Abstract
Background:
Cancer is a complex disease with a lucid etiology and in understanding the causation, we need to appreciate this complexity.
Objective:
Here we are aiming to gain insights into the genetic associations of prostate cancer through a network-based systems approach using the BC3Net algorithm.
Methods:
Specifically, we infer a prostate cancer Gene Regulatory Network (GRN) from a large-scale gene expression data set of 333 patient RNA-seq profiles obtained from The Cancer Genome Atlas (TCGA) database.
Results:
We analyze the functional components of the inferred network by extracting subnetworks based on biological process information and interpret the role of known cancer genes within each process. Fur-thermore, we investigate the local landscape of prostate cancer genes and discuss pathological associa-tions that may be relevant in the development of new targeted cancer therapies.
Conclusion:
Our network-based analysis provides a practical systems biology approach to reveal the collective gene-interactions of prostate cancer. This allows a close interpretation of biological activity in terms of the hallmarks of cancer.
Keywords: Gene regulatory network, Prostate cancer, Genomics, Systems biology, Network inference, Precision medicine, Data science
1. INTRODUCTION
Prostate cancer is among the most frequently diagnosed cancers and the 5th leading cause of cancer mortality worldwide for males [1]. Of the male population aged 50 and over, an estimated 40% have slow-growing prostate cancer. The incidence rate increases with age such that ageing is one of the several risk factors which also include: ethnicity, familial and genetic influences [2]. The survival rate of prostate cancer is improving which may be attributable to the increased monitoring of biomarkers such as prostate-specific antigen (PSA). PSA has allowed for the detection of prostate cancer in its primitive stage during which treatment may be most effective. However, the use of PSA as a diagnostic tool is far from ideal, mainly due to the over- detection of slow growing, non-fatal variants of the disease, the treatment of which causing more harm than good [3]. This slow growing phenotype typically results in death with prostate cancer rather than from the disease. Indeed this translates into the unique survival statistics of prostate cancer where in the UK, the one- five- and ten-year survival rates in 2011 have grown to 94%, 85% and 84% respectively [4].
As cancer progresses, oncogenes and tumour suppressor genes can undergo gain or loss in function, through changes in our genome. These changes are in the form of mutations and copy number variations. Many genes have recently been identified as contributing to the sporadic onset of prostate cancer along with hereditary predispositions to the disease. For instance, in a study [5] major genes associated with prostate cancer have been discussed, including ANX7, AR, ATBF1, BRCA1, BRCA2, CDKN1B, CHEK2, CYP17, CYP1B1, ELAC2, P53, PTEN and RAS. In general, cancer is understood to be a complex disease, because cancer is the result of mutations in many genes which may be involved in overlapping biological pathways that together form a complex cellular network. For this reason, cancer can be seen as a network disease that can be best interpreted by a systems perspective [6, 7]. Examples of such approaches for different cancer types can be found in some studies [8-11].
Owing to the fact that there is a poor separation between the lethal and non-lethal variants of prostate cancer, there is a growing need to determine reliable indicators that predict the best treatment for patients. With this in mind, our study aims to uncover the pathological associations of prostate cancer and to elucidate potential causes which may aid in biomarker discovery. Specifically, we infer a gene regulatory network (GRN) from a large-scale prostate cancer gene expression data set with the BC3Net algorithm [12]. Then, we apply a systems analysis of this GRN to provide insights into its functional and structural features by investigating, e.g., major hub genes and biological processes.
We would like to note that we call the network inferred from gene expression data a Gene Regulatory Network (GRN) and not, e.g., a gene transcriptomic network because it is known that a GRN contains aside from transcription regulations also information about protein bindings, and for this reason, the term is frequently used in the literature [13-17]. Hence, a GRN provides rich information about molecular regulations beyond transcription regulation.
This paper is organized as follows: First, we detail the methods and data used for the GRN inference and its analysis. Next, we present our results followed by their interpretation and discussions. Finally, the paper is concluded with a brief summary.
2. METHODS
2.1. Gene Expression Data
The most common form of prostate cancer is known as prostate adenocarcinoma that is present in 9 out of 10 cases. The remaining subtype is considered a rare form of the disease, and for this reason, we base our analysis on the most common prostate cancer form. To infer the prostate cancer GRN, we obtained data from The Cancer Genome Atlas (TCGA) containing 383 unique patient samples. Each patient sample is used to generate a gene expression profile on an Illumina next-generation sequencing (NGS) platform using the RNAseq_V2 protocol [18]. To quantify the reads RPKM (Reads per Kilobase per Million) mapped reads are used [18]. In total, each of the 383 patient samples consists of 20,531 gene expression values. On a technical note, we would like to remark that we repeated our analysis using TPM but found no differences in our results of the GRN.
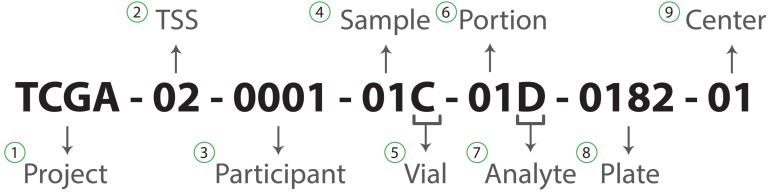
The TCGA has a comprehensive system in place to identify biospecimen data of samples. Specifically, each sample is assigned a unique barcode detailing specific data elements. Fig. (1) provides an example. Each of these 9 elements can vary depending on the patient hence, it is vital to use utilise this information.
Fig. (1).
Example of a TCGA barcode. 1) Project title, 2) Tissue source site, 3) Study patient identifier, 4) Type of sample, 5) Order of sample in a sequence of samples, 6) Order of portion in a sequence of sample portions, 7) Molecular type of material for analysis, 8) Order of plate in 96-well plates and 9) Centre analysing the material.
2.2. Preprocessing
Before the GRN can be inferred from the data, several preprocessing steps are needed. First, only samples obtained from solid tumours would be used. The TCGA barcodes are assigned to each expression profile and can be used to specify the type of sample used in acquiring the genetic material needed for NGS. Therefore the 4th element of these barcodes should be ‘01’ as this signifies the sample was taken from a solid tumour. This step reduced the number of patients from 383 to 333.
Networks are a description which provide a unique balance between simplicity and complexity. To maintain this harmony, we reduce the gene count, the next step in preprocessing. To achieve this, the average gene expression levels were calculated for each gene. The lower quartile was then removed. A large portion of these genes had a mean intensity level of 0 and as a result. were likely to be not expressed at all. This step reduced the number of genes in the data set from 20,502 to 15,376.
The final step in preprocessing was to log-transform the data. This step should always be employed in gene expression data as the intensity values are usually drastically skewed on a linear scale [19].
2.3. BC3NET
To infer the prostate cancer network from the gene expression dataset, the BC3NET algorithm [12] is employed. This GRN inference method is a bagging version of C3NET [20] used to construct a mutual information based network. The C3NET algorithm works as follows: For each gene pair in the TCGA dataset, the mutual information is estimated. Then, for each gene in the dataset,, the maximal mutual information pair is selected and tested for significance controlling the family-wise error rate by a Bonferroni correction for multiple testing.
