Abstract
Background:
Hepatitis B Viral (HBV) infection is one of the major causes of Hepatocellular Carcinoma (HCC). Mounting evidence had provided that the HBV integration might be a critical con-tributor of HCC carcinogenesis.
Objective and Methods:
To explore the profile of HBV integration in the plasma DNA, the method of next-generation sequencing, HBV capture and bioinformatics had been employed to screen for HBV in-tegration sites in the plasma samples.
Results:
In the initial experiment, a total of 87 breakpoints were detected in the 20 plasma samples. The distribution of breakpoints showed that there was significant enrichment of breakpoints in the region of intron. Furthermore, the HBV breakpoints were prone to occur in the region of X protein (1,700-2,000bp) in the plasma samples. The pathway analysis had revealed that the HBV integrations sites were specifically enriched in the cancer pathway.
Conclusion:
Altogether, our results had provided direct evidence for the HBV integration in plasma DNA, and they might be potentially useful for future HCC prognosis and diagnosis.
Keywords: Hepatocellular carcinoma, Plasma, HBV integration, Cell free DNA, HBV genome, Breakpoints
1. INTRODUCTION
HBV infection is an epidemic in Asia, Africa, Southern Europe and Latin America, and HBV consists of at least eight genotypes (A-H) [1]. Several factors have been known to be related to higher HCC risk among HBV carriers: demographic (male gender, older age, ethnicity), genetic (family history of HCC), viral (high viral load, viral genotype, duration of infection, co-infection with HCV, HIV or HDV), and environmental (exposure to aflatoxin, alcohol abuse or cigarette smoking) [2, 3]. Generally, HBV is the main causative agent in the high incidence HCC areas, while HCV is the major etiological factor related to HCC in low incidence HCC areas [2, 4-6]. In addition, HDV chronic infection has been found to be associated with a worsening of HBV infection. Usually, it increased the risk of liver decompensation and Hepatocellular Carcinoma (HCC) occurrence [7]. Furthermore, HBV DNA and HBeAg were detected less frequently in anti-HD-positive than in anti-HD-negative subjects among patients with severe liver disease [8]. These findings indicated that HBV infection was closely related to HDV infection, although the mechanism kept unclear.
Among HBV genotypes, HBV/B and HBV/C are more restricted to east/south-east Asia [9]. HBV infection of these two types has been known as a leading cause for chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) in China [10]. The integration events of HBV/B and HBV/C are also frequent to be detected in the cancer tissues of HCC patients.
Massive Parallel Sequencing (MPS) technology has provided an efficient mean to detect HBV integration through whole genome. Jiang et al. investigated the effect of HBV integration by whole genome sequencing. They found that HBV integration could trigger the copy number variation of genome and induce the abnormal expression [11]. Sung et al. had surveyed the hotspot genes of HBV integration through the whole genome and determined that there was a significant relationship between HBV integration and survival time [12]. Furthermore, the researcher found that the viral-human chimeric transcript may trigger the Wnt signaling pathways and had close relation with the development and progression of liver cancer [13]. Based on the above studies, it suggested that HBV integration is an important event in the process of tumorigenesis. HBV integration can not only cause genetic damage and chromosomal instability but also cause disorder to the host gene expression [14]. In addition, expression of viral proteins such as X protein and S antigen may further induce the tumorigenesis [13]. Indeed, the first descriptions of HBV integration events were based on primary HCC tissues and HCC-derived cell lines, prompting suggestions that HBV integration events might be causative in tumorigenesis [15, 16]. Hitherto, the reported mechanisms include (1) HBV integration mediated insertional mutagenesis of HCC-associated genes; (2) induction of chromosomal instability by HBV integration; (3) the expression of HBV genes from the HBV integration. However, the mechanism of HBV-induced HCC carcinogenesis still remains unclear so far [17].
However, the samples in these studies were collected via invasive procedures. In order to facilitate the clinical utilization, HBV integration events in plasma were urgently needed to be surveyed. The characteristic of HBV integration in the plasma might promote the clinical utilization potential of HBV integration.
