Abstract
Mammalian epithelial cells use a pair of parental centrioles and numerous deuterosomes as platforms for efficient basal body production during multiciliogenesis. How deuterosomes form and function, however, remain controversial. They are proposed to arise either spontaneously for massive de novo centriole biogenesis or in a daughter centriole‐dependent manner as shuttles to carry away procentrioles assembled at the centriole. Here, we show that both parental centrioles are dispensable for deuterosome formation. In both mouse tracheal epithelial and ependymal cells (mTECs and mEPCs), discrete deuterosomes in the cytoplasm are initially procentriole‐free. They emerge at widely dispersed positions in the cytoplasm and then enlarge, concomitant with their increased ability to form procentrioles. More importantly, deuterosomes still form efficiently in mEPCs whose daughter centriole or even both parental centrioles are eliminated through shRNA‐mediated depletion or drug inhibition of Plk4, a kinase essential to centriole biogenesis in both cycling cells and multiciliated cells. Therefore, deuterosomes can be assembled autonomously to mediate de novo centriole amplification in multiciliated cells.
Keywords: basal body, centriole, deuterosome, multicilia, Plk4
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Cell Cycle
Introduction
Centriole biogenesis in cycling cells is tightly controlled 1, 2, 3, 4. In G1 phase of the cell cycle, each cell contains a centrosome with a pair of parental centrioles: the mother and daughter centrioles. In S phase, the daughter centriole matures into a mother centriole. Both the older and younger mother centrioles are then allowed to assemble one nascent daughter centriole from a Cep63‐ and Cep152‐containing platform 5, 6, located around their basolateral walls, by restricting the levels and location of a critical protein kinase Plk4 7, 8, 9, 10. Plk4 recruits and phosphorylates Stil, which then recruits SAS6 to initiate centriole biogenesis 4, 11, 12, 13. The SAS6‐containing cartwheel further primes the assembly of other components such as the centriolar microtubules and Centrin into a procentriole 7, 14, 15, 16, 17, 18, 19. In G2 phase, the daughter centriole assembly is complete. Following mitosis, the two pairs of mother–daughter centrioles are partitioned in the form of the centrosome into two progeny cells so that constant centriole number can be maintained 3, 20. Overexpressing key regulators such as Plk4, Cep152, and SAS6 have been shown to potentiate the centriole biogenesis ability of the parental centrioles 7, 11, 21, 22, 23.
Epithelial tissues such as those in mammalian trachea, ependyma, and oviduct are abundant in terminally differentiated cells with dense motile cilia. These multiciliated cells each require up to hundreds of centrioles to serve as basal bodies of their cilia 24, 25, 26. To achieve this, the cells express high levels of proteins important for the massive basal body production through transcriptional regulators such as multicilin and E2f4 27, 28, 29, 30, 31, 32. As a result, both parental centrioles produce multiple daughter centrioles. Furthermore, dozens of spherical structures termed deuterosomes emerge to generate the majority of the basal bodies 33, 34, 35. The deuterosome adapts Deup1, a paralog of Cep63, and Ccdc78 to create a similar but parental centriole‐free platform of centriole biogenesis 27, 36, 37, 38. Its formation requires cytoplasmic E2f4 32. Fully assembled centrioles are eventually released from their “cradles” by APC/C‐activated proteolysis and mature into basal bodies 39, 40.
The origin and functions of deuterosomes, however, are still controversial. Based mainly on electron microscopic studies, deuterosomes have been proposed to either form autonomously and mediate parental centriole‐independent, or de novo, centriole biogenesis 3, 33, 34, 41 or form and support procentriole biogenesis in a parental centriole‐dependent manner 42. Our recent identification of Deup1 enables further examinations of these hypotheses using fluorescent microscopy. Our results in cultured mTECs strengthen the first model because deuterosomes are found to emerge massively, initially carrying fewer procentrioles (Fig 1A, stage II), and then grow into larger ones with more associated procentrioles (Fig 1A, stage III) 36. A later study mainly using cultured mEPCs, on the other hand, elaborates the second model by proposing that all the deuterosome and procentrioles are initially nucleated from the daughter centrosomal centriole. Deuterosomes only function as shuttles to load and carry away these procentrioles in the form of “halos” to facilitate centriole amplification (Fig 1B) 43, 44.
Figure 1. Discrete deuterosomes in mTECs are initially free of procentrioles.
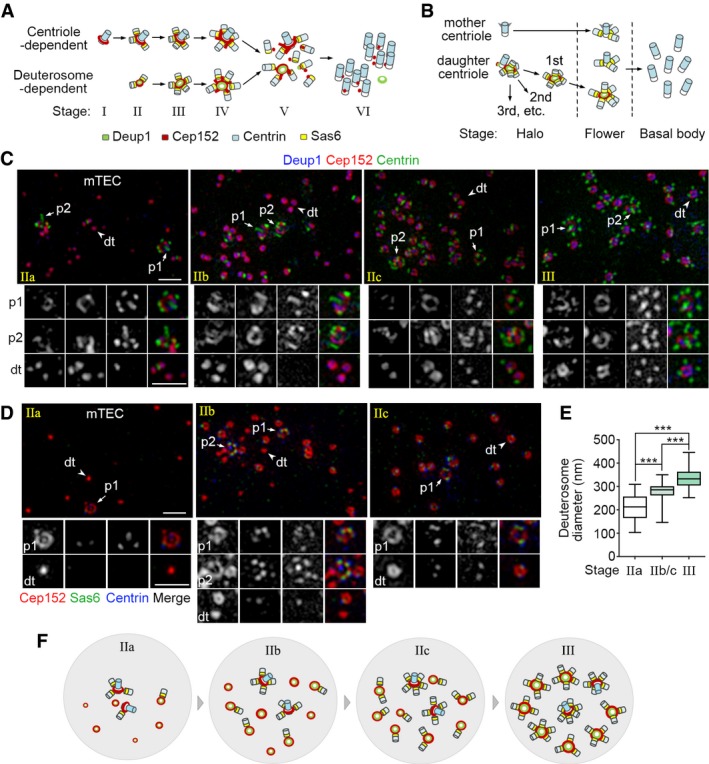
-
A, BTwo current models for the process of massive basal body formation based on studies in mTECs (A) and mEPCs (B). In (A), deuterosomes are proposed to form spontaneously in stage II and each supports the assembly of 1–2 procentrioles. They grow in size and nucleate more procentrioles in stage III. Each parental centriole also nucleates multiple procentrioles from stage II. Stage IV is characterized by the emergence of Cep152‐positive protrusions, stage V by the releasing of basal bodies from both deuterosomes and parental centrioles, and stage VI by the formation of basal body clusters under the apical side of the cell membrane 36. In (B), deuterosomes are assembled at the lateral wall of the daughter centriole and released one by one as procentriole‐occupied “halos” (the halo stage). During this stage, parental centrioles also start to generate their own procentrioles. When the last halo is released, procentrioles on both deuterosomes and parental centrioles start to mature simultaneously (the flower stage) and are eventually released as basal bodies (the basal body stage) 43. Deup1, Cep152, Centrin, and Sas6 are used as markers respectively for the deuterosome, both parental centriole and deuterosome, centriole (including procentriole), and procentriole.
-
C, DRepresentative mTECs containing early deuterosomes. mTECs cultured at an air–liquid interface for 2 days were immunostained for Cep152, Centrin, and Deup1 (C) or Sas6 (D) and subjected to 3D‐SIM. Parental centrioles (p1/p2; arrows) and typical deuterosomes (dt; arrowheads) are magnified 1.5× to show details. IIa: Parental centrioles contain procentrioles; most deuterosomes are procentriole‐free and usually small. IIb: A substantial portion of deuterosomes contains 1 procentriole; deuterosomes are usually medium in size. IIc: Most deuterosomes contain 1–2 procentrioles; deuterosomes are usually medium in size. A stage‐III cell, which contained larger deuterosomes associated with more procentrioles, is shown in (C) for comparison. Scale bar, 1 μm.
-
EDeuterosome‐size distributions in stage IIa, IIb/c, and III. For each group, the diameters of at least 348 deuterosomes were measured from 12 mTECs selected from three independent experiments according to the stages. The bottom and top of the box represent the 25th and 75th percentiles, respectively. The band is the median. The ends of the whiskers indicate the maximum and minimum of the data. Two‐tailed unpaired Student's t‐test: ***P < 0.001.
-
FAn illustration model for the progression of mTECs from stage IIa to III.
In this study, we sought to clarify the discrepancies using cultured mTECs and mEPCs.
Results and Discussion
Discrete deuterosomes in mTECs do not initially exist as halos
The global increase in the number of deuterosome‐associated procentrioles from stage II to stage III in mTECs 36 clearly argues against the mere “shuttle” role of the deuterosome. Nevertheless, the initial procentrioles on the stage‐II deuterosomes could still come from the centrosomal daughter centriole. To clarify this, we examined early events of deuterosome and procentriole formation. mTECs can be induced to differentiate into multiciliated cells efficiently by culturing at an air–liquid interface (ALI) 36, 45, 46. As mTECs thus cultured for 3 days were mainly at late stages of the basal body production 36, we examined those at day 2 with three‐dimensional structured illumination microscopy (3D‐SIM).