The above procedure is repeated for , bootstrap datasets resulting in networks. From this ensemble of networks, an aggregate network G is formed by testing the statistical significance of occurrences of edges in .
2.4. Cancer Gene Census
For our analysis, we use the tier 1 subset of the Cancer Gene Census [21] (obtained on 22-09-2018). The tier 1 subset consists of those genes with a documented and well-established link to its relevant cancer. Indeed, for each of the genes in this list, cancer-associated mutations, translocations, amplifications, inversions duplication’s, deletions or genes affected by Copy Number Variations (CNV) are known. At the time of downloading, there was a total of 574 cancer census genes, 534 of which were present within our TCGA prostate adenocarcinoma dataset.
2.5. Network Attributes
We use various methods of evaluating the network structure. Next, we describe each technique used in our analysis.
2.5.1. Degree Distribution
First, we evaluate the degree distribution, which simply measures the number of connections each node has. For an adjacency matrix A, of an undirected network, the degree of a node i equals the sum of the i row, i.e., . From this, the degree distribution of the entire network can be obtained and tested if it follows a power law, such that:, where is the percentage of genes of degree in the GRN and is the exponent of the power law distribution.
2.5.2. Shortest Path Length
For the prostate cancer GRN, the shortest path length between pairs of genes, i.e., , is calculated using the Dijkstra distance [22]. The average shortest path length is calculated by:
Here, is a graph and is the number of nodes in .
2.5.3. Edge Density
The edge density of a network is the number of edges in a network divided by the total number of possible edges. The number of possible edges within an undirected network is , where n is the total number of genes within the network. The edge density is a further quantitative measure of the connected nature of a network and useful in comparisons between subnetworks.
2.6. Gene Pair Enrichment Analysis
To investigate the functionality of the interconnected structure of the inferred prostate cancer GRN, we use a Gene Pair Enrichment Analysis (GPEA) [23]. The GPEA uses a hypergeometric test based on the edges of the network. This analysis is performed with the help of two annotation packages downloaded from Bioconductor [24], GO.db (version 2.14) and org.Hs.eg.db (version 2.14).
3. RESULTS
3.1. Inferring the Prostate Cancer Gene Regulatory Network
Using the TCGA gene expression dataset, we infer the prostate cancer GRN. In the following, we call this network . The inferred network consists of 15,376 nodes (synonymous with genes) and 82,579 edges. The network is almost fully connected because 99.62% (15,318/15,376) of the genes can be reached from any gene. Hence, the Giant Connected Component (GCC) consists of 15,318 genes and 82,544 edges. The average shortest path length between genes is and the edge density of is . Analysing the degrees of the network reveals that the maximal degree is 96. Table 1 denotes the 9 highest degree genes, termed hub genes.
Table 1.
Topmost degrees of genes within the prostate cancer gene regulatory network, .
| - | Gene Symbols | Entrez ID | Degree |
|---|---|---|---|
| 1 | PIK3C2A | 5286 | 96 |
| 2 | SCAF11 | 9169 | 90 |
| 3 | AURKAIP1 | 54998 | 86 |
| 4 | RIF1 | 55183 | 85 |
| 5 | NDUFA13 | 51079 | 84 |
| 6 | NOSIP | 51070 | 79 |
| 7 | NAA10 | 8260 | 78 |
| 8 | CLTB | 1212 | 75 |
| 9 | ASXL2 | 55252 | 75 |
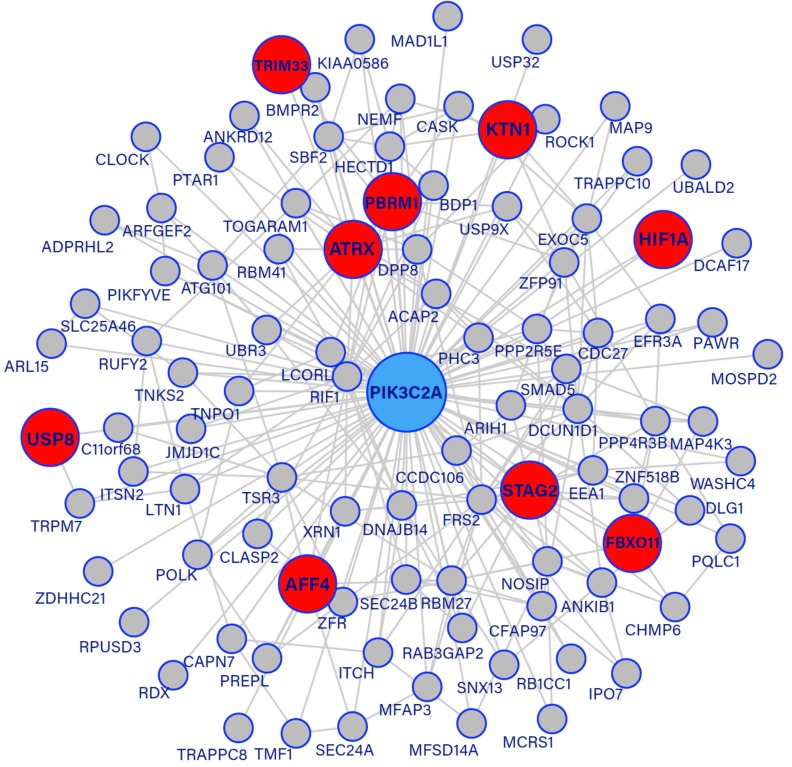
The topmost connected gene, PIK3C2A, functions in cellular proliferation, growth, survival and chemotaxis. In addition, it has been shown that actively lowering the levels of PIK3C2A reduced the proliferative ability of cancer cells in a number of cell lines [25]. Fig. (2) provides the subnetwork of the highest connected gene, PIK3C2A (in blue), including its 96 direct neighbours. Among these genes, there are 9 known cancer genes, highlighted in red.
Fig. (2).
Subnetwork of the topmost connected gene, PIK3C2A, shown in light-blue. Known cancer genes have been highlighted in red. (The color version of the figure is available in the electronic copy of the article).
The functional product of PIK3C2A is a member of the phosphoinositide 3-kinase protein family (PI3K). PI3K kinases are known to be involved in: cellular proliferation, signalling pathways, cell survival, migration, tumour formation, the progression of cancer and inter-cellular protein trafficking [26, 27]. PI3K kinases are composed of regulatory subunits that catalyse the synthesis of phosphoinositides which are important mediators of signal transduction in the mentioned processes. Furthermore, these kinases interact with various growth factors including Epidermal Growth Factor Receptor (EGFR) and Insulin-like Growth Factors (IGF). It has been shown that there is strong evidence of the expression of EGFR in prostate cancer leading to disease relapse and progression to androgen independence [28]. Here 100% of patients with metastatic prostate cancer expressed EGFR where an increased expression of this growth factor correlates with disease progression. For this reason, EGFR targeting could be relevant in the treatment of prostate cancer. IGF is also associated with tumorigenesis in prostate cancer where patients with an increased concentration of the genes functional product had a heightened risk of prostate cancer [29, 30]. PIK3C2A has a fundamental role in cell survival such that an mRNA threshold has been identified, below which the apoptotic (programmed cell death) process is switched on through the intrinsic cell death pathway [25].
