Abstract
Purpose
The etiology of Inflammatory Bowel Disease (IBD) remains currently unknown but evidence would suggest that it results from a complex interplay between genetic susceptibility genes, the intestinal microbiome and the environment, resulting in an increased response towards microbial and self-antigens, followed by the development of pre-clinical intestinal inflammation as a precursor to overt clinical disease. Efforts are needed to provide insights into the characterization of the disease, the possible prediction of complications, and the detection of a pre-clinical disease state where, through early screening and intervention, disease course can be reversed, attenuated or even prevented. A consortium of academic, industry and governmental organization investigators initiated this study to enable an assessment of pre-disease biomarkers in patients newly diagnosed with Crohn's disease (CD) and ulcerative colitis (UC).
Participants
A retrospective cohort of 1000 UC and 1000 CD cases with 500 matched controls was drawn from an active duty US military personnel population with relevant inclusion criteria with three associated pre-disease and a single disease-associated archived serum samples.
Findings to date
The PREDICTS study has been established as a biorepository platform study to perform novel discovery and analysis efforts in the field of IBD and proteomic systems biology.
Future plans
This study is poised to enable the assessment of novel biomarkers within the serum compartment to be analyzed with the goal of identifying pre-disease signals that ultimately predict disease risk, and further elucidate disease pathogenesis in the early stages of the disease process, and identify novel exposures that increase disease risk.
Highlights
•The current paradigm for the detection and treatment of IBD is dependent upon the development of symptoms and the subsequent confirmation of the diagnosis by biological, endoscopic, histological and/or radiologic exams, often at a time when the disease is already well-established and severe.
•"Furthermore, our understanding of inflammatory bowel disease including the trigger events, pathogenesis and variable phenotype in populations is not completely understood.
•To shift this paradigm, efforts are needed to provide insights into the characterization of the biological processes of disease, triggering events, the possible prediction of complications, and the detection of a pre-clinical disease state where, through early screening and intervention, disease course can be reversed, attenuated or even prevented.
•To address this gap, a consortium of academic, industry and governmental organization investigators was established and the PRoteomic Evaluation and Discovery in an IBD Cohort of Tri-service Subjects (PREDICTS) study was initiated to enable an assessment of pre-disease biomarkers in patients newly diagnosed with Crohn's disease (CD) and ulcerative colitis (UC).
•This publication describes a blueprint for the creation of public-private pre-competitive partnerships to investigate disease triggers and lay the foundation for disease interception and prevention.
1. Introduction
Inflammatory bowel diseases (IBD), including Crohn's disease (CD) and ulcerative colitis (UC), are chronic relapsing and remitting diseases, principally appearing in young adults, with peak onset between the ages of 15 and 30 years [1,2]. They are prominent in most developed countries, but it is becoming a global health problem as rising incidence is being observed in the developing world [3]. Incidence ranges in the United States (US) from approximately 2 to 15 cases per 100,000 person-years and rising for both forms with an estimated prevalence of 3.1 million [[4], [5], [6]]. Both types of IBD are clinically characterized by chronic disease courses, variability in disease severity and diversity in clinical manifestations including abdominal pain, diarrhea, hematochezia, weight loss, profound fatigue and frequent occurrence of serious intestinal and systemic disease complications, often leading to surgery and irreversible bowel damage. Consequently, IBD has a major impact on quality of life [7,8]. Disability attributed to IBD further increases the economic burden associated with these diseases, especially when it leads to partial or total unemployment in young people [9].
2. Study rationale
Basic research in IBD during the last few years has made significant advances in unveiling genetic loci of susceptibility, [10,11] revealing fundamental insights into enteric microbiota structures and their interaction with the innate and adaptive immune systems, [12] as well as discovering novel functions for and the regulation of previously recognized innate immune cells [13]. Additional research efforts have been initiated to predict treatment outcome, [14,15] and define risk stratification for disease progression in addition to shedding light on potential environmental triggers and their role in initiating post-infectious functional gastrointestinal disorders such as irritable bowel syndrome and IBD [[16], [17], [18], [19], [20]].
Despite such achievements in improving the understanding of IBD, and in part due to such advancements, several challenges and opportunities in IBD research remain [21]. These include: (1) identifying IBD patient subsets using ‘omics’ to predict aggressiveness of disease, complications, and response to treatment; understanding how environmental factors enhance the risk of IBD; (2) determining which environmental triggers initiate, perpetuate, and/or reactivate disease; and (3) exploration of the reciprocal interactions and functional pathways that lead to homeostasis versus inflammation with an ultimate goal of identifying novel disease interception strategies. The concept of disease interception from chronic immune-associated diseases is an emerging paradigm in chronic disease management where at-risk individuals are identified in a pre-disease state, or the window of time preceding clinical diagnosis and symptoms during which pathological molecular changes can be detected, and interventions can be targeted to prevent the overt irreversible damage that are associated with the disease process [22]. Overcoming these recognized challenges requires resources and unique settings whereby serial biospecimens can be obtained from incident IBD patients and in which data and biospecimens are available before disease onset and following treatment.
US military personnel constitute a large cohort of young healthy people particularly affected by IBD [19,23]. Unique to this population is an integrated database which links medical, deployment, and other exposure data, in addition to a serum repository which includes pre-disease and interval serum samples that are collected as part of a long-standing and ongoing HIV surveillance program [24]. Such a setting provides an opportunity to implement a systems proteomic approach to address critical discovery and translational research gaps in a well-characterized population. Coupled with the availability of this unique population data and sample resource, scientific and technologic innovation in recent years has led to high-throughput methodologies exploring proteomic measurements in complex biological systems [25]. As a consequence, primary discovery endeavors have been built upon and expanded beyond the reductionist approach to the "holistic" approach of comprehensively examining the globally interacting elements of biological systems [26]. The development of this systems approach has become an impetus for research by which large amounts of data are amassed, analyzed, and applied to complex questions of biology that were previously unsolvable. While another IBD pre-disease cohort study has initiated, [27] it is among genetically high risk individuals (first-degree relatives), and the population in our study represents an opportunity to understand disease processes and biomarkers of risk in a broader, more general population. However, there are some potential limitations given the demographics of this study population.
Thus, the alignment of emerging technologies and the unique data and specimen repository, led to a multinational, academic, industry and governmental partnership entitled “PRoteomic Evaluation and Discovery in an IBD Cohort of Tri-service Subjects (PREDICTS)” study to join resources and expertise on which novel discovery and translational research could be built. Furthermore, this partnership was built with the understanding that it represents a unique resource from which others, outside the founding partnership, could benefit and thus the study and opportunity is further described.
3. Cohort desciption
3.1. Study design and participants
This is a nested case-control study of subjects with incident IBD (UC and CD). Two-thousand IBD cases (1000 each with CD or UC) and 500 healthy controls were identified from the Defense Medical Surveillance System (DMSS). This is the main data repository for all US armed forces and contains relevant data from just over 10 million members having served in the armed forces since 1990, documenting their military and medical experiences throughout their career [28,29]. Subjects included in this study were active duty US military personnel. Our data (extracted from the DMSS) are limited to the years 1998–2013 (limited to 1998–2011 for CD).