Consistent with our previous report 36, both parental centrioles carried procentrioles in stage‐II mTECs when they were both detected (Fig 1C and D). By contrast, the size and number of the deuterosomes and the status of their procentrioles, which were better defined by using both Sas6 and Centrin as markers (Fig 1D) 47, 48, varied dramatically. For instance, a portion of the cells contained sparse, small (ϕ = 210 ± 60 nm), and usually procentriole‐free deuterosomes (Fig 1C–E: IIa), suggesting that they are in early stage II (Fig 1F: IIa). In the remaining stage‐II cells, deuterosomes were generally larger (ϕ = 280 ± 50 nm; Fig 1E: IIb/c) and more abundant (Fig 1C and D: IIb and IIc). Those mingled with deuterosomes either without procentriole or with only one procentriole were presumably in middle stage II (Fig 1C, D, and F: IIb). Sometimes, deuterosomes of both small and large sizes were observed in these cells (Fig 1D: IIb). By contrast, those with deuterosomes that were commonly associated with 1–2 procentrioles were in late stage II (Fig 1C, D, and F: IIc). In comparison, in stage‐III mTECs deuterosomes were 340 ± 50 nm in diameter (Fig 1E) and frequently associated with 3–5 procentrioles (Fig 1C and F). Therefore, in mTECs discrete deuterosomes do not initially carry procentrioles as indicated by Al Jord and colleagues 43. Neither does the marked increase in their associated procentrioles during the progression from stage II to stage III (Fig 1C) 36 fit the proposed shuttle function of the deuterosome (Fig 1B) 43.
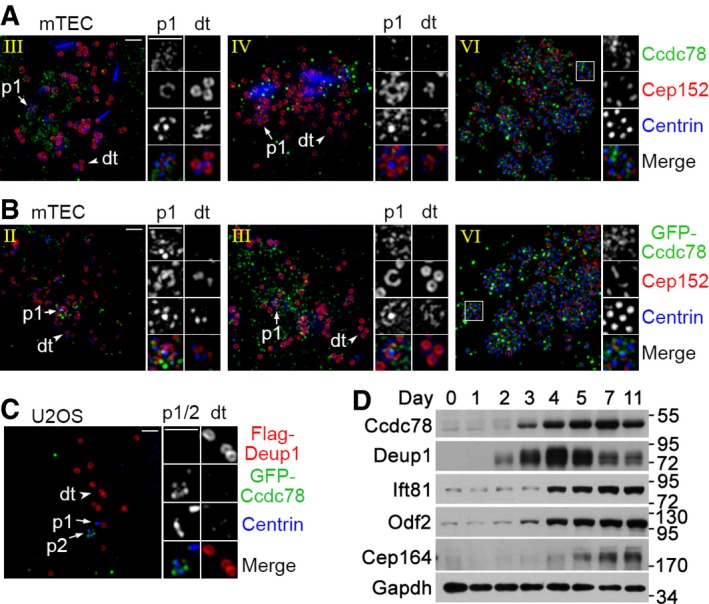
Murine Ccdc78, whose Xenopus orthologue is identified as a deuterosome protein 38, was also used by Al Jord and colleagues as a deuterosome marker in addition to Deup1 43. We thus examined its subcellular localization but were unable to detect deuterosome localization of endogenous or exogenous murine Ccdc78 in mTECs. Both endogenous Ccdc78 and exogenous GFP‐Ccdc78 formed numerous puncta irrelevant to deuterosomes in stages II–IV (Fig EV1A and B). In stage VI, they both showed correlation with basal bodies (Fig EV1A and B). As exogenous Deup1 can induce deuterosome‐like structures in cycling cells 36, we co‐expressed Flag‐Deup1 and GFP‐Ccdc78 in U2OS cells and still found no GFP‐Ccdc78 on the deuterosome‐like structures in the human cells (Fig EV1C). Instead, GFP‐Ccdc78 mostly decorated a parental centriole (Fig EV1C). We have previously shown that in differentiating mTECs, the expression patterns of proteins involved in centriole amplification, such as Deup1, Plk4, Cep152, and Sas6, are similar, but distinct from those involved in ciliary biogenesis or functions, such as Odf2 and Ift57 36. Ccdc78, however, was different from Deup1 in expression patterns but analogous to centriolar appendage proteins Odf2 and Cep164 and ciliary protein Ift81 (Fig EV1D) 25, 49, 50. These results do not suggest mammalian Ccdc78 as a deuterosome protein. We therefore only used Deup1 and Cep152 as deuterosome markers in the following experiments.
Figure EV1. Mammalian Ccdc78 is not a deuterosome protein.
-
A, BNeither endogenous nor exogenous murine Ccdc78 localized to deuterosomes in mTECs. Cultured mTECs were fixed at day 3 post‐ALI (A) or transfected with lentivirus at day −1 to express GFP‐Ccdc78, followed by fixation at day 3 (B). After immunostaining to visualize the indicated proteins, the samples were imaged with SIM. Parental centrioles (p1; arrows), typical deuterosomes (dt; arrowheads), and typical regions with basal bodies (framed) are magnified 2× to show details. Scale bar, 1 μm.
-
CGFP‐Ccdc78 did not localize to Flag‐Deup1‐induced deuterosome‐like structures in U2OS cells. U2OS cells were co‐transfected to express Flag‐Deup1 and GFP‐Ccdc78 for 48 h and fixed for immunostaining. Parental centrioles (p1/p2; arrows) and typical deuterosome‐like structures (dt; arrowheads) are magnified 2× to show details. Scale bar, 1 μm.
-
DCcdc78 and Deup1 had different expression patterns in differentiating mTECs. Cultured mTECs were induced to undergo multiciliation from day 0 and collected at the indicated time. Gapdh served as loading control.
Centriole amplification in mEPCs resembles mTECs
Next, we investigated whether the centriole amplification process in mEPCs is different. Cultured progenitor cells isolated from P0 mouse brain tissues can be induced to differentiate into multiciliated mEPCs through serum starvation 51, 52. We examined the mEPCs at day 2 or 3 postserum starvation because they were undergoing active centriole amplification 43, 53. As Centrin‐positive aggregates were frequently observed in mEPCs, especially in the area around the parental centrioles 54, 55, we also used Sas6 as additional procentriole marker 47, 48.
We found that mEPCs could also be grouped into six stages (Fig 2A and B), similar to mTECs (Fig 1A) 36: (i) Those containing only a pair of Cep152‐positive, Deup1‐negative parental centrioles were in stage I; (ii) those containing small deuterosomes with mostly 0–2 procentrioles were in stage II; (iii) those in the “halo” or “flower” stages (see Fig 1B) 43, containing larger deuterosomes commonly with 3–7 procentrioles, could be assigned to stages III and IV, respectively. The stage‐IV cells were also distinguished from those in stage III by the emergence of multiple Cep152‐positive protrusions from both deuterosomes and parental centrioles. Their Centrin staining was also elongated as compared to the punctate staining in the stage‐III cells; (iv) those with partially released basal bodies from their cradles were in stage V. Their basal bodies still contained the Sas6‐positive puncta; and (v) those with fully released basal bodies negative for the Sas6 staining were in stage VI (Fig 2A and B). Similar to mTECs 36, 53, stage‐VI mEPCs also underwent multiciliogenesis (Fig 2C). Only their basal bodies did not group into clusters (Fig 2A–C) as those do in mTECs 36.
Figure 2. mEPCs resemble mTECs in centriole amplification.
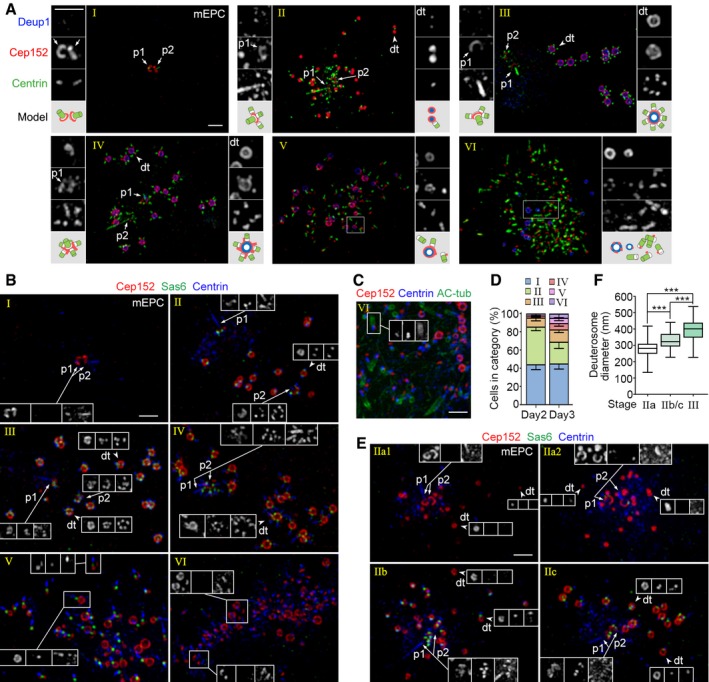
- Typical mEPCs representing different stages of centriole amplification. mEPCs cultured under serum starvation for 3 days were immunostained for Deup1, Cep152, and Centrin, followed by imaging with 3D‐SIM. Parental centrioles (p1/p2; arrows), typical deuterosomes (dt; arrowheads), and typical regions with basal bodies (framed) are magnified 2× to show details. An illustration is provided for each set of the magnified images. Scale bar, 1 μm.