Finally, it was found that actively reducing the level of PIK3C2A within tumour cells resulted in decreased proliferation and cell viability.
3.2. Functional Analysis of Biological Processes Using GPEA
The GPEA is performed using the R package, gpeaNet (version 1.0). This analysis highlights gene ontology terms with an enriched count of edges (interactions) among genes from the same gene ontology category. As multiple hypotheses are tested, the p-values have been adjusted by employing a Bonferroni correction. For the GPEA, we considered a total of 7,792 gene ontology terms from the category biological processes of which 670 (8.60%) are significant, for 0.05. To ensure only meaningful GO terms are tested, each term must have at least a size greater than 2 and less than 1,000 genes. The 35 most significant terms from this analysis are shown in Table 2. Each term is described by its biological process, the number of genes enriched for that term and corresponding interactions, an adjusted p-value and the number of known cancer genes present for that subnetwork. These GO terms describe important pathological processes such as the mRNA metabolic process (661 edges), mitotic cell cycle (710 edges), translation (485 edges), viral transcription (266 edges), cellular component disassembly (318 edges), protein localisation to membrane (305 edges) and protein targeting (359 edges).
Table 2.
Top most significant GO terms from a GPEA.
| GOID | Term | p-value | Size | Edges | CG |
|---|---|---|---|---|---|
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 0 | 117.00 | 259.00 | 5 |
| GO:0006414 | translational elongation | 0 | 105.00 | 268.00 | 3 |
| GO:0006415 | translational termination | 0 | 90.00 | 258.00 | 3 |
| GO:0006613 | cotranslational protein targeting to membrane | 0 | 106.00 | 267.00 | 4 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 0 | 104.00 | 267.00 | 4 |
| GO:0045047 | protein targeting to ER | 0 | 107.00 | 269.00 | 4 |
| GO:0070972 | protein localization to endoplasmic reticulum | 0 | 121.00 | 273.00 | 4 |
| GO:0072599 | establishment of protein localization to endoplasmic reticulum | 0 | 108.00 | 269.00 | 4 |
| GO:0006413 | translational initiation | 4.37e-316 | 149.00 | 283.00 | 8 |
| GO:0006612 | protein targeting to membrane | 1.94e-291 | 153.00 | 271.00 | 6 |
| GO:0019083 | viral transcription | 6.17e-290 | 149.00 | 266.00 | 12+ |
| GO:0043624 | cellular protein complex disassembly | 6.31e-278 | 153.00 | 262.00 | 6 |
| GO:0019080 | viral gene expression | 3.37e-275 | 159.00 | 266.00 | 12+ |
| GO:0043241 | protein complex disassembly | 3.63e-272 | 157.00 | 262.00 | 6 |
| GO:0000956 | nuclear-transcribed mRNA catabolic process | 1.91e-252 | 177.00 | 267.00 | 10+ |
| GO:0032984 | macromolecular complex disassembly | 5.74e-247 | 177.00 | 263.00 | 15+ |
| GO:0090150 | establishment of protein localization to membrane | 1.48e-244 | 213.00 | 294.00 | 13+ |
| GO:0006402 | mRNA catabolic process | 6.46e-243 | 188.00 | 270.00 | 10 |
| GO:0016071 | mRNA metabolic process | 4.26e-232 | 597.00 | 661.00 | 33+ |
| GO:0006401 | RNA catabolic process | 2.81e-222 | 213.00 | 276.00 | 12+ |
| GO:0019058 | viral life cycle | 4.87e-212 | 238.00 | 289.00 | 17+ |
| GO:0006412 | translation | 3.37e-207 | 459.00 | 485.00 | 26+ |
| GO:0072657 | protein localization to membrane | 6.30e-206 | 263.00 | 305.00 | 17+ |
| GO:0000278 | mitotic cell cycle | 1.57e-144 | 776.00 | 710.00 | 84+ |
| GO:0022411 | cellular component disassembly | 5.04e-140 | 364.00 | 318.00 | 33+ |
| GO:0022904 | respiratory electron transport chain | 4.44e-133 | 97.00 | 120.00 | 5 |
| GO:0045333 | cellular respiration | 6.56e-133 | 146.00 | 153.00 | 10+ |
| GO:0022900 | electron transport chain | 5.63e-131 | 99.00 | 120.00 | 5 |
| GO:0044764 | multi-organism cellular process | 8.93e-128 | 637.00 | 537.00 | 64+ |
| GO:0016032 | viral process | 3.64e-127 | 635.00 | 534.00 | 64+ |
| GO:0072594 | establishment of protein localization to organelle | 1.65e-122 | 427.00 | 345.00 | 49+ |
| GO:0044403 | symbiosis, encompassing mutualism through parasitism | 1.90e-117 | 678.00 | 557.00 | 64+ |
| GO:0044419 | interspecies interaction between organisms | 1.90e-117 | 678.00 | 557.00 | 64+ |
| GO:0044265 | cellular macromolecule catabolic process | 2.58e-114 | 704.00 | 576.00 | 53+ |
| GO:0006605 | protein targeting | 2.57e-110 | 467.00 | 359.00 | 48+ |
Interestingly, several of the 670 significant terms (not shown in Table 2) are known to be affected in prostate cancer while many are recognised as pathological trademarks of cancer. These terms include those associated with transcription where several transcription factors are known to initiate prostate cancer. These genes include NKX3.1, a mutation in which may predispose to prostate carcinogenesis [31] and MXI1 that regulates MYC (a known cancer gene) and is also associated with prostate cancer [32]. Cell adhesion processes are also linked to prostate carcinogenesis involving genes such as E-cadherin, decreased expression of which is associated with poor prognosis [33], c-Cam vital for prostate development and consistently found to have low expression levels in prostate cancer patients [34]. Finally, another important process is the growth factor (GF) response, e.g., involving IGF, TGF1 and EGF all of which are associated with advanced carcinoma and metastatic prostate cancer [30, 35-37].
3.3. Cancer Gene Census
To further study the functional modules of the prostate cancer GRN, the manually curated cancer gene census list was used to specify those genes in which mutations are directly linked to cancer. At the time of downloading this list consisted of a total of 574 genes, 534 of which were present
Size corresponds to the number of genes, edges the interactions, p-value is the adjusted value (Bonferroni correction), CG which lists the number of known cancer genes for that term. The “+” denotes a significant test for enrichment of the GO term with cancer genes. in our inferred . This list is used to highlight known cancer genes for each gene ontology term in the GPEA analysis. A hypergeometric test (one-sided Fishers exact test) is preformed to test for enrichment of cancer genes. In Table 2, the significant terms () are indicated by a “+” adjacent to the entry.
