 100 μm. Each assay was performed in triplicate. *p < 0.05; **p < 0.01.
miR-134-5p Suppresses Media Degeneration and TAD Progression In Vivo
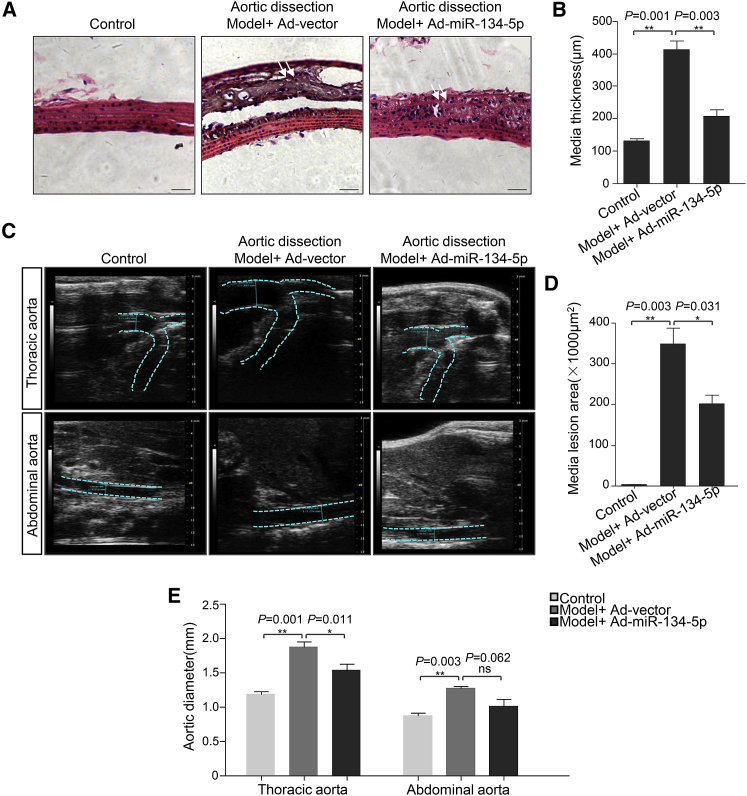
To explore whether miR-134-5p repressed vascular lesion formation in vivo, we constructed TAD mouse models to observe the vascular media degeneration in Ad-vector and Ad miR-134-5p group. After treatment of Angiotensin II (Ang II) peritoneal injection and high-fat diet for 4 weeks, the TAD models were constructed. At 2 weeks in model construction, we began the Ad-vector and Ad-miR-134-5p transduction simultaneously. While the model construction finished, the TAD mice were sacrificed and thoracic aortas were subjected to H&E staining. As shown in Figure 7A, no vascular lesion formation was found in control group. The morphous of VSMCs was normal, and they were in well-alignment. In contrast, there was a substantially vascular media degeneration with mussy filament assembly, bigger and round VSMCs, deficiency of normal VSMC contractile apparatus, and dissection formation in aortic dissection model + Ad-vector group. Transduction of thoracic aortas with miR-134-5p, however, significantly repressed the vascular media degeneration by ∼39%, which was indicated by decreases of both vascular media thickness and lesion area (Figures 7B and 7D). In compliance with the histochemical data of aortas, we observed that Ad-miR-134-5p strikingly inhibited the progressive dilatation of thoracic aortas by echocardiography examination (Figures 7C and 7E). In summary, miR-134-5p repressed the Ang II-induced vascular remodeling and progressive dilatation in vivo, which evidently contributed to TAD.