- Typical mEPCs at day 3, immunostained for Cep152, Sas6, and Centrin. The insets are arranged in the same sequence from left to right. Scale bar, 1 μm.
- Multicilia formation in a typical stage‐VI mEPC. Acetylated tubulin (AC‐tub) was used as ciliary marker. Note that the ciliogenesis is asynchronous. Scale bar, 1 μm.
- Stage distributions of mEPCs at day 2 and 3. The histograms represent mean values from three independent experiments. At least 108 cells were scored in each experiment and condition. Error bars represent SD.
- Discrete deuterosomes in early stage‐II mEPCs were also procentriole‐free. mEPCs at day 2 were immunostained for Cep152, Sas6, and Centrin. The insets are arranged in the same sequence from left to right. Scale bar, 1 μm.
- Deuterosome‐size distributions. For each group, the diameters of at least 496 deuterosomes were measured from 32 mEPCs selected from three independent experiments according to the stages. The bottom and top of the box represent the 25th and 75th percentiles, respectively. The band is the median. The ends of the whiskers indicate the maximum and minimum of the data. Two‐tailed unpaired Student's t‐test: ***P < 0.001.
Quantifications indicated that 44.3 ± 3.5% (day 2) and 45.3 ± 3.9% (day 3) of the cells were morphologically in stage I (Fig 2D). In addition to those that would soon undergo deuterosome formation, this population also contained cells of other fates because 30–60% of mEPCs were multiciliated at day 5 or later 51, 52, 53. The mEPCs at early stages were more abundant at day 2 than day 3. For instance, stage‐II cells occupied 41.3 ± 2.7% at day 2 but 23.8 ± 5.0% at day 3 (Fig 2D). On the other hand, 17.4% of the mEPCs were in stages IV–VI at day 3, whereas at day 2 only 4.1% of the cells were in stages IV–V and no stage‐VI cells were observed (Fig 2D). These results further support the conclusion that mEPCs progress from stage I to VI during their differentiation into multiciliated cells.
For further clues on early phases of the deuterosome formation, we examined the stage‐II mEPCs in detail. We found that, similar to mTECs (Fig 1), stage‐II mEPCs could also be grouped into those in IIa, IIb, and IIc (Fig 2E). The average deuterosome diameters were 280 ± 60 nm (IIa) and 330 ± 60 nm (IIb/c), which were smaller than those in stage‐III mEPCs (390 ± 80 nm; Fig 2F). Furthermore, when both parental centrioles were detected in the mEPCs at stage IIb or IIc, they all bore procentrioles (Fig 2E; also see Fig 2A and B). This was also true for the cells in stage III or IV (Fig 2A and B). In the stage‐IIa mEPCs, however, parental centrioles that were with or without procentrioles were observed (Fig 2E: IIa1 and IIa2). Therefore, even in mEPCs discrete deuterosomes do not initially exist as “halos” as reported (Fig 1B) 43, which makes it difficult to attribute all the procentrioles in the halos to the daughter centriole.
Nascent deuterosomes emerge from a wide variety of locations in mEPCs
To clarify the origin of the deuterosomes, we performed live cell imaging by using GFP‐Deup1 to directly label deuterosomes 36. To mimic the transient expression pattern of endogenous Deup1 36, we cloned the mouse Deup1 promoter and used it to drive GFP‐Deup1 expression (Fig 3A). We confirmed that GFP‐Deup1 was specifically expressed together with endogenous Deup1 and Plk4 in differentiating mEPCs at day 3, but not in the progenitor cells (Fig 3B and C). Its levels were low as compared to endogenous Deup1 (Fig 3C) and would thus minimize possible side effects of overexpression. 3D‐SIM also confirmed its specific expression in the cells undergoing centriole amplification and proper localization at the center of deuterosomes (Fig 3D).
Figure 3. Deuterosomes emerge in a dispersed fashion in mEPCs.
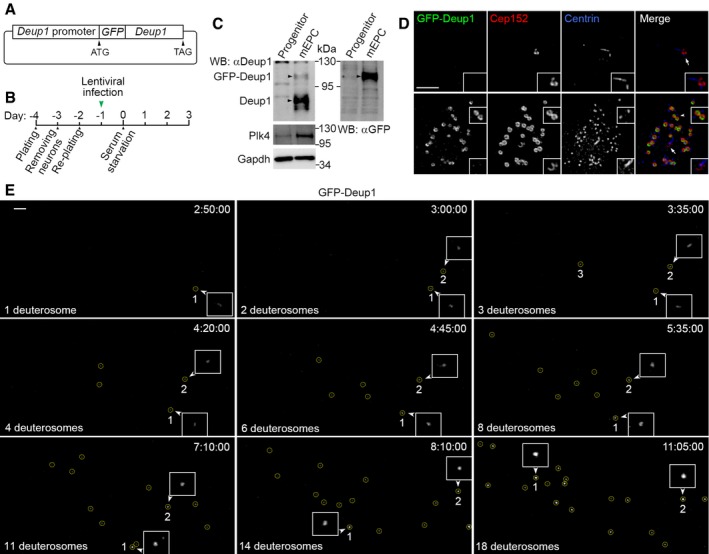
- Schematic illustration of the lentiviral construct used in the experiment. A 2‐kb mouse Deup1 genomic DNA fragment upstream of the first exon was used as the Deup1 promoter to drive the expression of GFP‐Deup1.
- Experimental scheme. Progenitors of mEPCs isolated from P0 mouse brain tissues were cultured for 6 days and infected with lentivirus described in (A) for 24 h, followed by serum starvation (day 0) to induce differentiation.
- Specific expression of GFP‐Deup1 in mEPCs. mEPCs treated as in (B) were collected at day 3 for immunoblotting to detect the indicated proteins (arrowheads). An aliquot of the infected progenitor cells was cultured to day 3 without serum starvation and used as control. Gapdh served as loading control.
- Specific expression and deuterosome localization of GFP‐Deup1 in mEPCs undergoing centriole amplification. mEPCs treated as in (B) were fixed at day 3; immunostained to visualize GFP, Cep152, and Centrin; and imaged with SIM. The insets are magnified images (1.5×) for deuterosomes (arrowhead) and parental centriole (arrow). Note that the top cell was not undergoing centriole amplification. Scale bar, 2 μm.
- Representative frames cropped from Movie EV2 to show the emergence of deuterosomes (encircled) in live imaging. Elapsed time is shown as h:min:s. The first two deuterosomes are marked, because they were traceable, and magnified 2× to show details. Scale bar, 2 μm.
We imaged the differentiating mEPCs for deuterosome formation events with spinning disk microscopy at 5‐min intervals for up to 12.5 h from day 2.5. We performed serial z‐stack sectioning at 0.5‐μm intervals to cover a depth of 20 μm and analyzed z‐projections of the images. Initially, we found that GFP‐positive puncta, presumably deuterosomes, moved rapidly and were difficult to trace over time (Movie EV1). Impairing microtubule‐dependent intracellular transport 56 by treating the cells with nocodazole (0.5 μg/ml), a microtubule‐destabilizing drug, markedly slowed down the deuterosome motilities. In the presence of nocodazole, we captured 15 cells that initiated their deuterosome biogenesis during the imaging (Fig 3E and Movie EV2) and 16 cells that already contained deuterosomes from the beginning and showed increased numbers of their deuterosomes over time (Movie EV3). Although clearly tracing every deuterosome was still difficult, the deuterosomes were found to initially emerge as tiny dim foci and enlarged dramatically over time with accordingly increased GFP‐Deup1 fluorescent intensity (Fig 3E, and [Link], [Link]). More importantly, nascent deuterosomes emerged at widely dispersed positions in these cells. Multiple deuterosomes were observed to appear from different locations within minutes or an hour (Fig 3E, and [Link], [Link]). These results are in sharp contrast to the observations by Al Jord and colleagues that deuterosomes require hours to form only at the daughter centriole and release from it 43. Therefore, they strongly suggest that deuterosomes self‐assemble efficiently.