3.4. Gene Ontology Terms
Next, we investigate two gene ontology terms extracted from the prostate cancer GRN and evaluate the genes which are present in these functional modules. Furthermore, the cancer gene census list is used to highlight known cancer genes within the networks. The investigated GO terms are GO:0000278 that represents the biological process of the mitotic cell cycle and GO:0002682 which describes the regulation of immune system process.
3.4.1. GO:0000278: Mitotic Cell Cycle
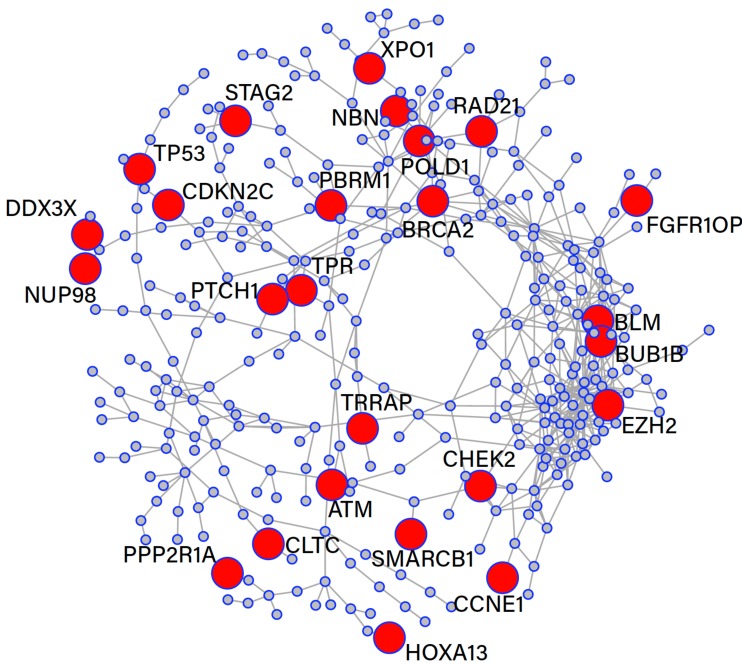
Cancer can be considered the result of a break down in function of the cell cycle, where proteins, transcription factors and regulatory components play an important role. If, through multiple mutations, this process malfunctions, there is a high likelihood of tumour formation. Fig. (3) shows a sub-graph of corresponding to the giant connected component of the gene ontology term GO:0000278 (Mitotic Cell Cycle). In total there are 776 genes with 710 interactions and the giant connected component contains 342 genes and 625 interactions. Interestingly, the network was found to contain a total of 84 known cancer genes for which Fisher’s exact test indicates a significant enrichment (p-value of ). Within the GCC, we found a total of 25 known cancer genes (highlighted in red) and the network has an average degree of 3.65. Interestingly, there appeared a cluster of 3 cancer genes BLM, BUB1B and EZH2 which have a significantly higher average degree of 10.
Fig. (3).
Gene Ontology term GO:0000278 corresponding to the mitotic cell cycle. (The color version of the figure is available in the electronic copy of the article).
Contained within this subnetwork there are several known cancer genes. For example: TP53, a tumour suppressor gene which under mutagenic transformation occurs in over 50% of cancers [38]. TP53 is considered a master regulator of diverse cellular processes that encodes for tumour protein p53, vital in the role of cancer prevention [39]. CLTC a gene which is in close proximity to TP53 currently has no association with prostate cancer. Although, Clathrin Heavy Chain (CHC), aliases of the CLTC gene, is required for p53 mediated transcription [40]. CHC, a cytosolic transporter found in both the cytosol and the nucleus has been shown to enhance p53 reliant trans-activation by binding to p53 forming a p53-CHC molecule. Furthermore, the reduction of CHC/CLTC via RNA interference mitigates the transcription of TP53. The p53-CHC molecule stabilises a communication between p53 and p300, this promotes the transcription of TP53 and translation to p53. For several decades TP53 has been studied for its potential in cancer therapies and several clinical trials using TP53 targeting have shown much promise [41, 42]. In prostate cancer, our subnetwork proposes a plausible targeting mechanism for TP53 through CLTC regulation.
Enhancer of Zeste Homolog 2 (EZH2) is also present in the GCC of this functional module and is known to be involved in the progression of prostate cancer by promoting proliferation and invasiveness of tumour cells [43, 44]. Furthermore, in a study of the effect of EZH2 in gastric cancer it was found that RNA interference of this gene up-regulated the p53 protein and down-regulated cyclin E, encoded by the CCNE1 gene [45]. CCNE1 is found in close proximity to the EZH2 gene and with the previous knowledge perhaps indicates involvement in EZH2 expression and prostate cancer progression. BRCA2 is another well characterised cancer gene that recently has traits linked to prostate cancer. It can be seen that a connection to RAD21 exists and together they play a major role in DNA repair by homologous recombination [46]. Recently, BRCA2 has proven to be a reliable biomarker for the prognosis of prostate cancer in that BRCA2 is the most strongly associated prostate cancer pre-disposition gene identified to date [47].
3.4.2. GO:0002682: Regulation of Immune System Process
The human immune system function is not limited by the war on infectious pathogens. In fact, its function extrapolates to maintaining tissue homeostasis and destroying damaged cells. As the immune system is usually highly effective in its purpose, it is strange that cancer has such a high incidence in humans [48]. In recent years tumourigenesis has been linked to mutations of genes involved in the immune response. Several mouse models have been investigated in which manipulation of the immune response has caused spontaneous tumours to form. We now know that the breakdown of the function of the immune system can play a major role in many cancers. Additionally, leukocytes have been identified as important regulators of cancer development. For this reason, we evaluate the regulation of the immune system process.
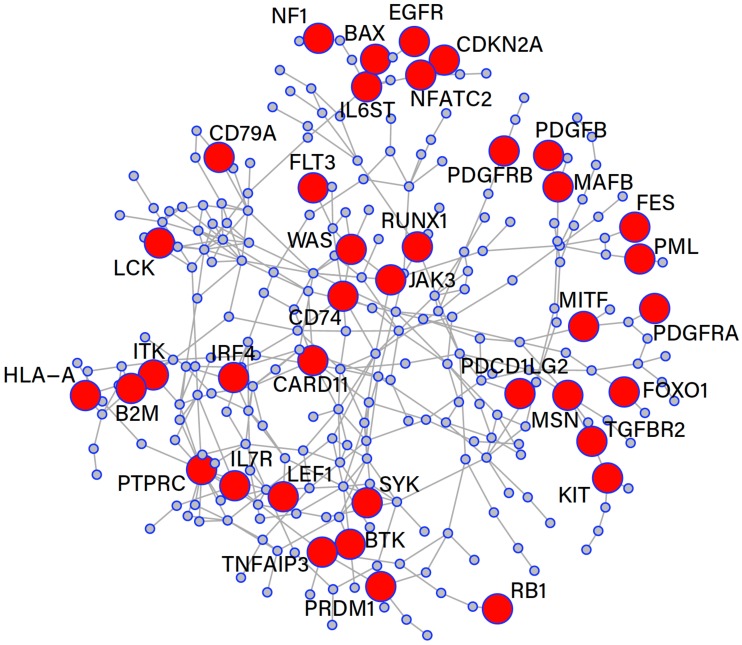
The giant connected component for this GO term is shown in Fig. (4). In total, this subnetwork contains 611 genes with 895 interactions, and there are 131 known cancer genes present in this subnetwork. A Fisher’s exact test for enrichment was performed and found that the cancer genes were significantly enriched for this GO term with a p-value of . The GCC contains 271 genes of which 38 are known cancer genes. The average degree for this network was 3.05.