3.2. Case selection
All selected cases had two or more medical encounters with an ICD-9 code for CD [555.0, 555.1 and 555.9 (including all subgroup codes)] or UC [556 (including all subgroup codes)] and available serum from the time of IBD diagnosis (±1 year) and from the preceding sampling points as per Fig. 1. Given that more than 1000 CD and UC cases were present in the database, we created an algorithm for case selection based on criteria as follows:
-
1)
≥ 2 medical encounters with an ICD-9 code for IBD;
-
2)
Available serum from the time of IBD diagnosis (±1 year) and from the preceding sampling points for that subject, 2, 4 and 6 + years before;
-
3)
≥ 1 medical encounters with a CPT code (45330-4, 45338, 45378, 45379, 45382, 45384, 45385) for lower gastrointestinal endoscopic procedure preceding ≥ 1 medical encounter with an ICD-9 code for IBD; and
-
4)Lack of medical encounters with an ICD-9 code for alternative form of IBD (e.g. if UC case, minimizing CD code visits)
-
a.No medical encounters with alternative IBD ICD-9 code
-
b.≤ 2 medical encounters with alternative IBD ICD-9 code
-
a.
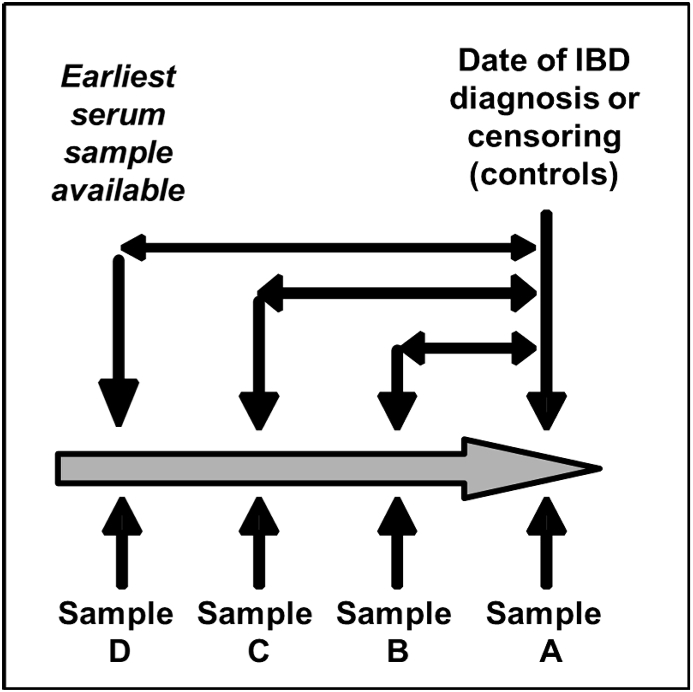
Fig. 1.
Serum Sample Selection concurrent with and prior to IBD diagnosis for PREDICTS subjects.
Legend: Sample A was the first sample available following the initial medical encounter in which an IBD diagnosis was made. In the absence of this sample, the last sample prior to that medical encounter was selected. Samples B and C represent the serum samples stored from the three preceding biennial (e.g. every two years) HIV screening test. Sample D represented the earliest serum sample available in the repository.
At a minimum, all subjects met criteria 1 and 2 above; however, preferential inclusion was given to subjects meeting criteria 3 and 4. The following algorithm outlines the order in which subjects were selected: 1,2,3,4a >1,2,3,4b > 1,2,3 > 1,2.
3.3. Control selection
Controls were matched on age, gender, race and timing of Sample A (±1 year) to UC cases. Control subjects were required to have no medical encounter evidence of Crohn's Disease [ICD9-CM: 555.0, 555.1, 555.9 (including all subgroup codes)], ulcerative colitis [ICD9-CM: 556 (including all subgroup codes)], rheumatoid arthritis (ICD9-CM: 714.0), celiac disease (ICD9-CM: 579.0) or colorectal cancer (ICD9-CM: 153.0–154.1).
3.4. Clinical and covariate data
Subjects for this study were identified from the DMSS from 1998 to 2013 (CD cases limited to 1998–2011). Medical encounter data were obtained from ambulatory and inpatient claims data for care obtained within the Military Health Services and the Tri-Service Reportable Events System data (Table 1). Demographic information including age, gender, race, education level, rank, marital status, and branch of service were obtained from personnel data records. Deployment data were derived from deployment rosters and deployment health assessments. All medical encounters with an IBD diagnosis at any diagnostic position were obtained from Armed Forces Health Surveillance Branch (AFHSB). Medical encounters associated with a procedural code for colonoscopy (CPT codes shown in Table 1) were also obtained. Infectious gastroenteritis (IGE), determined from ICD-9 codes for specific bacterial and viral pathogens, were also obtained (Table 1). All ICD-9 and CPT codes assigned in IBD-related medical encounters were also available, and allowed assessment of surgery rates, or other disease-associated complications. In an effort to assess exposures unique to this military population, deployment history was obtained to include the name of the operational deployment, duration of deployment and timing of deployment relative to disease onset. No information on smoking status, family history, medications, pathological results, radiographic studies, and/or clinical laboratory results was available.
Table 1.
List of variables available for study.
| All IBD Visits [CD - ICD9-CM: 555.0, 555.1 and 555.9 (including all subgroup codes) or UC [ICD9-CM: 556 (including all subgroup codes)] medical encounters |
|---|
| Date of medical encounter |
| All ICD-9 codes affiliated with medical encounter |
| Visit type (e.g., inpatient or outpatient) |
| All IBS (ICD9-CM: 564.1), or IGE (ICD9-CM: 001 [all subgroups], 003.0, 003.9, 004 [all subgroups], 008.0, 008.43, 008.44, 008.8, 005.4, 008.47, 008.49, 008.5, 009.0–3, 005.8, 005.9, 006.0, 006.1, 006.2, 006.9, 007 [all subgroups], 008.6 [all subgroups]) medical encounters prior and subsequent to initial IBD diagnosis |
| Date of medical encounter |
| All ICD-9 codes affiliated with medical encounter |
| Visit type (e.g., inpatient or outpatient) |
| All medical encounters in which a procedural code for colonoscopy was utilized |
| Date of medical encounter |
| All ICD-9 and CPT codes affiliated with medical encounter |
| Visit type (e.g., inpatient or outpatient) |
| All operational deployments |
| Date of deployment |
| Duration of deployment |
| Operation |
| Demographic Variables |
| Year of birth |
| Gender |
| Race |
| Education level |
| Military Rank |
| Marital status |
| Branch of service |
3.5. Serum sampling
For each IBD case and healthy control (HC), up to four serum samples were obtained from the DoD serum repository. Sample A was the first sample available following the initial medical encounter in which an IBD diagnosis was made (Fig. 1). In the absence of this sample, the last sample prior to that medical encounter was selected. Samples B and C represent the serum samples stored from the three preceding biennial (e.g. every two years) HIV screening test. In the absence of available serum, efforts were made to approximate two year intervals between serum samples. Sample A from HC subjects (frequency matched on age, race, gender and time) were matched to Sample A from UC cases based on year of collection (±1 year). Samples B and C represented samples obtained approximately two and four years prior to Sample A, respectively, without specific year-matching to the case samples. Sample D represented the earliest serum sample available in the repository. Serum samples were obtained in aliquots of 0.5 mL (from original 2.5 mL patient sample) for each subject and timepoint and labeled with a unique identifier which described both individual and sample timepoint. Serum at the DoD Serum Repository was archived at −30 °C, and upon transfer to the Naval Medical Research Center is held at −80 °C with continuous monitoring.
