Abstract
The current molecular understanding of Alzheimer’s disease (AD) has still not re-sulted in successful interventions. Mitochondrial dysfunction of the AD brain is currently emerging as a hallmark of this disease. One mitochondrial function often affected in AD is oxidative phosphorylation responsible for ATP production, but also for production of reactive oxygen species (ROS) and for the de novo synthesis of pyrimidines. This paper reviews the role of mitochondrial produced ROS and pyrimidines in the aetiology of AD and their pro-posed role in oxidative degeneration of macromolecules, synthesis of essential phospholipids and maintenance of mitochondrial viability in the AD brain.
Keywords: Mitochondria, DNA repair, dNTP pools, nucleotide metabolism, brain
1. Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disease that affects millions of people worldwide. Clinically, AD presents as a complex, heterogeneous disorder, roughly divided into two forms distinguished by latency: early onset / familial and the more common late onset / sporadic AD [1, 2]. Brain plaques, also called ‘senile’ plaques, containing insoluble Aβ and aggregates of Tau protein correlate with pathology of early and late onset AD, and these plaques have been suggested to play a fundamental role in disease pathology [3, 4]. However, despite a significant amount of research, the molecular mechanisms driving AD pathology remain poorly understood. This is reflected in the fact that successful interventions for treating or preventing AD remain elusive and the fact that clinical trials for interventions based on existing hypotheses about the role of Aβ and senile plaques in AD report limited or no success [5].
Recently, different aspects of mitochondrial dysfunction have been identified as novel components in the aetiology of AD. This is true for neuronal mito-chondria, but also for astroglial cells which have strong impact on neuronal function, neuronal development, and has been correlated with various neurodegenerative diseases, including AD and other forms of dementia [6, 7]. Astrocytes constitute the most abundant cell type in the human brain, and ensheath neurons and blood vessels. They maintain the brain extracellular milieu by buffering neurotransmitters and ions [8] and metabolize glucose to lactate, the major fuel for neurons [9], supplying neurons with substrates for oxidative phosphorylation [10]. These tasks are heavily dependent on mitochondrial function [11].
On a general scale, damaged mitochondria have been demonstrated to accumulate in brain tissue in both familial and sporadic forms of AD [12-15]. On a cellular scale, AD is demonstrated to be characterized by increased numbers of somatic mtDNA mutations [16], impairment of oxidative phosphorylation [17, 18], altered balance between mitochondrial fission and fusion [19] and changes in mitochondrial structure, dynamics, and motility [20, 21]. Furthermore, microarray data from hippocampal biopsies have revealed a significant decrease of nuclear and one mitochondrial encoded subunits of the mitochondrial electron transport chain in AD patients compared to age-matched controls [22].
The inverse Warburg hypothesis offers a bioenergetic model for AD, which postulates that AD is a consequence of mitochondrial deregulation inferring metabolic reprogramming as an initial attempt to maintain neuronal integrity [23, 24]. Once this compensatory mechanism is exhausted, bioenergetic deficits may lead to neuronal death and dementia. Thus, mitochondrial dysfunction may represent the missing link between aging and sporadic AD, and depicts attractive targets against neurodegeneration. Common to all mitochondrial dysfunctions associated with AD are the direct or indirect inhibition of the ability of neuronal and glial mitochondria to perform oxidative phosphorylation. One direct consequence of reduced oxidative phosphorylation is a decrease of the ATP produced and consequent changes of mitochondrial bioenergetics essential for the vitality of affected cells [25].
However, as will be reviewed here, other pathways important for neuronal viability are also affected and are therefore significant for the full understanding of the role of mitochondria in the aetiology of AD.
2. Reactive oxygen species in AD
A significant increase of oxidized biomolecules has been shown as a hallmark of AD in brain tissue: Oxidative base damage in both nuclear DNA (nDNA) and mtDNA has been reported to be increased in post-mortem brains of AD patients [26-29]. Also, lipid peroxidation of neuronal tissue and oxidative modifications of proteins has been demonstrated [30-32]. In a mouse model of AD, increase of oxidative stress precedes the appearance of Aβ plaques and neurofibrillary tangles, strongly indicating reactive oxygen species (ROS) as a very early event in AD [33]. This is supported by findings in human post-mortem AD brains, where oxidative damage is demonstrated to be quantitatively most predominant early in the disease and reduced with disease progression [34]. A mitochondrial origin of the increased levels of ROS in AD brain has often been suggested [12-14]. This hypothesis is based on the high oxygen consumption rate of neurons, utilized for oxidative phosphorylation, and on the observation that damaged mitochondria accumulate in the AD brain [12-14]. During the process of oxidative phosphorylation, ROS are continuously generated from up to 4% of the oxygen consumed by the process [35]. Complex I and especially complex III of the electron transport chain are the prime sites for electron leakage to molecular oxygen yielding the superoxide anion (·O2-) [36, 37], and the production of ROS is inversely correlated with the rate of electron transport, increasing exponentially when complex I or III are impaired [38, 39]. Mitochondrial produced ROS serve as a second messenger molecule proposed to report oxygen available for oxidative phosphorylation, affecting epigenetic marking of nDNA and regulating nuclear transcription factors, kinases, and phosphatases (reviewed in [40]). In the mitochondria, ·O2− is neutralized by intramitochondrial manganese (Mn)-dependent superoxide dismutase (SOD2) catalyzing the formation of H2O2, which within the mitochondria, in turn is inactivated by reaction with glutathione catalyzed by glutathione peroxidase [41-43]. If the amount of ROS produced exceeds the capacity of the mitochondrial antioxidant enzymes, ·O2− and H2O2 levels will rise. In the presence of transition metals, such as iron or copper, highly reactive OH· can be produced by Haber-Weiss or Fenton reactions. OH· can in turn give rise to a plethora of ROS, which has the potential to induce oxidative damage to lipids, proteins, RNA, and DNA [44-46].
The mitochondrion is not the only mediator of ROS in the brain, and several studies point to sources of ROS production that are completely independent of mitochondrial function. Microglia cells are such a source and increased production of ROS in the AD brain has been related to activation of microglia as a part of an inflammatory response in the brain [47-49]. In support of this, NADPH oxidase, the main mediator of microglial ROS, has been demonstrated to be activated in the AD brain, as evaluated by translocation of NADPH oxidase subunits [47]. In a mouse model of AD, where Aβ is overexpressed, removal of the active centre, gp91phox, of the NADPH oxidase, rescues the mouse from elevated levels of oxidative stress in neocortex and cerebrovascular dysfunction [50].
Rather than perceiving the two theories of origin of ROS in the AD brain to be mutually exclusive, recent studies suggest that the mitochondrial produced ROS can act as regulators of pro-inflammatory responses of microglia cells [51, 52]. In accordance, it has been demonstrated that an inhibition of mitochondrial produced ROS significantly prevented lipopolysaccharide (LPS)-induced activation of microglia [53]. Nevertheless, the hypothesis stating that mitochondrial produced ROS is solely responsible for the oxidative damage associated with AD is challenged with an emerging focus on the role of mitochondrial ROS as second messenger molecules in AD.
3. Base excision DNA repair of oxidative damage is decreased in AD
From a DNA repair perspective, neurons of AD pa-tients are much more exposed to and susceptible to in-creased levels of ROS, regardless of source, compared to controls [54]. One of the most frequent forms of oxidative DNA damage is 8oxoG [55]. 8oxoG mispairs are formed in vivo during DNA replication by two mechanisms, either by incorporation of an adenine nu-cleotide opposite an 8oxoG derived from the direct oxidation in the template strand [55] or by misincorporation of an 8oxoG that results from oxidation of GTP in the nucleotide pool [56]. Base ex-cision repair (BER) is the major repair pathway han-dling oxidative damage in DNA. BER involves the co-operative interaction of several proteins that work se-quentially to excise the target damage and restore DNA back to its original, unmodified form [57]. BER cor-rects DNA lesions through the action of DNA glycosy-lases that excise damaged bases, either by a monofunc-tional (uracil DNA glycosylase (UDG) or MPG) or a bifunctional (NTH1, OGG1, NEIL1 or NEIL2) DNA glycosylase leaving an AP site with an intact DNA phosphodiester backbone [58]. AP-endonuclase 1 (APE1) recognizes the AP site left by excision by the monofunctional DNA glycosylases and incise the DNA backbone adjacent to the AP site. Excision by a bifunc-tional DNA glycosylase is followed by an incision the DNA backbone facilitated by the AP lyase activity of these enzymes, leaving a DNA single strand break (SSB) [58]. The end processing is performed by either DNA polymerase β (Polβ), APE1 or PNKP depending on the terminus generated in the former step. The final step is ligation to seal the nick with a 3’-OH and a 5’-P terminus by the LIG3α-XRECC1 complex or LIG1 in association with PCNA [59].