Parental centrioles are dispensable for deuterosome formation
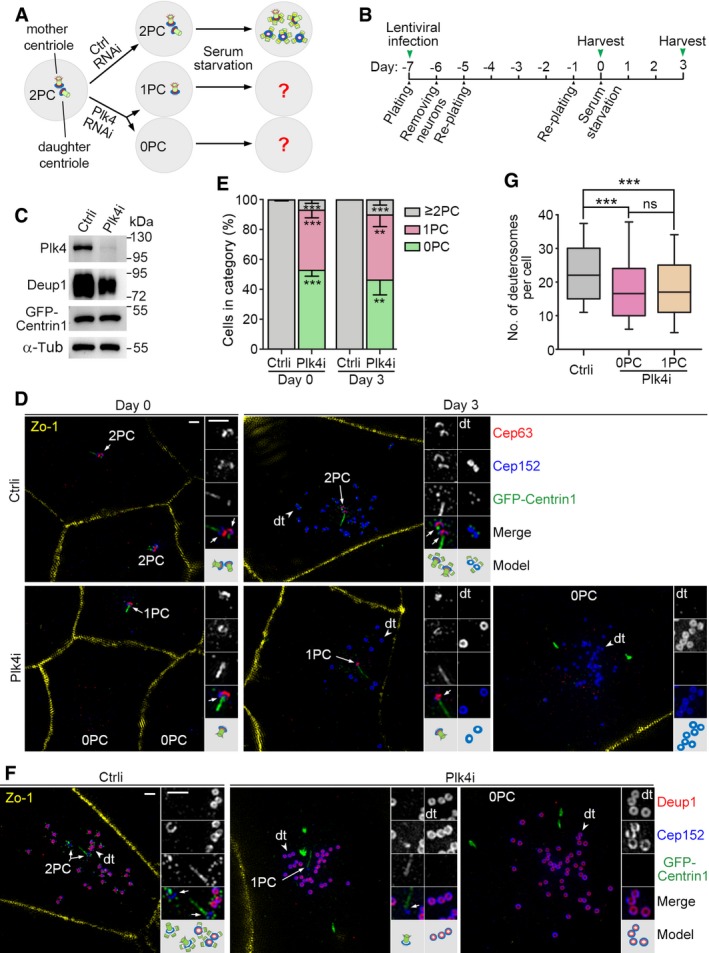
Next, we directly assessed the contribution of parental centrioles in deuterosome formation. We reasoned that, as Plk4 is essential for centriole biogenesis in both cycling and multiciliated cells 7, 8, 9, 36, its depletion in proliferating ependymal progenitors would result in the cells with either one parental centriole or no parental centriole (Fig 4A). Inducing their differentiation by serum starvation would allow us to examine how the deuterosome formation is affected by parental centrioles (Fig 4A).
Figure 4. Deuterosomes form efficiently in the absence of parental centrioles.
-
A, BExperimental design. Depletion of Plk4 will abolish centriole biogenesis and result in loss of one (1PC) or both (0PC) of the two parental centrioles (2PC) during the proliferation of ependymal progenitor cells, which can then be used to examine how parental centrioles contribute to the deuterosome formation (A). We transfected the progenitors prepared from P0 mouse brain tissues with lentiviral particles at day −7 to silence Plk4 expression and examined their progeny cells at day 0 and 3 (B).
-
CConfirmation of the Plk4 RNAi efficiency using mEPCs at day 3. The reduced expression of Deup1 in the Plk4‐depleted cells is attributed to reduced multiciliate cell differentiation.
-
DTypical cells immunostained for Zo‐1, Cep152, and Cep63. GFP‐Centrin1 expressed from the lentiviruses served as both infection and centriole markers. The green fluorescence was enhanced using anti‐GFP antibody and Alexa Fluor‐488‐conjugated secondary antibody. Parental centrioles (arrows) and representative deuterosomes (dt; arrowheads) are magnified twofold to show details. An illustration is provided for each set of the magnified images. Parental centrioles were identified based on the co‐staining patterns of Cep63, Cep152, and GFP‐Centrin1. The strong GFP‐Centrin1‐positive streaks or speckles in the Plk4‐depleted cells are not considered as centrioles because they did not co‐stain with other centriolar markers. Scale bar, 1 μm.
-
EParental centriole contents of the cells. The histograms represent mean values from three independent experiments. At least 186 cells at days 0 and 80 and deuterosome‐containing cells at day 3 were examined in each experiment and condition. Parental centrioles were identified based on the co‐staining patterns of at least two different centriolar markers. Error bars represent SD. Two‐tailed paired Student's t‐test, **P < 0.01; ***P < 0.001.
-
FConfirmation of deuterosome formation in Plk4i‐expressing mEPCs at day 3 by co‐staining for Deup1 and Cep152. Parental centrioles (arrows) and representative deuterosomes (dt; arrowheads) are magnified twofold to show details. Scale bar, 1 μm.
-
GBox plots for deuterosome numbers per cell. At least 156 deuterosome‐containing cells from three independent experiments were examined. Deuterosomes were scored as ring‐shaped structures decorated by both Deup1 and Cep152 (F) or by Cep152 but excluding parental centrioles (E). The bottom and top of the box represent the 25th and 75th percentiles, respectively. The band is the median. The ends of the whiskers indicate the 10th and 90th percentiles of the data. Two‐tailed unpaired Student's t‐test: ns, no significance; ***P < 0.001.
We have previously established a lentivirus that can stably co‐express an infection marker GFP‐Centrin1 with Plk4i, a short hairpin RNA (shRNA) against the murine Plk4 mRNA 36. We found that infecting ependymal progenitors with the virus for 7 days before serum starvation efficiently depleted Plk4 (Fig 4B and C). When Zo‐1, a tight junction protein 57, was used to mark cell boundaries, co‐staining Cep152 with a parental centriole‐specific marker, Cep63 5, 36, 58, indeed revealed that an average of 40.2% or 53.0% of the ependymal progenitors respectively lost one parental centriole (1PC) or both parental centrioles (0PC) immediately before serum starvation (day 0; Fig 4D and E). In comparison, 99.8% of the cells infected with a lentivirus co‐expressing a control shRNA (Ctrli) with GFP‐Centrin1 contained two or more parental centrioles (≥ 2PC; Fig 4E).
Notably, 33.6% of the Plk4‐depleted mEPCs at day 3 (n = 113 cells) were found to still contain deuterosomes (Fig 4D), which were further confirmed by co‐staining with Deup1 (Fig 4F). In comparison, deuterosome‐containing cells occupied 52.1% of the control virus‐infected mEPCs at day 3 (n = 71 cells). As expected, procentriole formation was abolished in the Plk4‐depleted mEPCs (n = 100 cells; Fig 4D and F). More importantly, 46.5% of the deuterosome‐producing cells had no parental centriole, whereas 43.5% of them contained a single parental centriole (Fig 4D–F). Only a small portion (10.0%) contained two or more parental centrioles (Fig 4E). Quantifications revealed that deuterosome numbers in the mEPCs with no or one parental centriole were very similar (Fig 4G). The average numbers were 19.3 ± 13.2 (n = 172 cells) and 19.2 ± 12.4 (n = 156 cells), respectively. Deuterosome numbers increased slightly in the control mEPCs (Fig 4G), with an average of 23.8 ± 11.5 (n = 316 cells). These results indicate that parental centriole is not important for deuterosome formation. The reduced percentage of the deuterosome‐containing Plk4‐depleted cells at day 3 correlated with the reduced protein levels of Deup1 (Fig 4C), suggesting a decreased differentiation potency of the progenitors, possibly due to Plk4 depletion‐induced self‐renewal defects 59.
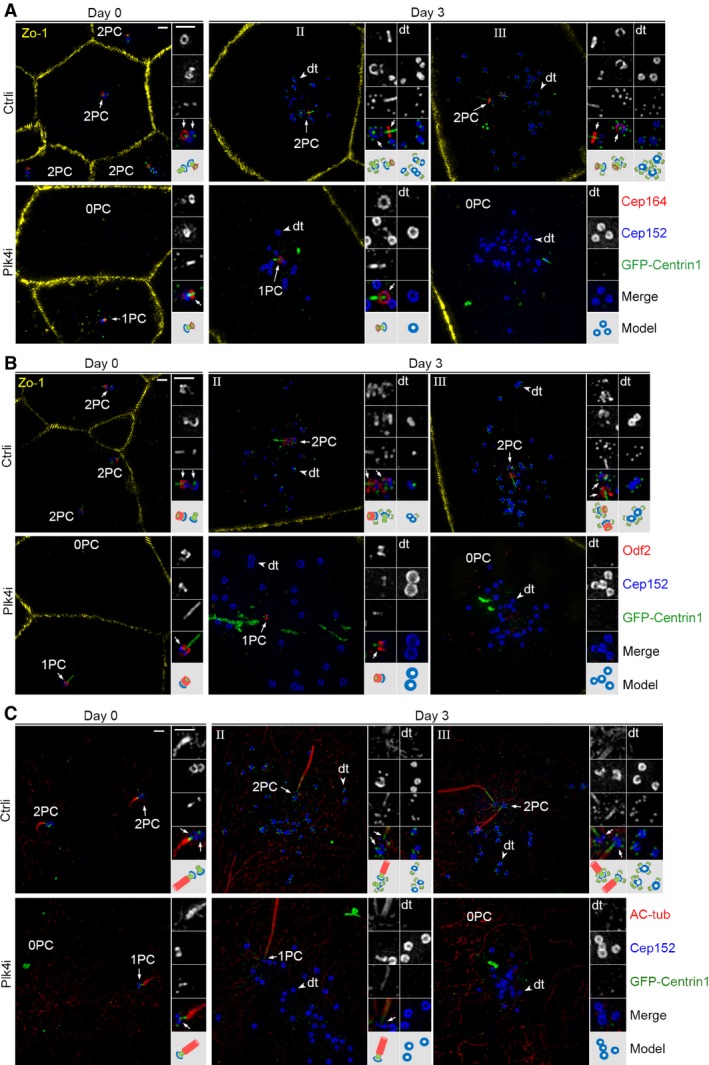
We also confirmed the parental centriole‐independent deuterosome formation by using Cep164 and Odf2, which localize to centriolar appendages of the mother centriole 32, 49, 50 or Centrobin, a daughter centriole‐specific protein 60, as markers, together with Cep152 and GFP‐Centrin1. In the control virus‐infected progenitors immediately before serum starvation (day 0), both parental centrioles were positive for Cep152 but only one of them (the mother centriole) was positive for Cep164 and Odf2 (Fig 5A and B). By contrast, in the Plk4‐depleted cells at day 0 or 3, when only one parental centriole was recognized, it was always double positive for Cep152‐Cep164 or Cep152‐Odf2, indicating that this centriole is the mother centriole (Fig 5A and B). Centrobin staining revealed that mother centriole accounted for approximately 92% of the Plk4‐depleted progenitors with one parental centriole at day 0 (Fig EV2A and B). At day 3, deuterosomes formed in the mEPCs with one or no recognizable parental centriole (Fig 5A and B).