3.6. Biorepository description
A centralized repository was established to collect, process, store, and distribute biospecimens and data to support scientific investigation. Accessions of samples were recorded as having arrived and maintained by unique sample ID. Upon thawing and sub-aliquoting a laboratory information management system is used to barcode each ‘daughter’ sample which links to the parent sample. Information on sample data and arrival are maintained. Parallel to the sample accession and archive system, data on subjects (e.g. demographics, diagnosis, deployments, and procedures) are received, curated and housed in a limited access central repository which maintains data integrity and links to specimen identification.
Processing of samples (subaliquoting) was initially performed manually to minimize repetitive freeze-thaws and subsequently transitioned to an automated process to minimize variations in sample handling. For each serum testing request and analysis plan, the parent serum is thawed once and multiple daughter aliquots are made which meet the volume requirement of the planned testing as well as anticipating future testing of retained daughter aliquots. Daughter samples are either immediately shipped if testing is to commence upon first thaw, or refrozen and sent to partner laboratories. Daughter samples are contained in bar-code labeled cryovials with a desiccation proof seal. No additives are used in the aliquoting and storage processes.
All samples are held at the Naval Medical Research Center prior to being requested via a distribution request. The inventory system is composed of sample holding boxes and the boxes are stored in monitored freezers at −80 °C. Accompanying data associated with samples are archived in a master database from which analytic subsets are generated. The process of requesting specimens and data from the repository are later described.
3.7. Sample size and statistical analysis
This study was designed to evaluate up to 1000 cases each of UC and CD, and 500 controls [age-, gender-, race -and time-frequency matched to the UC cases] with considerations of power to detect a significant difference in a single serologic marker being predicated on the seroprevalence of that marker in cases and controls through utilization of a variety of statistical techniques including traditional bivariate and multivariate analysis, as well as cluster analysis, ordination methods, and machine learning techniques. Given the exploratory nature of this study, we estimated that the proposed sample size would have over an 80% power to identify a significant difference in the presence of a single marker in cases and controls if the biomarker is present in as few as 10% of the cases with a seropositivity rate twice as in the control subjects (2-group continuity-adjusted chi-square test with two-sided alpha = 0.05). Additionally, among IBD cases, a single group repeated measures analysis of variance (alpha = 0.05) would have an 80% power to detect a change in specific parameter characterized by an effect size (an index of the separation expected between the observed means based on mean variance within and across subjects) of 0.0027. It was further anticipated that adjustments for multiple comparisons will be needed to control the Type I error rate which may yield a lower overall statistical power. Given the utilization of network analyses to maximize the probability of identifying statistical data patterns, it is anticipated that this study is powered adequately to identify significant differences in serum profiles across the study populations assuming there are in fact differences in the studied groups. Precise statistical methods and models to be used will be unique for each research objective; however, a general strategy has been proposed to include testing and evaluation on parallel subsets with validation on an independent subset.
3.8. Baseline description of the cohort
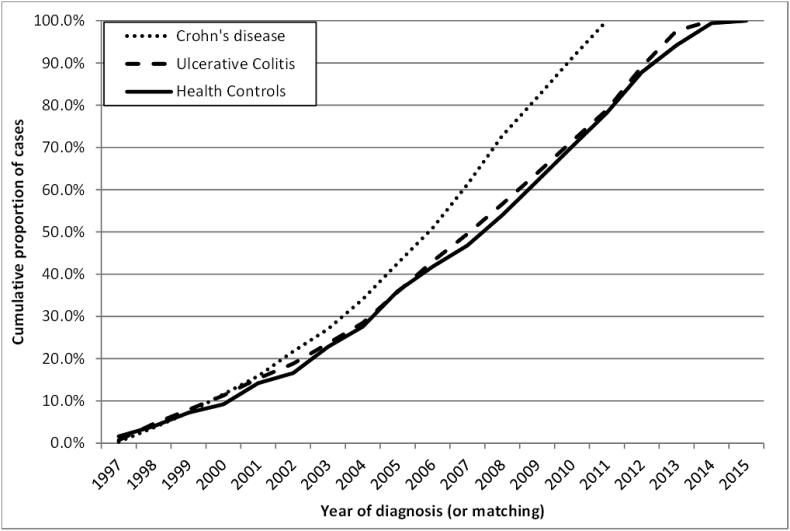
The demographics of the PREDICTS population are outlined in Table 2 and, in general, are reflective of the active duty US military population. Specifically, the majority of IBD subjects and healthy controls are male (82.5% and 92.0%, respectively) with no significant differences in IBD pathotypes (p = 0.7); however, control subjects are more predominantly male (p < 0.001). There are no significant differences in the age distribution of subjects (p = 0.5) with 84.8% under the age of 40 across all groups. As expected, less than 25% of the subjects are officers with a slightly higher proportion of enlisted personnel in the CD (82.1%) compared to UC (75.4%; p < 0.001) or HC cohorts (77.6%; p = 0.04). Approximately 70% of the CD subjects are classified as white, which is lower than UC (78.1%; p < 0.001) or HC subjects (82.8%; p < 0.001). The majority of subjects across all disease states had at least one operational deployment prior to the initial disease diagnosis (CD or UC) or matching HC. As shown in Fig. 3, incident cases (and HCs) were identified across the entire study period.
Table 2.
Demographics of PREDICTS study population.
