Abstract
Background:
Molecular epidemiological study of human immunodeficiency virus drug-resistant (HIVDR) markers is challenging in areas where the dominant subtype is non-B.
Objective:
Here we provide molecular data for HIVDR in the CRF01_AE subtype in Bali, Indonesia.
Method:
Seventy patients were enrolled in this study and grouped into treatment failure and treatment naïve groups. The full-length pol gene was amplified using nested reverse transcriptase polymerase chain reaction and the product was then sequenced. The readable sequence was then subjected to Stan-ford HIV Drug Resistance Database genotyping.
Results:
We found that clinical classification was in accordance with the presence of HIVDR markers in the pol gene. Independent of therapy history, the treatment failure group showed resistance markers against nucleoside reverse transcriptase inhibitors (NRTI) and non-nucleoside reverse transcriptase in-hibitors (NNRTI), ranging from 72%–100% of patients. Only a small proportion of naïve patients harbored HIV with drug resistance markers to NNRTI. No protease inhibitor-resistant marker was found in either patient group. Molecular marker mutations, which were found in more than 50% of treatment failure patients, were M184V (100%), T215A/Y/F (88.2%), D67N/G (76.5%), and M41L (58.8%).
Conclusion:
The protocol used in this study to determine genetic markers of HIVDR based on sub-type B can be applied for the rapid determination of resistance of the CRF01_AE subtype. All patients with progressive clinical signs and increased viral load should be recommended to undergo second-line treatment of the ARV regimen.
Keywords: Human Immunodeficiency Virus (HIV), CRF01_AE, treatment failure, naïve, Bali, nucleoside reverse transcriptase inhibitors (NRTI)
1. INTRODUCTION
Global surveillance of antiretroviral (ARV) drug-resistant human immunodeficiency virus (HIVDR) needs to be conducted. Effective ARV therapy is essential for HIV infected patients to reduce acquired immunodeficiency syndrome (AIDS) progression and prolong patient life expectancy. The pandemic emergence of HIVDR throughout the world has affected all age groups, including children [1, 2]. Drug resistance leads to treatment failure, resulting in patient death. The epidemiological impact can be disastrous as HIVDR may be transmitted to other patients and transmitted drug-resistant (TDR) strains have been reported in both developed and developing countries [3-5].
Mutations in the reverse transcriptase (RT) gene complex have been used to understand the genetic mechanism of HIVDR. This is particularly valid for the HIV-B subtype that predominantly circulates in America, Europe, and Australia [6]. However, the most globally prevalent subtype is HIV-1 C [7, 8], and the most widespread circulating subtype in Indonesia is CRF01_AE [9-11]. Molecular epidemiological study of HIV resistance is likely to be problematic in areas where the dominant subtype is non-B, as various reports have shown that molecular markers related to drug resistance are less valid for non-B subtypes [12].
Bali is a unique hotspot in understanding molecular markers of HIVDR subtype CRF01_AE. This major tourist destination is facing a rapidly growing HIV epidemic [13-15]. The island receives up to five million international visitors annually. Unfortunately, in Indonesia, knowledge of the disease is low and social stigma is high, meaning that only 5–10% of HIV/AIDS sufferers actually get diagnosed and treated [16, 17]. Additionally, HIVDR patients often have a tendency to maintain a high-risk lifestyle [18, 19], further contributing to TDR.
2. MATERIAL AND METHOD
2.1. Ethical Clearance
All subjects received a complete explanation of this study and agreed to sign an accompanying informed consent. Written informed consent was obtained from the patients or their families (in cases of patients under the age of 18 years) before the samples and patient data were collected. Ethical clearance for this study was granted by the Research Ethics Committee of the Faculty of Medicine, Udayana University, Denpasar, Bali, Indonesia. The inclusion criteria were patients who (1) underwent ARV therapy and showed increased viral load, or (2) were about to commence the ARV regimen, (3) the HIV-1 CRF01_AE subtype was confirmed, and (4) agreed to participate and signed the informed consent.
2.2. Study Design
This study was an observational cross-sectional analysis performed in 2008–2010. The target populations were HIV-AIDS patients undergoing ARV therapy who showed an increased viral load of >750 copies/ml (treatment failure group) and HIV patients who were about to start the ARV regimen (naïve group) at the Voluntary Consulting and Testing (VCT) clinic in Sanglah Hospital, Denpasar. Viral load was determined by reverse-transcriptase and polymerase chain reaction (RT-PCR) and DNA probe quantification of the product using the Amplicor HIV-1 Monitor kit (Roche Diagnostics GmbH, Mannheim, Germany).
2.3. Sample Collection
Five milliliters of venous blood were collected from all subjects who met the inclusion criteria, using Ethylene Diamine Tetra Acetic Acid as the anti-coagulant. Plasma was separated following clarification using centrifugation at 3000 rpm for 10 minutes.
2.4. RNA Isolation and Pol Gene Amplification
Total RNA was purified from plasma using a QIAamp Viral RNA Mini Kit (Qiagen Corporation, Germany) according to the manufacturer’s instructions. The whole RT gene in plasma was amplified using RT-PCR and nested PCR. In the first round, RT-PCR was conducted using Reverse Transcriptase-PCR SuperscriptTM III One-Step RT-PCR System with Platinum® Taq DNA Polymerase (Invitrogen® Carlsbad, CA) with an HIV-1F and HIV-1R primer set [20]. Nested PCR was conducted using Platinum® Taq DNA Polymerase High Fidelity (Invitrogen®) with an HIVGRT-2F2 and HIVGRT-2R primer set [20].
2.5. Sequencing
Sequencing of the nested PCR product was conducted at the Virology and Cancer Pathobiology Research Center (VCPRC) Faculty of Medicine Indonesia University, Jakarta, Indonesia. Sequencing was done using the BigDye Terminator (Thermo Fisher, Waltham, MA) following the manufacturer’s instructions, along with published primers [20]. Capillary electrophoresis of the reaction was performed in an Applied Biosystems 310 Sequencer (Applied Biosystem, Foster City, CA). Sequencing results were aligned using Viroseq® (Illinois, USA) or Mega4 [21] and the final sequence was subjected to Stanford HIV Drug Resistance Database genotyping (https://hivdb.stanford.edu).