Figure 7.
miR-134-5p Inhibited the Media Degeneration, Dissection Formation, and Dilatation of Thoracic Aortas In Vivo
(A) Representative graphs of H&E staining for thoracic aortas from mouse models in control, Ad-vector, or Ad-miR-134-5p groups. Arrows indicated bigger, round VSMCs in TAD model. Data are means ± SD. Scale bar, 100 μm. (B) Quantitative analysis of media thickness of thoracic aortas from different groups. (C) Representative echocardiography images of thoracic aortas or abdominal aortas from aortic dissection models treated with PBS, Ad-vector, or Ad-miR-134-5p, n = 10/group. (D) Quantification for media lesion area of aortic dissection models in different groups. (E) Plot for aortic diameter of thoracic aortas or abdominal aortas from aortic dissection models in different groups. *p < 0.05; **p < 0.01; ns, non-significant.
Discussion
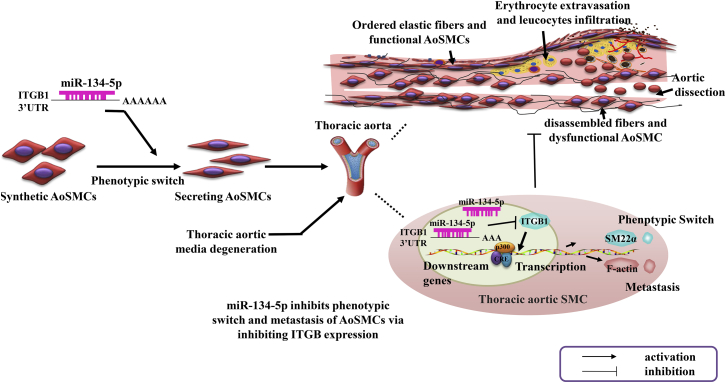
Despite essential progress in acknowledgment of pathogenesis of large vessel disease, the molecular mechanism underlying the phenotypic switch of human VSMCs remains unclear. In this study, we identified miR-134-5p as a novel regulator in human VSMC phenotypic switch, vascular remodeling, and TAD development. The data revealed that miR-134-5p expression is obviously decreased in TAD patients. Functional assay demonstrated that miR-134-5p facilitates VSMC phenotypic switch, proliferation, and migration though targeting STAT5B and ITGB1. In vivo experiments consolidated that miR-134-5p repressed the vascular remodeling and progressive dilatation, which evidently contributed to TAD (Figure 8).
Figure 8.
Schematic Diagram for the Potential Role of miR-134-5p in the Pathogenesis of Thoracic Aortic Dissection
miR-134-5p might suppress ITGB1 expression by affecting the binding of 3′ UTR. This process was involved in AoSMC phenotypic switch and metastasis. As a result, miR-134-5p repressed the vessel remodeling and thoracic aortic dissection via regulating ITGB1 expression.
Generous studies revealed that miR-134 was essential in cell proliferation, differentiation, and migration.22, 23, 24, 25, 26 In mouse embryonic stem cells, miR-134 was low in stem cells and significantly upregulated during cell differentiation to neurous.23 miR-134 was also reported to play a suppressor role in vascular endothelial cell growth, migration, and tuber formation, which occurred in atherosclerosis.27 However, the significance of miR-134-5p in VSMC function and TAD development has not been explored.
Mechanistically, we authenticated transcription factor STAT5B and cell motility-related molecule ITGB1 as pivotal targets of miR-134-5p in VSMCs. STAT5B is a member of STAT family. It is implicated in response to growth factors and cell proliferation.20 ITGB1 is reported as a crucial mediator in cell motility, metastasis, and invasion.21 Importantly, STAT5 has been confirmed as a key regulator which activation was markedly enhanced during vascular injury.28 In vascular cells, ITGB1 was pivotal for adhesion, migration, and survival during angiogenesis. Blocking ITGB1 expression might effectively impair neovascularization.29 Hypoxia-induced ITGB1 high expression also triggered vascular endothelial cell migration and angiogenesis via the SNRK/SP1-ITGB1 axis.30 Consistent with these studies, we found that STAT5B silence significantly inhibited human VSMC proliferation. ITGB1 deficiency obviously facilitated the expression of VSMC differentiation markers and inhibited VSMC migration. These functional experiments confirmed the bioinformatical prediction results, such as GO and pathway analysis regarding miRNA microarrays, that the targets were involved in vascular pathophysiology. The strikingly downregulated miRNA in TAD-miR-134-5p substantially repressed VSMC phenotypic switch, proliferation, and migration via the target genes—STAT5B and ITGB1. Then it further contributed to inhibiting vascular lesion formation in TAD progression. In accordance with these data, overexpression of miR-134-5p in TAD mouse models obviously suppressed pathological vascular remodeling such as vascular media degeneration and dissection formation.