3.1. BER Activity in AD
It has been hypothesized that neurons from AD patients have a genetic defect in BER [54]. In accordance, both a significant decrease in 8oxoG glycosylase activity [60, 61] and reduced DNA synthesis capacity by Polβ [61] have been demonstrated in AD brain. Our previous studies show a down-regulation of APE1 and an up-regulation of OGG1 in the prodromal phases of AD in the Tg-ArcSwe AD mouse model at the age of 4 months [62]. In support, we found lower APE1 mRNA levels in the blood and in the entorhinal cortex of AD patients than in healthy controls [63]. The entorhinal cortex is one of the first regions to be affected in AD [64] and alterations observed here may represent late changes in the progression of AD.
We have not been able to detect the OGG1 protein by mass spectrometry of AD patient frontal cortex and cerebellum brain tissue, suggesting low expression of OGG1, a highly catalytic enzyme [63]. In studies on mild cognitive impairment (MCI), OGG1 mRNA transcript abundance in blood was reduced in MCI, MCI/AD and AD patients compared to healthy controls as well as in patients with abnormal levels of cerebrospinal fluid (CSF) Aβ-42 and tau and in patients with normal CSF levels of Aβ-42 and tau [63]. The findings indicate that BER mRNA profile alterations occur independent of Aβ and tau pathology since the alterations are also seen in patients with no CSF pathology, however, not in healthy controls. This is consistent with findings from other studies [65-68]. It has been suggested that oxidative DNA damage increases only during the early stages of AD and then declines with the progression of the disease due to activation of a compensatory mechanism [34]. Other studies show increased OGG1 mRNA levels in brain tissue from the hippocampus, parahippocampal gyri and middle temporal gyri of patients with preclinical stages of AD compared to healthy controls [68]. This elevation of OGG1 may represent a compensatory increase in protein expression to moderate the loss of activity due to altered posttranslational modifications in response to increased oxidative DNA damage [68].
3.2. Base Excision Repair in Mitochondria in AD
The mitochondria only offers very simple DNA repair pathways to ensure the fidelity of mtDNA, and BER is the major DNA repair pathway for oxidative damage and the dominant repair pathway in mitochondria [69].
Several of the mitochondrial BER (mtBER) proteins are splice variants that localize to the mitochondria. Seven of the 10 human glycosylases have been detected in the mitochondria including UNG1/2, MPG, OGG1, MUTYH and NEIL1/2 [70, 71]. MUTYH-α-1 appears to be the primary MUTYH splice variant in mitochondria [70], and UNG1 excise uracil and oxidized cytosine [72]. NEIL1 excises oxidative modified pyrimidine bases [73] and primarily lesions in dsDNA, bubble, bulge and fork structures while NEIL2 excises lesions in ssDNA primarily [74]. APE1 performs incision of the AP site [75]. Polymerase γ (PolG) is the sole polymerase in mitochondria [76] and performs gap-filling and procession of the termini [77]. Ligation is carried out by LIG3β performing a nick ligation without requiring XRCC1 like the nuclear variant [78, 79]. PARP1 is also present in mitochondria in a complex with LIG3β and mtDNA [80]. Studies suggest that long patch-BER also occurs in mitochondria [81]
Studies in AD patients have demonstrated reduced mitochondrial levels of βOGG1 in the neuronal cytoplasm of affected AD tissues that were associated with neurofibrillary tangles, dystrophic neuritis and reactive astrocytes [82]. When comparing levels of oxidized bases in AD brains compared to control brain, significantly higher levels were demonstrated in the frontal, parietal, and temporal lobes, and the mtDNA of affected tissues contained approximately 10-fold higher levels of oxidized bases compared to nuclear DNA [29]. These findings indicate that the mitochondria are particularly sensitive to increased ROS levels and dysfunctional BER. In support of this, transcriptional profiling of frontal cortical tissue from a mouse model of common familial AD mutations (3xTgAD) with a 50% reduction in in Polβ expression, revealed a down-regulation of genes involved in mitochondrial bioenergetics [83].
4. Mitochondrial regulated nucleotide metabolism in AD
In eukaryotes, nucleotide levels are maintained by nucleotide salvage and/or de novo synthesis of ribo- and deoxyribonucleotide triphosphates (rNTPs and dNTPs). Imbalanced dNTP pools have been demonstrated in cells from AD individuals [84] and may be an early risk marker for AD [85]. Thus, it is possible that rNTP and/or dNTP pool imbalances play a role in the aetiology of AD, although the underlying mechanism remains unknown [85].
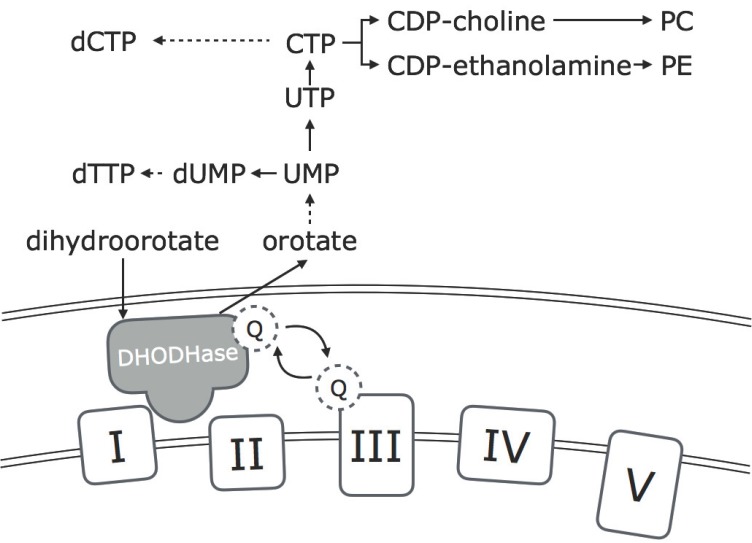
Mitochondrial respiration is linked to synthesis of rNTP and dNTP indirectly through production of ATP and directly through the enzyme dihydroorotate dehydrogenase (DHODHase). As in many other cellular processes, ATP is used as a substrate in several of the steps of rNTP and dNTP synthesis. Additionally, binding of ATP to the activity site of ribonucleotide reductase (RNR) is necessary for activation of the rate–limiting enzyme of the de novo synthesis of dNTP [86]. DHODHase is an integral protein of the inner mitochondrial membrane that faces the intermembrane space. The enzyme mediates the conversion of dihydroorotate to orotate, which is further converted to uridine monophosphates, pyrimidine ribonucleotides and pyrimidine deoxyribonucleotides (Fig. 1). DHODHase is functionally connected to the mitochondrial respiration by a flavin prosthetic group that couples dihydroorotate oxidation to respiratory ubiquinone reduction [87]. Inhibition of mitochondrial respiration, caused by lack of oxygen, presence of electron transport chain inhibitors or mutations of complex III and IV of the electron transport chain, cause impairments of the de novo UMP synthesis and a subsequent decrease in the de novo synthesis of pyrimidines [88, 89].
Fig. (1).