2.6. Statistical Analysis
Variable normality of the subject demographic data was tested using the Shapiro-Wilk normality test [22]. Quantitative statistics were calculated by the Mann-Whitney test. Non-parametric statistics were calculated using the Chi-Square test. All calculations were conducted using the Statistical Package for the Social Sciences 16.0 (SPSS Inc., Chicago, IL).
3. RESULT
Seventy HIV patients were enrolled in this study. Thirty-four (48.6%) subjects were in the treatment failure group and 36 (51.5%) were treatment-naïve patients. The HIV RT gene could be amplified and sequenced in 48 of the total number of patients. From the positive samples, one was confirmed as HIV subtype A and the remaining 47 (97.9%) as HIV-1 CRF01_AE subtype. Ultimately, the numbers of samples belonging to treatment failure and treatment naïve groups were 17 and 30, respectively. All sequences from this study have been deposited in GenBank with accession numbers of KY927941–KY927982. The sequences covered nucleotide no 1252 – 2953 based on reference CRF01_AE/B of CM240, GenBank Acc. No. AF516184 [23].
Demographic data of ARV treatment failure and naïve patients included in this study, incorporating risk factors, viral load, name and duration of the ARV received, and treatment failure discovery, are available in Supplementary Material 1 and 2. A summary of both data sets is presented in Table 1. The results show that the groups were not statistically different (p>0.05) in sex, age, and viral load parameters. However, the risk factor for the two groups differed significantly (p=0.018). The predominant at-risk group within the treatment-naïve group was heterosexuals (86.7%), while those present in the treatment failure group were heterosexuals (55.6%), intravenous drug users (27.8%), and perinatal infected infants (16.7%)
Table 1.
Summary of demographic data of ARV treatment failure and naïve of HIV-1-CRF01_AE positive patients in Bali, Indonesia 2008 – 2010*.
| Variable | ARV Treatment Failure Patients | Naïve Patients | P |
|---|---|---|---|
| Number | 18 | 30 | |
| Sex | 0.296 | ||
| Male | 14 (77.8%) | 19 (63.3%) | |
| Female | 4 (22.2%) | 11 (36.7%) | |
| Age | 0.56 | ||
| 5-15 years | 3 (16.7%) | - | |
| 16-44 years | 15 (83.3%) | 26 (86.7%) | |
| 45-65 years | - | 4 (13.3%) | |
| Age average value | 31.6 | 35.2 | |
| Age range | 5 – 48 | 27 – 50 | |
| Duration (days) | |||
| ARV initiation since date of positive detection | 1 – 870 | 1 – 2,370 | |
| Mean | 146.6 | 324.3 | 0.163 |
| ARV treatment duration to drug failure discovery | 420 – 2,130 | - | |
| Mean | 1,077.20 | - | |
| Viral Load (copy/ml) | 0.354 | ||
| Range | 6,700 – 750,000 | 2,345 – 750,000 | |
| Risk factor | 0.018 | ||
| Perinatal infection | 3 (16.7%) | - | |
| Tattoo + heterosexual | - | 1 (3.3%) | |
| IVDU** | 5 (27.8%) | 1 (3.3%) | |
| IVDU + heterosexual | - | 1 (3.3%) | |
| Heterosexual | 10 (55.6%) | 26 (86.7%) | |
| Heterosexual spouse | - | 1 (3.3%) |
*The data of patients, whom the sequences could not be obtained were excluded; **IVDU: Intra-venous drug user;
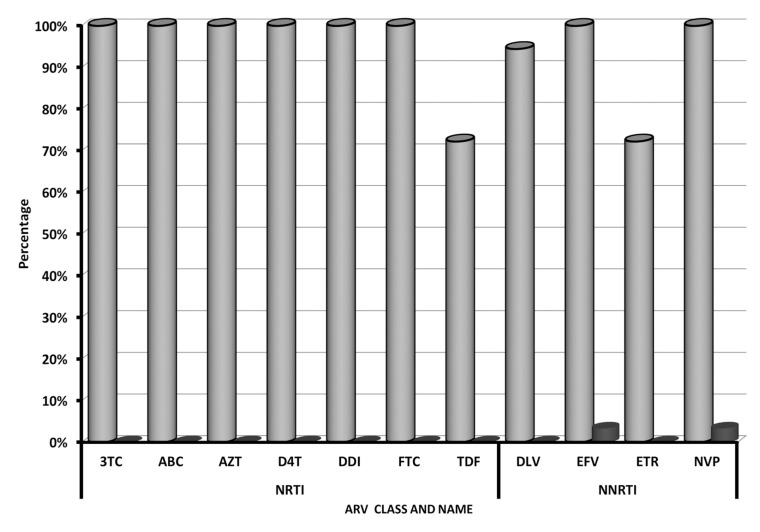
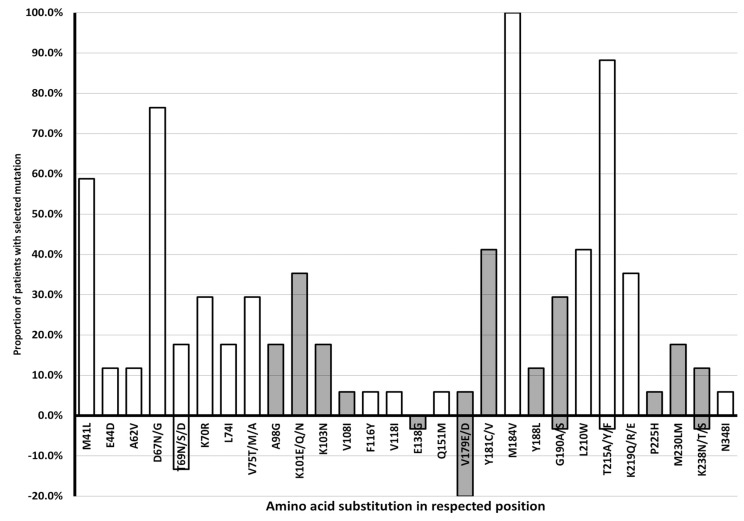
Percentage of HIVDR genotype of HIV-1 of CRF01_AE subtype to various Nucleoside Reverse Transcriptase Inhibitor (NRTI) and Non-Nucleoside Reverse Transcriptase Inhibitor (NRTI) in treatment failure and naïve patient groups is shown in Fig. (1). This result indicated that treatment failure patients harbor HIV with resistance markers to nucleoside reverse transcriptase inhibitors (NRTI) and non-nucleoside reverse transcriptase inhibitors (NNRTI), ranging from 72%–100% of patients, while only a small proportion of naïve patients harbor HIV with NNRTI drug-resistant markers. No protease inhibitor (PI) resistant marker was found in either patient group. Percentage of HIV-1 of CRF01_AE subtype in treatment failure and naïve patient groups harboring deduced amino-acid substitutions that generate HIVDR genotype to NRTI and NNRTI is presented in Fig. (2). The result shows that molecular marker mutations, which were found in more than 50% of treatment failure patients, were M184V (100%), T215A/Y/F (88.2%), D67N/G (76.5%), and M41L (58.8%).