A high throughput virus integration detection (HIVID) approach was adopted to investigate the HBV integration sites [18]. Overall, there were 15 samples with HBV integration among 20 plasma samples and 87 breakpoints were found in the 15 plasma samples. Furthermore, we determine the characteristic of HBV integration sites in the plasma samples. HBV integration was prone to the region of INTRON. Pathway analysis indicated that the HBV integration events were enriched in the pathway of cancer. Altogether, our results provided evidence for the HBV integration in these plasma samples and explored the characteristic of HBV integration in the plasma, which might be useful for HCC prognosis and diagnosis.
2. MATERIALS AND METHODS
2.1. Sample Collection and Plasma DNA Extraction
We obtained the plasma and tumor tissue samples from the Affiliated Hospital of Jining Medical University, Jining, China, and patients had been diagnosed with HCC. All patients signed the written informed consent form, and the study had been approved by the Ethics Review Committee in the Jining Medical University. Plasma was stored at 80°C and DNA was extracted from a 0.5-ml plasma aliquot with DNA Blood Midi Kit (Qiagen, Germany) according to the manufacturer’s instructions and stored at -20°C before further analysis. The inclusion criteria for this study included: (i) HBV-positive HCCs (HBV-B type) (ii) obtained from consenting patients and (iii) all patients were HCV-negative and HIV-negative (iv) negative for autoimmune hepatitis and metabolic and/or genetic disorders, such as Wilson’s disease, hemochromatosis.
2.2. HBV Fragments Enrichment and Sequencing
The construction of sequencing library strictly followed the standard instructions provided by Illumina. The cell free DNA purified, and their ends were blunted, “A” tailed, ligated to adaptors and then PCR. The DNA libraries were quantified using Bioanalyser 2100 (Agilent Technologies, Santa Clara, CA, USA). The hybridization procedures were carried out following MyGenostics’s GenCapTM Target Enrichment Protocol (GenCapTM Enrichment, MyGenostics, USA). The DNA libraries were hybridized with HBV probes at 65°C for 24 hours and subsequently subjected to washes to remove unbound. The eluted fragments were amplified by 12 PCR cycles in order to generate the sequencing library. Each library was further quantified and proceeded to 101 cycles of paired-end index sequencing in the Illumina HiSeq 2000 sequencer according to the manufacturer’s official instruction.
2.3. Breakpoints Detection and Annotation of HBV Integration Sites
Deploying an algorithm established by our team previously [18], the low-quality reads, duplication reads and adaptor contaminations were removed. Subsequently, the filtered clean reads were mapped to both the human (NCBI build 37, HG19) and the HBV genomes. The chimeric reads that partially aligned to the human genome and partially aligned to the HBV genome were remained as the reads of our interest. The selected chimeric reads were then subjected to paired-end reads assembly, which helped to reconstruct fragment sequences, and additionally increased the efficacy to locate the precise position of the breakpoints. The PE-assembled reads were re-mapped to the human and the HBV genome using BWA [19]. The HBV integration breakpoints were annotated using ANNOVAR [20].
3. RESULTS
3.1. The Distribution of Breakpoints in the Human Genome
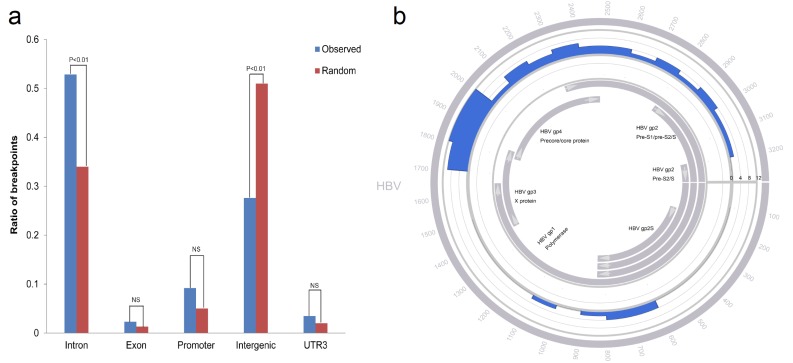
Plasma samples of 20 HCC patients were obtained in order to investigate the HBV integration sites (Table 1). The DNA of plasma was processed according to our innovative HIVID approach [18]. Among the samples, 15 samples showed positive for HBV integration, altogether 87 integration sites were determined (Table 2). Among all the integration sites, 46 of them were in the intron region, and 24 were in the intergenic region (Table 3). Therefore, it seems that the breakpoints were more prone to be enriched in the intron region (Fig. 1a; Intron observed ratio=0.53; Intron random ratio=0.34; Chisquare Test P<0.01).