Overview of the relationship of the electron transport chain (complex I-IV) to the dihydroorotate dehydrogenase (DHODHase). DHODHase is an integral enzyme in the de novo synthesis of pyrimidines. The enzyme is located in the inner mitochondrial membrane and its activity is dependent on an active electron transport chain. Pyrimidines synthesised are important as substrates for nuclear and mitochondrial DNA replication and repair, and for synthesis of the essential phospholipids: phosphatidylcholine (PC) and phosphatidylethanolamine (PE).
In dividing cells, dNTP levels are important as substrates for DNA replication. In post-mitotic cells such as neurons, dNTP levels are important as substrates for mitochondrial DNA replication and post replicative DNA repair processes. rNTP levels are substrates for synthesis of RNA, additionally pyrimidine ribonucleotides also serve as essential precursors in the synthesis of phospholipids, glycolipids and glycoproteins of the plasma membrane (reviewed in [90]), expression of genes encoding essential proteins for de novo synthesis of pyrimidines has been verified in rat brain using in situ hybridization [91]. In support, expression and activity of DHODHase has been determined in neuronal cell bodies of rat brain by western blot analysis and immunocytochemistry [92]. For both genes and proteins of DHODHase and other de novo constituents, expression was found to be highest in the neocortex and the in hippocampus [91, 92]. Two brain regions that are known to be severely affected in AD. The essential and many-facetted roles of mitochondrial produced pyrimidines in neurons can offer an explanation to the involvement of imbalanced dNTP levels in the aetiology of AD.
4.1. Balanced Levels of dNTP are Essential for mtDNA Replication
Replication of mitochondrial DNA is not regulated by mitosis and occurs continuously in post mitotic tissue including neurons. However, dNTP pool imbalance also affect replication of mtDNA and point mutations and deletions accumulate in mtDNA in cells [93] and tissue [94] that exhibit dNTP pool imbalance. This phenomenon has been observed in patients with mitochondrial neurogastrointestinal encephalopathy (MNGIE) [95] and hepatocerebral mitochondrial depletion disorders (MDS) [96]. The cytosolic pool of dNTP which supplies the nuclear genome, is cell cycle regulated where dNTP synthesis is initiated at the beginning of S phase of the cell cycle and shut off at G2 phase. This regulation is mediated by cell cycle regulated expression and degradation of RNR-R2, one of the two proteins constituting RNR [97] and by translational regulation of constituents of the nucleotide salvage pathway [98]. The dNTP pool of the mitochondrial compartment is separate from the much larger pool supplying the nuclear genome [99, 100], however, a cytosolic de novo synthesis of dNTP has also been demonstrated to be essential for mtDNA maintenance and repair in post mitotic cells and tissue [101-103]. P53R2 is a protein which substitutes RNR-R2 in post mitotic cells in response to DNA damage. P53R2 is transcriptionally regulated by P53 and results in an activation of RNR and de novo synthesis of dNTP intended as substrates for DNA repair [104]. Mutations of the RRM2B gene encoding P53R2 have in human fibroblasts been demonstrated to induce mtDNA replication and repair deficiency in post mitotic, but not dividing cells [101]. In humans, mutations of RRM2B have been correlated with severe mtDNA depletion in muscle tissue and Rrm2b-/- mice display a severe decrease of mtDNA content in liver, kidney and muscle [102]. Although some neurological conditions are reported in patients with P53R2 mutations, the effects of P53R2 deficiency on mitochondria in neural tissue have not been examined [102, 105]. Nevertheless, transcripts encoding RNR-R2 are undetectable in brain, whereas the RRM2B gene is ubiquitously expressed in human muscle [102], implying that neurons entirely rely on P53R2 for dNTP synthesis.
Here, we hypothesize that dNTP pool imbalance plays a role in the aetiology of AD, possibly mediated by an ability to induce mutation and depletion of mtDNA. Furthermore, we postulate a dNTP based vicious cycle where imbalance of the cytosolic dNTP pool will induce mutation and depletion of mtDNA, which in turn will impact on the de novo synthesis of dNTP through decreased function of the DHODHase and decreased activation of the RNR due to lower levels of ATP. Our hypotheses are consistent with the suggestion that altered abundance of cytosolic dNTPs may be an early marker of susceptibility to AD [85]
4.2. Pyrimidines and Phospholipid Synthesis in AD
Ribonucleotides and deoxyribonucleotides are not the final products of the de novo synthesis pathway. Rather, they are intimately involved in the synthesis of phospholipids that are essential for neuronal function and survival. Phosphatidylcholine (PC) and phosphatidylethanolamine (PE) are the most abundant phospholipids in the plasma membrane of neurons and AD or risk of AD has been correlated with significant decreases of PC in plasma [106, 107]. In addition significant decrease of PC and PE levels have been demonstrated in post mortem brains of AD patients [108-113].
Synthesis of PC and PE is dependent on presence of cytidine triphosphate (CTP). PC is synthesised in a process catalysed by the enzyme CTP:phosphocholine cytidylyltransferase (PCCT) where CTP and phosphocholine reacts and forms CDP-choline. CDP-choline subsequently reacts with diacylglycerol in a reaction catalysed by CDP-choline:1,2-diacylgylcerol cholinephosphotransferase (PECT) whereby PC is produced and the cytidine ribonucleotide is released in form of CMP. In a similar process to PC synthesis, CTP reacts with ethanolamine forming CDP-ethanolamine in a process mediated by CTP: phosphoethanolamine cytidylyltransferase. CDP-ethonalamine reacts with diacylglycerol and forms PE and CMP in a process catalysed by CDP-ethanolamine: 1,2-diacylglycerol ethanolaminephosphotransfease (reviewed by [114]).
For both PC and PE, CTP is required for the synthesis of the two phospholipids, however, CTP exceeds the role of an intermediate substrate. CTP has been suggested to serve as a universal signal for biosynthesis of all phospholipids and thereby control the rate of membrane synthesis to the energy state of the cell [115]. In support of this notion, both PECT and PCCT from human brain biopsies exhibit low affinity for CTP suggesting that the activities of these enzymes, and by implication, the rate of PC and PE synthesis, are highly dependent upon the cellular concentration of CTP [116]. Finally, it has in rat myoblasts been demonstrated that a specific inhibition of CTP synthesis results in a 50% reduction in synthesis of PC and PE [117]. Conversely, incubation of rat striatal slices with cytidine causes dose dependent increases of intracellular levels of CTP and in turn a significant increase of PC and PE synthesis [118].
As dysfunction of the ETC impairs the activity of DHODHase, and as consequence the de novo synthesis of CTP, this suggests a correlation between mitochondrial fitness and synthesis of phospholipids. This correlation has to some extent been documented. Treating differentiated rat pheochromocytoma PC12 cells with the uncoupler of the inner mitochondrial membrane carbonyl cyanide m-chlorophenylhydrazone (CCCP) caused an accelerated and significant synthesis of PC. The uncoupling used in this study was mild and did not cause depolarization of the mitochondrial membrane, as evidenced with the ATP levels remained unaltered [119]. Whether this change is due to increased activity of the DHODHase or other mitochondrial activities related to ETC, is unclear, as levels of CTP was not monitored. This necessitates further experiments to describe the correlation between mitochondrial function, the DHODHase and phospholipid synthesis.
Imbalances of the dNTP levels have been correlated with risk of AD [84, 85]. These imbalance events can be symptomatic of AD which in itself is an important realization and a fact that hopefully can be used for early AD risk assessment in the near future [85]. The mitochondrial relation to de novo synthesis of rNTP and dNTP, and in turn the strong impact of CTP in synthesis of essential phospholipids and the important role of de novo synthesized dNTP for maintenance of mtDNA, strongly indicates a causal role of mitochondrial impact on rNTP and dNTP synthesis in the aetiology of AD. An important realisation is that due to the importance of balanced dNTP for mtDNA maintenance, the initial dNTP imbalance does not need to be of mitochondrial origin. Imbalances of the dNTP pool resulting from genetic predisposition or even diet [120] have the potential to initiate a vicious cycle whereby failing maintenance of mtDNA results in decreased synthesis of PC and PE, but also other mitochondrial dysfunction phenotypes associated with AD.