| Crohn's disease |
Ulcerative colitis |
Healthy controls |
||||
|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |
| Gender | ||||||
| Male | 822 | 82.2% | 828 | 82.8% | 460 | 92.0% |
| Female | 178 | 17.8% | 172 | 17.2% | 40 | 8.0% |
| Age | ||||||
| 20-29 | 445 | 44.5% | 427 | 42.7% | 210 | 42.0% |
| 30-39 | 409 | 32.8% | 411 | 35.5% | 218 | 35.5% |
| 40-49 | 135 | 14.5% | 147 | 16.7% | 71 | 15.8% |
| >=50 | 11 | 1.2% | 15 | 1.6% | 1 | 0.2% |
| Education | ||||||
| High school (or equivalent) | 691 | 69.1% | 586 | 58.6% | 324 | 64.8% |
| Some college | 92 | 9.2% | 134 | 13.4% | 52 | 10.4% |
| College | 120 | 12.0% | 181 | 18.1% | 77 | 15.4% |
| Advanced degree | 75 | 7.5% | 75 | 7.5% | 36 | 7.2% |
| Unknown | 22 | 2.2% | 24 | 2.4% | 11 | 2.2% |
| Marital status | ||||||
| Married | 77 | na | 713 | 71.3% | 369 | 73.8% |
| Single/Other | 23 | na | 287 | 28.7% | 131 | 26.2% |
| Military Rank | ||||||
| Jr. Enlisted | 185 | 18.5% | 140 | 14.0% | 70 | 14.0% |
| Sr. Enlisted | 636 | 63.6% | 614 | 61.4% | 318 | 63.6% |
| Officer | 179 | 17.9% | 246 | 24.6% | 112 | 22.4% |
| Branch of Service | ||||||
| Army | na | na | 324 | 32.4% | 161 | 32.2% |
| Air Force | na | na | 263 | 26.3% | 114 | 22.8% |
| Marines | na | na | 109 | 10.9% | 52 | 10.4% |
| Navy/Coast Guard | na | na | 304 | 30.4% | 173 | 34.6% |
| Race/Ethnicity | ||||||
| White | 696 | 69.6% | 781 | 78.1% | 414 | 82.8% |
| Black | 156 | 15.6% | 171 | 17.1% | 71 | 14.2% |
| Other | 148 | 14.8% | 48 | 4.8% | 15 | 3.0% |
| Deployments | ||||||
| Deployed | 628 | 69.1% | 738 | 77.1% | 390 | 81.9% |
| Not deployed | 372 | 33.1% | 262 | 25.0% | 110 | 21.4% |
na = not available at time of publication.
Fig. 3.
Year of incident diagnosis (or matching).
4. Findings to date
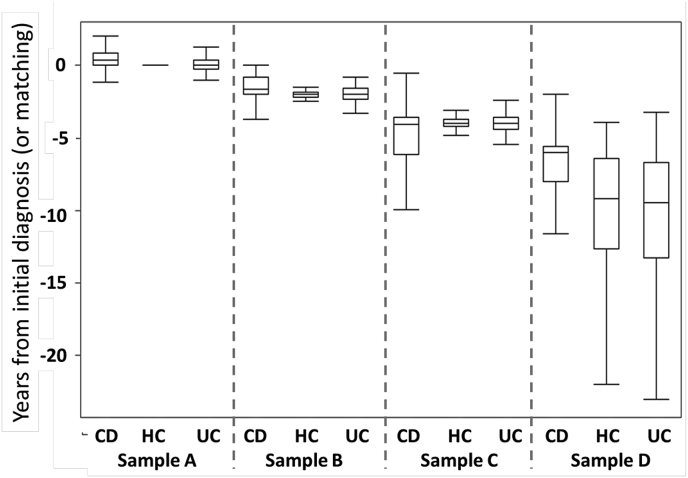
As shown in Fig. 4, disease-associated samples from CD and UC subjects were obtained a median of 145 (interquartile range (IQR): 16, 311) and 3 (IQR: −100, 230) days from the incident IBD diagnosis, respectively. All control samples were obtained at the time of censoring. Antecedent samples for CD were obtained at a median of 1.6 (IQR: 0.8, 2.0), 4.0, (3.6, 6.1) and 6.0 (5.6, 8.0) years prior to the initial medical encounter with the CD-associated ICD-CM encounter. Similar samples for the UC subjects were obtained at a median of 2.0 (IQR: 1.6, 2.3), 4.0, (IQR: 3.6, 4.4) and 9.5 (IQR: 6.7, 13.3) years prior to the incident diagnosis.
Fig. 4.
Box and whisker plots of serum sample timing stratified by CD, UC and healthy controls.
Legend. UC, ulcerative colitis; CD, Crohn's disease, HC, healthy controls. The box and whisker plots represent the median (mid-line) 25% and 75% quartiles (boxes) and the 1st and 3rd quartiles + 1.5 * the interquartile range (whiskers).
As shown in Table 3, among the CD cases, just over 50% of the subjects had evidence within the DMSS of being seen in a gastroenterology clinic concurrent with any CD-associated medical encounter. Gastroenterologist associated visits obtained outside of the Military Health System, which occurs commonly in areas that do not have specialist availability, lacked visit specification in the database. This proportion was significantly higher in the UC cases (%, p < 0.001).73.4%). Depending on IBD subtype, 11–15% of subjects had at least one inpatient IBD-related medical encounter (CD: 36.0%, UC: 26.6%) with a median of one inpatient encounter for each admitted subject. Procedural and ICD-9-CM-based codes for endoscopy highlight a significant difference in the prevalence of these procedures being documented in the DMSS with a significantly higher proportion of UC subjects (68.1%, p < 0.001)) with at least one concurrent endoscopy compared to CD subjects (36.8%). Throughout the medical encounter history, CD and UC subjects often received multiple visit-associated sub-codes. While the most common CD sub-code was ‘regional enteritis, unspecified site’ documented in just over 85% of subjects, small and large intestine sub-codes were also documented in 42.5% and 41.8% of subjects, respectively. Additionally, a small proportion of CD subjects received UC co-diagnoses during their IBD workup. Similarly to CD, UC subjects received a spectrum of UC-specific sub-codes, the most common being ‘ulcerative colitis, unspecified’ documented in 87.3% of the UC cases and identified in a median of 7 (IQR: 3, 16) separate encounters for each subject with that sub-code. Other sub-codes such as ‘universal ulcerative colitis’ (40.1%), ‘ulcerative proctitis’ (30.6%) and ‘left-sided ulcerative colitis’ (24.6%) were also common. Additional data to assist in classification of standard phenotype of disease severity based on disease location and behavior, and history of surgery are also available in the repository (data not shown).
Table 3.
Additional information related to medical CD- or UC-associated medical encounters.
| N (%) with CD/UC-associated medical encounters |
Median (IQR) visits per subject |
|||
|---|---|---|---|---|
| CD | UC | CD | UC | |
| Gastroenterology clinic | 555 (55.5) | 734 (73.4) | 4 (2, 9) | 5 (2, 9) |
| Inpatient encounter | 360 (36.0) | 266 (26.6) | 1 (1, 2) | 1 (1, 2) |
| Endoscopy | 368 (36.8) | 681 (68.1) | 1 (1, 2) | 2 (1, 2) |
| Regional enteritis, small intestine | 425 (42.5) | 0 (0.0) | 3 (1, 6) | – |
| Regional enteritis, large intestine | 418 (41.8) | 1 (0.1) | 2 (1, 5) | 1 (1, 1) |
| Regional enteritis, small/large intestine | 218 (21.8) | 0 (0.0) | 2 (1, 4) | – |
| Regional enteritis, unspecified site | 866 (86.6) | 2 (0.2) | 6 (2, 14) | 1 (1, 1) |
| Ulcerative enterocolitis | 0 (0.0) | 0 (0.0) | – | – |
| Ulcerative ileocolitis | 2 (0.2) | 41 (4.1) | 2 (1, 2) | 1 (1, 2) |
| Ulcerative proctitis | 6 (0.6) | 306 (30.6) | 2 (1, 2) | 3 (1, 6) |
| Ulcerative proctosigmoiditis | 3 (0.3) | 165 (16.5) | 1 (1, 1) | 2 (1, 4) |
| Pseudopolyposis colon | 14 (1.4) | 34 (3.4) | 1 (1, 1) | 1 (1, 2) |
| Left-sided ulcerative colitis | 1 (0.1) | 246 (24.6) | 1 (1, 1) | 2 (1, 4) |
| Universal ulcerative colitis | 13 (1.3) | 401 (40.1) | 1 (1, 2) | 3 (1, 7) |
| Other ulcerative colitis | 5 (0.5) | 281 (28.1) | 1 (1, 1) | 2 (1, 3) |
| Ulcerative colitis, unspecified | 33 (3.3) | 873 (87.3) | 1 (1, 1) | 7 (3, 16) |
| Intestinal resectiona | 70 (7.0) | 54 (5.4) | 2 (1, 4) | 3 (2, 9) |
| Infusion procedure (biological)b | 25 (2.5) | 0 (0.0) | 1 (1, 1) | – |
CPT codes: 44160, 44205, 44207 and 44120.