Table 1.
Demographic and clinicopathologic characteristics of 20 HCC patients.
| Variables | Mean ± SD / n (%) |
|---|---|
| Age, years | 49.5±7.2 |
| Sex | |
| Female | 5 (25%) |
| Male | 15 (75%) |
| Diagnosis | |
| Primary HCC | 20 (100%) |
| HCC Differentiation (Edmondson-Steiner) | |
| II | 2 (10%) |
| III | 18 (90%) |
| Tumor diameter, cm | 6.2±4.3 |
| Microvascular invasion | |
| Absence | 15 (75%) |
| Presence | 5 (25%) |
| ALB, g/L | 43.9±5.1 |
| TBIL, µmol/L | 16.1±5.7 |
| PT, seconds | 11.9±1.1 |
| AFP, µg/L | 579.8±564.8 |
| HBV type | |
| HBV_B | 20 (100%) |
| HBV DNA levels, IU/mL | 417825.9±856030.9 |
| HBeAg | |
| Positive | 6 (30%) |
| Negative | 14 (70%) |
| Anti-HBe status | |
| Positive | 17 (85%) |
| Negative | 3 (15%) |
| HBsAg status | |
| Positive | 20 (100%) |
| Anti-HBs status | |
| Negative | 20 (100%) |
Table 2.
Data production of samples. Breakpoint number in human genome and coverage of HBV genome were shown.
| Library | Total Bases | Q20 | HBV Type | HBV Coverage | Breakpoint Number |
|---|---|---|---|---|---|
| B001 | 5.42G | 88.42;79.57 | B | 99% | 3 |
| B002 | 5.01G | 86.90;78.08 | B | 98% | 4 |
| B0074 | 5.22G | 85.49;80.51 | B | 97% | 7 |
| B0068 | 5.31G | 86.97;79.88 | B | 99% | 2 |
| B0064 | 5.40G | 87.38;81.57 | B | 99% | 3 |
| B0061 | 5.81G | 86.08;78.67 | B | 100% | 9 |
| B0049 | 5.12G | 85.49;80.55 | B | 99% | 5 |
| B0037 | 5.16G | 88.97;81.86 | B | 96% | 6 |
| B0032 | 5.20G | 86.55;81.97 | B | 97% | 5 |
| B0075 | 5.25G | 87.89;79.72 | B | 98% | 7 |
| B007 | 5.86G | 83.50;81.52 | B | 99% | 5 |
| B0034 | 5.27G | 87.87;80.78 | B | 97% | 8 |
| B0021 | 5.68G | 86.78;82.47 | B | 99% | 5 |
| B0012 | 5.09G | 88.77;79.88 | B | 98% | 6 |
| B0010 | 5.50G | 84.52;81.63 | B | 99% | 12 |
| B0016 | 5.91G | 85.87;79.25 | B | 96% | 0 |
| B0011 | 5.53G | 89.67;81.98 | B | 98% | 0 |
| B008 | 5.57G | 86.98;78.66 | B | 99% | 0 |
| B009 | 5.61G | 85.59;80.79 | B | 99% | 0 |
| B005 | 5.55G | 86.49;79.74 | B | 99% | 0 |
Table 3.