Conclusion
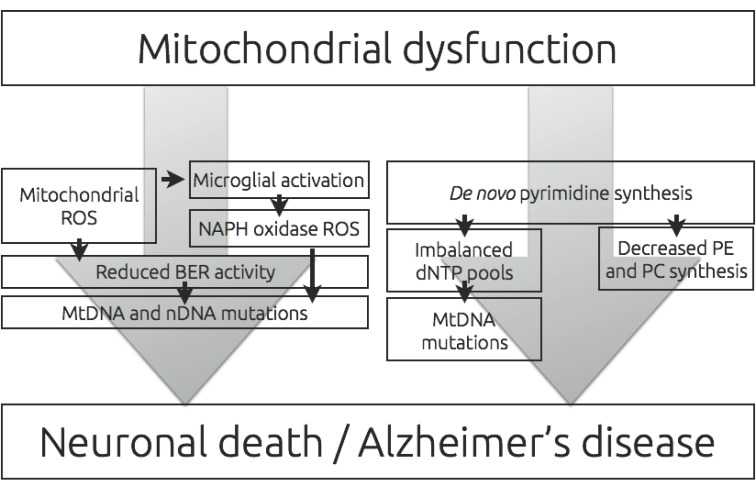
Despite a significant amount of research on AD, interventions to treat or prevent AD are not yet available. This reflects the poor understanding of the molecular mechanisms underlying AD development and that targeting of existing well-validated markers of AD, including Aβ and tau protein so far only have resulted in no effect on disease progression or only a slowing of clinical decline. [121-123]. Recent studies imply an increasing role of dysfunctional neuronal and glial mitochondria in the aetiology of AD. As the majority of observed mitochondrial dysfunctions affect the ability to perform oxidative phosphorylation it is important to review the downstream effects of such an inhibition in AD (Fig. 2). Previously, we have reviewed the effects of a decreased capacity to produce ATP in mitochondria of neurons [25], and in this study we have reviewed the role of an increased ROS production and a decreased pyrimidine production resulting from mitochondrial dysfunction. ROS produced in mitochondria has in several studies been associated with aging of a wide array of tissues, caused by oxidative damage to DNA, lipids and proteins. However, recent literature suggest that oxidative damage events in the AD brain, at least in part, can be related to inflammatory responses rather than directly from mitochondrial dysfunction. A correlation between mitochondrial function and propagation of an inflammatory response mediated by mitochondrial ROS functioning as a second messenger molecule, suggest a more advanced role of mitochondrial produced ROS in the aetiology of AD.
Fig. (2).
Reduced level and decreased efficiency of oxidative phosphorylation is a hallmark of Alzheimer’s disease (AD). In turn, a decreased efficiency of oxidative phosphorylation has the potential to induce an increased production of mitochondrial reactive oxygen species (ROS). Mitochondrial ROS can activate the microglial oxidative response and the combined production of ROS has the potential of induce nuclear and mitochondrial DNA mutations. This deleterious effect of ROS is often additive in the AD brain with a reduced activity of the base excision repair (BER) pathway. Reduced level of oxidative phosphorylation also has the potential to decrease de novo synthesis of pyrimidines, which in turn can result in imbalance of dNTP levels and a resulting increased mutation load on mitochondrial DNA. Decreased levels of pyrimidines also affect synthesis of essential phopsholipids: phosphatidylcholine (PC) and phosphatidylethanolamine (PE). Together and separately, these effects of altered oxidative phosphorylation increase the risk of neuronal death.
Nucleotide synthesis of neurons is not a widely researched topic. This is most likely due to the fact that neurons are post-mitotic, and therefore not need abundant pools of nucleotides. Nevertheless, several pathways of nucleotide synthesis has been demonstrated to be expressed in brain tissue [91, 92]. Synthesis of nucleotides is important for mtDNA synthesis, but also for the synthesis of essential phospholipids demonstrated to be down regulated in AD.
With this review, the hope is to raise attention to the downstream effects of mitochondrial dysfunction, as they, in theory, can explain several aspects of AD which current hypothesis on the pathogenesis of AD cannot. It is hoped that additional studies of both normal aging and the pathogenesis of AD and other neurological diseases will eventually provide insight into the molecular basis of AD, and lay the groundwork for effective treatment and/or prevention of AD.
Acknowledgements
This research was supported by a grant from Nordea fonden CD and LJR. From the South-Eastern Norway Regional Health Authority: Project 2014020 to MSL and TT and project 2014050 to TT. From the Olav Thon Foundation to LJR and TT.
Consent for Publication
Not applicable.
conflict of interest
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Pritchard S.M., Dolan P.J., Vitkus A., Johnson G.V.W. The toxicity of tau in Alzheimer disease: Turnover, targets and potential therapeutics. J. Cell. Mol. Med. 2011;15:1621–1635. doi: 10.1111/j.1582-4934.2011.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serrano-Pozo A., Frosch M.P., Masliah E., Hyman B.T. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2011;1:a006189–a006189. doi: 10.1101/cshperspect.a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., Mohs R.C., Morris J.C., Rossor M.N., Scheltens P., Carrillo M.C., Thies B., Weintraub S., Phelps C.H. The Diagnosis of Dementia Due to Alzheimer’s Disease: Recommendations From the National Institute on Aging-Alzheimer. Vol. 7. S Association Workgroups on Diagnostic Guidelines for Alzheimer’s Disease; 2011. pp. 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Möller H.J., Graeber M.B. The Case Described by Alois Alzheimer in 1911. Historical and Conceptual Perspectives Based on the Clinical Record and Neurohistological Sections. Eur. Arch. Psychiatry Clin. Neurosci. 1998;248(3):111–122. doi: 10.1007/s004060050027. [DOI] [PubMed] [Google Scholar]
- 5.Cummings J.L., Morstorf T., Zhong K. Alzheimer’s disease drug-development pipeline: Few candidates, frequent failures. Alzheimers Res. Ther. 2014;6:37. doi: 10.1186/alzrt269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verkhratsky A., Olabarria M., Noristani H.N., Yeh C-Y., Rodriguez J.J. Astrocytes in Alzheimer’s disease. Neurotherapeutics. 2010;7:399–412. doi: 10.1016/j.nurt.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen L.J., Shiloh Y., Bergersen L.H., Sander M., Bohr V.A., Tønjum T. DNA damage response, bioenergetics, and neurological disease: The challenge of maintaining brain health in an aging human population. Mech. Ageing Dev. 2013;134:427–433. doi: 10.1016/j.mad.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubik L.L., Philbert M.A. The role of astrocyte mitochondria in differential regional susceptibility to environmental neurotoxicants: Tools for understanding neurodegeneration. Toxicol. Sci. 2015;144:7–16. doi: 10.1093/toxsci/kfu254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stobart J.L., Anderson C.M. Multifunctional role of astrocytes as gatekeepers of neuronal energy supply. Front. Cell. Neurosci. 2013;7:38. doi: 10.3389/fncel.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellerin L., Pellegri G., Bittar P.G., Charnay Y., Bouras C., Martin J.L., Stella N., Magistretti P.J. Evidence supporting the existence of an activity-dependent astrocyte-neuron lactate shuttle. Dev. Neurosci. 1998;20:291–299. doi: 10.1159/000017324. [DOI] [PubMed] [Google Scholar]