The regulation of VSMC phenotype is pivotal for vascular remodeling and lesion formation.10 SMC phenotypic switch plays a pivotal role in TAD.31 Increasing evidence suggested that miRNAs, e.g., miR-31, miR-143/145, and miR-221, were critical effector molecules in response to growth factor signaling for VSMC phenotype regulation.9, 32 The molecular mechanisms underlying the modulation of VSMC phenotypic alteration by miRNAs are discriminate but invariably involved in regulating the expression of transcription factors or key factors in signaling, for instance, serum response factor and Kruppel-like zinc finger proteins.32, 33 In the current study, we evaluated the impact of miR-134-5p on its target key factors related to vascular pathophysiology and found that miR-134-5p did not notably interfere with the expression of SMAD6, MKNK1, COL2A1, and VEGFA, whereas ITGB1 was confirmed as a mediator in the effect of miR-134-5p on VSMC phenotypic switch. This process might occur in VSMC through a mechanism that needs to be further investigated.
In summary, this is the first time that miR-134-5p was identified as a pivotal regulator in human AoSMC phenotypic switch and migration via targeting the ITGB1 pathway. The expression of miR-134-5p was substantially downregulated in AoSMCs from TAD patients, indicating a pathological role of miR-134-5p in SMC dysfunctional vascular diseases. Intriguingly, miR-134-5p ectopic expression was shown to prevent mice from experiencing development of aorta wall media degeneration and its associated adaptive responses, such as aorta dilatation. Taken together, miR-134-5p/ITGB1 signaling might be a novel therapeutic target for treatment of macrovascular disease such as TAD.
Materials and Methods
Subjects
Twenty thoracic aorta samples from focus of TAD patient were collected during Bentall procedures in the Department of Cardiovascular Surgery, the First Affiliated Hospital of Soochow University (December 2016–December 2017). Among them, 12 samples were used in miRNA microarray detection, and eight samples were applied in validation quantitative real-time PCR. Twelve normal aorta samples were harvested from age-gender-matched patients receiving valve replacement. The aorta status of subjects were evaluated using at least one imaging examination, e.g., echocardiography, MRI, or angiography. All the subjects have signed the informed consent form. The tests using human or animal aorta specimens were approved by the Institutional Review Board from the First Affiliated Hospital of Soochow University.
RNA Extraction and miRNA Microarray Analysis
Total RNA from 12 TAD patients was extracted using mirVana miRNA isolation kit (Ambion, Austin, TX, USA) according to the manufacturer’s protocol. RNA quantification was conducted with NanoDrop ND-2000 (Thermo Scientific, USA), and the RNA integrity was examined using Agilent Bioanalyzer 2100 (Agilent Technologies, USA). The miRNA expression profile of aorta tissues was analyzed by Agilent Human miRNA array (8 × 60k; Design ID: 070156). In brief, total RNA were amplified, dephosphorylated, denatured, and labeled with cyanine-3-CTP (Cy3). After purifying and washing, the labeled RNA were hybridized onto the microarray and scanned using the Agilent Scanner G2505C (Agilent Technologies, USA). Raw data was normalized, and the basic analysis was performed by Genespring GX 12.5 software (Agilent Technologies, USA). The threshold of screening differentiated expressed miRNAs was a fold change ≥ 2.0 and p value ≤ 0.05. Target genes of miRNAs were intersection predicted using three different algorithms—Targetscan (http://targetscan.org/), microRNAorg (http://microrna.org), PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_data.html). The further GO and KEGG analysis were conducted with R software (https://www.r-project.org/) to study the potential roles of target genes.