The breakpoints of plasma samples. The breakpoints detected in the plasma samples.
| Sample | Chr | Position | Support | Element | Gene |
|---|---|---|---|---|---|
| B0074 | chr4 | 8977829 | 4 | intergenic | LOC650293/USP17L10 |
| B0074 | chr8 | 103211319 | 2 | downstream | RRM2B |
| B0074 | chr8 | 30611415 | 2 | intronic | UBXN8 |
| B0074 | chr8 | 103119207 | 2 | intronic | NCALD |
| B0074 | chr4 | 125111322 | 2 | intergenic | LINC01091/ANKRD50 |
| B0074 | chr4 | 65178643 | 2 | intronic | TECRL |
| B0074 | chr20 | 47656089 | 2 | promoter | CSE1L |
| B0068 | chr4 | 118968708 | 2 | intronic | NDST3 |
| B0068 | chr17 | 21903348 | 2 | promoter | FLJ36000 |
| B0064 | chr11 | 65986345 | 8 | intronic | PACS1 |
| B0064 | chr1 | 249121201 | 7 | promoter | SH3BP5L |
| B0064 | chr15 | 34708490 | 4 | intergenic | GOLGA8A |
| Sample | Chr | Position | Support | Element | Gene |
| B0061 | chr18 | 22570340 | 7 | intergenic | RP11-449D8.1/ZNF521 |
| B0061 | chr6 | 169471301 | 4 | intergenic | SMOC2/THBS2 |
| B0061 | chr3 | 110721179 | 4 | intergenic | LINC01205/PVRL3-AS1 |
| B0061 | chr7 | 74141512 | 3 | ncRNA_intronic | LOC101926943 |
| B0061 | chr21 | 39816650 | 3 | intronic | ERG |
| B0061 | chr4 | 86519608 | 2 | intronic | ARHGAP24 |
| B0061 | chr2 | 45757229 | 2 | intronic | SRBD1 |
| B0061 | chr1 | 225407594 | 2 | intronic | DNAH14 |
| B0061 | chr1 | 10649629 | 2 | intronic | PEX14 |
| B0049 | chr7 | 55273463 | 4 | UTR3 | EGFR |
| B0049 | chr4 | 92355373 | 3 | intronic | CCSER1 |
| B0049 | chr6 | 31003961 | 2 | downstream | MUC22 |
| B0049 | chr4 | 31901689 | 2 | intergenic | PCDH7 |
| B0049 | chr3 | 49089016 | 2 | intronic | QRICH1 |
| B0037 | chrX | 112015468 | 2 | downstream | AMOT |
| B0037 | chr2 | 28331047 | 2 | intronic | BRE |
| B0037 | chr17 | 33939459 | 2 | intronic | AP2B1 |
| B0037 | chr14 | 100715229 | 2 | intronic | YY1 |
| B0037 | chr10 | 57261124 | 2 | intergenic | PCDH15/MTRNR2L5 |
| B0037 | chr1 | 38451027 | 2 | intronic | SF3A3 |
| B0032 | chr17 | 21906061 | 4 | ncRNA_intronic | FLJ36000 |
| B0032 | chr17 | 21906355 | 3 | ncRNA_intronic | FLJ36000 |
| B0032 | chr11 | 191802 | 3 | promoter | LOC653486 |
| B0032 | chr4 | 49649534 | 2 | intergenic | CWH43 |
| B0032 | chr17 | 21903348 | 2 | promoter | FLJ36000 |
| B0075 | chr8 | 30611415 | 3 | intronic | UBXN8 |
| B0075 | chr5 | 7349039 | 3 | intergenic | LOC442132/ADCY2 |
| B0075 | chr3 | 73158614 | 3 | intergenic | PPP4R2/PDZRN3 |
| B0075 | chr8 | 124890891 | 2 | intronic | FER1L6 |
| B0075 | chr4 | 100009507 | 2 | intronic | ADH5 |
| B0075 | chr18 | 22570340 | 2 | intergenic | RP11-449D8.1/ZNF521 |
| B0075 | chr12 | 27136775 | 2 | intronic | TM7SF3 |
| B007 | chr10 | 89954886 | 6 | intergenic | PTEN/RNLS |
| B007 | chr10 | 89954930 | 4 | intergenic | PTEN/RNLS |
| B007 | chr11 | 47814540 | 3 | intronic | NUP160 |
