Watch a video presentation of this article
Abbreviations
- cccDNA
covalently closed circular DNA
- CHB
chronic HBV
- HBeAg
HBV secreted e antigen
- HBsAg
HBV surface antigen
- HBV
hepatitis B virus
- IFN
interferon
- NA
nucleos(t)ide analogue
- pgRNA
pregenomic RNA
- Pol
polymerase
- rcDNA
relaxed circular DNA
Clinicians rely on hepatitis B virus (HBV) biomarkers in patient serum to monitor infection in the liver. Serum HBV DNA is used to track viral replication within infected hepatocytes. Development of antibodies to HBV surface antigen (HBsAg) and the secreted e antigen (HBeAg) mark important immune control milestones and inform treatment decisions in chronic HBV (CHB) infections. Recently, it has been appreciated that HBV pregenomic RNA (pgRNA) is also found in serum1, 2; the potential relevance of pgRNA as a biomarker during CHB infection is currently the subject of investigation.
The ability of HBV to sustain chronic infection is due in part to the maintenance of the HBV DNA genome within the hepatocyte nucleus in the form of a covalently closed circular DNA (cccDNA) mini‐chromosome. cccDNA drives infection by serving as the template for most HBV RNAs, including pgRNA and mRNAs (Fig. 1). pgRNA is required for synthesis of the HBV relaxed circular DNA (rcDNA) genome found in the mature virus. Within the cytoplasm, the HBV capsid protein (Core) assembles around an HBV polymerase (Pol) and a single pgRNA copy to form the nascent virion. Within the capsid, HBV DNA is reverse transcribed by Pol (with concomitant pgRNA degradation). Following envelopment, the DNA viruses are released from the cell. Serum HBV DNA level is indicative of the extent of viral replication and risk for transmission, because each copy of DNA represents an infectious virus.
Figure 1.
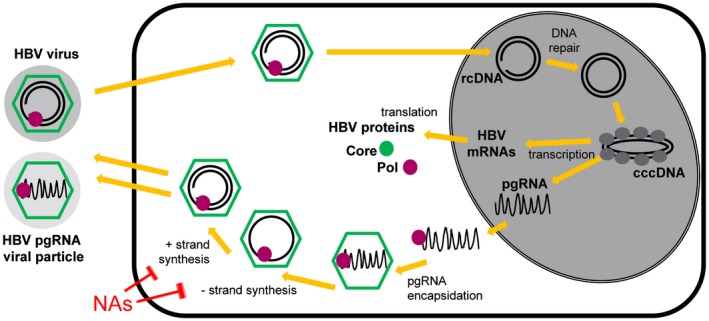
HBV life cycle. HBV is uncoated on entry and the partially double‐stranded rcDNA enters the nucleus. Host DNA repair machinery completes the partial strand. Host histones (gray circles) associate with cccDNA. HBV mRNAs and pgRNA are transcribed from cccDNA and are sent to the cytoplasm. HBV proteins, including Core (green) and Pol (mauve), are translated from mRNAs. Pol associates with the pgRNA; the complex is encapsidated by Core. Within the Core capsid, Pol reverse transcribes the minus (−) strand DNA from pgRNA, then the plus (+) strand. The completed virion is enveloped (not shown) and exits the cell. It is unknown how pgRNA‐containing encapsidated particles exit. NAs block Pol activity (red text).
Encapsidated pgRNA is also found in the serum. The presence of pgRNA‐containing viral particles in serum is part of the HBV life cycle as demonstrated in studies of longitudinal samples from patients with untreated acute and chronic hepatitis B3 and may be influenced by HBV genotype.4 During acute infection, HBV DNA is detectable first and pgRNA becomes detectable after a short lag. pgRNA levels correlate with viral load4 and are consistently ~2 log lower than DNA in the absence of treatment regardless of whether infection resolves or persists (Fig. 2A and 2B).3 This suggests a model wherein exit of pgRNA‐containing viral particles from the hepatocyte consistently occurs in parallel with DNA‐containing particles. Whether pgRNA‐containing particles are infectious has not been fully established, but recently published evidence suggests that these particles lack replicative proficiency.5
Figure 2.