miRNA Molecular Beacon Probe and Immunofluorescence
A custom miRNA detection probe miR-134-5p was labeled with TYE665 (Focobio, Guangzhou, China) and used in the immunofluorescence experiment. For stress fiber staining, AoSMCs were incubated with Alexa Fluor 488 phalloidine (1:1000, Abcam, Cambridge, MA, USA) in 0.17 mm coverglass-bottom dishes (30 min, room temperature). Cell nuclei were stained by DAPI. Fluorescence images were digitally captured applying Carl Zeiss LSM880 laser scanning confocal microscope (Carl Zeiss, Jena, Germany). Average integral optical density of stress fiber was quantified from 10 visual fields by Image Pro Plus 6.0 software (Media Cybernetics, Silver Spring, MD, USA).
Dual-Luciferase Reporter Assay
To validate the binding of miR-134-5p to 3′ UTR of target genes, the reporter construct containing two copies of ∼100-bp WT or mutant 3′ UTR sequences of STAT5B/ITGB1 were created. Two tandem copies of ∼100-bp DNA sequences around miR-134-5p binding site or mutant site were synthesized by GENEWIZ (Suzhou, China); then, the sequences were inserted into pGL-3 Basic vector (Promega, Madison, WI, USA), following the luciferase gene with BamHI and SalI enzyme sites. Transfection of reporter constructs and pRL-TK control plasmids into AoSMCs was performed according to the instructions of Dual-luciferase Reporter Assay System (Promega, Madison, WI, USA). The luciferase activity was tested by Synergy H1 microplate reader (BioTek Instruments, Winooski, VT, USA). Relative luciferase activity was calculated via normalization to Renilla luciferase activity of pRL-TK.
AoSMC Scratch-Wound Healing Assay
Primary human AoSMCs were isolated from thoracic aorta media of patients who received aortic valve replacements. These cells were cultured in human SMC medium (Sciencell, San Diego, CA, USA) and identified using VSMC markers (Figure S1). In this test, AoSMCs were used at passages 3–8 and a purity over 95%. First, cells were seeded in 35-mm dishes and starved in DMEM + 0.5% fetal bovine serum (FBS) for 48 h. Then a linear scratch was quickly generated using 200-μL tip in the center of a dish. Cells were treated with or without PDGF-BB (20 ng/mL, PeproTech, NJ, USA) for 12 h. The images of migrated AoSMCs were captured with an inversion fluorescence microscope (Olympus IX51, Tokyo, Japan). Cells/group in 10 independent visual fields were counted, and the average number was calculated for statistical analysis.
Quantitative Real-Time PCR Analysis and Western Blot Assay
The miRNA expression was validated using RNA-tailing quantitative real-time PCR in accordance with the protocol of the all-in-one miRNA detection kit (Genecopoeia, Guangzhou, China). miR-3713, miR-4769-5p, miR-6832-5p, miR-4754, miR-134-5p, and miR-597 forward primer and universal adaptor PCR primer were obtained from Genecopoeia (Guangzhou, China). The primers of contractile markers, matrix metalloproteinases, and vascular development target genes in quantitative real-time PCR assay were listed in Table S1. For western blot assay, proteins were extracted from AoSMCs with different treatment and subjected to immunoblotting using antibodies against α-SMA, SM22α, STAT5B, ITGB1, or β-actin (1:1000, Abcam, Cambridge, MA, USA). β-actin was used as internal control, and Gel-Pro analyzer 4.0 software was applied in protein quantitative densitometric analysis (Media Cybernetics, Silver Spring, MD, USA).