| B007 | chr7 | 72974855 | 2 | promoter | BCL7B |
| B007 | chr18 | 42356727 | 2 | intronic | SETBP1 |
| Sample | Chr | Position | Support | Element | Gene |
| B0034 | chr12 | 30265504 | 5 | intergenic | TMTC1/IPO8 |
| B0034 | chr2 | 119052347 | 4 | intergenic | INSIG2/LOC101927709 |
| B0034 | chr15 | 67917548 | 4 | intronic | MAP2K5 |
| B0034 | chr12 | 33057564 | 3 | promoter | PKP2 |
| B0034 | chr9 | 98598152 | 2 | ncRNA_intronic | LINC00476 |
| B0034 | chr6 | 32546766 | 2 | UTR3 | HLA-DRB1 |
| B0034 | chr6 | 32546685 | 2 | UTR3 | HLA-DRB1 |
| B0034 | chr4 | 42527080 | 2 | intronic | ATP8A1 |
| B0021 | chr7 | 39673979 | 9 | intronic | RALA |
| B0021 | chr7 | 129485990 | 2 | intronic | UBE2H |
| B0021 | chr7 | 40234460 | 2 | intronic | SUGCT |
| B0021 | chr17 | 43211456 | 2 | intronic | ACBD4 |
| B0021 | chr17 | 43211378 | 2 | intronic | ACBD4 |
| B0012 | chr5 | 58123101 | 3 | intronic | RAB3C |
| B0012 | chr2 | 80380053 | 3 | intronic | CTNNA2 |
| B0012 | chr17 | 39084514 | 3 | exonic | KRT23 |
| B0012 | chr1 | 173786892 | 3 | intronic | CENPL |
| B0012 | chrX | 61684765 | 2 | intergenic | SPIN4 |
| B0012 | chr7 | 158778486 | 2 | intergenic | WDR60/LINC00689 |
| B0010 | chr4 | 184204577 | 3 | intronic | WWC2 |
| B0010 | chr3 | 69838919 | 3 | intronic | MITF |
| B0010 | chr17 | 7267199 | 3 | downstream | TMEM95 |
| B0010 | chr16 | 1723993 | 3 | exonic | CRAMP1L |
| B0010 | chr14 | 69887597 | 3 | intronic | SLC39A9 |
| B0010 | chr10 | 56134248 | 3 | intronic | PCDH15 |
| B0010 | chr1 | 111605674 | 3 | intergenic | LRIF1/DRAM2 |
| B0010 | chr9 | 20278555 | 2 | intergenic | SLC24A2/MLLT3 |
| B0010 | chr9 | 68429933 | 2 | ncRNA_intronic | LOC642236 |
| B0010 | chr9 | 6881037 | 2 | intronic | KDM4C |
| B0010 | chr8 | 88368708 | 2 | intronic | CNBD1 |
| B0010 | chr8 | 124052577 | 2 | intronic | DERL1 |
| B002 | chr5 | 52674905 | 2 | intergenic | LOC257396/FST |
| B002 | chr3 | 42691176 | 2 | promoter | ZBTB47 |
| B002 | chr12 | 38638139 | 4 | intergenic | ALG10B |
| B002 | chr2 | 28693259 | 2 | intergenic | FOSL2/PLB1 |
| B001 | chr2 | 219543519 | 2 | intronic | STK36 |
| B001 | chr2 | 219543501 | 4 | intronic | STK36 |
| B001 | chr18 | 11550976 | 11 | intergenic | PIEZO2/SLC35G4 |
Fig. (1).
The distribution of breakpoints in gene elements and HBV genome. (a) Observed represented the observed ratio of breakpoints; Random represented the random ratio of breakpoints. NS represented no significant. (b) Histograms were constructed for 100-bp intervals. HBV genes with different functions are marked. The number of breakpoints located in the HBV genome was shown.
3.2. The Distribution of Breakpoints in the HBV Genome
The distribution of breakpoints in the HBV genome was then analysed. It was also revealed that the breakpoints from plasma samples were enriched in the region of 1700-2000 bp of the HBV genome specifically (Fig. 1b).