Figure 5. Status of parental centrioles and deuterosome formation in the Plk4‐depleted mEPCs.
-
A, BThe remaining single parental centriole (1PC) was usually positive for centriolar appendage proteins Cep164 and Odf2. Cultured ependymal progenitors treated as described in Fig 4B were immunostained to visualize Zo‐1, Cep152, and Cep164 or Odf2. GFP‐Centrin1, whose green fluorescence was enhanced using anti‐GFP antibody and Alexa Fluor‐488‐conjugated secondary antibody, served as both infection and centriole markers. Parental centrioles were identified based on the co‐staining patterns of Cep152, GFP‐Centrin1, and Cep164 (A) or Odf2 (B). Representative parental centrioles (arrows) and deuterosomes (dt; arrowheads) were magnified twofold to show details. An illustration is provided for each set of the magnified images. Scale bar, 1 μm.
-
CCilium formation in progenitors (day 0) and mEPCs (day 3). Ependymal progenitors treated as described in Fig 4B were immunostained to visualize Cep152, GFP, and acetylated tubulin (AC‐tub; cilia marker). Note that the bottom region of the cilium is Centrin‐positive. Scale bar, 1 μm.
Figure EV2. The daughter centriole accounts for a small portion of the single parental centriole in Plk4‐depleted ependymal progenitors.
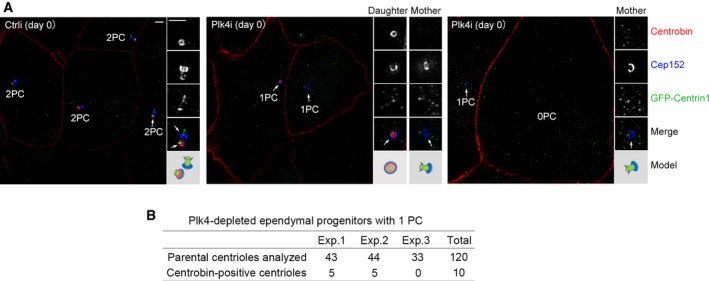
- Identification of the daughter centriole through Centrobin. Ependymal progenitors treated as in Fig 4B were fixed at day 0 and immunostained for Centrobin and Cep152. GFP‐Centrin1 served as both infection and centriole markers. Zo‐1 was visualized in the same channel with Centrobin to label the cell boundaries because their antibodies were from mouse and their staining patterns did not overlap. Parental centrioles are denoted by arrows. An illustration is provided for each set of the magnified (2×) images. Scale bar, 1 μm.
- Quantification for Centrobin‐positive parental centrioles in three independent experiments.
Immunostaining using the ciliary marker acetylated tubulin 61 confirmed that a cilium (1.2 ± 0.5 μm in length; n = 424) already existed in the progenitors at day 0 (Figs 5C, and EV3A and B) as reported 51. The cilium became elongated at day 3 (3.9 ± 1.4 μm in length; n = 436; Figs 5C, and EV3A and B). Interestingly, some of control virus‐infected mEPCs at day 3 contained two cilia (Fig 5C). Consistently, in some of the stage‐III control mEPCs, both parental centrioles were positive for Cep164 and Odf2 (Fig 5A and B), suggesting maturation of the daughter centriole into a basal body. This is interesting because in cycling cells the daughter centriole needs to go through mitosis to mature into the mother centriole, which possesses appendages and can serve as a basal body 2, 15. Possibly, this unexpected centriole maturation is rendered by the recently reported mitosis‐like program in the differentiating mEPCs 39. In addition, the bottom region of these cilia was usually Centrin‐positive (Fig 5C) 43. This explains the frequent observation of a Centrin‐positive stick over the appendages of the mother centriole (Figs 2, 4, and 5), though its physiological significance remains to be clarified.
Figure EV3. Ciliary length increases dramatically during the differentiation of ependymal progenitors.
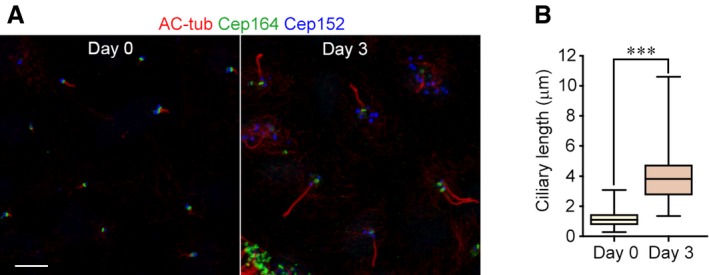
- Cilia in the progenitors (day 0) and mEPCs (day 3). Ependymal progenitors treated as in Fig 4B were fixed and immunostained to visualize acetylated tubulin (AC‐tub; a cilia marker), Cep164 (a marker for mother centriole or basal body), and Cep152 (a marker for parental centriole and deuterosome). Scale bar, 5 μm.
- Quantification for the ciliary length. The results were from three independent experiments. At least 105 cilia were measured in each experiment and condition. The bottom and top of the box represent the 25th and 75th percentiles, respectively. The band is the median. The ends of the whiskers indicate the maximum and minimum of the data. Two‐tailed unpaired Student's t‐test: ***P < 0.001.
To further corroborate the parental centriole‐independent deuterosome assembly, we treated ependymal progenitors with centrinone, a chemical inhibitor of Plk4 62 (Fig EV4A). We tested different concentrations and found that 1.5 μM of centrinone provided the best centriole depletion efficiency with low mortality in ependymal progenitors (Fig EV4B). At 3.0 μM concentration, cell death was prominent. We thus performed the subsequent experiments using 1.5 μM concentration. When the DMSO‐treated cells were immunostained with Cep164, Cep152, and Centrin, they did not display centriole loss at both day 0 and 3 (Fig EV4C and D). By contrast, 51.6% of the centrinone‐treated progenitors at day 0 contained no or one parental centriole (0PC + 1PC; n = 488 cells). At day 3, 65.7% of the deuterosome‐containing cells treated with centrinone (n = 134 cells) contained no or one parental centriole (Fig EV4C and D). Quantification indicated that the deuterosome numbers in the cells with no parental centriole (34.2 ± 16.2; n = 66 cells) or one parental centriole (35 ± 12.9; n = 31 cells) were similar but increased as compared to the DMSO‐treated cells (20.7 ± 11.1; n = 102 cells; Fig EV4E). The increase could be attributed to the limited sample size or other unknown effects of the drug. In addition, procentrioles were observed on deuterosomes in most centrinone‐treated cells at day 3 (97%; n = 97 cells; Fig EV4D), possibly due to the failure of the centrinone to inhibit the markedly elevated levels of Plk4 in these cells (Fig 3C) 36. Taken together, we demonstrate that parental centrioles are not important for deuterosome formation.
Figure EV4. Depletion of parental centrioles with centrinone does not affect deuterosome formation.
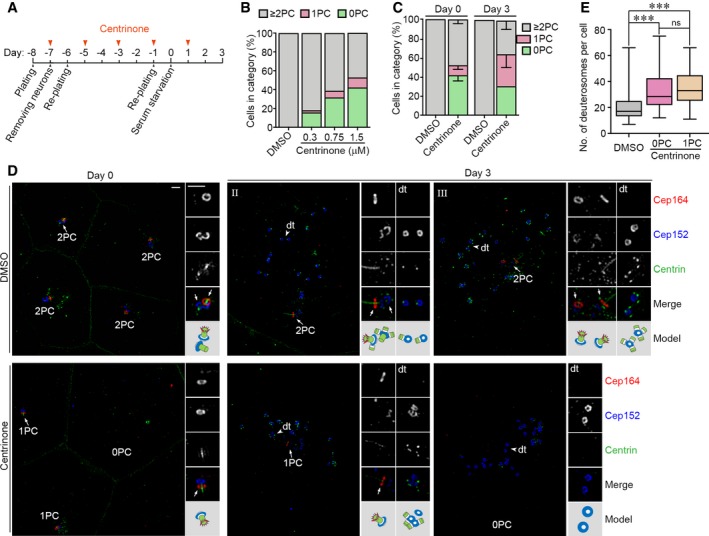
- Experimental design.
- Effects of different centrinone concentrations on parental centrioles. Mouse ependymal progenitors treated as in (A) with 0.3–3.0 μM centrinone were fixed at day 0 and immunostained as in (D). Parental centrioles were identified based on the co‐staining patterns of Cep152, Centrin, and Cep164. The histograms represent mean values from one experiment, and at least 89 cells were scored in each condition. The cells treated with 3.0 μM of centrinone died massively and were not analyzed.