Fig. (4).
Gene Ontology term GO:0002682, regulation of immune system process.
In prostate cancer, there is a wealth of literature that points towards an association of chronic inflammation and carcinogenesis [49]. Several genes found in our subnetwork play a role in the pathology of the inflammatory response, e.g., Interleukin 6 Signal Transducer (IL6ST) which has an interesting association with many cytokines involved in prostate cancer. IL6ST is a signal transducer of interleukin 6 (IL6) and oncostatin M (OSM), which are growth factors for prostate cancer. Both of these GF’s have been shown to significantly up-regulate Vascular Endothelial Growth Factor (VEGF) in prostate cancer cells which promotes the vasculature to support tumour cell growth [50]. An important concept in cancer pathology is that inflammation will promote the growth of new vasculature which tumour cells are completely dependant upon for survival. This is usually mediated through GF’s such as VEGF which is a well-known target for cancer therapies [51]. It has also been observed that higher levels of IL6 and OSM are present in metastatic prostate cancer. Therefore with this understanding IL6ST regulates IL6 and OSM within a cell which in turn up-regulates
VEGF, where high levels correlate to a well-defined vascular network, symptomatic of aggressive tumours.
In our subnetwork, there are several other important observations such as the presence of CD74, a receptor for macrophage migration inhibitory factor (MIF). MIF is commonly found to be over-expressed in prostate cancer and with an over-expression of CD74 in tumour cells, has been shown to attenuate growth and invasion of prostate cancer cells [52]. FLT3, codes for a receptor tyrosine kinase that is activated by the FLT3-ligand. Targeting of FLT3 in immunotherapy treatments for prostate cancer has shown great promise in animal models. Here treatment of 30 microgram/day dose of FLT3-ligand resulted in significantly decreased tumour size (p 0.0001) [53]. Another gene known to be associated with cancer and present in the GCC is Runt-related transcription factor (RUNX1). RUNX1 is associated with a 2.76 fold increased risk of advanced stage prostate cancer and a 9.52 fold greater risk of metastasis [54].
3.5. Local Landscape of Prostate Cancer Genes
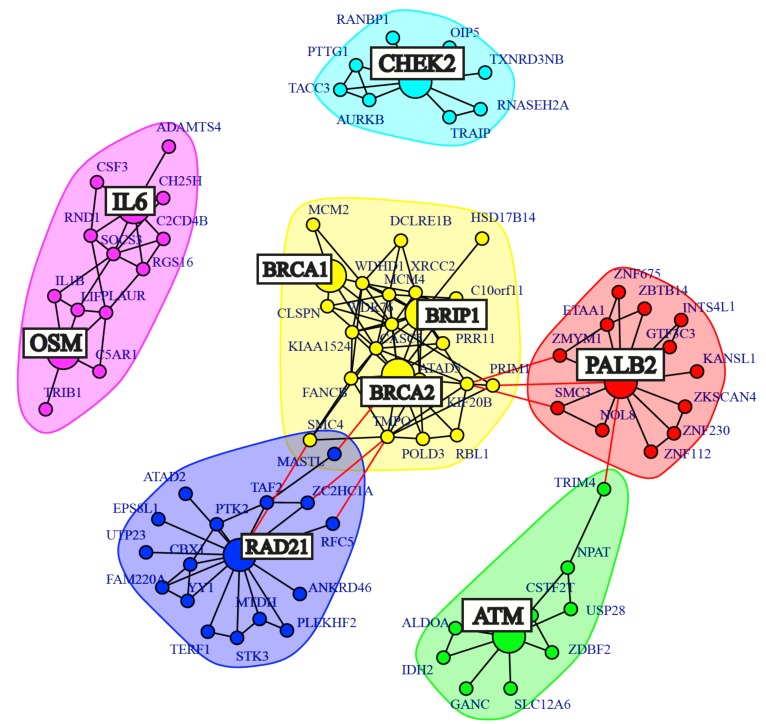
Next, we examine 6 genes that are frequently observed in prostate cancer and 3 genes for which previous results indicate an association. Specifically, the 6 genes known to be associated with prostate cancer are: ATM, BRCA1, BRCA2, BRIP1, CHEK2, PALB2 and the further 3 genes are: IL6, OSM and RAD21. Out of these 9 genes, 7 are known cancer genes contained within the cancer gene census list (BRCA1, BRCA2, BRIP1, RAD21, ATM, PALB2 and CHEK2).
For these genes, a subnetwork was constructed by determining all shortest paths that connect these genes, plus the first degree neighbours of the genes. For future reference, this network will be referred to as , shown in Fig. (5), with the known cancer genes highlighted as large nodes. Overall contains 86 genes (9 specific cancer genes and 77 neighbours) and 164 interactions.
Fig. (5).
First degree neighbour network of 9 genes known to be implemented in prostate cancer.
Interestingly, BRCA1, BRCA2 and BRIP1 are in grouped together. BRIP1 is involved in DNA repair by homologous recombination in a process that is dependent on the interaction with BRCA1 and therefore BRCA2. Furthermore to investigate the discovered interaction between RAD21 and BRCA2, (Fig. 3), the RAD21 gene was included in our analysis. There are a number of interactions linking RAD21 to BRCA2, the most strongly associated prostate cancer pre-disposition gene [47]. RAD21 over-expression is commonly observed in BRCA2 present prostate cancer [55], where RAD21 is a key component of a cohesion complex needed for several cellular processes. In fact, RAD21 over-expression was present in 53% of the prostate tumours and is perhaps an equivalently useful biomarker in prostate cancer as BRCA2. It is also important to note that all of the mentioned genes are well known hallmarks of cancer as indicated by the cancer gene census list.
Two more purposely extracted genes, IL6 and OSM were used from the prostate cancer GRN. IL6 and OSM are regulated by IL6ST, a gene that was present in the sub-network shown in Fig. (4). Both IL6 and OSM are growth factors associated with prostate cancer that together significantly up-regulate the expression of VEGF [50]. In a meta-analysis of prostate cancer, VEGF is strongly associated with poor survival for patients (hazard ratio = 1.54) [56]. In recent years VEGF has been a hot topic in cancer research such that there is resounding evidence of its association with cancer progression, poor prognosis and survival in prostate cancer [50, 57, 58]. Interestingly, both IL6 and OSM are interconnected in . An additional analysis showed that VEGFA, VEGFB, VEGFC, IL6, OSM and IL6ST are connected indirectly through a 2nd degree neighbour network (results not shown).