CPT codes: J1745, S9359.
In addition to the IBD-related medical encounters, subjects received ICD-9-CM codes associated with other diseases concurrent with all pre- and post-diagnosis IBD-related medical encounters (Table 4). The most common co-diagnoses were diseases of the digestive system (occurring in 58.9% of the CD cases and 63.0% of the UC cases) and symptoms, signs and ill-defined conditions (occurring in 53.9% of CD cases and 55.4% of the UC cases) with no significant difference in the proportion of cases (p = 0.5 and 0.06, respectively). ICD-9-CM codes for diseases of the digestive system were assigned on a total of over 2500 (CD: 2714; UC: 2555) separate IBD-related encounters for a median of three encounters per CD case (IQR: 1, 6) and two for each UC case (IQR: 1, 5). Diseases of the musculoskeletal system and connective tissue were also common among cases (CD: 25.7%, UC: 30.5%) with a median of two visits per case. Diseases of the circulatory system were more common among UC cases (25.2%) than CD cases (17.6%), as were endocrine, nutritional and metabolic diseases and immunity (p < 0.001 and p = 0.002, respectively). Among the HC subjects, the most commonly identify medical encounters were similar to the co-diagnosed conditions among the IBD cases with symptoms, signs and ill-defined conditions being most common (44.2%) followed by mental disorders (28.4%) and diseases of the digestive system (17.6%). The median IBD related medical follow-up following incident diagnosis was 373 days (IQR: 151, 977) and 852 days (IQR: 350, 1967) for CD and UC, respectively.
Table 4.
ICD-9-CM codes corresponding with IBD-related medical encounters and all HC-associated encounters.
| ICD-9-CM description | ICD-9-CM code | Number (%) of subjects with co-Diagnoses |
Total number, median (IQR) of separate medical encounters |
||||
|---|---|---|---|---|---|---|---|
| CD | UC | HC | CD | UC | HC | ||
| Infectious and parasitic diseases | 001–139 | 120 (12.0) | 145 (14.5) | 61 (12.2) | 2451 (1,2) | 3401 (1, 3) | 881 (1, 2) |
| Neoplasms | 140–239 | 77 (7.7) | 158 (15.8) | 26 (5.2) | 1231 (1, 2) | 2961 (1, 2) | 811 (1, 2) |
| Endocrine, nutritional and metabolic diseases and immunity disorders | 240–279 | 202 (20.2) | 263 (26.3) | 55 (11.0) | 4822 (1, 3) | 8442 (1, 4) | 991 (1, 2) |
| Diseases of the blood and blood forming organs | 280–289 | 155 (15.5) | 206 (20.6) | 29 (5.8) | 4822 (1, 4) | 7452 (1, 4) | 1202 (1, 4) |
| Mental disorders | 290–319 | 207 (20.7) | 199 (19.9) | 142 (28.4) | 6192 (1, 4) | 6542 (1, 3) | 8133 (1, 6) |
| Diseases of the nervous system | 320–359 | 99 (9.9) | 119 (11.9) | 19 (3.8) | 2231 (1, 2) | 2871 (1, 3) | 221 (1, 1) |
| Diseases of the sense organs | 360–389 | 47 (4.7) | 88 (8.8) | 11 (2.2) | 691 (1, 2) | 1441 (1, 1) | 141 (1, 2) |
| Diseases of the circulatory system | 390–459 | 176 (17.6) | 252 (25.2) | 40 (8.0) | 3671 (1, 2) | 7482 (1, 3) | 651 (1, 2) |
| Diseases of the respiratory system | 460–519 | 117 (11.7) | 154 (15.4) | 55 (11.0) | 2191 (1, 2) | 3001 (1, 2) | 1181 (1, 2) |
| Diseases of the digestive system | 520–579 | 589 (58.9)a | 630 (63.0)b | 88 (17.6) | 2714a3 (1,6) | 2555b2 (1, 5) | 1981 (1, 2) |
| Diseases of the genitourinary system | 580–629 | 97 (9.7) | 96 (9.6) | 39 (7.8) | 2071 (1, 2) | 1721 (1, 2) | 691 (1, 2) |
| Complications of pregnancy, childbirth and the puerperium | 630–679 | 17 (1.7) | 23 (2.3) | 10 (2.0) | 1256 (4, 10) | 1633 (2, 6) | 303 (2, 4) |
| Diseases of the skin and subcutaneous tissue | 680–709 | 136 (13.6) | 155 (15.5) | 19 (3.8) | 2331 (1, 2) | 2981 (1, 2) | 281 (1, 2) |
| Diseases of the musculoskeletal system and connective tissue | 710–739 | 257 (25.7) | 305 (30.5) | 73 (14.6) | 7002 (1, 3) | 9932 (1, 4) | 1881 (1, 3) |
| Congenital anomalies | 740–759 | 30 (3.0) | 13 (1.3) | 5 (1.0) | 451 (1, 2) | 181 (1, 2) | 81 (1, 2) |
| Certain conditions originating in the perinatal period | 760–779 | 2 (0.2) | 1 (0.1) | 0 (0.0) | 21 (1, 1) | 8 (-, -) | 0 (-, -) |
| Symptoms, signs and ill-defined conditions | 780–799 | 539 (53.9) | 554 (55.4) | 221 (44.2) | 22152 (1, 5) | 22982 (1, 5) | 8392 (1, 4) |
| Injury and poisoning | 800–999 | 119 (11.9) | 110 (11.0) | 51 (10.2) | 2521 (1, 2) | 2191 (1, 2) | 1081 (1, 2) |
Does not include CD diagnoses.
Does not include UC diagnoses.