TAD Model Construction
Apoe−/− mice were purchased from Vital River Company (Beijing, China). All animal experiments performed conform to institutional guidelines for laboratory animals. To construct the TAD models, 8- to 10-week-old Apoe−/− mice were treated with Angiotensin II (Ang II) (20 mg/kg.d, Sangon, Shanghai, China) and high-fat diet (21% butterfat, Vital River, Beijing, China) for 4 weeks. At 2 weeks of model construction, 20 μL Ad-vector or Ad-miR-134-5p (1010 pfu/mL, Genechem, Shanghai, China) were injected around thoracic aorta arch three times (n = 10 per group). At the end of model construction, mice were anesthetized and echocardiographic detected using a Vevo2100 cardiovascular ultrasound system with B mode (VisualSonics, Toronto, Canada). Thoracic aortas were harvested and histochemistry stained.
Statistical Analysis
Data were presented as mean ± SD. Differences between groups were evaluated by Student’s t test (two groups) or one-way ANOVA. All the statistical tests were two-sided and conducted using Statistical Program for Social Science (SPSS 18.0, Chicago, IL, USA). p values lower than 0.05 is considered as statistically significant.
Author Contributions
Y.W. designed the experiments and analyzed data. Other researchers performed the experiments. Y.W. and G.-Y.P. wrote the manuscript and prepared the figures. All authors reviewed and approved the final manuscript.
Conflicts of Interest
The authors have no competing interests in this work.
Acknowledgments
This work is supported by the National Clinical Key Specialty of Cardiovascular Surgery, Jiangsu Province’s Key Discipline/Laboratory of Medicine (XK201118), and the Jiangsu Clinical Research Center for Cardiovascular Surgery (no. BL201451). The funding had no role in study design, data analysis, or article writing.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.02.021.
Supplemental Information
References
- 1.Jones J.A., Beck C., Barbour J.R., Zavadzkas J.A., Mukherjee R., Spinale F.G., Ikonomidis J.S. Alterations in aortic cellular constituents during thoracic aortic aneurysm development: myofibroblast-mediated vascular remodeling. Am. J. Pathol. 2009;175:1746–1756. doi: 10.2353/ajpath.2009.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis-Dusenbery B.N., Wu C., Hata A. Micromanaging vascular smooth muscle cell differentiation and phenotypic modulation. Arterioscler. Thromb. Vasc. Biol. 2011;31:2370–2377. doi: 10.1161/ATVBAHA.111.226670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rensen S.S., Doevendans P.A., van Eys G.J. Regulation and characteristics of vascular smooth muscle cell phenotypic diversity. Neth. Heart J. 2007;15:100–108. doi: 10.1007/BF03085963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali M.S., Starke R.M., Jabbour P.M., Tjoumakaris S.I., Gonzalez L.F., Rosenwasser R.H., Owens G.K., Koch W.J., Greig N.H., Dumont A.S. TNF-α induces phenotypic modulation in cerebral vascular smooth muscle cells: implications for cerebral aneurysm pathology. J. Cereb. Blood Flow Metab. 2013;33:1564–1573. doi: 10.1038/jcbfm.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ignatieva E., Kostina D., Irtyuga O., Uspensky V., Golovkin A., Gavriliuk N., Moiseeva O., Kostareva A., Malashicheva A. Mechanisms of Smooth Muscle Cell Differentiation Are Distinctly Altered in Thoracic Aortic Aneurysms Associated with Bicuspid or Tricuspid Aortic Valves. Front. Physiol. 2017;8:536. doi: 10.3389/fphys.2017.