Fig. (1).
Percentage of HIVDR genotype of HIV-1 of CRF01_AE subtype to various Nucleoside Reverse Transcriptase Inhibitor (NRTI) and Non-Nucleoside Reverse Transcriptase Inhibitor (NRTI) in treatment failure and naïve patient groups in Bali, Indonesia 2008 – 2010. Grey blocks: treatment failure group; Black blocks: naïve group; NRTI: lamivudine (3TC), Abacavir (ABC), Zidovudine (AZT), Stavudine (D4T), Didanosine (DDI), Emtricitabine (FTC), and Tenofovir (TDF); NNRTI: Delavirdine (DLV), Efavirenz (EFV), Etravirine (ETR), Nevirapine (NVP).
Fig. (2).
Percentage of HIV-1 of CRF01_AE subtype in treatment failure and naïve patient groups in Bali, Indonesia 2008 – 2010, with deduced amino-acid substitutions that generate HIVDR genotype to NRTI and NNRTI. Treatment failure: above the axis; naïve: below the axis; HIVDR genotype to NRTI: unfilled bars; HIVDR genotype to NNRTI: filled bars. The numbering was annotated by The Stanford HIV Drug Resistance Database genotyping (https://hivdb.stanford.edu).
Comparison of the percentage of amino acid substitutions in CRF01_AE subtype in treatment failure group in Bali – Indonesia and global CRF01, B, and C subtypes n NRTI±NNRTI Treated Patients available in HIV Drug Resistance Database of Stanford University (https://hivdb.stanford.edu/pages/surveillance.html; accessed January 26, 2019) is presented in Table 2. The result shows that the major substitutions (percentage >50%) of M41, D67, M184, and T215 were higher than all subtype data, while the K103 substitution was less frequent in our study.
Table 2.
Percentage of amino acid substitutions in CRF01_AE subtype in the treatment failure group in Bali – Indonesia and Global CRF01, B, and C subtypes available in HIV Drug Resistance Database of Stanford University.*
| Pos. | CR | Subtype | |||
|---|---|---|---|---|---|
| CRF01_AE (This Study) | Global CRF01 | B | C | ||
| 41 | M | 58.8 | 19.0 | 40.0 | 9.2 |
| 44 | E | 11.8 | NA | NA | NA |
| 62 | A | 11.8 | NA | NA | NA |
| 67 | D | 76.5 | 33.0 | 37.1 | 18.8 |
| 69 | T | 17.6 | 3.3 | 10.2 | 3.1 |
| 70 | K | 29.4 | 21.7 | 22.6 | 13.7 |
| 74 | L | 17.6 | 6.8 | 16.6 | 6.4 |
| 75 | V | 29.4 | 17.2 | 8.4 | 5.7 |
| 98 | A | 17.6 | 10.1 | 6.3 | 5.9 |
| 101 | K | 35.3 | 18.6 | 9.5 | 10.5 |
| 103 | K | 17.6 | 42.6 | 51.0 | 76.1 |
| 108 | V | 5.9 | NA | NA | NA |
| 116 | F | 5.9 | 4.4 | 2.2 | 1.4 |
| 118 | V | 5.9 | NA | NA | NA |
| 138 | E | NA | 5.4 | 4.4 | 9.7 |
| 151 | Q | 5.9 | 6.8 | 3.0 | 2.1 |
| 179 | V | 5.9 | 4.4 | 3.6 | 6.1 |
| 181 | Y | 41.2 | 41.4 | 21.0 | 15.8 |
| 184 | M | 100 | 78.7 | 77.1 | 69.6 |
| 188 | Y | 11.8 | 5.9 | 5.5 | 9.0 |
| 190 | G | 29.4 | 41.0 | 25.4 | 24.0 |
| 210 | L | 41.2 | 11.9 | 27.5 | 3.3 |
| 215 | T | 88.2 | 32.7 | 53.6 | 16.6 |
| 219 | K | 35.3 | 22.6 | 27.8 | 12.3 |
| 225 | P | 5.9 | 3.9 | 3.2 | 7.7 |
| 230 | M | 17.6 | 3.4 | 1.1 | 3.5 |
| 238 | K | 11.8 | 2.3 | 2.6 | 2.3 |
| 348 | N | 5.9 | 6.5 | 13.0 | 15.1 |
* In NRTI±NNRTI Treated Patients Available in https://hivdb.stanford.edu/pages/surveillance.html; NA: not available; CR: consensus residue
DISCUSSION
As expected, almost all of the HIV-1 identified individuals in this study harbored CRF01_AE subtype, which is the most common circulating subtype in Indonesia [9, 11, 24]. Only one was of A subtype. Along the RT gene, we found unambiguous sequence reads in many positions with a very clean background. The primer set used for sequencing allowed us to identify nucleotides with multiple signals. In some cases, we also repeated the sequencing to confirm the signals. This is a hallmark of the viral quasi-species concept
[25-34]. The reads covered nucleotide no 1252 – 2953 based on reference CRF01_AE/B of CM240, GenBank Acc. No. AF516184 [23]. This should include viral protease, reverse transcriptase, and RT-thumb domain of HIV-1 [35], calculated from the ribosomal slippage site of TTTTTTAG [36].