3.6. Connectivity and Edge Weight Comparison
For the construction of the prostate cancer GRN, , an ensemble of 100 were generated and then aggregated to form the main network. An edge weight can be estimated for each gene pair that interacts with one another. This weight signifies the fraction of how often the edge was observed between two genes in the ensemble and, hence, represents a statistical interrelation of the data, showing which gene pairs have the strongest association.
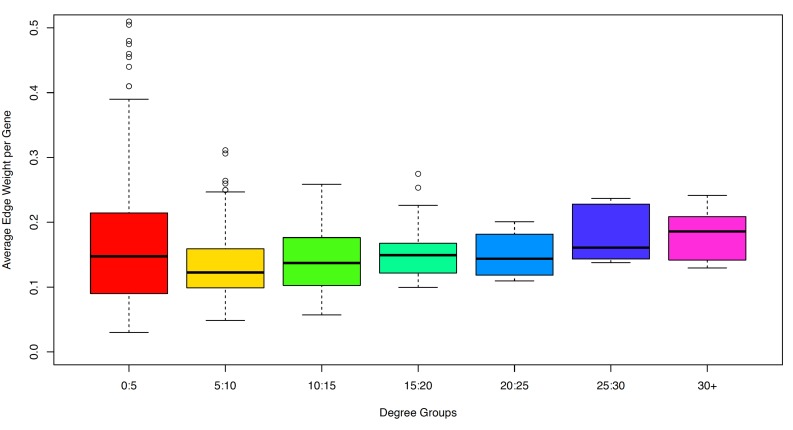
The degree for each gene in was classified into groups with increments of 5, for instance, those genes which had a degree between 0 and 5 were assigned to group 1. This was then plotted as the average edge weight per gene pair (y axis) against the Degree Groups (x axis) in the form of a boxplot in Fig. (6). A one way analysis of variance (ANOVA) was performed and found to be significant with a p-value of . Indeed, from Fig. (6), we see that this significance is due to increasing values in the average edge weight. As a consequence of this, our results indicate that hub genes, for example PIK3C2A, have several interactions which show a strong association. Potentially, this could mean the interactions observed for high degree genes may be more reliant.
Fig. (6).
Boxplot of average edge-weight versus degree groups. Degree groups represent discrete groups of genes in with increments of 5.
CONCLUSION
Of the many observable interactions within our prostate cancer GRN, we focused here on discussing those that may be of clinical relevance. That is to say, several of these molecular associations present plausible mechanisms that may be used for targeted cancer therapies. For instance, the hub gene PIK3C2A can be inhibited by PI3K inhibitors acting on the alpha subunit of this protein. Furthermore, the IL6ST-[IL6-OSM]-VEGF pathway presents a mechanism for actively reducing the VEGF abundance within prostate cancer cells. Here it was found that IL6 and OSM increased VEGF expression that is mediated by IL6ST. Importantly, the increased expression of VEGF within tumour cells is often attributed to poor prognosis and aggressive cancers.
In the current research, there is much focus on this growth factor and its role in cancer. The over-expression of VEGF in tumour cells has an angiogenic effect where the blood vessels allow the delivery of nutrients maintaining the continued cell division and survival, one of the hallmarks of cancer [59, 60].
Acknowledgements
For all calculations, data processing and network inference, the programming language ’R’ has been utilized [60]. Matthias Dehmer would like to thank the Austrian Science Funds for supporting this work (project P 30031).
Ethics Approval and Consent to Participate
Not applicable.
Human and Animal Rights
No Animals/Humans were used for studies that are the basis of this research.
Consent for Publication
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: IARC Cancer Base No. 11. IARC Publications; 2013. [DOI] [PubMed] [Google Scholar]
- 2.Patel A.R., Klein E.A. Risk factors for prostate cancer. Nat. Clin. Pract. Urol. 2009;6(2):87–95. doi: 10.1038/ncpuro1290. [DOI] [PubMed] [Google Scholar]
- 3.Chodak G. Prostate cancer: Epidemiology, screening, and biomarkers. Rev. Urol. 2006;8(2):S3–S8. [PMC free article] [PubMed] [Google Scholar]
- 4.Quaresma M., Coleman M.P., Rachet B. 40-year trends in an index of survival for all cancers combined and survival adjusted for age and sex for each cancer in England and Wales, 1971-2011: A population-based study. Lancet. 2015;385(9974):1206–1218. doi: 10.1016/S0140-6736(14)61396-9. [DOI] [PubMed] [Google Scholar]
- 5.Dong J.T. Prevalent mutations in prostate cancer. J. Cell. Biochem. 2006;97(3):433–447. doi: 10.1002/jcb.20696. [DOI] [PubMed] [Google Scholar]
- 6.Wang E., Lenferink A., O’Connor-McCourt M. Cancer systems biology: Exploring cancer-associated genes on cellular networks. Cell. Mol. Life Sci. 2007;64(14):1752–1762. doi: 10.1007/s00018-007-7054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreeger P.K., Lauffenburger D.A. Cancer systems biology: A network modeling perspective. Carcinogenesis. 2010;31(1):2–8. doi: 10.1093/carcin/bgp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Matos Simoes R., Tripathi S., Emmert-Streib F. Organizational structure of the peripheral gene regulatory network in B-cell lymphoma. BMC Syst. Biol. 2012;6:38. doi: 10.1186/1752-0509-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Matos Simoes R., Dehmer M., Emmert-Streib F. B-cell lymphoma gene regulatory networks: Biological consistency among inference methods. Front. Genet. 2013;4:281. doi: 10.3389/fgene.2013.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emmert-Streib F., de Matos Simoes R., Glazko G., McDade S., Haibe-Kains B., Holzinger A., Dehmer M., Campbell F. Functional and genetic analysis of the colon cancer network. BMC Bioinformatics. 2014;15(Suppl. 6):S6. doi: 10.1186/1471-2105-15-S6-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emmert-Streib F., de Matos Simoes R., Mullan P., Haibe-Kains B., Dehmer M. The gene regulatory network for breast cancer: Integrated regulatory landscape of cancer hallmarks. Front. Genet. 2014;5:15. doi: 10.3389/fgene.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Matos Simoes R., Emmert-Streib F. Bagging statistical network inference from large-scale gene expression data. PLoS One. 2012;7(3):e33624. doi: 10.1371/journal.pone.0033624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hecker M., Lambeck S., Toepfer S., van Someren E., Guthke R. Gene regulatory network inference: Data integration in dynamic models - A review. Biosystems. 2009;96(1):86–103. doi: 10.1016/j.biosystems.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Hartemink A.J. Reverse engineering gene regulatory networks. Nat. Biotechnol. 2005;23(5):554–555. doi: 10.1038/nbt0505-554. [DOI] [PubMed] [Google Scholar]