- 11.Voloboueva L.A., Suh S.W., Swanson R.A., Giffard R.G. Inhibition of mitochondrial function in astrocytes: Implications for neuroprotection. J. Neurochem. 2007;102:1383–1394. doi: 10.1111/j.1471-4159.2007.4634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swerdlow R.H., Burns J.M., Khan S.M. The Alzheimer’s disease mitochondrial cascade hypothesis. J. Alzheimers Dis. 2010;20(Suppl. 2):S265–S279. doi: 10.3233/JAD-2010-100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du H., Guo L., Yan S.S. Synaptic mitochondrial pathology in Alzheimer’s disease. Antioxid. Redox Signal. 2012;16:1467–1475. doi: 10.1089/ars.2011.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy P.H. Is the mitochondrial outermembrane protein VDAC1 therapeutic target for Alzheimer’s disease? Biochim. Biophys. Acta. 2013;1832:67–75. doi: 10.1016/j.bbadis.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swerdlow R.H. Bioenergetics and metabolism: A bench to bedside perspective. J. Neurochem. 2016;139(Suppl. 2):126–135. doi: 10.1111/jnc.13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin M.T., Simon D.K., Ahn C.H., Kim L.M., Beal M.F. High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer’s disease brain. Hum. Mol. Genet. 2002;11:133–145. doi: 10.1093/hmg/11.2.133. [DOI] [PubMed] [Google Scholar]
- 17.Rhein V., Song X., Wiesner A., Ittner L.M., Baysang G., Meier F., Ozmen L., Bluethmann H., Dröse S., Brandt U., Savaskan E., Czech C., Götz J., Eckert A. Amyloid-beta and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer’s disease mice. Proc. Natl. Acad. Sci. USA. 2009;106:20057–20062. doi: 10.1073/pnas.0905529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krako N., Magnifico M.C., Arese M., Meli G., Forte E., Lecci A., Manca A., Giuffrè A., Mastronicola D., Sarti P., Cattaneo A. Characterization of mitochondrial dysfunction in the 7PA2 cell model of Alzheimer’s disease. J. Alzheimers Dis. 2013;37:747–758. doi: 10.3233/JAD-130728. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Su B., Lee H-G., Li X., Perry G., Smith M.A., Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J. Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baloyannis S.J. Mitochondrial alterations in Alzheimer’s disease. J. Alzheimers Dis. 2006;9:119–126. doi: 10.3233/jad-2006-9204. [DOI] [PubMed] [Google Scholar]
- 21.Chen H., Chan D.C. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum. Mol. Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mastroeni D., Khdour O.M., Delvaux E., Nolz J., Olsen G., Berchtold N., Cotman C., Hecht S.M., Coleman P.D. Nuclear but not mitochondrial-encoded OXPHOS genes are altered in aging, mild cognitive impairment, and Alzheimer’s disease. Alzheimers Dement. 2016;13:510–519. doi: 10.1016/j.jalz.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demetrius L.A., Driver J. Alzheimer’s as a metabolic disease. Biogerontology. 2013;14:641–649. doi: 10.1007/s10522-013-9479-7. [DOI] [PubMed] [Google Scholar]
- 24.Demetrius L.A., Magistretti P.J., Pellerin L. Alzheimer’s disease: The amyloid hypothesis and the inverse warburg effect. Front. Physiol. 2014;5:522. doi: 10.3389/fphys.2014.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desler C., Hansen T.L., Frederiksen J.B., Marcker M.L., Singh K.K., Juel Rasmussen L. Is there a link between mitochondrial reserve respiratory capacity and aging? J. Aging Res. 2012;2012:192503–192509. doi: 10.1155/2012/192503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullaart E., Boerrigter M.E., Ravid R., Swaab D.F., Vijg J. Increased levels of DNA breaks in cerebral cortex of Alzheimer’s disease patients. Neurobiol. Aging. 1990;11:169–173. doi: 10.1016/0197-4580(90)90542-8. [DOI] [PubMed] [Google Scholar]
- 27.Mecocci P., MacGarvey U., Beal M.F. Oxidative damage to mitochondrial DNA Is increased in Alzheimer’s disease. Ann. Neurol. 1994;36:747–751. doi: 10.1002/ana.410360510. [DOI] [PubMed] [Google Scholar]
- 28.Lyras L., Cairns N.J., Jenner A., Jenner P., Halliwell B. An assessment of oxidative damage to proteins, lipids, and DNA in brain from patients with Alzheimer’s disease. J. Neurochem. 1997;68:2061–2069. doi: 10.1046/j.1471-4159.1997.68052061.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang J., Xiong S., Xie C., Markesbery W.R., Lovell M.A. Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer’s disease. J. Neurochem. 2005;93:953–962. doi: 10.1111/j.1471-4159.2005.03053.x. [DOI] [PubMed] [Google Scholar]
- 30.Hensley K., Hall N., Subramaniam R., Cole P., Harris M., Aksenov M., Aksenova M., Gabbita S.P., Wu J.F., Carney J.M. Brain regional correspondence between Alzheimer’s disease histopathology and biomarkers of protein oxidation. J. Neurochem. 1995;65:2146–2156. doi: 10.1046/j.1471-4159.1995.65052146.x. [DOI] [PubMed] [Google Scholar]
- 31.Butterfield D.A., Kanski J. Brain protein oxidation in age-related neurodegenerative disorders that are associated with aggregated proteins. Mech. Ageing Dev. 2001;122:945–962. doi: 10.1016/s0047-6374(01)00249-4. [DOI] [PubMed] [Google Scholar]
- 32.Markesbery W.R. Oxidative stress hypothesis in Alzheimer’s disease. Free Radic. Biol. Med. 1997;23:134–147. doi: 10.1016/s0891-5849(96)00629-6. [DOI] [PubMed] [Google Scholar]
- 33.Hauptmann S., Scherping I., Dröse S., Brandt U., Schulz K.L., Jendrach M., Leuner K., Eckert A., Müller W.E. Mitochondrial dysfunction: An early event in Alzheimer pathology accumulates with age in AD transgenic mice. Neurobiol. Aging. 2009;30:1574–1586. doi: 10.1016/j.neurobiolaging.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Nunomura A., Perry G., Aliev G., Hirai K., Takeda A., Balraj E.K., Jones P.K., Ghanbari H., Wataya T., Shimohama S., Chiba S., Atwood C.S., Petersen R.B., Smith M.A. Oxidative damage is the earliest event in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2001;60:759–767. doi: 10.1093/jnen/60.8.759. [DOI] [PubMed] [Google Scholar]
- 35.O’Donovan D.J., Fernandes C.J. Mitochondrial glutathione and oxidative stress: Implications for pulmonary oxygen toxicity in premature infants. Mol. Genet. Metab. 2000;71:352–358. doi: 10.1006/mgme.2000.3063. [DOI] [PubMed] [Google Scholar]
- 36.Ide T., Tsutsui H., Kinugawa S., Utsumi H., Kang D., Hattori N., Uchida K., Arimura K.I., Egashira K., Takeshita A. Mitochondrial electron transport complex I is a potential source of oxygen free radicals in the failing myocardium. Circ. Res. 1999;85:357–363. doi: 10.1161/01.res.85.4.357. [DOI] [PubMed] [Google Scholar]
- 37.Chen Q. Production of reactive oxygen species by mitochondria: Central role of complex III. J. Biol. Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 38.Lesnefsky E.J., Gudz T.I., Moghaddas S., Migita C.T., Ikeda-Saito M., Turkaly P.J., Hoppel C.L. Aging decreases electron transport complex III activity in heart interfibrillar mitochondria by alteration of the cytochrome C binding site. J. Mol. Cell. Cardiol. 2001;33:37–47. doi: 10.1006/jmcc.2000.1273. [DOI] [PubMed] [Google Scholar]
- 39.Indo H.P., Davidson M., Yen H-C., Suenaga S., Tomita K., Nishii T., Higuchi M., Koga Y., Ozawa T., Majima H.J. Evidence of ROS generation by mitochondria in cells with impaired electron transport chain and mitochondrial DNA damage. Mitochondrion. 2007;7:106–118. doi: 10.1016/j.mito.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 40.Weinberg F., Chandel N.S. Mitochondrial metabolism and cancer. Ann. N. Y. Acad. Sci. 2009;1177:66–73. doi: 10.1111/j.1749-6632.2009.05039.x. [DOI] [PubMed] [Google Scholar]