Global genotyping seems valid for the HIV-1 CRF01_AE subtype. There have been very few reports regarding polymorphisms and molecular drug resistance markers in this subtype, although protease polymorphism has been described [37, 38]. Patterns of point mutations associated with ARV treatment failure in the CRF01_AE subtype likely differ from that of B subtype [39]. Despite this, we found that there was a strong concordance of molecular markers that were initially established for type B with the clinical signs of the patients in our study. All patients exhibiting signs of treatment failure expressed resistance marker mutations following drug resistance genotyping in the Stanford database.
However, some patterns seem unique to our study. Comparison of incidence of amino acid substitutions in our study to global CFR, B, and C subtypes data shows that the major substitutions (percentage of 50% or higher) of M41, D67, M184, and T215 were higher than all subtype data, while the K103 substitution was less frequent in our study. Substitution of 138E was not found in our study, while others of 44E, 62A, 108V, and 118V were not available in the database. Unique host factors might contribute in replication rate of the HIV-1, therefore its mutation pattern varies. There has been yet no clear evidence to prove this possibility. Host factors have been indicated to contribute to HIV-1 replication [40-42]. The influence of host factors in susceptibility to HIV infection and AIDS progression, but not in HIV mutation pattern, has been described elsewhere [43-46].
All treatment failure subjects harbored resistance to NRTI and NNRTI, which are first-line ARV drugs. The pattern is concordance to a recent report from North Sulawesi, Indonesia [47]. The figure is higher than that reported in South Africa [48]. The group included three pediatric patients who underwent 6.5 years of ARV therapy. The presence of HIVDR strains in children has been reported elsewhere, such as Brazil [49, 50] and France [51], with varying degrees of resistance to NRTI and NNRTI, reaching between 30–80% of patients. The low number of children enrolled in this study might cause the observed difference.
Cross-resistance in HIV-1 subtypes is obvious from this study. Independent of therapy history, the pol gene sequences found in all treatment failure subjects contained all NRTI and NNRTI resistant markers (Fig. 1), with the prevalence of each molecular marker being 72–100%. All subjects within the treatment failure group received two NRTI and one NNRTI drugs, available during the study period. The NRTIs used were lamivudine and zidovudine, although stavudine was given to zidovudine-allergic patients. The NNRTI was either nevirapine or efavirenz. Of note, no patient was given abacavir, didanosine, emtricitabine or tenofovir, yet genotypic testing showed that all treatment failure subjects were 100% resistant to these drugs despite never having received them, with the exception of resistance to tenofovir, which reached 72.2%. These data demonstrate that cross-resistant HIV is not uncommon [52-59].
We found no TDR in our study. Only a small proportion of naïve patients harbored HIV with drug-resistant markers to NNRTI. This may be unique to this study or to the subtype in question. Transmitted drug resistant strains have been observed elsewhere [4, 60-65], including in Indonesia [66, 67]. The TDR prevalence ranges from 9–25% in Europe and the United States [7]. This might be related to the length of implementation of the ARV program [12]. The program was introduced in 2002–2004 in developing countries, including Indonesia. The resistant markers found in the treatment naïve group are most likely not TDR and seem merely due to genetic variation of HIV through the high mutation rate of the virus [68-72]. The three cases of HIVDR in children were not TDR either, as all of them underwent ARV treatment during the previous five years (Table 1).
Thymidine analogue mutations (TAMs) of cluster 1 were dominant in our study. Codon polymorphisms were found throughout the RT gene in both groups. Significant differences exist in codon number 41, 67, 70, 75, 184, 210, 215 and 219. Except for codon 75, all others belong to TAM codons, while codon 75 is a nucleoside analogue mutation (NAM) [73, 74]. Molecular markers of TAMs, M184V (100%), T215A/Y/F (88.2%), D67N/G (76.5%), and M41L (58.8%), were found in more than 50% of treatment failure patients. These TAMs were found in the treatment failure group but not in any member of the treatment naïve group. The emergence of TAMs correlates with the use of NRTIs. There are two clusters of TAMs; cluster 1 includes substitutions of M41L, L210W and T215Y, while cluster 2 of D67N, K70R, T125F and K219Q/E/N [75]. The phenotypic picture of cluster 2 mutations is less resistant than cluster 1 [76]. Therefore, the treatment failure in this study is mostly due to cluster 1 TAMs.
The pattern of resistance-associated mutations found in this study differs to that found in Maumere, East Nusa Tenggara, except for M184V [11]. This discrepancy might relate to the subject of the genotypic study. We were targeting naïve as well as treatment failure patients, while the other study did not explain if the subjects were naïve or treatment failure patients [11]. The different time frame of sampling might also cause this dissimilar observation.
There are two major drawbacks of this study. Firstly, the success rate of RT-PCR amplification and sequencing is only around 70% of the samples. This might be because of the quality and quantity of RNA as well as the great variation in primer binding sites. A better approach could be the sequencing of pro-viral DNA [77] and application of next-generation sequencing [78]. The second limitation is the lack of facilities to conduct phenotypic testing to provide in vitro evidence. Different approaches to HIVDR should be applied and in-country HIVDR phenotyping infrastructure should be established, including in Indonesia.
That the international database applied in this study is in agreement with the clinical manifestations of subjects with treatment failure demonstrates that the protocol can be recommended for immediate determination of resistance genotype, and ideally, the HIVDR genotyping should combine pro-viral DNA and viral RNA amplification. All patients with progressive clinical signs and increased viral load should be recommended to undergo second line ARV therapy, as no protease inhibitor (PI) resistant marker was found in the treatment failure group.
In conclusion, clinical classification was in concordance with the presence of HIVDR markers in the pol gene. Independent of therapy history, the treatment failure group showed resistance markers against nucleoside reverse transcriptase inhibitors (NRTI) and non-nucleoside reverse transcriptase inhibitors (NNRTI), ranging from 72%–100% of patients. Only a small proportion of naïve patients harbored HIV with drug resistance markers to NNRTI. Molecular marker mutations in more than 50% of treatment failure patients were M184V, T215A/Y/F, D67N/G, and M41L. The protocol used in this study to determine genetic markers of HIVDR based on subtype B can be applied for the rapid determination of resistance the CRF01_AE subtype. All patients with progressive clinical signs and increased viral load should be recommended to undergo second-line treatment of the ARV regimen.