- 15.Emmert-Streib F., Dehmer M., Haibe-Kains B. Untangling statistical and biological models to understand network inference: The need for a genomics network ontology. Front. Genet. 2014;5:299. doi: 10.3389/fgene.2014.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emmert-Streib F., Dehmer M., Haibe-Kains B. Gene regulatory networks and their applications: Understanding biological and medical problems in terms of networks. Front. Cell Dev. Biol. 2014;2:38. doi: 10.3389/fcell.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Joshi T., Zhang X-S., Xu D., Chen L. Inferring gene regulatory networks from multiple microarray datasets. Bioinformatics. 2006;22(19):2413–2420. doi: 10.1093/bioinformatics/btl396. [DOI] [PubMed] [Google Scholar]
- 18.Li B., Ruotti V., Stewart R.M., Thomson J.A., Dewey C.N. RNA-seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26(4):493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newton M.A., Kendziorski C.M., Richmond C.S., Blattner F.R., Tsui K.W. On differential variability of expression ratios: Improving statistical inference about gene expression changes from microarray data. J. Comput. Biol. 2001;8(1):37–52. doi: 10.1089/106652701300099074. [DOI] [PubMed] [Google Scholar]
- 20.Altay G., Emmert-Streib F. Structural influence of gene networks on their inference: Analysis of c3net. Biol. Direct. 2011;6(1):31. doi: 10.1186/1745-6150-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nat. Rev. Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dijkstra E.W. A note on two problems in connexion with graphs. Numer. Math. 1959;1(1):269–271. [Google Scholar]
- 23.de Matos Simoes R., Dehmer M., Emmert-Streib F. Interfacing cellular networks of S. cerevisiae and E. coli: Connecting dynamic and genetic information. BMC Genomics. 2013;14(1):324. doi: 10.1186/1471-2164-14-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A.J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J.Y., Zhang J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elis W., Triantafellow E., Wolters N.M., Sian K.R., Caponigro G., Borawski J., Gaither L.A., Murphy L.O., Finan P.M., Mackeigan J.P. Down-regulation of class ii phosphoinositide 3-kinase α expression below a critical threshold induces apoptotic cell death. Mol. Cancer Res. 2008;6(4):614–623. doi: 10.1158/1541-7786.MCR-07-0262. [DOI] [PubMed] [Google Scholar]
- 26.Vivanco I., Sawyers C.L. The phosphatidylinositol 3-kinase-akt pathway in human cancer. Nat. Rev. Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 27.Cantley L.C. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 28.Di Lorenzo G., Tortora G., D’Armiento F.P., De Rosa G., Staibano S., Autorino R., D’Armiento M., De Laurentiis M., De Placido S., Catalano G., Bianco A.R., Ciardiello F. Expression of epidermal growth factor receptor correlates with disease relapse and progression to androgen-independence in human prostate cancer. Clin. Cancer Res. 2002;8(11):3438–3444. [PubMed] [Google Scholar]
- 29.Platz E.A., Pollak M.N., Leitzmann M.F., Stampfer M.J., Willett W.C., Giovannucci E. Plasma insulin- like growth factor-1 and binding protein-3 and subsequent risk of prostate cancer in the PSA era. Cancer Causes Control. 2005;16(3):255–262. doi: 10.1007/s10552-004-3484-8. [DOI] [PubMed] [Google Scholar]
- 30.Chan J.M., Stampfer M.J., Giovannucci E., Gann P.H., Ma J., Wilkinson P., Hennekens C.H., Pollak M. Plasma insulin-like growth factor-i and prostate cancer risk: a prospective study. Science. 1998;279(5350):563–566. doi: 10.1126/science.279.5350.563. [DOI] [PubMed] [Google Scholar]
- 31.Bhatia-Gaur R., Donjacour A.A., Sciavolino P.J., Kim M., Desai N., Young P., Norton C.R., Gridley T., Cardiff R.D., Cunha G.R., Abate-Shen C., Shen M.M. Roles for nkx3.1 in prostate development and cancer. Genes Dev. 1999;13(8):966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eagle L.R., Yin X., Brothman A.R., Williams B.J., Atkin N.B., Prochownik E.V. Mutation of the mxi1 gene in prostate cancer. Nat. Genet. 1995;9(3):249–255. doi: 10.1038/ng0395-249. [DOI] [PubMed] [Google Scholar]
- 33.Umbas R., Isaacs W.B., Bringuier P.P., Schaafsma H.E., Karthaus H.F., Oosterhof G.O., Debruyne F.M., Schalken J.A. Decreased e-cadherin expression is associated with poor prognosis in patients with prostate cancer. Cancer Res. 1994;54(14):3929–3933. [PubMed] [Google Scholar]
- 34.Kleinerman D.I., Troncoso P., Lin S.H., Pisters L.L., Sherwood E.R., Brooks T., von Eschenbach A.C., Hsieh J.T. Consistent expression of an epithelial cell adhesion molecule (c-cam) during human prostate development and loss of expression in prostate cancer: Implication as a tumor suppressor. Cancer Res. 1995;55(6):1215–1220. [PubMed] [Google Scholar]
- 35.Wolk A., Mantzoros C.S., Andersson S.O., Bergström R., Signorello L.B., Lagiou P., Adami H.O., Trichopoulos D. Insulin-like growth factor 1 and prostate cancer risk: A population-based, case-control study. J. Natl. Cancer Inst. 1998;90(12):911–915. doi: 10.1093/jnci/90.12.911. [DOI] [PubMed] [Google Scholar]
- 36.Wikström P., Stattin P., Franck-Lissbrant I., Damber J.E., Bergh A. Transforming growth factor β1 is associated with angiogenesis, metastasis, and poor clinical outcome in prostate cancer. Prostate. 1998;37(1):19–29. doi: 10.1002/(sici)1097-0045(19980915)37:1<19::aid-pros4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 37.Olapade-Olaopa E.O., Moscatello D.K., MacKay E.H., Horsburgh T., Sandhu D.P., Terry T.R., Wong A.J., Habib F.K. Evidence for the differential expression of a variant EGF receptor protein in human prostate cancer. Br. J. Cancer. 2000;82(1):186–194. doi: 10.1054/bjoc.1999.0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oren M. Decision making by p53: Life, death and cancer. Cell Death Differ. 2003;10(4):431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 39.Farnebo M., Bykov V.J., Wiman K.G. The p53 tumor suppressor: a master regulator of diverse cellular processes and therapeutic target in cancer. Biochem. Biophys. Res. Commun. 2010;396(1):85–89. doi: 10.1016/j.bbrc.2010.02.152. [DOI] [PubMed] [Google Scholar]