- 41.Arai M., Imai H., Koumura T., Yoshida M., Emoto K., Umeda M., Chiba N., Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase plays a major role in preventing oxidative injury to cells. J. Biol. Chem. 1999;274:4924–4933. doi: 10.1074/jbc.274.8.4924. [DOI] [PubMed] [Google Scholar]
- 42.Simmons T.W., Jamall I.S. Relative importance of intracellular glutathione peroxidase and catalase in vivo for prevention of peroxidation to the heart. Cardiovasc. Res. 1989;23:774–779. doi: 10.1093/cvr/23.9.774. [DOI] [PubMed] [Google Scholar]
- 43.Radi R., Turrens J.F., Chang L.Y., Bush K.M., Crapo J.D., Freeman B.A. Detection of catalase in rat heart mitochondria. J. Biol. Chem. 1991;266:22028–22034. [PubMed] [Google Scholar]
- 44.Lu C.Y., Lee H.C., Fahn H.J., Wei Y.H. Oxidative damage elicited by imbalance of free radical scavenging enzymes is associated with large-scale mtDNA deletions in aging human skin. Mutat. Res. 1999;423:11–21. doi: 10.1016/s0027-5107(98)00220-6. [DOI] [PubMed] [Google Scholar]
- 45.Tabatabaie T., Floyd R.A. Inactivation of glutathione peroxidase by benzaldehyde. Toxicol. Appl. Pharmacol. 1996;141:389–393. doi: 10.1006/taap.1996.0304. [DOI] [PubMed] [Google Scholar]
- 46.Mello Filho A.C., Meneghini R. In vivo formation of single-strand breaks in DNA by hydrogen peroxide is mediated by the Haber-Weiss reaction. Biochim. Biophys. Acta. 1984;781:56–63. doi: 10.1016/0167-4781(84)90123-4. [DOI] [PubMed] [Google Scholar]
- 47.Shimohama S., Tanino H., Kawakami N., Okamura N., Kodama H., Yamaguchi T., Hayakawa T., Nunomura A., Chiba S., Perry G., Smith M.A., Fujimoto S. Activation of NADPH oxidase in Alzheimer’s disease brains. Biochem. Biophys. Res. Commun. 2000;273:5–9. doi: 10.1006/bbrc.2000.2897. [DOI] [PubMed] [Google Scholar]
- 48.Bianca V.D., Dusi S., Bianchini E., Dal Prà I., Rossi F. Beta-amyloid activates the O-2 forming NADPH oxidase in microglia, monocytes, and neutrophils. A possible inflammatory mechanism of neuronal damage in Alzheimer’s disease. J. Biol. Chem. 1999;274:15493–15499. doi: 10.1074/jbc.274.22.15493. [DOI] [PubMed] [Google Scholar]
- 49.Abramov A.Y., Duchen M.R. The role of an astrocytic NADPH oxidase in the neurotoxicity of amyloid beta peptides. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:2309–2314. doi: 10.1098/rstb.2005.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park L., Anrather J., Zhou P., Frys K., Pitstick R., Younkin S., Carlson G.A., Iadecola C. NADPH-oxidase-derived reactive oxygen species mediate the cerebrovascular dysfunction induced by the amyloid beta peptide. J. Neurosci. 2005;25:1769–1777. doi: 10.1523/JNEUROSCI.5207-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu Z., Yu F., Gong P., Qiu Y., Zhou W., Cui Y., Li J., Chen H. Subneurotoxic copper(II)-induced NF-κB-dependent microglial activation is associated with mitochondrial ROS. Toxicol. Appl. Pharmacol. 2014;276:95–103. doi: 10.1016/j.taap.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 52.Voloboueva L.A., Emery J.F., Sun X., Giffard R.G. Inflammatory response of microglial BV-2 cells includes a glycolytic shift and is modulated by mitochondrial glucose-regulated protein 75/mortalin. FEBS Lett. 2013;587:756–762. doi: 10.1016/j.febslet.2013.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park J., Min J-S., Kim B., Chae U-B., Yun J.W., Choi M-S., Kong I-K., Chang K-T., Lee D-S. Mitochondrial ROS govern the LPS-induced pro-inflammatory response in microglia cells by regulating MAPK and NF-κB pathways. Neurosci. Lett. 2015;584:191–196. doi: 10.1016/j.neulet.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 54.Boerrigter M.E., Wei J.Y., Vijg J. DNA repair and Alzheimer’s disease. J. Gerontol. 1992;47:B177–B184. doi: 10.1093/geronj/47.6.b177. [DOI] [PubMed] [Google Scholar]
- 55.Demple B., Harrison L. Repair of oxidative damage to DNA: Enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 56.Maki H., Sekiguchi M., Mut T. Protein Specifically Hydrolyses a Potent Mutagenic Substrate for DNA Synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 57.Seeberg E., Eide L., Bjørås M. The base excision repair pathway. Trends Biochem. Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 58.Jeppesen D.K., Bohr V.A., Stevnsner T. DNA repair deficiency in neurodegeneration. Prog. Neurobiol. 2011;94:166–200. doi: 10.1016/j.pneurobio.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ranalli T.A., Tom S., Bambara R.A. AP endonuclease 1 coordinates flap endonuclease 1 and DNA ligase I activity in long patch base excision repair. J. Biol. Chem. 2002;277:41715–41724. doi: 10.1074/jbc.M207207200. [DOI] [PubMed] [Google Scholar]
- 60.Lovell M.A., Xie C., Markesbery W.R. Decreased base excision repair and increased helicase activity in Alzheimer’s disease brain. Brain Res. 2000;855:116–123. doi: 10.1016/s0006-8993(99)02335-5. [DOI] [PubMed] [Google Scholar]
- 61.Weissman L., Jo D-G., Sørensen M.M., de Souza-Pinto N.C., Markesbery W.R., Mattson M.P., Bohr V.A. Defective DNA base excision repair in brain from individuals with Alzheimer’s disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007;35:5545–5555. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lillenes M.S., Støen M., Gómez-Muñoz M., Torp R., Günther C-C., Nilsson L.N.G., Tønjum T. Transient OGG1, APE1, PARP1 and Polβ expression in an Alzheimer’s disease mouse model. Mech. Ageing Dev. 2013;134:467–477. doi: 10.1016/j.mad.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 63.Lillenes M.S., Rabano A., Støen M., Riaz T., Misaghian D., Møllersen L., Esbensen Y., Günther C-C., Selnes P., Stenset V.T.V., Fladby T., Tønjum T. Altered DNA base excision repair profile in brain tissue and blood in Alzheimer’s disease. Mol. Brain. 2016;9:61. doi: 10.1186/s13041-016-0237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khan U.A., Liu L., Provenzano F.A., Berman D.E., Profaci C.P., Sloan R., Mayeux R., Duff K.E., Small S.A. Molecular drivers and cortical spread of lateral entorhinal cortex dysfunction in preclinical Alzheimer’s disease. Nat. Neurosci. 2014;17:304–311. doi: 10.1038/nn.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee S.L., Thomas P., Fenech M. Genome instability biomarkers and blood micronutrient risk profiles associated with mild cognitive impairment and Alzheimer’s disease. Mutat. Res. 2015;776:54–83. doi: 10.1016/j.mrfmmm.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 66.Ding Q., Markesbery W.R., Cecarini V., Keller J.N. Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer’s disease. Neurochem. Res. 2006;31:705–710. doi: 10.1007/s11064-006-9071-5. [DOI] [PubMed] [Google Scholar]