3.3. The Pathway Analysis of Breakpoints
Using DAVID pathway enrichment software, the genes integrated by HBV were analysed [21, 22], and the results indicated that cancer pathway was particularly targeted (P<0.01). These five genes CTNNA2, EGFR, MITF, STK36 and RALA were located in the cancer pathway.
4. DISCUSSION
HBV integration had previously been demonstrated having a close association with the tumorigenesis of HCC. In the studies led by Sung et al., the authors had identified several genes preferentially integrated by HBV [12]. In this study, our team had analysed the HBV integration sites in 20 plasma samples of HCC patients. The results suggested that the genes in the cancer pathway were particularly targeted by HBV integration. In recent years, the use of plasma DNA sample in clinical diagnosis has become increasingly important [23], this is due to the presence of circulating DNA originated from the degenerating tumor cells [24]. According to the established data, one of the major sources of plasma or serum DNA may be from the apoptotic cells [25], though the entire mechanism of DNA being released into circulating blood still remains to be thoroughly investigated.
Generally, there was a significant portion of the cf-DNA from the tumor tissues. Many researchers have identified that common genetic alterations exist in both tumor tissue and paired plasma samples [26]. The level and characteristic of cf-DNA in human plasma have been affected by the dynamic balance between cellular DNA release and DNA degradation. Thus, stability of distinct cf-DNA forms [27, 28], activity of blood nucleases [29], adsorption of cf-DNA on blood cells [30, 31], as well as degradation of cf-DNA by phagocytes should also be considered as factors regulating the characteristic and level of cf-DNA in cancer patients [32].
Moreover, it is curious to find a number of breakpoints located in the intergenic regions. In recent years, an increasing trend of research interests had drawn to resolve the usefulness of breakpoints located in the intergenic region. For instances, MYC activation was driven by an upstream integration of HPV-18 genome [33]; β-catenin transactivity could be modulated by HBV integration in Long Interspersed Nuclear Element (LINE) [13]. Thus the importance and significance of HBV integration sites in the intergenic region remain elusive.
The distribution of breakpoints in the HBV genome was also investigated. It was revealed that the breakpoints were particularly enriched in the region of HBV X and core genes, which is in line with the previous findings by others and also our group.
CONCLUSION
Our study had adopted an effective method to seek HBV integration sites in the plasma samples. The results provided evidence for HBV integration in the plasma samples, which could be potentially useful for future HCC prognosis and diagnosis.
Acknowledgements
The study was funded by Doctoral Setup Foundation of Jining Medical University (No. 600491001), Technology Development Project of Medical and Health Science in Shandong Province (No. 2017WS516), Supporting Fund for Teachers Research of Jining Medical University (No. JY2017JS004), Natural Science Foundation of Shandong (No. ZR2018PH018), Jining Science and Technology Project for Benefit People (No. 10).
AUTHORS’ CONTRIBUTIONS
WYL, QSK, and CXF conceived and designed the experiments. QH, MHT, YWQ and APZ performed the experiments. YHS and QSK analyzed the data and HQ, CXF, WYL wrote the paper.
Ethics Approval and Consent to Participate
The study has been approved by the Ethics Review Committee of Jining Medical University (No. 2018KY003), Jining, China.
Human and Animal Rights
No animals were used in this study, Reported experiments on humans were in accordance with the ethical standards of the committee responsible for human experimentation (institutional national), and with the Helsinki Declaration of 1975, as revised in 2008 (http://www.wma.net/en/20activities/10ethics/10helsinki/).