- 40.Enari M., Ohmori K., Kitabayashi I., Taya Y. Requirement of clathrin heavy chain for p53-mediated transcription. Genes Dev. 2006;20(9):1087–1099. doi: 10.1101/gad.1381906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimada H., Matsubara H., Shiratori T., Shimizu T., Miyazaki S., Okazumi S., Nabeya Y., Shuto K., Hayashi H., Tanizawa T., Nakatani Y., Nakasa H., Kitada M., Ochiai T. Phase i/ii adenovi- ral p53 gene therapy for chemoradiation resistant advanced esophageal squamous cell carcinoma. Cancer Sci. 2006;97(6):554–561. doi: 10.1111/j.1349-7006.2006.00206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventura A., Kirsch D.G., McLaughlin M.E., Tuveson D.A., Grimm J., Lintault L., Newman J., Reczek E.E., Weissleder R., Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445(7128):661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 43.Varambally S., Dhanasekaran S.M., Zhou M., Barrette T.R., Kumar-Sinha C., Sanda M.G., Ghosh D., Pienta K.J., Sewalt R.G., Otte A.P., Rubin M.A., Chinnaiyan A.M. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 44.Bryant R.J., Cross N.A., Eaton C.L., Hamdy F.C., Cunliffe V.T. EZH2 promotes proliferation and invasiveness of prostate cancer cells. Prostate. 2007;67(5):547–556. doi: 10.1002/pros.20550. [DOI] [PubMed] [Google Scholar]
- 45.Choi J.H., Song Y.S., Yoon J.S., Song K.W., Lee Y.Y. Enhancer of zeste homolog 2 expression is associated with tumor cell proliferation and metastasis in gastric cancer. APMIS. 2010;118(3):196–202. doi: 10.1111/j.1600-0463.2009.02579.x. [DOI] [PubMed] [Google Scholar]
- 46.Yan M., Xu H., Waddell N., Shield-Artin K., Haviv I., kCon Fab, McKay M.J., Fox, S.B. Enhanced RAD21 cohesin expression confers poor prognosis in BRCA2 and BRCAX, but not BRCA1 familial breast cancers. Breast Cancer Res. 2012;14(2):R69. doi: 10.1186/bcr3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitra A.V., Bancroft E.K., Barbachano Y., Page E.C., Foster C.S., Jameson C., Mitchell G., Lindeman G.J., Stapleton A., Suthers G., Evans D.G., Cruger D., Blanco I., Mercer C., Kirk J., Maehle L., Hodgson S., Walker L., Izatt L., Douglas F., Tucker K., Dorkins H., Clowes V., Male A., Donaldson A., Brewer C., Doherty R., Bulman B., Osther P.J., Salinas M., Eccles D., Axcrona K., Jobson I., Newcombe B., Cybulski C., Rubinstein W.S., Buys S., Townshend S., Friedman E., Domchek S., Ramon Y., Cajal T., Spigelman A., Teo S.H., Nicolai N., Aaronson N., Ardern-Jones A., Bangma C., Dearnaley D., Eyfjord J., Falconer A., Grönberg H., Hamdy F., Johannsson O., Khoo V., Kote-Jarai Z., Lilja H., Lubinski J., Melia J., Moynihan C., Peock S., Rennert G., Schröder F., Sibley P., Suri M., Wilson P., Bignon Y.J., Strom S., Tischkowitz M., Liljegren A., Ilencikova D., Abele A., Kyriacou K., van Asperen C., Kiemeney L., IMPACT Study Collaborators Easton, D.F.; Eeles, R.A. Targeted prostate cancer screening in men with mutations in BRCA1 and BRCA2 detects aggressive prostate cancer: Preliminary analysis of the results of the IMPACT study. BJU Int. 2011;107(1):28–39. doi: 10.1111/j.1464-410X.2010.09648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Visser K.E., Eichten A., Coussens L.M. Paradoxical roles of the immune system during cancer development. Nat. Rev. Cancer. 2006;6(1):24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 49.Vasto S., Carruba G., Candore G., Italiano E., Di Bona D., Caruso C. Inflammation and prostate cancer. Future Oncol. 2008;4(5):637–645. doi: 10.2217/14796694.4.5.637. [DOI] [PubMed] [Google Scholar]
- 50.Weiss T.W., Simak R., Kaun C., Rega G., Pflüger H., Maurer G., Huber K., Wojta J. Oncostatin M and IL-6 induce u-PA and VEGF in prostate cancer cells and correlate in vivo. Anticancer Res. 2011;31(10):3273–3278. [PubMed] [Google Scholar]
- 51.Ellis L.M., Hicklin D.J. VEGF-targeted therapy: Mechanisms of anti-tumour activity. Nat. Rev. Cancer. 2008;8(8):579–591. doi: 10.1038/nrc2403. [DOI] [PubMed] [Google Scholar]
- 52.Meyer-Siegler K.L., Iczkowski K.A., Leng L., Bucala R., Vera P.L. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J. Immunol. 2006;177(12):8730–8739. doi: 10.4049/jimmunol.177.12.8730. [DOI] [PubMed] [Google Scholar]
- 53.Ciavarra R.P., Somers K.D., Brown R.R., Glass W.F., Consolvo P.J., Wright G.L., Schellhammer P.F. Flt3-ligand induces transient tumor regression in an ectopic treatment model of major histocompatibility complex-negative prostate cancer. Cancer Res. 2000;60(8):2081–2084. [PubMed] [Google Scholar]
- 54.Huang S.P., Lan Y.H., Lu T.L., Pao J.B., Chang T.Y., Lee H.Z., Yang W.H., Hsieh C.J., Chen L.M., Huang L.C., Ting W.C., Bao B.Y. Clinical significance of runt-related transcription factor 1 polymorphism in prostate cancer. BJU Int. 2011;107(3):486–492. doi: 10.1111/j.1464-410X.2010.09512.x. [DOI] [PubMed] [Google Scholar]
- 55.Deb S., Huiling X., Thorne H., Willems-Jones A., Clouston D., Bolton D., Ramsay R., Fox S.B. Rad21 overexpression is frequently observed in BRCA-X prostate cancers. Hered. Cancer Clin. Pract. 2012;10(Suppl. 2):A59. [Google Scholar]
- 56.Zhan P., Ji Y.N., Yu L.K. VEGF is associated with the poor survival of patients with prostate cancer: A meta-analysis. Transl. Androl. Urol. 2013;2(2):99–105. doi: 10.3978/j.issn.2223-4683.2013.06.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y.Z., Wong Y.C. Sex hormone-induced prostatic carcinogenesis in the noble rat: The role of insulin-like growth factor-1 (IGF-1) and vascular endothelial growth factor (VEGF) in the development of prostate cancer. Prostate. 1998;35(3):165–177. doi: 10.1002/(sici)1097-0045(19980515)35:3<165::aid-pros2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 58.Zhong H., Semenza G.L., Simons J.W., De Marzo A.M. Up-regulation of hypoxia-inducible factor 1α is an early event in prostate carcinogenesis. Cancer Detect. Prev. 2004;28(2):88–93. doi: 10.1016/j.cdp.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 59.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 60.R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]