5. Discussion
Herein we have described a large cohort of active duty US military personnel with incident Crohn's disease or ulcerative colitis along with matched healthy control subjects in which serum samples have been periodically collected prior to and immediately after IBD onset. This cohort study, called PREDICTS, is designed as a platform study that may revolutionize our understanding of IBD pathoetiology and facilitate movement toward individualized medicine and disease risk prediction and ultimately prevention [22,30,31]. Ongoing serologic assays include those focused on unique proteomic markers, serologic evidence of infectious exposures, cytokine profiles and responses to commensal intestinal microbiota will serve to generate new hypotheses for future in and ex vivo IBD research [16]. We recognize that other cohort studies exist including the Nurses Health Study I and II, and the GEM (Genetics, Environmental, Microbial) Project that promise to provide unique opportunities to address current research gaps. However, the PREDICTS study is unique given the availability of multiple samples in the pre-clinical period and the opportunity this affords in discerning factors that are associated with initial triggers and progression of subclinical immuno-inflammatory processes, before overt disease has been established [22,32].
This cohort of IBD patients represents a well-characterized group of subjects whose initial sample collections were obtained during periods of time in which the subjects were sufficiently healthy to actively serve in the US military. While these outcomes have not been validated by chart review, a significant proportion of both UC and CD cases have ‘confirmatory’ medical encounter data that provide confidence in the diagnoses, such as codes associated with gastroenterology specialists visits and endoscopic procedures. It is important to note that the absence of these data does not reduce the likelihood of these being actual IBD cases. The primary reasons for missing ‘confirmatory’ data are likely multifactorial, and may include the absence of an available Department of Defense specialty clinic at the duty site and subsequent civilian care (specialist and endoscopy procedures) received which are not recorded into the DMSS, or simply incomplete or inaccurate records.
We also fully recognize that an ideal study of this type would involve multiple specimen types including genetic, tissue biopsy, fecal and whole blood specimens in a prospectively designed study where detailed diagnostic, treatment, and exposure data would be collected. Such studies are underway and promise to yield high quality data on which similar hypotheses could be tested [33]. The limitation of this study to serum is recognized but does not preclude important and rigorous research possibilities. While genetics clearly play an important role in IBD, it only explains a proportion of IBD risk [34]. Poor correlation between the regulation of transcripts and actual protein quantities may confound associations between genetic polymorphisms disease likely due to the fact that genomic analysis does not account for post-translational processes such as protein modifications and protein degradation or epigenetic changes. Therefore, methods employed in the disease biomarker process have expanded to include proteomics which has allowed for interrogation of these systems indirectly through the serum compartment [35]. In addition, the readily accessible sample type (e.g., blood draw) may have practical considerations in the future development of clinically applicable screening and evaluation tools. In addition to lack of complementary specimen types, this study is limited by available medical and demographic data which are obtainable in the DMSS as well as the inability to link with medical encounter data upon discharge from Active Duty. Clinical laboratory data, endoscopic and histological data, pharmacy data, as well as smoking status and other behavioral risk factor information are unavailable. While other prospective studies of active duty military populations have included post-discharge follow-up data, the de-identified nature of PREDICTS precludes linking with those ongoing studies, and does not allow for prospectively following participants who may be beneficiaries in the Veteran Affairs system.
Despite these limitations, the number of clinical cases, accessible clinical and exposure data in the data archive, as well as serial serum time points provide a robust platform to inform relevant gaps in our knowledge and understanding of IBD. At present, one study has been published from the PREDICTS study in which antimicrobial biomarkers tested many years prior to disease diagnosis were found to predict complicated disease phenotype at clinical presentation [16]. Efforts are underway to expand these initial findings. In addition, a variety of protein and antibody panels are currently being evaluated to explore additional aims of biomarker prevalence at clinical onset, biomarker conversion associated with disease onset, enteric infection and disease/biomarker conversion, novel disease biomarkers and pathogenesis, biomarker results compared to historical populations, demographic and deployment factors and association with disease biomarkers, as well as biomarkers predicting disease progression after diagnosis. The potential value and utility of PREDICTS has yet to be fully revealed, and we hope that through future collaborations, advances in technology, and accumulation of a data set rich in multiple biomarker results, and promises of advances in our understanding of IBD that translate into new solutions and practice changes will be fulfilled. Finally, with this initiative we feel that we have provided a blueprint for the creation of public-private pre-competitive partnerships to investigate disease triggers and lay the foundation for disease interception and prevention.
Collaboration
-
•
Participating Institutions: This study was conceived, developed and funded through a partnership between the Naval Medical Research Center (Silver Spring, MD, US), the Mayo Clinic (Rochester, MN, US), Prometheus Laboratories, Inc. (San Diego, CA, US), the Icahn School of Medicine at Mount Sinai (New York, NY, US) and Janssen Pharmaceuticals (Springhouse, PA, US).
Author contributions
Study concept and design: Mark S. Riddle, Chad K. Porter, Francisco Leon, Joseph. A. Murray, Fred Princen and Jean Frederic Colombel;
Acquisition of data: Chad K. Porter, Ramiro L. Gutierrez, and Mark S. Riddle;
Analysis and data interpretation: Mark S. Riddle, Chad K. Porter, Ramiro L. Gutierrez.
Sample repository archiving: Renee M. Laird, Chad K. Porter.
Drafting of manuscript: Mark S. Riddle, Chad K. Porter.
Critical revision of manuscript for important intellectual content: Chad K. Porter, Mark S. Riddle, Ramiro L. Gutierrez, Fred Princen, Rick Strauss, Shannon E. Telesco, Joana Torres, Rok Seon Choung, Renee M. Laird, Jean Fred Colombel, Joseph A. Murray.
Administrative, technical, or material support: Chad K. Porter, Ramiro L. Gutierrez, and Mark S. Riddle, Renee M. Laird.
Study supervision: Mark S. Riddle, Joseph. A. Murray, Fred Princen, Rick Straus, and Jean-Frederic Colombel.
Conflicts of interest
Dr Murray has received grant funding from Alba Therapeutics and Alvine Pharmaceuticals, Inc., has served on an advisory board for Alvine Pharmaceuticals, Inc., and has served as a consultant for AMAG Pharmaceuticals, BioLineRx, Glenmark, UCB Pharma, Takeda, Celimmune and GlaxoSmithKline. J-F Colombel has served as consultant or advisory board member for Abbvie, ABScience, Amgen, Bristol-Myers Squibb, Celltrion, Danone, Ferring, Genentech, Giuliani SPA, Given Imaging, Janssen, Immune Pharmaceuticals, Eli Lilly, Medimmune, Merck & Co., Millenium Pharmaceuticals Inc., Neovacs, Nutrition Science Partners Ltd., Pfizer Inc. Prometheus Laboratories, Protagonist, Receptos, Sanofi, Schering Plough Corporation, Second Genome, Shire, Takeda, Teva Pharmaceuticals, Tigenix, UCB Pharma, Vertex, Dr. August Wolff GmbH & Co. J-F Colombel has served as a speaker for Abbvie, Ferring, Janssen, Merck & Co., Nutrition Science Partners Ltd., Takeda. Joana Torres received consulting fees from Takeda and support from Abbvie to travel to conferences. Fred Princen, Thomas P. Stockfisch, and Sharat Singh are employees of Prometheus Laboratories. Francisco Leon was employee of Janssen R&D. The remaining author discloses no conflicts.