CONCLUSION
Clinical classification of patients to treatment failure was in concordance with the presence of HIVDR markers in the pol gene. Independent of therapy history, the treatment failure group showed resistance markers against nucleoside reverse transcriptase inhibitors (NRTI) and non-nucleoside reverse transcriptase inhibitors (NNRTI), ranging from 72%–100% of patients. Only a small proportion of naïve patients harbored HIV with drug resistance markers to NNRTI. Molecular marker mutations in more than 50% of treatment failure patients were M184V, T215A/Y/F, D67N/G, and M41L.
Acknowledgements
This study was part of the doctoral dissertation of Nyoman Sri Budayanti at the Post Graduate Program of Medicine of Udayana University, Bali, Indonesia. The late Prof. Dr. Ketut Suata supervised the study. The funding was partly from the TREAT Asia Studies to Evaluate Resistance (TASER) project and Minister of Health Indonesia funding to the Virology and Cancer Pathobiology Research Center (VCPRC) Faculty of Medicine Indonesia University, Jakarta, Indonesia. We thank Mrs. Aroem Naroeni, PhD. and Mrs. Silvia Tri Midyaningtyas of VCPRC for lab assistance. The manuscript has been through professional English editing at Edanz Editing (https://www.edanzediting.com).
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers web site along with the published article.
1. Demography of ARV treatment failure, including risk factors, viral load, name and duration of the ARV, and treatment failure discovery of HIV-1 of CRF01_AE subtype positive patients in Bali, Indonesia 2008 – 2010.
2. Demography of ARV-naïve, including risk factors and viral load of HIV-1 of CRF01_AE positive patients in Bali, Indonesia 2008 – 2010.
Ethics Approval and Consent to Participate
The study was approved by the Research Ethics Committee of the Faculty of Medicine, Udayana University, Denpasar, Bali, Indonesia.
Human and Animal Rights
No animals were used in the study. All humans research procedures followed were in accordance with the standards set forth in the Declaration of Helsinki principles of 1975, as revised in 2008 (http://www.wma.net/en/20activities/ 10 ethics/10helsinki/).
Consent for Publication
Written informed consent was obtained from the patients or their families (in cases of patients under the age of 18 years) before the samples and patient data were collected.
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Rojas Sanchez P., Holguin A. Drug resistance in the HIV-1-infected paediatric population worldwide: a systematic review. J. Antimicrob. Chemother. 2014;69(8):2032–2042. doi: 10.1093/jac/dku104. [DOI] [PubMed] [Google Scholar]
- 2.Hamers R.L., Kityo C., Lange J.M., et al. Global threat from drug resistant HIV in sub-Saharan Africa. BMJ. 2012;344:e4159. doi: 10.1136/bmj.e4159. [DOI] [PubMed] [Google Scholar]
- 3.Rhee S.Y., Blanco J.L., Jordan M.R., et al. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis. PLoS Med. 2015;12(4):e1001810. doi: 10.1371/journal.pmed.1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferreira A.C.G., Coelho L.E., Grinsztejn E., et al. Transmitted drug resistance in patients with acute/recent HIV infection in Brazil. Braz. J. Infect. Dis. 2017;21(4):396–401. doi: 10.1016/j.bjid.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jayaraman G.C., Archibald C.P., Kim J., et al. A population-based approach to determine the prevalence of transmitted drug-resistant HIV among recent versus established HIV infections: results from the Canadian HIV strain and drug resistance surveillance program. J. Acquir. Immune Defic. Syndr. 2006;42(1):86–90. doi: 10.1097/01.qai.0000196666.16616.fe. [DOI] [PubMed] [Google Scholar]
- 6.Abecasis A.B., Wensing A.M., Paraskevis D., et al. HIV-1 subtype distribution and its demographic determinants in newly diagnosed patients in Europe suggest highly compartmentalized epidemics. Retrovirology. 2013;10:7. doi: 10.1186/1742-4690-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch M.S., Brun-Vezinet F., Clotet B., et al. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA Panel. Clin. Infect. Dis. 2003;37(1):113–128. doi: 10.1086/375597. [DOI] [PubMed] [Google Scholar]
- 8.Buonaguro L., Tornesello M.L., Buonaguro F.M. Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: pathogenetic and therapeutic implications. J. Virol. 2007;81(19):10209–10219. doi: 10.1128/JVI.00872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merati T.P., Ryan C.E., Spelmen T., et al. CRF01_AE dominates the HIV-1 epidemic in Indonesia. Sex. Health. 2012;9(5):414–421. doi: 10.1071/SH11121. [DOI] [PubMed] [Google Scholar]
- 10.Sahbandar I.N., Takahashi K., Djoerban Z., et al. Current HIV type 1 molecular epidemiology profile and identification of unique recombinant forms in Jakarta, Indonesia. AIDS Res Hum. 2009;25(7):637–646. doi: 10.1089/aid.2008.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Indriati D.W., Kotaki T., Khairunisa S.Q., et al. Appearance of drug resistance mutations among the dominant HIV-1 subtype, CRF01_AE in maumere, Indonesia. Curr. HIV Res. 2018;16(2):158–166. doi: 10.2174/1570162X16666180502114344. [DOI] [PubMed] [Google Scholar]
- 12.Chan P.A., Kantor R. Transmitted drug resistance in nonsubtype B HIV-1 infection. HIV Ther. 2009;3(5):447–465. doi: 10.2217/hiv.09.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Januraga P.P., Wulandari L.P., Muliawan P., et al. Sharply rising prevalence of HIV infection in Bali: a critical assessment of the surveillance data. Int. J. STD AIDS. 2013;24(8):633–637. doi: 10.1177/0956462413477556. [DOI] [PubMed] [Google Scholar]
- 14.Ford K., Wirawan D.N., Sumantera G.M., Sawitri A.A., Stahre M. Voluntary HIV testing, disclosure, and stigma among injection drug users in Bali, Indonesia. AIDS Educ. Prev. 2004;16(6):487–498. doi: 10.1521/aeap.16.6.487.53789. [DOI] [PubMed] [Google Scholar]