Consent for Publication
Informed consent was obtained from all individual participants included in the study.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Lin C.L., Liao L.Y., Wang C.S., Chen P.J., Lai M.Y., Chen D.S., Kao J.H. Basal core-promoter mutant of hepatitis B virus and progression of liver disease in hepatitis B e antigen-negative chronic hepatitis B. Liver Int. 2005;25(3):564–570. doi: 10.1111/j.1478-3231.2005.01041.x. [DOI] [PubMed] [Google Scholar]
- 2.Petruzziello A. Epidemiology of Hepatitis B virus (HBV) and Hepatitis C virus (HCV) related hepatocellular carcinoma. Open Virol. J. 2018;12(1):26–32. doi: 10.2174/1874357901812010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sagnelli E., Felaco F.M., Rapicetta M., Stroffolini T., Petruzziello A., Annella T., Chionne P., Pasquale G., Filippini P., Peinetti P. Interaction between HDV and HBV infection in HBsAg-chronic carriers. Infection. 1991;19(3):155–158. doi: 10.1007/BF01643238. [DOI] [PubMed] [Google Scholar]
- 4.Perz J.F., Armstrong G.L., Farrington L.A., Hutin Y.J., Bell B.P. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol. 2006;45(4):529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Sherman M. Hepatocellular carcinoma: Epidemiology, risk factors, and screening. Semin. Liver Dis. 2005;25(2):143–154. doi: 10.1055/s-2005-871194. [DOI] [PubMed] [Google Scholar]
- 6.Aghemo A., Colombo M. Hepatocellular carcinoma in chronic hepatitis C: From bench to bedside. Semin. Immunopathol. 2013;35(1):111–120. doi: 10.1007/s00281-012-0330-z. [DOI] [PubMed] [Google Scholar]
- 7.Romeo R., Petruzziello A., Pecheur E.I., Facchetti F., Perbellini R., Galmozzi E., Khan N.U., Di Capua L., Sabatino R., Botti G., Loquercio G. Hepatitis delta virus and hepatocellular carcinoma: An update. Epidemiol. Infect. 2018;146(13):1612–1618. doi: 10.1017/S0950268818001942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stroffolini T., Sagnelli E., Rapicetta M., Felaco F.M., Filippini P., Annella T., Petruzziello A., Chionne P., Sarrecchia B., Piccinino F. Hepatitis B virus DNA in chronic HBsAg carriers: Correlation with HBeAg/anti-HBe status, anti-HD and liver histology. Hepatogastroenterology. 1992;39(1):62–65. [PubMed] [Google Scholar]
- 9.Araujo N.M., Waizbort R., Kay A. Hepatitis B virus infection from an evolutionary point of view: How viral, host, and environmental factors shape genotypes and subgenotypes. Infect. Genet. Evol. 2011;11(6):1199–1207. doi: 10.1016/j.meegid.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka M., Katayama F., Kato H., Tanaka H., Wang J., Qiao Y.L., Inoue M. Hepatitis B and C virus infection and hepatocellular carcinoma in China: A review of epidemiology and control measures. J. Epidemiol. 2011;21(6):401–416. doi: 10.2188/jea.JE20100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang Z., Jhunjhunwala S., Liu J., Haverty P.M., Kennemer M., Guan Y., Lee W., Carnevali P., Stinson J., Johnson S., Diao J., Yeung S., Jubb A., Ye W., Wu T.D., Kapadia S.B., de Sauvage F.J., Gentleman R.C., Stern H.M., Seshagiri S., Pant K.P., Modrusan Z., Ballinger D.G., Zhang Z. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res. 2012;22(4):593–601. doi: 10.1101/gr.133926.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sung W.K., Zheng H., Li S., Chen R., Liu X., Li Y., Lee N.P., Lee W.H., Ariyaratne P.N., Tennakoon C., Mulawadi F.H., Wong K.F., Liu A.M., Poon R.T., Fan S.T., Chan K.L., Gong Z., Hu Y., Lin Z., Wang G., Zhang Q., Barber T.D., Chou W.C., Aggarwal A., Hao K., Zhou W., Zhang C., Hardwick J., Buser C., Xu J., Kan Z., Dai H., Mao M., Reinhard C., Wang J., Luk J.M. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 2012;44(7):765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 13.Lau C.C., Sun T., Ching A.K., He M., Li J.W., Wong A.M., Co N.N., Chan A.W., Li P.S., Lung R.W., Tong J.H., Lai P.B., Chan H.L., To K.F., Chan T.F., Wong N. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25(3):335–349. doi: 10.1016/j.ccr.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Edman J.C., Gray P., Valenzuela P., Rall L.B., Rutter W.J. Integration of hepatitis B virus sequences and their expression in a human hepatoma cell. Nature. 1980;286(5772):535–538. doi: 10.1038/286535a0. [DOI] [PubMed] [Google Scholar]