Chad K. Porter, Ramiro L. Gutierrez, Renee Laird, Mark Riddle - The views expressed in this article are those of the author and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the U.S. Government.
Human Subjects Research and Data Protections: The human subjects' research protocols (NMRC.2012.0007 and NMRC.2014.0019) under which the data and samples were obtained were approved as ‘Exempt’ research by the Naval Medical Research Center Institutional Review Board in compliance with all applicable Federal regulations governing the protection of human subjects. In addition, this study is conducted under a support agreement with the AFHSB. All data are were de-identified by personnel at AFHSB who removed the list of 18 identifiers outlined in 45 CFR 164.514(b)(2) and DOD 6025.18-R (DoD Health Information Privacy Regulation) prior to providing the data to the investigators.
Study governance
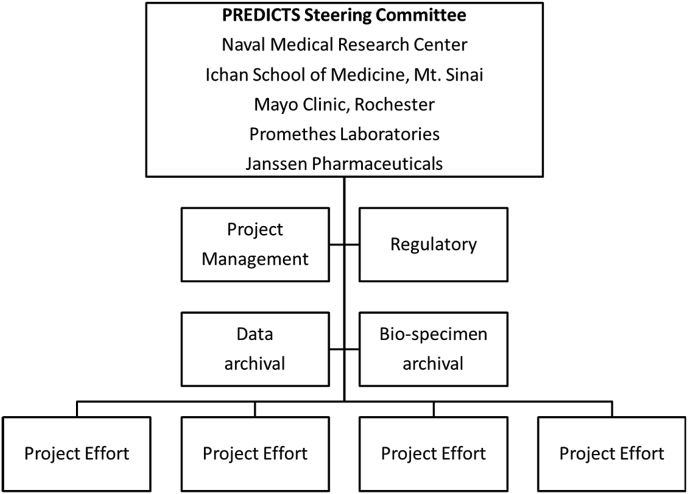
This study is coordinated and executed through a Steering Committee (SC) and supported by project management, regulatory, biospecimen and data archival support functions under a Navy Cooperative Research and Development Agreement (CRADA# NMR-11-3920, Antimicrobial Antibodies as Predictors of Inflammatory Bowel Diseases) (Fig. 2). The primary function of the SC is to oversee research efforts conducted under the PREDICTS protocols and CRADA. The SC and PREDICTS Study is governed by a written charter (Supplemental Files) which includes specific guidance, policies and procedures for handling data and serum utilization requests, publication and presentation clearance, data sharing, as well as appeal processes to handle disputes. In addition to managing these procedural functions, the SC reviews and monitors all research efforts and provides guidance and direction for future research. The SC provides a stabilizing influence among vested stakeholders so organizational concepts and directions are established and maintained with a mission-oriented view. The SC provides insight on long-term strategies in support of research objectives. Members of the SC ensure objectives are adequately addressed and projects remain under appropriate control and oversight. The SC is also charged with the responsibility of advocating for the Study and identifying new partnerships and opportunities.
Fig. 2.
Predicts study Governance structure.
Procedure for application and use of study data and additional testing
The SC welcomes submissions requesting the utilization of existing data and serum resources. A concept submission process involves four steps: 1) an invitation to develop and submit a Data Analysis Proposal (Supplemental Files) by one or more members of the SC; 2) submission of the Data Analysis Proposal and review by the SC; 3) Decision by SC on project (or request for modifications; 4) Proposal execution. Once a concept is approved upon review, the SC provides ongoing oversight to ensure applicability to the PREDICTS objectives, as well as regulatory, data sharing, publication, and intellectual property requirements. All results and data interpretation are reported to the SC and, if applicable, published in the peer reviewed literature.
Copyright statement
This work was prepared as part of the official duties of US Government personnel and service members (CKP, MSR, RLG). Title 17 U.S C. §105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person's official duties.
Funding
Funding and support of the PREDICTS study platform was provided through a Cooperative and Research Development Agreement with direct contributions by Janssen Pharmaceuticals, Prometheus Laboratories and the Naval Medical Research Center (NCRADA number NMR-11-3920).
Acknowledgements
The authors would like to thank the staff at the Armed Forces Health Surveillance Branch for their assistance in compiling and providing the data and sera utilized for this study. Additionally, the authors would like to thank all the men and women in uniform who have dedicated their service and lives to our country and through the simple act of providing blood allow the potential for discoveries to help millions.
We are indebted to the PREDICTS study team of program coordinators, data managers, affiliated scientists, and administrative support personnel for their tireless hours to ensure the success of this project. The PREDICTS study team: Christina Hill, Vicky Chapman, Ashley Alcala, Alex Maue (Naval Medical Research Center); Takihiro Soto, Joseph Russell, Scott Plevy, Carrie Brodmerkel (Janssen Pharmaceuticals); Bénédicte De Vroey, Corinne Gower-Rousseau (Center Hospitalier Régional Universitaire, Lille, France); Sharat Singh, Tharak Rao, Xiaojun Li (Prometheus Laboratories Inc), Francesca Petralia (Mount Sinai).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.conctc.2019.100345.
Contributor Information
Chad K. Porter, Email: chad.k.porter2.civ@mail.mil.