- 15.Sagung Sawitri A.A., Sumantera G.M., Wirawan D.N., Ford K., Lehman E. HIV testing experience of drug users in Bali, Indonesia. AIDS Care. 2006;18(6):577–588. doi: 10.1080/09540120500275015. [DOI] [PubMed] [Google Scholar]
- 16.Commision I.N.A. 2014 http://www. unaids.org/sites/default/files/country/documents/IDN_narrative_report_2014.pdf
- 17.Waluyo A., Culbert G.J., Levy J., Norr K.F. Understanding HIV-related stigma among Indonesian nurses. JANAC. 2015;26(1):69–80. doi: 10.1016/j.jana.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozal M.J., Amico K.R., Chiarella J., et al. Antiretroviral resistance and high-risk transmission behavior among HIV-positive patients in clinical care. AIDS. 2004;18(16):2185–2189. doi: 10.1097/00002030-200411050-00011. [DOI] [PubMed] [Google Scholar]
- 19.Culbert G.J., Earnshaw V.A., Wulanyani N.M., et al. Correlates and Experiences of HIV Stigma in Prisoners Living With HIV in Indonesia: A Mixed-Method Analysis. JANAC. 2015;26(6):743–757. doi: 10.1016/j.jana.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J.H., Wong K.H., Li P.C., et al. In-house human immunodeficiency virus-1 genotype resistance testing to determine highly active antiretroviral therapy resistance mutations in Hong Kong. Hong Kong Med. J. 2012;18(1):20–24. [PubMed] [Google Scholar]
- 21.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 22.Ghasemi A., Zahediasl S. Normality tests for statistical analysis: a guide for non-statisticians. Int. J. Endocrinol. Metab. 2012;10(2):486–489. doi: 10.5812/ijem.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viputtijul K., de Souza M., Trichavaroj R., et al. Heterosexually acquired CRF01_AE/B recombinant HIV type 1 found in Thailand. AIDS Res. Hum. Retroviruses. 2002;18(16):1235–1237. doi: 10.1089/08892220260387986. [DOI] [PubMed] [Google Scholar]
- 24.Foley B., Donegan E., Silitonga N., et al. Importation of multiple HIV type 1 strains into West Papua, Indonesia (Irian Jaya). AIDS Res. Hum. Retroviruses. 2001;17(17):1655–1659. doi: 10.1089/088922201753342068. [DOI] [PubMed] [Google Scholar]
- 25.Yahi N., Tamalet C., Tourres C., et al. Mutation patterns of the reverse transcriptase and protease genes in human immunodeficiency virus type 1-infected patients undergoing combination therapy: survey of 787 sequences. J. Clin. Microbiol. 1999;37(12):4099–4106. doi: 10.1128/jcm.37.12.4099-4106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinones-Mateu M.E., Albright J.L., Mas A., Soriano V., Arts E.J. Analysis of pol gene heterogeneity, viral quasispecies, and drug resistance in individuals infected with group O strains of human immunodeficiency virus type 1. J. Virol. 1998;72(11):9002–9015. doi: 10.1128/jvi.72.11.9002-9015.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Rourke S.M., Sutthent R., Phung P., et al. Glycans flanking the hypervariable connecting peptide between the A and B strands of the V1/V2 domain of HIV-1 gp120 confer resistance to antibodies that neutralize CRF01_AE viruses. PLoS One. 2015;10(3):e0119608. doi: 10.1371/journal.pone.0119608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paydary K., Khaghani P., Emamzadeh-Fard S., Alinaghi S.A., Baesi K. The emergence of drug resistant HIV variants and novel anti-retroviral therapy. Asian Pac. J. Trop. Biomed. 2013;3(7):515–522. doi: 10.1016/S2221-1691(13)60106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosn J., Galimand J., Raymond S., et al. Cohort ACP: X4 tropic multi-drug resistant quasi-species detected at the time of primary HIV-1 infection remain exclusive or at least dominant far from PHI. PLoS One. 2011;6(8):e23301. doi: 10.1371/journal.pone.0023301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dybowski J.N., Heider D., Hoffmann D. Structure of HIV-1 quasi-species as early indicator for switches of co-receptor tropism. AIDS Res. Ther. 2010;7:41. doi: 10.1186/1742-6405-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aasa-Chapman M.M., Aubin K., Williams I., McKnight A. Primary CCR5 only using HIV-1 isolates does not accurately represent the in vivo replicating quasi-species. Virology. 2006;351(2):489–496. doi: 10.1016/j.virol.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Matsushita S., Takahama S., Shibata J., et al. Ex vivo neutralization of HIV-1 quasi-species by a broadly reactive humanized monoclonal antibody KD-247. Hum. Antibodies. 2005;14(3-4):81–88. [PubMed] [Google Scholar]
- 33.Sankale J.L., De La Tour R.S., Marlink R.G., et al. Distinct quasi-species in the blood and the brain of an HIV-2-infected individual. Virology. 1996;226(2):418–423. doi: 10.1006/viro.1996.0671. [DOI] [PubMed] [Google Scholar]
- 34.Sabino E., Pan L.Z., Cheng-Mayer C., Mayer A. Comparison of in vivo plasma and peripheral blood mononuclear cell HIV-1 quasi-species to short-term tissue culture isolates: an analysis of tat and C2-V3 env regions. AIDS. 1994;8(7):901–909. [PubMed] [Google Scholar]
- 35.Nagata S., Imai J., Makino G., Tomita M., Kanai A. Evolutionary Analysis of HIV-1 Pol Proteins Reveals Representative Residues for Viral Subtype Differentiation. Front. Microbiol. 2017;8:2151. doi: 10.3389/fmicb.2017.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuiken C., Korber B., Shafer R.W. HIV sequence databases. AIDS Rev. 2003;5(1):52–61. [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J., Yue J., Wu S., Yan Y. Polymorphisms and drug resistance analysis of HIV-1 CRF01_AE strains circulating in Fujian Province, China. Arch. Virol. 2007;152(10):1799–1805. doi: 10.1007/s00705-007-1019-9. [DOI] [PubMed] [Google Scholar]