- Parental centriole contents of the cells treated with 1.5 μM centrinone, based on the co‐staining patterns of Cep152, Centrin, and Cep164 (D). The histograms represent mean values from three independent experiments, and at least 482 cells at days 0 and 100 deuterosome‐containing cells at day 3 were scored in each experiment and condition. Error bars represent SD.
- Typical cells treated with DMSO or centrinone, immunostained for Cep152, Cep164, Centrin, and Zo‐1. Zo‐1 and Centrin were immunostained in the same channel because their antibodies were both from mouse and their staining patterns did not overlap. Magnified images (2×) show details of representative parental centrioles (arrows) and deuterosomes (dt; arrowheads). An illustration is provided for each set of the magnified images. Scale bar, 1 μm.
- Box plots for deuterosome numbers per cell at day 3. Deuterosomes were scored as ring‐shaped structures decorated by Cep152 but excluding parental centrioles (D). At least 100 deuterosome‐containing cells from three independent experiments were scored. The bottom and top of the box represent the 25th and 75th percentiles, respectively. The band is the median. The ends of the whiskers indicate the maximum and minimum of the data. Two‐tailed unpaired Student's t‐test: ns, no significance; ***P < 0.001.
During the revision of the manuscript, two preprints posted online in bioRxiv also report similar parental centriole‐independent deuterosome formation in centrinone‐treated mTECs and mEPCs, respectively [preprint: 63, preprint: 64]. One of the preprints [preprint: 64] is posted by the same group that has previously proposed the daughter centriole‐dependent model of deuterosome formation 43. It is known that in cycling cells, centrioles can be assembled de novo in the absence of parental centrioles 65, 66, 67, 68. The presence of parental centriole inhibits the activation of the de novo pathway to avoid deregulated centriole formation 65. Apparently, deuterosomes are able to efficiently form and fully function in the absence of parental centrioles. Moreover, our live imaging results (Fig 3; [Link], [Link]) indicate that their spontaneous assembly is not repressed by parental centrioles.
The efficient, spontaneous formation of deuterosomes (Figs 3, 4, 5 and EV4, and [Link], [Link]) [preprint: 63, preprint: 64] accordingly suggests that the daughter centriole does not possess a unique mechanism to induce deuterosome assembly as proposed 43, 44. Deup1 is sometimes observed to co‐localize with Cep152 at both the mother and the daughter centrioles (Fig 1C) 27, 36. The parental centriole‐localized Deup1 can even replace Cep63 to sustain the centriole‐dependent procentriole assembly 36. Deuterosomes may thus form from the centriolar Deup1, Cep152, Plk4, and other unknown proteins, if any, through mechanisms identical or similar to those that govern their autonomous assembly. Such deuterosomes keep associated with parental centrioles until they are released as halos 43. Consistently, Mercey and colleagues also provide evidence in their preprint to suggest that the mother centriole is capable of deuterosome formation as well [preprint: 64].
The results to date also strengthen the idea that the deuterosome indeed functions as a platform for massive de novo procentriole biogenesis (Fig 1A) 27, 36. In centrinone‐treated, parental centriole‐ablated mEPCs, procentriole‐associated deuterosomes were still observed (Fig EV4) [preprint: 64]. When centrinone was removed at day 0, the deuterosomes exhibit similar ability to associate with procentrioles as those in mEPCs with intact parental centrioles [preprint: 64]. Nor are the total numbers of centrioles produced in the parental centriole‐ablated mEPCs reduced as compared to the control cells [preprint: 64]. Similar situations occur in mTECs [preprint: 63]. Therefore, deuterosomes are still fully functional in procentriole biogenesis in the absence of parental centrioles.
Taken together, we propose that in differentiating multiciliated cells, deuterosomes form mainly in the cytosol and less frequently at the parental centrioles. Both the deuterosomes and the parental centrioles induce procentriole biogenesis to maximize the efficiency of the centriole amplification. Although multiple studies suggest that the centriole amplification processes are similar in multiciliated tissues and in vitro cultured multiciliated cells 43, 45, 46, 69, future studies are still required to clarify whether deuterosomes are generated in the same way in vivo.
Materials and Methods
Cell culture, lentivirus production, and infection
mTECs were isolated from 4‐week C57BL/6J mice and cultured as described previously 36. mEPCs were cultured as described 51, 52 with some modifications. Briefly, the telencephalon was dissected from P0 mice, followed by careful removal of the meninx, choroid plexus, hippocampus, and olfactory bulb in dissection buffer (161 mM NaCl, 5 mM KCl, 1 mM MgSO4, 3.7 mM CaCl2, 5 mM HEPES, and 5.5 mM glucose, pH 7.4) on ice. The remaining tissues were cut into small pieces and digested in freshly prepared digestion buffer (10 U/ml papain, 0.2 mg/ml l‐cysteine, 0.5 mM EDTA, 1 mM CaCl2, 1.5 mM NaOH, and 5 U/ml DNase I in the dissection buffer) for 30 min at 37°C. The digestion was then stopped by adding 10% FBS. After gentle pipetting with a P1000 tip, the cells were collected by centrifugation at 400 × g for 5 min at room temperature. The pelleted cells were re‐suspended in the culture medium and seeded in a laminin‐coated flask. Neurons were shaken off after a 1‐day culture, and the remaining cells were further cultured to reach confluency. The cells were then transferred into the wells of laminin‐coated 29‐mm glass‐bottomed dishes (Cellvis, D29‐14‐1.5‐N) at a density of 2 × 105 cells per well and were maintained in serum‐free medium to induce multiciliate mEPCs.
Lentiviral productions for the RNAi experiments were performed as described previously 36. Eighteen 10‐cm dishes of HEK293T cells transfected for 48 h were used to produce the lentiviral particles, which were further concentrated to 1 ml. Ependymal progenitor‐enriched brain cells isolated from three P0 mice were re‐suspended into 10 ml of the culture medium 51, 52 containing 60 μl of the concentrated lentiviral particles and seeded into a 75‐cm2 flask (day −7). To suppress the p53‐dependent apoptosis associated with centriole loss 70, 10 μM of the p53 inhibitor, pifithrin‐α (S2929, Selleckchem), was always included in the culture medium to sustain cell viability 71. After 24 h of culture, neurons were shaken off and fresh culture medium was added (day −6). After additional 6 days (day 0), the cells were serum‐starved to induce differentiation and assayed at day 3.
To deplete parental centrioles using centrinone (a gift from Dr. Karen Oegema, UCSD), the drug dissolved in DMSO was added to ependymal progenitors at day −7, together with the p53 inhibitor pifithrin‐α (10 μM). The culture medium was changed every 2 days, with supplemented centrinone and pifithrin‐α. We initially tested different concentrations (0.3, 0.75, 1.5, and 3 μM) of centrinone and finally used 1.5 μM as the optimal concentration. The cells were assayed at day 0 or day 3.
Experiments involving mouse tissues were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Institute of Biochemistry and Cell Biology.
Plasmid constructs
The cDNAs of full‐length mouse Deup1 (NM_181816) and Ccdc78 (NM_001165929) were PCR‐amplified and subcloned into the lentiviral expression vector, pLV‐GFP, to generate pLV‐GFP‐Deup1 and pLV‐GFP‐Ccdc78. To express GFP‐Deup1 under the control of its own promoter, a 2‐kb genomic DNA sequence of mouse Deup1 upstream of the first exon was PCR‐amplified and used to replace the CMV promoter of pLV‐GFP‐Deup1.
Antibodies
Secondary antibodies used for immunofluorescence (IF) were as follows: donkey anti‐rabbit conjugated with Cy3 or DyLight 405, anti‐chicken conjugated with Alexa Fluor‐488 or DyLight‐405, anti‐mouse conjugated with Cy3 or DyLight‐405, anti‐rat conjugated with Alexa Fluor‐488, anti‐guinea pig conjugated with Cy3 (Jackson ImmunoResearch), and anti‐mouse conjugated with Alexa Fluor‐647 (Thermo Fisher Scientific). The DyLight 405‐conjugated antibodies were used at 1:200, and the remaining antibodies were used at 1:1,000. Secondary antibodies used for Western blotting (WB) were HRP‐conjugated goat anti‐mouse and anti‐rabbit antibodies (Thermo Fisher; 1:5,000).
Commercial primary antibodies used were as follows: mouse anti‐Sas6 (sc‐81431, Santa Cruz; IF 1:50), mouse anti‐Centrin (04‐1624(20H5), Millipore; IF 1:200), mouse anti‐α‐Tubulin (T5168, Sigma‐Aldrich; WB 1:5,000), mouse anti‐acetylated tubulin (T7451, Sigma‐Aldrich; IF 1:1,000), mouse anti‐Centrobin (ab70448, Abcam; IF 1:200), mouse anti‐Zo‐1 (33‐9100, Thermo Fisher; IF 1:1,000), rabbit anti‐Centrin1 (12794‐1‐AP, Proteintech; IF 1:200), rat anti‐GFP (sc‐101536, Santa Cruz; IF 1:50), rabbit anti‐GFP (A‐11122, Thermo Fisher; WB 1:3,000), and rabbit anti‐Cep63 (06‐1292, Millipore; IF 1:200).