- 16.Brechot C., Pourcel C., Louise A., Rain B., Tiollais P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature. 1980;286(5772):533–535. doi: 10.1038/286533a0. [DOI] [PubMed] [Google Scholar]
- 17.Tu T., Budzinska M.A., Shackel N.A., Jilbert A.R. Conceptual models for the initiation of hepatitis B virus-associated hepatocellular carcinoma. Liver Int. 2015;35(7):1786–1800. doi: 10.1111/liv.12773. [DOI] [PubMed] [Google Scholar]
- 18.Li W., Zeng X., Lee N.P., Liu X., Chen S., Guo B., Yi S., Zhuang X., Chen F., Wang G., Poon R.T., Fan S.T., Mao M., Li Y., Li S., Wang J., Jianwang; Xu X., Jiang H., Zhang X. HIVID: An efficient method to detect HBV integration using low coverage sequencing. Genomics. 2013;102(4):338–344. doi: 10.1016/j.ygeno.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahr S., Hentze H., Englisch S., Hardt D., Fackelmayer F.O., Hesch R.D., Knippers R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–1665. [PubMed] [Google Scholar]
- 24.Anker P., Mulcahy H., Chen X.Q., Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev. 1999;18(1):65–73. doi: 10.1023/a:1006260319913. [DOI] [PubMed] [Google Scholar]
- 25.Elshimali Y.I., Khaddour H., Sarkissyan M., Wu Y., Vadgama J.V. The clinical utilization of circulating cell free DNA (CCFDNA) in blood of cancer patients. Int. J. Mol. Sci. 2013;14(9):18925–18958. doi: 10.3390/ijms140918925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleischhacker M., Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim. Biophys. Acta. 2007;1775(1):181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Holdenrieder S., Von Pawel J., Nagel D., Stieber P. Long-term stability of circulating nucleosomes in serum. Anticancer Res. 2010;30(5):1613–1615. [PubMed] [Google Scholar]
- 28.Thakur B.K., Zhang H., Becker A., Matei I., Huang Y., Costa-Silva B., Zheng Y., Hoshino A., Brazier H., Xiang J., Williams C., Rodriguez-Barrueco R., Silva J.M., Zhang W., Hearn S., Elemento O., Paknejad N., Manova-Todorova K., Welte K., Bromberg J., Peinado H., Lyden D. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24(6):766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamkovich S.N., Cherepanova A.V., Kolesnikova E.V., Rykova E.Y., Pyshnyi D.V., Vlassov V.V., Laktionov P.P. Circulating DNA and DNase activity in human blood. Ann. N. Y. Acad. Sci. 2006;1075(1):191–196. doi: 10.1196/annals.1368.026. [DOI] [PubMed] [Google Scholar]
- 30.Tamkovich S.N., Vlasov V.V., Laktionov P.P. Circulating deoxyribonucleic acids in blood and their using in medical diagnostics. Mol. Biol. 2008;42(1):12–23. [PubMed] [Google Scholar]
- 31.Tamkovich S.N., Litviakov N.V., Bryzgunova O.E., Dobrodeev A.Y., Rykova E.Y., Tuzikov S.A., Zav’ialov A.A., Vlassov V.V., Cherdyntseva N.V., Laktionov P.P. Cell-surface-bound circulating DNA as a prognostic factor in lung cancer. Ann. N. Y. Acad. Sci. 2008;1137(1):214–217. doi: 10.1196/annals.1448.042. [DOI] [PubMed] [Google Scholar]
- 32.Choi J.J., Reich C.F., III, Pisetsky D.S. The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology. 2005;115(1):55–62. doi: 10.1111/j.1365-2567.2005.02130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adey A., Burton J.N., Kitzman J.O., Hiatt J.B., Lewis A.P., Martin B.K., Qiu R., Lee C., Shendure J. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013;500(7461):207–211. doi: 10.1038/nature12064. [DOI] [PMC free article] [PubMed] [Google Scholar]