- 38.Manosuthi W., Butler D.M., Perez-Santiago J., et al. Protease polymorphisms in HIV-1 subtype CRF01_AE represent selection by antiretroviral therapy and host immune pressure. AIDS. 2010;24(3):411–416. doi: 10.1097/QAD.0b013e3283350eef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ariyoshi K., Matsuda M., Miura H., et al. Patterns of point mutations associated with antiretroviral drug treatment failure in CRF01_AE (subtype E) infection differ from subtype B infection. J. Acquir. Immune Defic. Syndr. 2003;33(3):336–342. doi: 10.1097/00126334-200307010-00007. [DOI] [PubMed] [Google Scholar]
- 40.Santa-Marta M., de Brito P.M., Godinho-Santos A., Goncalves J. Host factors and HIV-1 replication: Clinical evidence and potential therapeutic approaches. Front. Immunol. 2013;4:343. doi: 10.3389/fimmu.2013.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salgado M., Swanson M.D., Pohlmeyer C.W., et al. HLA-B*57 elite suppressor and chronic progressor HIV-1 isolates replicate vigorously and cause CD4+ T cell depletion in humanized BLT mice. J. Virol. 2014;88(6):3340–3352. doi: 10.1128/JVI.03380-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ndzinu J.K., Takeuchi H., Saito H., Yoshida T., Yamaoka S. eIF4A2 is a host factor required for efficient HIV-1 replication. Microbes Infect. 2018;20(6):346–352. doi: 10.1016/j.micinf.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Sharma G., Kaur G., Mehra N. Genetic correlates influencing immunopathogenesis of HIV infection. Indian J. Med. Res. 2011;134(6):749–768. doi: 10.4103/0971-5916.92623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lama J., Planelles V. Host factors influencing susceptibility to HIV infection and AIDS progression. Retrovirology. 2007;4:52. doi: 10.1186/1742-4690-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Candore G., Romano G.C., D’Anna C., et al. Biological basis of the HLA-B8, DR3-associated progression of acquired immune deficiency syndrome. Pathobiology. 1998;66(1):33–37. doi: 10.1159/000027992. [DOI] [PubMed] [Google Scholar]
- 46.Pinching A.J. Factors affecting the natural history of human immunodeficiency virus infection. Immunodefic. Rev. 1988;1(1):23–38. [PubMed] [Google Scholar]
- 47.Ueda S., Witaningrum A.M., Khairunisa S.Q., et al. Genetic Diversity and Drug Resistance of HIV-1 Circulating in North Sulawesi, Indonesia. AIDS Res. Hum. Retroviruses. 2018 doi: 10.1089/AID.2018.0221. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 48.El-Khatib Z., Ekstrom A.M., Ledwaba J., et al. Viremia and drug resistance among HIV-1 patients on antiretroviral treatment: a cross-sectional study in Soweto, South Africa. AIDS. 2010;24(11):1679–1687. doi: 10.1097/QAD.0b013e32833a097b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Almeida F.J., Berezin E.N., Rodrigues R., et al. Diversity and prevalence of antiretroviral genotypic resistance mutations among HIV-1-infected children. J. Pediatr. (Rio J.) 2009;85(2):104–109. doi: 10.2223/JPED.1877. [DOI] [PubMed] [Google Scholar]
- 50.Machado D.M., Fernandes S.C., Succi R.C., et al. Analysis of HIV- type 1 protease and reverse transcriptase in Brazilian children failing highly active antiretroviral therapy (HAART). Rev. Inst. Med. Trop. São Paulo. 2005;47(1):1–5. doi: 10.1590/s0036-46652005000100001. [DOI] [PubMed] [Google Scholar]
- 51.Delaugerre C., Warszawski J., Chaix M.L., et al. Prevalence and risk factors associated with antiretroviral resistance in HIV-1-infected children. J. Med. Virol. 2007;79(9):1261–1269. doi: 10.1002/jmv.20940. [DOI] [PubMed] [Google Scholar]
- 52.Riemenschneider M., Senge R., Neumann U., Hullermeier E., Heider D. Exploiting HIV-1 protease and reverse transcriptase cross-resistance information for improved drug resistance prediction by means of multi-label classification. BioData Min. 2016;9:10. doi: 10.1186/s13040-016-0089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller V., Larder B.A. Mutational patterns in the HIV genome and cross-resistance following nucleoside and nucleotide analogue drug exposure. Antivir. Ther. 2001;6(Suppl. 3):25–44. [PubMed] [Google Scholar]
- 54.Ma L., Huang J., Xing H., et al. Genotypic and phenotypic cross-drug resistance of harboring drug-resistant HIV type 1 subtype B′ strains from former blood donors in central Chinese provinces. AIDS Res. Hum. Retroviruses. 2010;26(9):1007–1013. doi: 10.1089/aid.2009.0252. [DOI] [PubMed] [Google Scholar]
- 55.Heider D., Senge R., Cheng W., Hullermeier E. Multilabel classification for exploiting cross-resistance information in HIV-1 drug resistance prediction. Bioinformatics. 2013;29(16):1946–1952. doi: 10.1093/bioinformatics/btt331. [DOI] [PubMed] [Google Scholar]
- 56.Groschel B., Cinatl J., Perigaud C., et al. S-acyl-2-thioethyl (SATE) pronucleotides are potent inhibitors of HIV-1 replication in T-lymphoid cells cross-resistant to deoxycytidine and thymidine analogs. Antiviral Res. 2002;53(2):143–152. doi: 10.1016/s0166-3542(01)00205-4. [DOI] [PubMed] [Google Scholar]
- 57.Van Laethem K., Witvrouw M., Balzarini J., et al. Patient HIV-1 strains carrying the multiple nucleoside resistance mutations are cross-resistant to abacavir. AIDS. 2000;14(4):469–471. doi: 10.1097/00002030-200003100-00027. [DOI] [PubMed] [Google Scholar]
- 58.Palmer S., Shafer R.W., Merigan T.C. Highly drug-resistant HIV-1 clinical isolates are cross-resistant to many antiretroviral compounds in current clinical development. AIDS. 1999;13(6):661–667. doi: 10.1097/00002030-199904160-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu Z., Fletcher R.S., Arts E.J., Wainberg M.A., Parniak M.A. The K65R mutant reverse transcriptase of HIV-1 cross-resistant to 2′, 3′-dideoxycytidine, 2′,3′-dideoxy-3′-thiacytidine, and 2′,3′-dideoxyinosine shows reduced sensitivity to specific dideoxynucleoside triphosphate inhibitors in vitro. J. Biol. Chem. 1994;269(45):28118–28122. [PubMed] [Google Scholar]