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inamoto S., Kwartler C.S., Lafont A.L., Liang Y.Y., Fadulu V.T., Duraisamy S., Willing M., Estrera A., Safi H., Hannibal M.C. TGFBR2 mutations alter smooth muscle cell phenotype and predispose to thoracic aortic aneurysms and dissections. Cardiovasc. Res. 2010;88:520–529. doi: 10.1093/cvr/cvq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukherjee K., Gitlin J.M., Loftin C.D. Effectiveness of cyclooxygenase-2 inhibition in limiting abdominal aortic aneurysm progression in mice correlates with a differentiated smooth muscle cell phenotype. J. Cardiovasc. Pharmacol. 2012;60:520–529. doi: 10.1097/FJC.0b013e318270b968. [DOI] [PubMed] [Google Scholar]
- 8.Riches K., Clark E., Helliwell R.J., Angelini T.G., Hemmings K.E., Bailey M.A., Bridge K.I., Scott D.J.A., Porter K.E. Progressive Development of Aberrant Smooth Muscle Cell Phenotype in Abdominal Aortic Aneurysm Disease. J. Vasc. Res. 2018;55:35–46. doi: 10.1159/000484088. [DOI] [PubMed] [Google Scholar]
- 9.Rangrez A.Y., Massy Z.A., Metzinger-Le Meuth V., Metzinger L. miR-143 and miR-145: molecular keys to switch the phenotype of vascular smooth muscle cells. Circ Cardiovasc Genet. 2011;4:197–205. doi: 10.1161/CIRCGENETICS.110.958702. [DOI] [PubMed] [Google Scholar]
- 10.Owens G.K., Kumar M.S., Wamhoff B.R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 11.Tallquist M., Kazlauskas A. PDGF signaling in cells and mice. Cytokine Growth Factor Rev. 2004;15:205–213. doi: 10.1016/j.cytogfr.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Small E.M., Olson E.N. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boettger T., Beetz N., Kostin S., Schneider J., Krüger M., Hein L., Braun T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J. Clin. Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Y., Liu X., Yang J., Lin Y., Xu D.Z., Lu Q., Deitch E.A., Huo Y., Delphin E.S., Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ. Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordes K.R., Sheehy N.T., White M.P., Berry E.C., Morton S.U., Muth A.N., Lee T.H., Miano J.M., Ivey K.N., Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao N., Koenig S.N., Trask A.J., Lin C.H., Hans C.P., Garg V., Lilly B. MicroRNA miR145 regulates TGFBR2 expression and matrix synthesis in vascular smooth muscle cells. Circ. Res. 2015;116:23–34. doi: 10.1161/CIRCRESAHA.115.303970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J., Zou Y., Liu X., Zhu J., Zhang J., Zhang R., Yang T., Xia H. Association of Common Genetic Variants in Pre-microRNAs and Neuroblastoma Susceptibility: A Two-Center Study in Chinese Children. Mol. Ther. Nucleic Acids. 2018;11:1–8. doi: 10.1016/j.omtn.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B., Zhang J.N., Pu P.Y. Expressions of PDGF-B and collagen type III in the remodeling of experimental saccular aneurysm in rats. Neurol. Res. 2008;30:632–638. doi: 10.1179/174313208X291595. [DOI] [PubMed] [Google Scholar]
- 19.Ren P., Zhang L., Xu G., Palmero L.C., Albini P.T., Coselli J.S., Shen Y.H., LeMaire S.A. ADAMTS-1 and ADAMTS-4 levels are elevated in thoracic aortic aneurysms and dissections. Ann. Thorac. Surg. 2013;95:570–577. doi: 10.1016/j.athoracsur.2012.10.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Liu K., Zhang Y., Qi J., Lu B., Shi C., Yin Y., Cai W., Li W. ABL-N may induce apoptosis of human prostate cancer cells through suppression of KLF5, ICAM-1 and Stat5b, and upregulation of Bax/Bcl-2 ratio: An in vitro and in vivo study. Oncol. Rep. 2015;34:2953–2960. doi: 10.3892/or.2015.4293. [DOI] [PubMed] [Google Scholar]