Rabbit anti‐Deup1 (IF 1:200; WB 1:4,000), chicken anti‐Cep152 (IF 1:300), and rabbit anti‐Plk4 (IF 1:200; WB 1:2,000) antibodies were homemade 36. To generate antibodies against murine Cep164, Odf2, or Ccdc78, cDNA fragments of mouse Cep164 (NM_001081373; encoding 1–400 aa), Odf2 (NM_001177659; encoding 401–826 aa), and Ccdc78 (NM_001165929; encoding 1–437 aa) were PCR‐amplified from the total cDNAs of mTECs. The cDNA fragments of Cep164 and Odf2 were subcloned into pET32a to express His‐tagged fusion proteins. The cDNA of Ccdc78 was subcloned into pGEX‐4T‐1 to express GST‐fusion protein. The proteins were purified by using Ni‐NTA beads (Qiagen) or glutathione‐agarose beads (Sigma) and used as antigens. Rabbit anti‐Cep164 (IF 1:200; WB 1:1,000), guinea pig anti‐Odf2 (IF 1:200; WB 1:2,000), and rabbit anti‐Ccdc78 (IF 1:200; WB 1:2,000) antibodies were generated through contracted services (ABclonal) and affinity‐purified.
Immunofluorescent microscopy
Immunostaining and immunofluorescent microscopy were performed as described 36. Briefly, mEPCs grown on glass‐bottomed dishes and mTECs on Transwells were pre‐extracted with 0.5% Triton X‐100 in PBS for 40 s or for 3 min, respectively, followed by fixation with 4% fresh paraformaldehyde in PBS for 15 min at room temperature. After fixation, the cells were permeabilized with 0.5% Triton X‐100 in PBS for 15 min and blocked with blocking buffer (4% BSA in TBST) for 1 h at room temperature. Primary and secondary antibodies were diluted into the blocking buffer and applied to cells at room temperature for 2 and 1 h, respectively, interspaced with three rounds of washing. The samples were imaged with a structured illumination microscope (GE OMX V3) with a 100×/1.40 NA oil‐immersion objective lens (Olympus). Serial z‐stack sectioning was performed at 125‐nm intervals. Raw images were processed for maximum intensity projection with SoftWoRx software.
Confocal images were captured using Leica TCS SP8 system with a 63×/1.40 oil‐immersion objective lens. Serial z‐stack sectioning was set at 125‐nm intervals. Images were processed with maximum intensity projections.
Live cell imaging
Mouse ependymal progenitors were infected with lentivirus at day −1 for the expression of GFP‐Deup1 under Deup1 promoter. Live cell imaging was performed at day 2.5. The images in Fig 3E, and [Link], [Link] were captured with an Olympus SpinSR10 spinning disk confocal super‐resolution microscope equipped with an APON 60 × OTIRF/1.49 NA oil objective (Olympus) and ORCA‐Flash 4.0 V3 Digital CMOS Camera (Hamamatsu). The laser power (488 nm) was set to 10% to reduce cell toxicity. The images in Movie EV1 were recorded using an Andor Dragonfly high‐speed confocal microscope equipped with a Plan Apo λ 60×/1.40 NA oil objective (Nikon) and a Zyla sCMOS camera (Andor). The laser power (488 nm) was set to 3%. The exposure time was 100 ms. The images were recorded at 5‐min intervals for 12.5 h. z‐stack sectioning was performed at 0.5‐μm intervals to cover a depth of 20 μm. The images and movies were processed with Imaris (Bitplane) and ImageJ (Fiji) softwares.
Quantification and statistical analysis
Deuterosome diameters were measured from 3D‐SIM images of Deup1 using the “automatic bright objects” mode of the “count/size” function of Image‐Pro Plus 6.0 software (Media Cybernetics). Cells in different categories were scored manually using available original 3D‐SIM images (1,024 × 1,024 pixels), each of which contained multiple cells, from three independent experiments. Parental centrioles were identified and scored based on at least two different centriolar markers. Ciliary length of ependymal progenitors at day 0 and mEPCs at day 3 was measured using the “measurements” module of Image‐Pro Plus 6.0. Quantification results are presented as mean ± SD unless otherwise stated. Differences are considered significant when P was < 0.05 in a two‐tailed unpaired Student's t‐test using GraphPad Prism software (GraphPad Software).
Author contributions
XZ and XY conceived and directed the project; HZ and QC performed major experiments; CF did the quantification of the ciliary length; QH generated the homemade antibodies; JZ provided the 3D‐SIM imaging system; XZ, XY, and HZ designed experiments, interpreted data, and wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Review Process File
Acknowledgements
The authors thank Dr Nathalie Spassky (CNRS, France) for mEPC culture protocol, Dr. Karen Oegema (University of California, San Diego) for providing centrinone, the Centre for Biological Imaging (Institute of Biophysics, CAS) for support on 3D‐SIM imaging, Dr. Wenjuan Cai and Kefeng Wang (Olympus Corporation) for support on spinning disk imaging, and Yanli Zhao (Bitplane) for support on Imaris. This work was supported by National Natural Science Foundation of China (31330045 to X.Z. and 31501092 to H.Z.), National Key R&D Program of China (2017YFA0503500), and Chinese Academy of Sciences (XDB19000000).
EMBO Reports (2019) 20: e46735
Contributor Information
Xiumin Yan, Email: yanx@sibcb.ac.cn.
Xueliang Zhu, Email: xlzhu@sibcb.ac.cn.
References
- 1. Banterle N, Gonczy P (2017) Centriole biogenesis: from identifying the characters to understanding the plot. Annu Rev Cell Dev Biol 33: 23–49 [DOI] [PubMed] [Google Scholar]
- 2. Nigg EA, Holland AJ (2018) Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat Rev Mol Cell Biol 19: 297–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nigg EA, Stearns T (2011) The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 13: 1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ito D, Bettencourt‐Dias M (2018) Centrosome remodelling in evolution. Cells 7: 71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown NJ, Marjanovic M, Luders J, Stracker TH, Costanzo V (2013) Cep63 and cep152 cooperate to ensure centriole duplication. PLoS One 8: e69986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sir JH, Barr AR, Nicholas AK, Carvalho OP, Khurshid M, Sossick A, Reichelt S, D'Santos C, Woods CG, Gergely F (2011) A primary microcephaly protein complex forms a ring around parental centrioles. Nat Genet 43: 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kleylein‐Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA (2007) Plk4‐induced centriole biogenesis in human cells. Dev Cell 13: 190–202 [DOI] [PubMed] [Google Scholar]
- 8. Bettencourt‐Dias M, Rodrigues‐Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM (2005) SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol 15: 2199–2207 [DOI] [PubMed] [Google Scholar]
- 9. Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA (2005) The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 7: 1140–1146 [DOI] [PubMed] [Google Scholar]
- 10. Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T (2010) Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol 191: 721–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arquint C, Sonnen KF, Stierhof YD, Nigg EA (2012) Cell‐cycle‐regulated expression of STIL controls centriole number in human cells. J Cell Sci 125: 1342–1352 [DOI] [PubMed] [Google Scholar]
- 12. Arquint C, Gabryjonczyk AM, Imseng S, Bohm R, Sauer E, Hiller S, Nigg EA, Maier T (2015) STIL binding to Polo‐box 3 of PLK4 regulates centriole duplication. Elife 4: e07888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hilbert M, Noga A, Frey D, Hamel V, Guichard P, Kraatz SH, Pfreundschuh M, Hosner S, Fluckiger I, Jaussi R et al (2016) SAS‐6 engineering reveals interdependence between cartwheel and microtubules in determining centriole architecture. Nat Cell Biol 18: 393–403 [DOI] [PubMed] [Google Scholar]
- 14. Lane HA, Nigg EA (1996) Antibody microinjection reveals an essential role for human polo‐like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol 135: 1701–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loncarek J, Bettencourt‐Dias M (2018) Building the right centriole for each cell type. J Cell Biol 217: 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA (2012) 3D‐structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open 1: 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moyer TC, Clutario KM, Lambrus BG, Daggubati V, Holland AJ (2015) Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J Cell Biol 209: 863–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ohta M, Ashikawa T, Nozaki Y, Kozuka‐Hata H, Goto H, Inagaki M, Oyama M, Kitagawa D (2014) Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat Commun 5: 5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pelletier L, O'Toole E, Schwager A, Hyman AA, Muller‐Reichert T (2006) Centriole assembly in Caenorhabditis elegans . Nature 444: 619–623 [DOI] [PubMed] [Google Scholar]
- 20. Kong D, Farmer V, Shukla A, James J, Gruskin R, Kiriyama S, Loncarek J (2014) Centriole maturation requires regulated Plk1 activity during two consecutive cell cycles. J Cell Biol 206: 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha‐Ferreira I, Riparbelli M, Rodrigues‐Martins A, Bettencourt‐Dias M, Callaini G, Glover DM (2010) Asterless is a scaffold for the onset of centriole assembly. Nature 467: 714–718 [DOI] [PubMed] [Google Scholar]
- 22. Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P (2007) Regulated HsSAS‐6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell 13: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vulprecht J, David A, Tibelius A, Castiel A, Konotop G, Liu F, Bestvater F, Raab MS, Zentgraf H, Izraeli S et al (2012) STIL is required for centriole duplication in human cells. J Cell Sci 125: 1353–1362 [DOI] [PubMed] [Google Scholar]