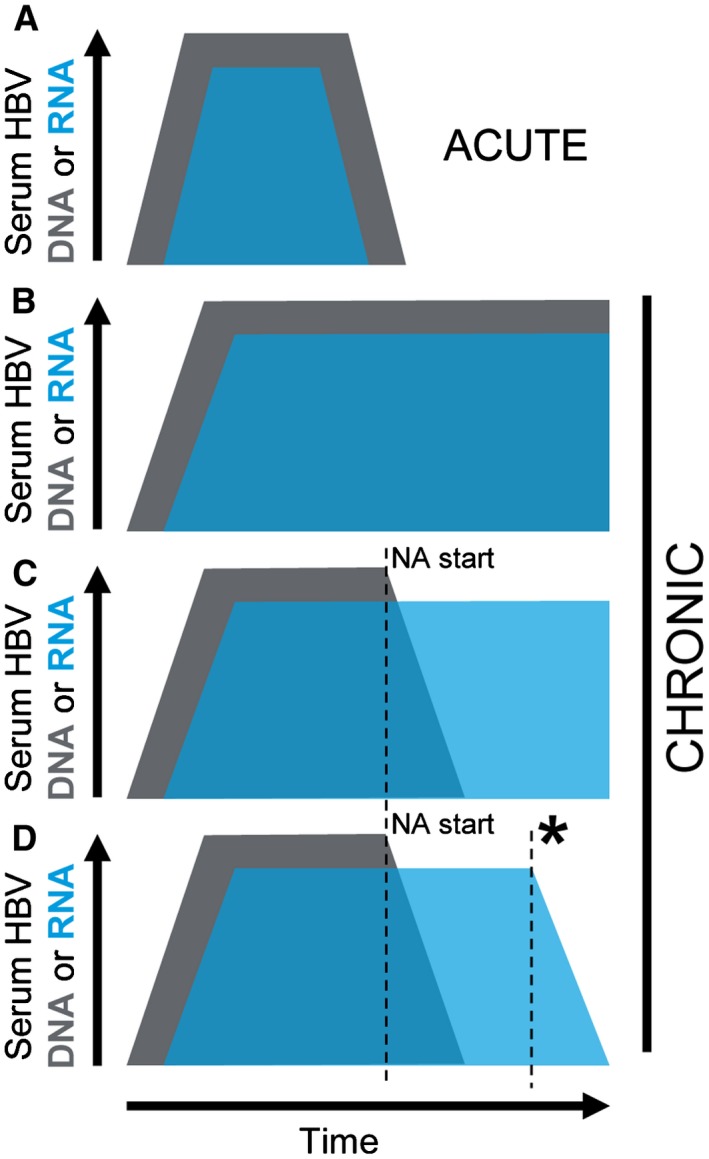
Schematic of HBV DNA and RNA kinetics during acute or chronic infection. Relative DNA (gray) and pgRNA (blue) concentrations in the serum during HBV infection. In acute infection (A), increasing viral replication results in elevated HBV DNA until spontaneous resolution and loss of DNA detection. pgRNA follows the same kinetics but remains consistently lower than the DNA viral load. During chronic infection, HBV DNA and pgRNA levels can remain detectable over time (B). Commencing treatment with NAs (NA start) can rapidly reduce DNA (C, D) while pgRNA remains detectable. Decreasing cccDNA activity (asterisk) would result in reduced pgRNA levels (D).
