Abstract
Introduction
Emergence of resistance determinants of bla NDM and mcr-1 has undermined the antimicrobial effectiveness of the last line drugs carbapenems and colistin.
Aim
This work aimed to assess the prevalence of bla NDM and mcr-1 in E. coli strains collected from food in Shenzhen, China, during the period 2015 to 2017.
Methods
Multidrug-resistant E. coli strains were isolated from food samples. Plasmids encoding mcr-1 or bla NDM genes were characterised and compared with plasmids found in clinical isolates.
Results
Among 1,166 non-repeated cephalosporin-resistant E. coli strains isolated from 2,147 food samples, 390 and 42, respectively, were resistant to colistin and meropenem, with five strains being resistant to both agents. The rate of resistance to colistin increased significantly (p < 0.01) from 26% in 2015 to 46% in 2017, and that of meropenem resistance also increased sharply from 0.3% in 2015 to 17% in 2017 (p < 0.01). All meropenem-resistant strains carried a plasmid-borne bla NDM gene. Among the colistin-resistant strains, three types of mcr-1-bearing plasmids were determined. Plasmid sequencing indicated that these mcr-1 and bla NDM-bearing plasmids were structurally similar to those commonly recovered from clinical isolates. Interestingly, both mcr-1-bearing and bla NDM-bearing plasmids were transferrable to E. coli strain J53 under selection by meropenem, yet only mcr-1-bearing plasmids were transferrable under colistin selection.
Conclusion
These findings might suggest that mobile elements harbouring mcr-1 and bla NDM have been acquired by animal strains and transmitted to our food products, highlighting a need to prevent a spike in the rate of drug resistant food-borne infections.
Keywords: blaNDM, mcr-1, E. coli, food, plasmid resistance
Introduction
Antimicrobial resistance poses an increasing risk to human and animal health worldwide. In particular, carbapenem resistance mediated by serine β-lactamases and metallo-β-lactamases (MBLs), such as the OXA enzymes produced by Acinetobacter baumannii and Klebsiella pneumoniae carbapenemase (KPC-1) and New Delhi metallo-β-lactamase (NDM-1) produced by Enterobacteriaceae, is associated with a high mortality rate among hospitalised patients [1,2]. NDM-1, a type of Ambler class B metallo-β-lactamases (MBLs), exhibits high hydrolytic activity against almost all known β-lactam antimicrobials (except aztreonam), including the last-line carbapenems [3,4]. It was first found to be produced by K. pneumoniae and Escherichia coli strains isolated from a Swedish patient of Indian origin who was admitted to hospital in New Delhi, India [5]. Thereafter, the bla NDM-1 gene disseminated in various countries and regions such as China, the Middle East, South East Asia and Europe [4]. This multidrug resistance gene, which may be located on either plasmids or chromosome [3,6,7], leaves few therapeutic options for infected patients. In China, Ho et al. reported the first isolation of bla NDM-1-positive E. coli from a 1-year-old infant and its mother in 2011 [8]. NDM-1-producing Enterobacteriaceae have since disseminated to various provinces in China, with the majority of such strains isolated from stool samples [9]. However, reports of isolation of carbapenem-resistant Enterobacteriaceae (CRE) from food samples remain scarce around the world.
Tigecycline and colistin have been regarded as last-line antibiotics used to treat serious infections caused by carbapenemase-producing Enterobacteriaceae [10]. Colistin (polymyxin E) belongs to the family of polymyxins which act against Gram-negative bacteria, including most species of Enterobacteriaceae, with broad-spectrum activity. Until 2015, almost all reported polymyxin resistance mechanisms were coded on the chromosome and mediated by mutations in two component regulatory systems (PmrAB, PhoPQ), loss of lipopolysaccharide, or mgrB inactivation in the case of K. pneumoniae [10,11]. Importantly, a report by Liu et al. in November 2015 described the emergence of a conjugative plasmid-mediated colistin resistance gene, mcr-1, which encodes an enzyme (MCR-1) that belongs to the phosphoethanolamine transferase enzyme family and leads to altered bacterial lipid A in Enterobacteriaceae through modification of the phosphoethanolamine moiety [10-12]. Identification of mcr-1 as a plasmid-mediated resistance mechanism highlights its potential to act as a transmissible colistin resistance determinant. Following this discovery, several reports have documented the presence of mcr-1 in strains of different species of Enterobacteriaceae that exhibited multidrug resistance phenotypes and were recovered from farm animals, food samples, human faecal samples, environmental samples and samples collected in clinical settings [11-14]. Plasmid types including IncX4, IncI2, IncP, IncFII and IncHI2, which harbour the mcr-1 gene, have been reported [12,14-16].