- 67.Lovell M.A., Markesbery W.R. Oxidatively modified RNA in mild cognitive impairment. Neurobiol. Dis. 2008;29:169–175. doi: 10.1016/j.nbd.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lovell M.A., Soman S., Bradley M.A. Oxidatively modified nucleic acids in preclinical Alzheimer’s disease (PCAD) brain. Mech. Ageing Dev. 2011;132:443–448. doi: 10.1016/j.mad.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Larsen N.B., Rasmussen M., Rasmussen L.J. Nuclear and mitochondrial DNA repair: Similar pathways? Mitochondrion. 2005;5:89–108. doi: 10.1016/j.mito.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 70.Prakash A., Doublié S. Base excision repair in the mitochondria. J. Cell. Biochem. 2015;116:1490–1499. doi: 10.1002/jcb.25103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hashiguchi K., Stuart J.A., de Souza-Pinto N.C., Bohr V.A. The C-Terminal alphaO helix of human Ogg1 is essential for 8-oxoguanine DNA glycosylase activity: The mitochondrial beta-Ogg1 lacks this domain and does not have glycosylase activity. Nucleic Acids Res. 2004;32:5596–5608. doi: 10.1093/nar/gkh863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nilsen H., Otterlei M., Haug T., Solum K., Nagelhus T.A., Skorpen F., Krokan H.E. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vartanian V., Lowell B., Minko I.G., Wood T.G., Ceci J.D., George S., Ballinger S.W., Corless C.L., McCullough A.K., Lloyd R.S. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc. Natl. Acad. Sci. USA. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prakash A., Doublié S., Wallace S.S. The Fpg/Nei family of dna glycosylases: Substrates, structures, and search for Damage. Prog. Mol. Biol. Transl. Sci. 2012;110:71–91. doi: 10.1016/B978-0-12-387665-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tomkinson A.E., Bonk R.T., Linn S. Mitochondrial endonuclease activities specific for Apurinic/Apyrimidinic sites in DNA from mouse cells. J. Biol. Chem. 1988;263:12532–12537. [PubMed] [Google Scholar]
- 76.Yakubovskaya E., Chen Z., Carrodeguas J.A., Kisker C., Bogenhagen D.F. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J. Biol. Chem. 2006;281:374–382. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- 77.Kaguni L.S. DNA polymerase gamma, the mitochondrial replicase. Annu. Rev. Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 78.Lakshmipathy U., Campbell C. Double strand break rejoining by mammalian mitochondrial extracts. Nucleic Acids Res. 1999;27:1198–1204. doi: 10.1093/nar/27.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lakshmipathy U., Campbell C. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 2000;28:3880–3886. doi: 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rossi M.N., Carbone M., Mostocotto C., Mancone C., Tripodi M., Maione R., Amati P. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J. Biol. Chem. 2009;284:31616–31624. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Akbari M., Visnes T., Krokan H.E., Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst.) 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 82.Iida T., Furuta A., Nishioka K., Nakabeppu Y., Iwaki T. Expression of 8-oxoguanine DNA glycosylase is reduced and associated with neurofibrillary tangles in Alzheimer’s disease brain. Acta Neuropathol. 2002;103:20–25. doi: 10.1007/s004010100418. [DOI] [PubMed] [Google Scholar]
- 83.Sykora P., Misiak M., Wang Y., Ghosh S., Leandro G.S., Liu D., Tian J., Baptiste B.A., Cong W-N., Brenerman B.M., Fang E., Becker K.G., Hamilton R.J., Chigurupati S., Zhang Y., Egan J.M., Croteau D.L., Wilson D.M., Mattson M.P., Bohr V.A. DNA polymerase B deficiency leads to neurodegeneration and exacerbates Alzheimer disease phenotypes. Nucleic Acids Res. 2015;43:943–959. doi: 10.1093/nar/gku1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maynard S., Hejl A-M., Dinh T., Keijzers G., Hansen M., Desler C., Moreno-Villanueva M., Bürkle A., Rasmussen L.J., Bohr V.A. Defective mitochondrial respiration, altered dNTP pools and reduced AP endonuclease 1 activity in peripheral blood mononuclear cells of Alzheimer’s disease patients. Aging (Albany N.Y.) 2015;7:793–815. doi: 10.18632/aging.100810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Desler C., Frederiksen J.H., Angleys M., Maynard S., Keijzers G., Fagerlund B., Mortensen E.L., Osler M., Lauritzen M., Bohr V.A., Rasmussen L.J. Increased deoxythymidine triphosphate levels is a feature of relative cognitive decline. Mitochondrion. 2015;25:34–37. doi: 10.1016/j.mito.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kashlan O.B., Scott C.P., Lear J.D., Cooperman B.S. A comprehensive model for the allosteric regulation of mammalian ribonucleotide reductase. Functional consequences of ATP- and dATP-induced oligomerization of the large subunit. Biochemistry. 2002;41:462–474. doi: 10.1021/bi011653a. [DOI] [PubMed] [Google Scholar]
- 87.Bader B., Knecht W., Fries M., Löffler M. Expression, purification, and characterization of histidine-tagged rat and human flavoenzyme dihydroorotate dehydrogenase. Protein Expr. Purif. 1998;13:414–422. doi: 10.1006/prep.1998.0925. [DOI] [PubMed] [Google Scholar]
- 88.Löffler M., Jöckel J., Schuster G., Becker C. Dihydroorotat-ubiquinone oxidoreductase links mitochondria in the biosynthesis of pyrimidine nucleotides. Mol. Cell. Biochem. 1997;174:125–129. [PubMed] [Google Scholar]
- 89.Beuneu C., Auger R., Löffler M., Guissani A., Lemaire G., Lepoivre M. Indirect inhibition of mitochondrial dihydroorotate dehydrogenase activity by nitric oxide. Free Radic. Biol. Med. 2000;28:1206–1213. doi: 10.1016/s0891-5849(00)00239-2. [DOI] [PubMed] [Google Scholar]
- 90.Micheli V., Camici M., Tozzi M.G., Ipata P.L., Sestini S., Bertelli M., Pompucci G. Neurological disorders of purine and pyrimidine metabolism. Curr. Top. Med. Chem. 2011;11:923–947. doi: 10.2174/156802611795347645. [DOI] [PubMed] [Google Scholar]
- 91.Gerlach J., Löffler M., Schäfer M.K-H. Gene expression of enzymes required for the de novo synthesis and degradation of pyrimidines in rat peripheral tissues and brain. Nucleosides Nucleotides Nucleic Acids. 2011;30:1147–1154. doi: 10.1080/15257770.2011.603712. [DOI] [PubMed] [Google Scholar]
- 92.Schaefer C.M., Schäfer M.K.H., Löfflerr M. Region-specific distribution of dihydroorotate dehydrogenase in the rat central nervous system points to pyrimidine de novo synthesis in neurons. Nucleosides Nucleotides Nucleic Acids. 2010;29:476–481. doi: 10.1080/15257771003730128. [DOI] [PubMed] [Google Scholar]
- 93.Song S., Wheeler L.J., Mathews C.K. Deoxyribonucleotide pool imbalance stimulates deletions in HeLa cell mitochondrial DNA. J. Biol. Chem. 2003;278:43893–43896. doi: 10.1074/jbc.C300401200. [DOI] [PubMed] [Google Scholar]
- 94.López L.C., Akman H.O., García-Cazorla A., Dorado B., Marti R., Nishino I., Tadesse S., Pizzorno G., Shungu D., Bonilla E., Tanji K., Hirano M. Unbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum. Mol. Genet. 2009;18:714–722. doi: 10.1093/hmg/ddn401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nishino I., Spinazzola A., Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283:689–692. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- 96.Mandel H., Szargel R., Labay V., Elpeleg O., Saada A., Shalata A., Anbinder Y., Berkowitz D., Hartman C., Barak M., Eriksson S., Cohen N. The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nat. Genet. 2001;29:337–341. doi: 10.1038/ng746. [DOI] [PubMed] [Google Scholar]