- 24. Brooks ER, Wallingford JB (2014) Multiciliated cells. Curr Biol 24: R973–R982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ishikawa H, Marshall WF (2011) Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol 12: 222–234 [DOI] [PubMed] [Google Scholar]
- 26. Spassky N, Meunier A (2017) The development and functions of multiciliated epithelia. Nat Rev Mol Cell Biol 18: 423–436 [DOI] [PubMed] [Google Scholar]
- 27. Yan X, Zhao H, Zhu X (2016) Production of Basal Bodies in bulk for dense multicilia formation. F1000Res 5: 1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stubbs JL, Vladar EK, Axelrod JD, Kintner C (2012) Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol 14: 140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma L, Quigley I, Omran H, Kintner C (2014) Multicilin drives centriole biogenesis via E2f proteins. Genes Dev 28: 1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoh RA, Stowe TR, Turk E, Stearns T (2012) Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS One 7: e52166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim S, Ma L, Shokhirev MN, Quigley I, Kintner C (2018) Multicilin and activated E2f4 induce multiciliated cell differentiation in primary fibroblasts. Sci Rep 8: 12369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mori M, Hazan R, Danielian PS, Mahoney JE, Li H, Lu J, Miller ES, Zhu X, Lees JA, Cardoso WV (2017) Cytoplasmic E2f4 forms organizing centres for initiation of centriole amplification during multiciliogenesis. Nat Commun 8: 15857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dirksen ER (1971) Centriole morphogenesis in developing ciliated epithelium of the mouse oviduct. J Cell Biol 51: 286–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sorokin SP (1968) Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci 3: 207–230 [DOI] [PubMed] [Google Scholar]
- 35. Anderson RG, Brenner RM (1971) The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. J Cell Biol 50: 10–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao H, Zhu L, Zhu Y, Cao J, Li S, Huang Q, Xu T, Huang X, Yan X, Zhu X (2013) The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nat Cell Biol 15: 1434–1444 [DOI] [PubMed] [Google Scholar]
- 37. Tang TK (2013) Centriole biogenesis in multiciliated cells. Nat Cell Biol 15: 1400–1402 [DOI] [PubMed] [Google Scholar]
- 38. Klos Dehring DA, Vladar EK, Werner ME, Mitchell JW, Hwang P, Mitchell BJ (2013) Deuterosome‐mediated centriole biogenesis. Dev Cell 27: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Al Jord A, Shihavuddin A, Servignat d'Aout R, Faucourt M, Genovesio A, Karaiskou A, Sobczak‐Thepot J, Spassky N, Meunier A (2017) Calibrated mitotic oscillator drives motile ciliogenesis. Science 358: 803–806 [DOI] [PubMed] [Google Scholar]
- 40. Revinski DR, Zaragosi LE, Boutin C, Ruiz‐Garcia S, Deprez M, Thome V, Rosnet O, Gay AS, Mercey O, Paquet A et al (2018) CDC20B is required for deuterosome‐mediated centriole production in multiciliated cells. Nat Commun 9: 4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Beisson J, Wright M (2003) Basal body/centriole assembly and continuity. Curr Opin Cell Biol 15: 96–104 [DOI] [PubMed] [Google Scholar]
- 42. Kalnins VI, Porter KR (1969) Centriole replication during ciliogenesis in the chick tracheal epithelium. Z Zellforsch Mikrosk Anat 100: 1–30 [DOI] [PubMed] [Google Scholar]
- 43. Al Jord A, Lemaitre AI, Delgehyr N, Faucourt M, Spassky N, Meunier A (2014) Centriole amplification by mother and daughter centrioles differs in multiciliated cells. Nature 516: 104–107 [DOI] [PubMed] [Google Scholar]
- 44. Meunier A, Spassky N (2016) Centriole continuity: out with the new, in with the old. Curr Opin Cell Biol 38: 60–67 [DOI] [PubMed] [Google Scholar]
- 45. You Y, Richer EJ, Huang T, Brody SL (2002) Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol 283: L1315–L1321 [DOI] [PubMed] [Google Scholar]
- 46. Vladar EK, Stearns T (2007) Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol 178: 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P (2005) SAS‐6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol 7: 115–125 [DOI] [PubMed] [Google Scholar]
- 48. Nakazawa Y, Hiraki M, Kamiya R, Hirono M (2007) SAS‐6 is a cartwheel protein that establishes the 9‐fold symmetry of the centriole. Curr Biol 17: 2169–2174 [DOI] [PubMed] [Google Scholar]
- 49. Yang TT, Chong WM, Wang WJ, Mazo G, Tanos B, Chen Z, Tran TMN, Chen YD, Weng RR, Huang CE et al (2018) Super‐resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat Commun 9: 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA (2007) Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 179: 321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Delgehyr N, Meunier A, Faucourt M, Bosch Grau M, Strehl L, Janke C, Spassky N (2015) Ependymal cell differentiation, from monociliated to multiciliated cells. Methods Cell Biol 127: 19–35 [DOI] [PubMed] [Google Scholar]
- 52. Spassky N, Merkle FT, Flames N, Tramontin AD, Garcia‐Verdugo JM, Alvarez‐Buylla A (2005) Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J Neurosci 25: 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zheng J, Liu H, Zhu L, Chen Y, Zhao H, Zhang W, Li F, Xie L, Yan X, Zhu X (2019) Microtubule‐bundling protein Spef1 enables mammalian ciliary central apparatus formation. J Mol Cell Biol 11: 67–77 [DOI] [PubMed] [Google Scholar]
- 54. Dammermann A, Merdes A (2002) Assembly of centrosomal proteins and microtubule organization depends on PCM‐1. J Cell Biol 159: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hori A, Peddie CJ, Collinson LM, Toda T (2015) Centriolar satellite‐ and hMsd1/SSX2IP‐dependent microtubule anchoring is critical for centriole assembly. Mol Biol Cell 26: 2005–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barlan K, Gelfand VI (2017) Microtubule‐based transport and the distribution, tethering, and organization of organelles. Cold Spring Harb Perspect Biol 9: a025817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA (1986) Identification of ZO‐1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol 103: 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lukinavicius G, Lavogina D, Orpinell M, Umezawa K, Reymond L, Garin N, Gonczy P, Johnsson K (2013) Selective chemical crosslinking reveals a Cep57‐Cep63‐Cep152 centrosomal complex. Curr Biol 23: 265–270 [DOI] [PubMed] [Google Scholar]
- 59. Martin CA, Ahmad I, Klingseisen A, Hussain MS, Bicknell LS, Leitch A, Nurnberg G, Toliat MR, Murray JE, Hunt D et al (2014) Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat Genet 46: 1283–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zou C, Li J, Bai Y, Gunning WT, Wazer DE, Band V, Gao Q (2005) Centrobin: a novel daughter centriole‐associated protein that is required for centriole duplication. J Cell Biol 171: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Piperno G, Fuller MT (1985) Monoclonal antibodies specific for an acetylated form of alpha‐tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol 101: 2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, Seo CP, Hsia JE, Kim SK, Mitchell JW et al (2015) Cell biology. Reversible centriole depletion with an inhibitor of Polo‐like kinase 4. Science 348: 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nanjundappa R, Kong D, Shim K, Stearns T, Brody S, Loncarek J, Mahjoub M (2018) Regulation of cilia abundance in multiciliated cells. bioRxiv 10.1101/478297 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mercey O, Al Jord A, Rostaing P, Mahuzier A, Fortoul A, Boudjema A, Faucourt M, Spassky N, Meunier A (2018) Dynamics of centriole amplification in centrosome‐depleted brain multiciliated progenitors. bioRxiv 10.1101/503730 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. La Terra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A (2005) The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J Cell Biol 168: 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL (2002) De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol 158: 1171–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Szollosi D, Ozil JP (1991) De novo formation of centrioles in parthenogenetically activated, diploidized rabbit embryos. Biol Cell 72: 61–66 [DOI] [PubMed] [Google Scholar]
- 68. Calarcogillam PD, Siebert MC, Hubble R, Mitchison T, Kirschner M (1983) Centrosome development in early mouse embryos as defined by an autoantibody against pericentriolar material. Cell 35: 621–629 [DOI] [PubMed] [Google Scholar]
- 69. Guirao B, Meunier A, Mortaud S, Aguilar A, Corsi JM, Strehl L, Hirota Y, Desoeuvre A, Boutin C, Han YG et al (2010) Coupling between hydrodynamic forces and planar cell polarity orients mammalian motile cilia. Nat Cell Biol 12: 341–350 [DOI] [PubMed] [Google Scholar]
- 70. Bazzi H, Anderson KV (2014) Acentriolar mitosis activates a p53‐dependent apoptosis pathway in the mouse embryo. Proc Natl Acad Sci USA 111: E1491–E1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Djuzenova CS, Fiedler V, Memmel S, Katzer A, Hartmann S, Krohne G, Zimmermann H, Scholz CJ, Polat B, Flentje M et al (2015) Actin cytoskeleton organization, cell surface modification and invasion rate of 5 glioblastoma cell lines differing in PTEN and p53 status. Exp Cell Res 330: 346–357 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Review Process File