Mark S. Riddle, Email: markriddlemd@gmail.com.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Loftus E.V., Jr., Silverstein M.D., Sandborn W.J. Ulcerative colitis in Olmsted County, Minnesota, 1940-1993: incidence, prevalence, and survival. Gut. 2000;46:336–343. doi: 10.1136/gut.46.3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vermeire S., van Assche G., Rutgeerts P. Review article: altering the natural history of Crohn's disease--evidence for and against current therapies. Aliment Pharmacol. Ther. 2007;25:3–12. doi: 10.1111/j.1365-2036.2006.03134.x. [DOI] [PubMed] [Google Scholar]
- 3.M'Koma A.E. Inflammatory bowel disease: an expanding global health problem. Clin. Med. Insights Gastroenterol. 2013;6:33–47. doi: 10.4137/CGast.S12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loftus C.G., Loftus E.V., Jr., Harmsen W.S. Update on the incidence and prevalence of Crohn's disease and ulcerative colitis in Olmsted County, Minnesota, 1940-2000. Inflamm. Bowel Dis. 2007;13:254–261. doi: 10.1002/ibd.20029. [DOI] [PubMed] [Google Scholar]
- 5.Molodecky N.A., Soon I.S., Rabi D.M. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54. doi: 10.1053/j.gastro.2011.10.001. e42; quiz e30. [DOI] [PubMed] [Google Scholar]
- 6.Dahlhamer J.M., Zammitti E.P., Ward B.W. Prevalence of inflammatory bowel disease among adults aged >/=18 Years - United States, 2015. MMWR Morb. Mortal. Wkly. Rep. 2016;65:1166–1169. doi: 10.15585/mmwr.mm6542a3. [DOI] [PubMed] [Google Scholar]
- 7.Russell A.S., Gulliver W.P., Irvine E.J. Quality of life in patients with immune-mediated inflammatory diseases. J. Rheumatol. Suppl. 2011;88:7–19. doi: 10.3899/jrheum.110899. [DOI] [PubMed] [Google Scholar]
- 8.van der Have M., van der Aalst K.S., Kaptein A.A. Determinants of health-related quality of life in Crohn's disease: a systematic review and meta-analysis. J. Crohns. Colitis. 2014;8:93–106. doi: 10.1016/j.crohns.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Floyd D.N., Langham S., Severac H.C. The economic and quality-of-life burden of Crohn's disease in Europe and the United States, 2000 to 2013: a systematic review. Dig. Dis. Sci. 2015;60:299–312. doi: 10.1007/s10620-014-3368-z. [DOI] [PubMed] [Google Scholar]
- 10.Jostins L., Ripke S., Weersma R.K. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkes M. The genetics universe of Crohn's disease and ulcerative colitis. Dig. Dis. 2012;30(Suppl 1):78–81. doi: 10.1159/000341130. [DOI] [PubMed] [Google Scholar]
- 12.Magrone T., Jirillo E. The interplay between the gut immune system and microbiota in health and disease: nutraceutical intervention for restoring intestinal homeostasis. Curr. Pharmaceut. Des. 2013;19:1329–1342. doi: 10.2174/138161213804805793. [DOI] [PubMed] [Google Scholar]
- 13.Scharl M., Rogler G. Inflammatory bowel disease: dysfunction of autophagy? Dig. Dis. 2012;30(Suppl 3):12–19. doi: 10.1159/000342588. [DOI] [PubMed] [Google Scholar]
- 14.Barnes E.L., Burakoff R. New biomarkers for diagnosing inflammatory bowel disease and assessing treatment outcomes. Inflamm. Bowel Dis. 2016;22(12):2956–2965. doi: 10.1097/MIB.0000000000000903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heier C.R., Fiorillo A.A., Chaisson E. Identification of pathway-specific serum biomarkers of response to glucocorticoid and infliximab treatment in children with inflammatory bowel disease. Clin. Transl. Gastroenterol. 2016;7:e192. doi: 10.1038/ctg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choung R.S., Princen F., Stockfisch T.P. Serologic microbial associated markers can predict Crohn's disease behaviour years before disease diagnosis. Aliment Pharmacol. Ther. 2016;43:1300–1310. doi: 10.1111/apt.13641. [DOI] [PubMed] [Google Scholar]
- 17.Dubinsky M.C., Lin Y.C., Dutridge D. Serum immune responses predict rapid disease progression among children with Crohn's disease: immune responses predict disease progression. Am. J. Gastroenterol. 2006;101:360–367. doi: 10.1111/j.1572-0241.2006.00456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zholudev A., Zurakowski D., Young W. Serologic testing with ANCA, ASCA, and anti-OmpC in children and young adults with Crohn's disease and ulcerative colitis: diagnostic value and correlation with disease phenotype. Am. J. Gastroenterol. 2004;99:2235–2241. doi: 10.1111/j.1572-0241.2004.40369.x. [DOI] [PubMed] [Google Scholar]
- 19.Porter C.K., Cash B.D., Pimentel M. Risk of inflammatory bowel disease following a diagnosis of irritable bowel syndrome. BMC Gastroenterol. 2012;12:55. doi: 10.1186/1471-230X-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verdu E.F., Riddle M.S. Chronic gastrointestinal consequences of acute infectious diarrhea: evolving concepts in epidemiology and pathogenesis. Am. J. Gastroenterol. 2012;107:981–989. doi: 10.1038/ajg.2012.65. [DOI] [PubMed] [Google Scholar]
- 21.Denson L.A., Long M.D., McGovern D.P. Challenges in IBD research: update on progress and prioritization of the CCFA's research agenda. Inflamm. Bowel Dis. 2013;19:677–682. doi: 10.1097/MIB.0b013e31828134b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres J., Burisch J., Riddle M. Preclinical disease and preventive strategies in IBD: perspectives, challenges and opportunities. Gut. 2016;65:1061–1069. doi: 10.1136/gutjnl-2016-311785. [DOI] [PubMed] [Google Scholar]
- 23.Porter C.K., Tribble D.R., Aliaga P.A. Infectious gastroenteritis and risk of developing inflammatory bowel disease. Gastroenterology. 2008;135:781–786. doi: 10.1053/j.gastro.2008.05.081. [DOI] [PubMed] [Google Scholar]
- 24.Moore M., Eiseman E., Fisher G. The RAND Corporation; 2010. Harnessing Full Value from the DoD Serum Repository and the Defense Medical Surveillance System. [PMC free article] [PubMed] [Google Scholar]
- 25.Shi T., Song E., Nie S. Advances in targeted proteomics and applications to biomedical research. Proteomics. 2016;16:2160–2182. doi: 10.1002/pmic.201500449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fontana J.M., Alexander E., Salvatore M. Translational research in infectious disease: current paradigms and challenges ahead. Transl. Res. 2012;159:430–453. doi: 10.1016/j.trsl.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kevans D., Silverberg M.S., Borowski K. IBD genetic risk profile in healthy first-degree relatives of crohn's disease patients. J. Crohns. Colitis. 2016;10:209–215. doi: 10.1093/ecco-jcc/jjv197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubertone M.V., Brundage J.F. The Defense Medical Surveillance System and the Department of Defense serum repository: glimpses of the future of public health surveillance. Am. J. Public Health. 2002;92:1900–1904. doi: 10.2105/ajph.92.12.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Defense Medical Surveillance System Armed Forces Health Surveillance Branch. https://health.mil/Military-Health-Topics/Health-Readiness/Armed-Forces-Health-Surveillance-Branch/Data-Management-and-Technical-Support/Defense-Medical-Surveillance-System: Defense Health Agency last accessed 23 October 2016.
- 30.Cayer D.M., Nazor K.L., Schork N.J. Mission critical: the need for proteomics in the era of next-generation sequencing and precision medicine. Hum. Mol. Genet. 2016;25:R182–r189. doi: 10.1093/hmg/ddw214. [DOI] [PubMed] [Google Scholar]
- 31.Jain K.K. Role of proteomics in the development of personalized medicine. Adv. Protein Chem. Struct. Biol. 2016;102:41–52. doi: 10.1016/bs.apcsb.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Torres J., Colombel J.F., Riddle M.S. Evidence for life before inflammatory bowel disease. Clin. Gastroenterol. Hepatol. 2016;14:825–828. doi: 10.1016/j.cgh.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Kevans D., Turpin W., Madsen K. Determinants of intestinal permeability in healthy first-degree relatives of individuals with Crohn's disease. Inflamm. Bowel Dis. 2015;21:879–887. doi: 10.1097/MIB.0000000000000323. [DOI] [PubMed] [Google Scholar]
- 34.Elson C.O., Cong Y., McCracken V.J. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol. Rev. 2005;206:260–276. doi: 10.1111/j.0105-2896.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- 35.Okerberg E.S., Wu J., Zhang B. High-resolution functional proteomics by active-site peptide profiling. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4996–5001. doi: 10.1073/pnas.0501205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.