CHB infection cannot be fully cured because of the persistence of HBV cccDNA in the hepatocyte nuclei even after establishment of host immune control.6 Thus, treatment of patients with CHB aims to attain a “functional” cure of infection wherein immune control is attained and cccDNA activity is minimal. Directly monitoring cccDNA activity within the hepatocyte would provide a direct assessment of infection status during treatment of CHB but would require repeat liver biopsies. Treatment options for CHB are currently limited to two classes of drugs: (1) pegylated interferon (IFN) or (2) nucleos(t)ide analogues (NAs). IFN is efficacious in inducing immune control; however, long‐term (often lifelong) treatment with NAs is often favored because of relative lack of side effects. NAs suppress viral replication by blocking DNA synthesis from the pgRNA template (Fig. 1). Whereas NA treatment reduces HBV DNA to undetectable levels, pgRNA is generally still present in the serum because NAs do not impact pgRNA transcription or encapsidation (Figs. 1 and 2C).1, 7
Serum HBV RNA levels correlate to intrahepatic pgRNA, liver histopathology,8, 9 and cccDNA,9 supporting a potential utility for serum pgRNA in indirectly monitoring cccDNA activity during NA treatment. However, the use of this marker during NA treatment in predicting clinical outcomes remains undemonstrated, although the following model of pgRNA utility during NA treatment could be extrapolated: a subject with undetectable HBV DNA but consistently elevated pgRNA presumably has active cccDNA transcription occurring (Fig. 2C); such a subject would be expected to respond poorly to termination of treatment. In contrast, decreasing cccDNA activity would likely manifest as decreased detection of serum pgRNA (Fig. 2D). Two studies have demonstrated that HBeAg‐positive status is linked with elevated levels of serum HBV RNA4 both off and on treatment,3 consistent with higher levels of cccDNA transcriptional activity. HBV pgRNA has shown potential in predicting HBeAg loss.10 Treatment guidelines allow NA treatment termination where HBsAg loss occurs or, in some cases, HBeAg loss and/or sustained loss of HBV DNA detection.11 Loss of pgRNA detection in subjects with undetectable HBV DNA could potentially serve as a new NA treatment endpoint alone or in conjunction with other markers. In HBV‐related hepatocellular carcinoma tumor biopsies, elevated intracellular pgRNA was correlated with better prognosis12; whether pgRNA serum levels have predictive utility in this type of patient cohort should also be examined. In conclusion, monitoring of pgRNA during treatment of patients with CHB may have clinical value for stratifying NA treatment responses in patients with CHB when HBV DNA synthesis is suppressed. Studies are needed to define the potential predictive utility of pgRNA during treatment with NA, as well as with newer, novel therapies and treatment regimens.
Potential conflict of interest: All authors are employees and shareholders of Abbott Laboratories.
References
- 1. Wang J, Shen T, Huang X, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol 2016;65:700‐710. [DOI] [PubMed] [Google Scholar]
- 2. Jansen L, Kootstra NA, van Dort KA, et al. Hepatitis B virus pregenomic RNA is present in virions in plasma and is associated with a response to pegylated interferon alfa‐2a and nucleos(t)ide analogues. J Infect Dis 2016;213:224‐232. [DOI] [PubMed] [Google Scholar]
- 3. Butler E, Gersch J, McNamara A, et al. Hepatitis B virus serum DNA and RNA levels in nucleos(t)ide analog‐treated or untreated patients during chronic and acute infection. Hepatology. 10.1002/hep.30082 [DOI] [PubMed] [Google Scholar]
- 4. van Campenhout MJH, van Bömmel F, Pfefferkorn M, et al. Host and viral factors associated with serum hepatitis B virus RNA levels among patients in need for treatment. Hepatology 2018;68:839‐847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J, Sheng Q, Ding Y, et al. HBV RNA virion‐like particles produced under nucleos(t)ide analogues treatment are mainly replication‐deficient. J Hepatol 2018;68:847‐849. [DOI] [PubMed] [Google Scholar]
- 6. Kumar R, Pérez‐Del‐Pulgar S, Testoni B, et al. Clinical relevance of the study of hepatitis B virus covalently closed circular DNA. Liver Int 2016;36(Suppl. 1):72‐77. [DOI] [PubMed] [Google Scholar]
- 7. Lok AS, Zoulim F, Dusheiko G, et al. Hepatitis B cure: From discovery to regulatory approval. J Hepatol 2017;67:847‐861. [DOI] [PubMed] [Google Scholar]
- 8. Wang J, Yu Y, Li G, et al. Relationship between serum HBV‐RNA levels and intrahepatic viral as well as histologic activity markers in entecavir‐treated patients. J Hepatol. 10.1016/j.jhep.2017.08.021 [DOI] [PubMed] [Google Scholar]
- 9. Wang J, Yu Y, Li G, et al. Natural history of serum HBV‐RNA in chronic HBV infection. J Viral Hepat 2018;25:1038‐1047. [DOI] [PubMed] [Google Scholar]
- 10. van Bömmel F, Bartens A, Mysickova A, et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology 2015;61:66‐76. [DOI] [PubMed] [Google Scholar]
- 11. Terrault NA, Bzowej NH, Chang KM, et al. American Association for the Study of Liver Diseases. AASLD guidelines for treatment of chronic hepatitis B. Hepatology 2016;63:261‐283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Halgand B, Desterke C, Rivière L, et al. Hepatitis B virus pregenomic RNA in hepatocellular carcinoma: A nosological and prognostic determinant. Hepatology 2018;67:86‐96. [DOI] [PubMed] [Google Scholar]