- 97.Chabes A.L., Pfleger C.M., Kirschner M.W., Thelander L. Mouse ribonucleotide reductase R2 protein: A new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc. Natl. Acad. Sci. USA. 2003;100:3925–3929. doi: 10.1073/pnas.0330774100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Munch-Petersen B., Cloos L., Jensen H.K., Tyrsted G. Human thymidine kinase 1. Regulation in normal and malignant cells. Adv. Enzyme Regul. 1995;35:69–89. doi: 10.1016/0065-2571(94)00014-t. [DOI] [PubMed] [Google Scholar]
- 99.Pontarin G., Gallinaro L., Ferraro P., Reichard P., Bianchi V. Origins of mitochondrial thymidine triphosphate: Dynamic relations to cytosolic pools. Proc. Natl. Acad. Sci. USA. 2003;100:12159–12164. doi: 10.1073/pnas.1635259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Desler C., Munch-Petersen B., Rasmussen L.J. The role of mitochondrial dNTP levels in cells with reduced TK2 activity. Nucleosides Nucleotides Nucleic Acids. 2006;25:1171–1175. doi: 10.1080/15257770600894501. [DOI] [PubMed] [Google Scholar]
- 101.Pontarin G., Ferraro P., Bee L., Reichard P., Bianchi V. Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc. Natl. Acad. Sci. USA. 2012;109:13302–13307. doi: 10.1073/pnas.1211289109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bourdon A., Minai L., Serre V., Jais J-P., Sarzi E., Aubert S., Chrétien D., de Lonlay P., Paquis-Flucklinger V., Arakawa H., Nakamura Y., Munnich A., Rötig A. Mutation of RRM2B, encoding P53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 103.Pontarin G., Ferraro P., Rampazzo C., Kollberg G., Holme E., Reichard P., Bianchi V. Deoxyribonucleotide metabolism in cycling and resting human fibroblasts with a missense mutation in p53R2, a subunit of ribonucleotide reductase. J. Biol. Chem. 2011;286:11132–11140. doi: 10.1074/jbc.M110.202283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakamura Y., Tanaka H., Arakawa H., Yamaguchi T., Shiraishi K., Fukuda S., Matsui K., Takei Y. A ribonucleotide reductase gene involved in a P53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 105.Wang D.B., Kinoshita C., Kinoshita Y., Morrison R.S. P53 and mitochondrial function in neurons. Biochim. Biophys. Acta. 2014;1842:1186–1197. doi: 10.1016/j.bbadis.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Whiley L., Sen A., Heaton J., Proitsi P., García-Gómez D., Leung R., Smith N., Thambisetty M., Kloszewska I., Mecocci P., Soininen H., Tsolaki M., Vellas B., Lovestone S., Legido-Quigley C. AddNeuroMed Consortium. Evidence of altered phosphatidylcholine metabolism in Alzheimer’s disease. Neurobiol. Aging. 2014;35:271–278. doi: 10.1016/j.neurobiolaging.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schaefer E.J., Bongard V., Beiser A.S., Lamon-Fava S., Robins S.J., Au R., Tucker K.L., Kyle D.J., Wilson P.W.F., Wolf P.A. Plasma phosphatidylcholine docosahexaenoic acid content and risk of dementia and Alzheimer disease: The framingham heart study. Arch. Neurol. 2006;63:1545–1550. doi: 10.1001/archneur.63.11.1545. [DOI] [PubMed] [Google Scholar]
- 108.Nitsch R.M., Blusztajn J.K., Pittas A.G., Slack B.E., Growdon J.H., Wurtman R.J. Evidence for a membrane defect in Alzheimer disease brain. Proc. Natl. Acad. Sci. USA. 1992;89:1671–1675. doi: 10.1073/pnas.89.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prasad M.R., Lovell M.A., Yatin M., Dhillon H., Markesbery W.R. Regional membrane phospholipid alterations in Alzheimer’s disease. Neurochem. Res. 1998;23:81–88. doi: 10.1023/a:1022457605436. [DOI] [PubMed] [Google Scholar]
- 110.Guan Z., Wang Y., Cairns N.J., Lantos P.L., Dallner G., Sindelar P.J. Decrease and structural modifications of phosphatidylethanolamine plasmalogen in the brain with Alzheimer disease. J. Neuropathol. Exp. Neurol. 1999;58:740–747. doi: 10.1097/00005072-199907000-00008. [DOI] [PubMed] [Google Scholar]
- 111.Wells K., Farooqui A.A., Liss L., Horrocks L.A. Neural membrane phospholipids in Alzheimer disease. Neurochem. Res. 1995;20:1329–1333. doi: 10.1007/BF00992508. [DOI] [PubMed] [Google Scholar]
- 112.Pettegrew J.W., Panchalingam K., Hamilton R.L., McClure R.J. Brain membrane phospholipid alterations in Alzheimer’s disease. Neurochem. Res. 2001;26:771–782. doi: 10.1023/a:1011603916962. [DOI] [PubMed] [Google Scholar]
- 113.Gottfries C.G., Karlsson I., Svennerholm L. Membrane components separate early-onset Alzheimer’s disease from senile dementia of the Alzheimer type. Int. Psychogeriatr. 1996;8:365–372. doi: 10.1017/s1041610296002736. [DOI] [PubMed] [Google Scholar]
- 114.Vance D.E., Vance J.E. Physiological consequences of disruption of mammalian phospholipid biosynthetic genes. J. Lipid Res. 2009;50(Suppl.):S132–S137. doi: 10.1194/jlr.R800048-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vance D.E., Choy P.C. How is phosphatidylcholine biosynthesis regulated? Trends Biochem. Sci. 1979;4:145–148. [Google Scholar]
- 116.Ross B.M., Moszczynska A., Blusztajn J.K., Sherwin A., Lozano A., Kish S.J. Phospholipid biosynthetic enzymes in human brain. Lipids. 1997;32:351–358. doi: 10.1007/s11745-997-0044-x. [DOI] [PubMed] [Google Scholar]
- 117.Hatch G.M., McClarty G. Regulation of cardiolipin biosynthesis in H9c2 cardiac myoblasts by cytidine 5′-triphosphate. J. Biol. Chem. 1996;271:25810–25816. doi: 10.1074/jbc.271.42.25810. [DOI] [PubMed] [Google Scholar]
- 118.Savci V., Wurtman R.J. Effect of cytidine on membrane phospholipid synthesis in rat striatal slices. J. Neurochem. 1995;64:378–384. doi: 10.1046/j.1471-4159.1995.64010378.x. [DOI] [PubMed] [Google Scholar]
- 119.Farber S.A., Slack B.E., Blusztajn J.K. Acceleration of phosphatidylcholine synthesis and breakdown by inhibitors of mitochondrial function in neuronal cells: A model of the membrane defect of Alzheimer’s disease. FASEB J. 2000;14:2198–2206. doi: 10.1096/fj.99-0853. [DOI] [PubMed] [Google Scholar]
- 120.James S.J., Miller B.J., McGarrity L.J., Morris S.M. the effect of folic acid and/or methionine deficiency on deoxyribonucleotide pools and cell cycle distribution in mitogen‐stimulated rat lymphocytes. Cell Prolif. 1994;27:395–406. [Google Scholar]
- 121.Sevigny J., Chiao P., Bussière T., Weinreb P.H., Williams L., Maier M., Dunstan R., Salloway S., Chen T., Ling Y., O’Gorman J., Qian F., Arastu M., Li M., Chollate S., Brennan M.S., Quintero-Monzon O., Scannevin R.H., Arnold H.M., Engber T., Rhodes K., Ferrero J., Hang Y., Mikulskis A., Grimm J., Hock C., Nitsch R.M., Sandrock A. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature. 2016;537:50–56. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- 122.Doody R.S., Thomas R.G., Farlow M., Iwatsubo T., Vellas B., Joffe S., Kieburtz K., Raman R., Sun X., Aisen P.S., Siemers E., Liu-Seifert H., Mohs R. Alzheimer’s disease cooperative study steering committee; solanezumab study group. Phase 3 trials of solanezumab for mild-to-moderate Alzheimer’s disease. N. Engl. J. Med. 2014;370:311–321. doi: 10.1056/NEJMoa1312889. [DOI] [PubMed] [Google Scholar]
- 123.Anand K., Sabbagh M. Early investigational drugs targeting tau protein for the treatment of Alzheimer’s disease. Expert Opin. Investig. Drugs. 2015;24:1355–1360. doi: 10.1517/13543784.2015.1075002. [DOI] [PMC free article] [PubMed] [Google Scholar]