- 21.Chen S., Wu D.D., Sang X.B., Wang L.L., Zong Z.H., Sun K.X., Liu B.L., Zhao Y. The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis. 2017;8:e3118. doi: 10.1038/cddis.2017.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y., Zhang M., Qian J., Bao M., Meng X., Zhang S., Zhang L., Zhao R., Li S., Cao Q. miR-134 functions as a tumor suppressor in cell proliferation and epithelial-to-mesenchymal Transition by targeting KRAS in renal cell carcinoma cells. DNA Cell Biol. 2015;34:429–436. doi: 10.1089/dna.2014.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L., Zheng Y., Sun Y., Zhang Y., Yan J., Chen Z., Jiang H. MiR-134-Mbd3 axis regulates the induction of pluripotency. J. Cell. Mol. Med. 2016;20:1150–1158. doi: 10.1111/jcmm.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y.H., Zhao H., Zhou L.P., Zhao C.X., Wu Y.F., Zhen L.X., Li J., Ge D.X., Xu L., Lin L. miR-134 Modulates the Proliferation of Human Cardiomyocyte Progenitor Cells by Targeting Meis2. Int. J. Mol. Sci. 2015;16:25199–25213. doi: 10.3390/ijms161025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zha R., Guo W., Zhang Z., Qiu Z., Wang Q., Ding J., Huang S., Chen T., Gu J., Yao M., He X. Genome-wide screening identified that miR-134 acts as a metastasis suppressor by targeting integrin β1 in hepatocellular carcinoma. PLoS ONE. 2014;9:e87665. doi: 10.1371/journal.pone.0087665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y., Meng L., Yu Q., Dong D., Tan G., Huang X., Tan Y. The miR-134 attenuates the expression of transcription factor FOXM1 during pluripotent NT2/D1 embryonal carcinoma cell differentiation. Exp. Cell Res. 2015;330:442–450. doi: 10.1016/j.yexcr.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 27.Miao C., Cao H., Zhang Y., Guo X., Wang Z., Wang J. LncRNA DIGIT Accelerates Tube Formation of Vascular Endothelial Cells by Sponging miR-134. Int. Heart J. 2018;59:1086–1095. doi: 10.1536/ihj.17-290. [DOI] [PubMed] [Google Scholar]
- 28.Janmaat M.L., Heerkens J.L., de Bruin A.M., Klous A., de Waard V., de Vries C.J. Erythropoietin accelerates smooth muscle cell-rich vascular lesion formation in mice through endothelial cell activation involving enhanced PDGF-BB release. Blood. 2010;115:1453–1460. doi: 10.1182/blood-2009-07-230870. [DOI] [PubMed] [Google Scholar]
- 29.Carlson T.R., Hu H., Braren R., Kim Y.H., Wang R.A. Cell-autonomous requirement for beta1 integrin in endothelial cell adhesion, migration and survival during angiogenesis in mice. Development. 2008;135:2193–2202. doi: 10.1242/dev.016378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Q., Xie Z., Yan C., Ding Y., Ma Z., Wu S., Qiu Y., Cossette S.M., Bordas M., Ramchandran R., Zou M.H. SNRK (Sucrose Nonfermenting 1-Related Kinase) Promotes Angiogenesis In Vivo. Arterioscler. Thromb. Vasc. Biol. 2018;38:373–385. doi: 10.1161/ATVBAHA.117.309834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J., Wang L., Fu W., Wang C., Guo D., Jiang J., Wang Y. Smooth muscle cell phenotypic diversity between dissected and unaffected thoracic aortic media. J. Cardiovasc. Surg. (Torino) 2013;54:511–521. [PubMed] [Google Scholar]
- 32.McDonald R.A., Hata A., MacLean M.R., Morrell N.W., Baker A.H. MicroRNA and vascular remodelling in acute vascular injury and pulmonary vascular remodelling. Cardiovasc. Res. 2012;93:594–604. doi: 10.1093/cvr/cvr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nazari-Jahantigh M., Wei Y., Schober A. The role of microRNAs in arterial remodelling. Thromb. Haemost. 2012;107:611–618. doi: 10.1160/TH11-12-0826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.