- 60.Clutter D.S., Zhou S., Varghese V., et al. Prevalence of drug-resistant minority variants in untreated HIV-1-infected individuals with and those without transmitted drug resistance detected by sanger sequencing. J. Infect. Dis. 2017;216(3):387–391. doi: 10.1093/infdis/jix338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li T., Qian F., Yuan T., et al. Drug resistance mutation profiles of the drug-naive and first-line regimen-treated HIV-1-infected population of Suzhou, China. Virol. Sin. 2017;32(4):271–279. doi: 10.1007/s12250-017-4002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andreis S., Basso M., Scaggiante R., et al. Drug resistance in B and non-B subtypes amongst subjects recently diagnosed as primary/recent or chronic HIV-infected over the period 2013-2016: Impact on susceptibility to first-line strategies including integrase strand-transfer inhibitors. J. Glob. Antimicrob. Resist. 2017;10:106–112. doi: 10.1016/j.jgar.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 63.Paraskevis D., Kostaki E., Magiorkinis G., et al. Prevalence of drug resistance among HIV-1 treatment-naive patients in Greece during 2003-2015: Transmitted drug resistance is due to onward transmissions. Infect. Genet. Evol. 2017;54:183–191. doi: 10.1016/j.meegid.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 64.Ghafari S., Memarnejadian A., Samarbaf-Zadeh A., et al. Prevalence of HIV-1 transmitted drug resistance in recently infected, treatment-naive persons in the Southwest of Iran, 2014-2015. Arch. Virol. 2017;162(9):2737–2745. doi: 10.1007/s00705-017-3431-0. [DOI] [PubMed] [Google Scholar]
- 65.Socias M.E., Nosova E., Kerr T., et al. Patterns of transmitted drug resistance and virological response to first-line antiretroviral treatment among HIV-positive people who use illicit drugs in a Canadian setting. Clin. Infect. Dis. 2017;65(5):796–802. doi: 10.1093/cid/cix428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kotaki T., Khairunisa S.Q., Witaningrum A.M., et al. HIV-1 transmitted drug resistance mutations among antiretroviral therapy-Naive individuals in Surabaya, Indonesia. AIDS Res. Ther. 2015;12:5. doi: 10.1186/s12981-015-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Witaningrum A.M., Kotaki T., Khairunisa S.Q., et al. Genotypic Characterization of Human Immunodeficiency Virus Type 1 Derived from Antiretroviral Therapy-Naive Individuals Residing in Sorong, West Papua. AIDS Res. Hum. Retroviruses. 2016;32(8):812–817. doi: 10.1089/AID.2016.0054. [DOI] [PubMed] [Google Scholar]
- 68.Roberts J.D., Bebenek K., Kunkel T.A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242(4882):1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 69.Cuevas J.M., Geller R., Garijo R., Lopez-Aldeguer J., Sanjuan R. Extremely High Mutation Rate of HIV-1 In Vivo. PLoS Biol. 2015;13(9):e1002251. doi: 10.1371/journal.pbio.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Luca A., Sidumo Z.J., Zanelli G., et al. Accumulation of HIV-1 drug resistance in patients on a standard thymidine analogue-based first line antiretroviral therapy after virological failure: implications for the activity of next-line regimens from a longitudinal study in Mozambique. BMC Infect. Dis. 2017;17(1):605. doi: 10.1186/s12879-017-2709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moscona R., Ram D., Wax M., et al. Comparison between next-generation and Sanger-based sequencing for the detection of transmitted drug-resistance mutations among recently infected HIV-1 patients in Israel, 2000-2014. J. Int. AIDS Soc. 2017;20(1):1–9. doi: 10.7448/IAS.20.1.21846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Golmohammadi R., Baesi K., Moradi A., et al. The first characterization of HIV-1 subtypes and drug resistance mutations among antiretrovirally treated patients in Kermanshah, Iran. Intervirology. 2017;60(1-2):33–37. doi: 10.1159/000478701. [DOI] [PubMed] [Google Scholar]
- 73.Cozzi-Lepri A., Ruiz L., Loveday C., et al. Thymidine analogue mutation profiles: factors associated with acquiring specific profiles and their impact on the virological response to therapy. Antivir. Ther. 2005;10(7):791–802. [PubMed] [Google Scholar]
- 74.Rezende L.F., Prasad V.R. Nucleoside-analog resistance mutations in HIV-1 reverse transcriptase and their influence on polymerase fidelity and viral mutation rates. Int. J. Biochem. Cell Biol. 2004;36(9):1716–1734. doi: 10.1016/j.biocel.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 75.Johnson V.A., Brun-Vezinet F., Clotet B., et al. Update of the drug resistance mutations in HIV-1. Top. HIV Med. 2008;16(5):138–145. [PubMed] [Google Scholar]
- 76.Boyer P.L., Sarafianos S.G., Arnold E., Hughes S.H. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 2001;75(10):4832–4842. doi: 10.1128/JVI.75.10.4832-4842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Demeter L.M., D’Aquila R., Weislow O., et al. Interlaboratory concordance of DNA sequence analysis to detect reverse transcriptase mutations in HIV-1 proviral DNA. ACTG Sequencing Working Group. AIDS Clinical Trials Group. J. Virol. Methods. 1998;75(1):93–104. doi: 10.1016/s0166-0934(98)00100-1. [DOI] [PubMed] [Google Scholar]
- 78.Gall A., Ferns B., Morris C., et al. Universal amplification, next-generation sequencing, and assembly of HIV-1 genomes. J. Clin. Microbiol. 2012;50(12):3838–3844. doi: 10.1128/JCM.01516-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publishers web site along with the published article.
1. Demography of ARV treatment failure, including risk factors, viral load, name and duration of the ARV, and treatment failure discovery of HIV-1 of CRF01_AE subtype positive patients in Bali, Indonesia 2008 – 2010.
2. Demography of ARV-naïve, including risk factors and viral load of HIV-1 of CRF01_AE positive patients in Bali, Indonesia 2008 – 2010.