Several recent studies have reported the recovery of strains that harboured both the bla NDM and mcr-1 genes, including bla NDM-1/bla NDM-5/bla NDM-7 and mcr-1 in Enterobacteriaceae species, especially in E. coli [17-19], as well as bla NDM-9 and mcr-1 in Cronobacter sakazakii [20]. These reports focused on the epidemiological features, evolutionary origin and dissemination routes of resistance genes in different countries. However, few of these studies involved surveillance of prevalence of the bla NDM and mcr-1 genes for a prolonged period. Here, we report the presence of bla NDM and mcr-1 in E. coli isolated from food samples in Shenzhen, China, during the period 2015 to 2017 and assessed the prevalence of both genes in these E. coli food isolates. We investigated the genomic structures of the mobile genetic elements that encoded bla NDM and/or mcr-1 in these strains to elucidate the transferability of these important resistance determinants.
Methods
Bacterial isolation
E. coli isolates were obtained from food samples, including pork, chicken, beef and shrimps, collected from wet markets and supermarkets in Shenzhen, Guangdong Province, China during the period from 10 August 2015 to 17 April 2017. Three isolation methods were employed to increase the yield. Details of these are described in the Supplement.
Antimicrobial susceptibility testing
All E. coli isolates recovered from food samples were subjected to determination of their antimicrobial susceptibilities using the agar dilution method according to the Clinical and Laboratory Standards Institute (CLSI) except for colistin and tigecycline that was tested using the broth-microdilution method [21]. We used resistance breakpoints published by the CLSI [21]. E. coli strain ATCC 25922 was tested as the quality control strain. Further E. coli isolates from the same food sample and exhibiting identical MIC profile were considered as similar clones and eliminated from further analysis. Only non-duplicated E. coli isolates from each sample were used for further analysis.
Characterisation of bla NDM and mcr-1 positive Escherichia coli strains
All E. coli strains were screened for the presence of the bla NDM and mcr-1 gene by PCR using primers as previously described [10,22]. PCR products were purified and subjected to Sanger sequencing to confirm the genetic identity.
Methodological details for the conjugation experiments, XbaI-PFGE, S1-PFGE and Southern hybridisation are given in the Supplement.
Plasmid sequencing and bioinformatics analyses
Plasmid sequencing was performed as previously described using the Illumina and PacBio RS II platforms [14]. All plasmid sequences were submitted to the RAST tool for annotations and modified manually by BLAST [23]. The Easyfig software was used in comparative plasmid analysis [24].
Statistical analysis
The statistical analysis was performed by Minitab16 software (Minitab, version 16.2.3, Minitab Inc., State College, United States). The resistance rates of E. coli to colistin and meropenem in 2015, 2016 and 2017 were determined by the chi-squared test. The threshold for significant difference was p < 0.05.
Results
Prevalence of cephalosporin-resistant Escherichia coli in food products
A total of 1,166 (55%) non-repeated cephalosporin-resistant E. coli isolates were recovered from 2,137 food samples purchased during the period from 2015 to 2017 in Shenzhen, China, for the Food-borne Pathogen Surveillance Programme conducted by the Shenzhen Key Laboratory for Food Biological Food Safety Control. No food sample was found to contain more than one type of E. coli isolate. Among the 2,137 food samples, 1,368 were purchased from wet markets, and 769 samples were purchased from supermarkets of different regions in Shenzhen. These food samples included 1,182 pork, 230 beef, 383 chicken and 342 shrimp samples (Supplementary Table S1). Among the 1,166 ceftaxidime-resistant E. coli isolates, 385 (52%) E. coli were isolated from 747 food samples in 2015, 570 (56%) from 1,019 food samples in 2016, and 211 (57) from 371 food samples in 2017 (Table 1). Among these E. coli isolates, 710 were isolated from the 1,182 pork samples (60%), 107 were recovered from the 230 beef samples (47%), 315 from the 383 chicken samples (82%) and 34 from the 342 shrimp samples (10%). The detailed sampling and isolation information is provided in Supplementary Table S1.
Table 1. Isolation rate of Escherichia coli strains resistant to ceftaxime, mcr-1 bearing strains, bla NDM bearing strains and strains harbouring both mcr-1 and bla NDM genes, Shenzhen, 2015–2017 (n = 2,137).
| Year | Number of food samples | Cefotaxime-resistant E. coli isolates | mcr-1-bearing E. coli | bla NDM-bearing E. coli, with or without mcr-1 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Isolation rate (%)a | Pork | Beef | Chicken | Shrimp | Total | mcr-1 positivity rate (%)a | Total | bla NDM positivity rate (%)a | Number of mcr-1-positive strains | bla NDM/mcr-1 positivity rate (%) | ||||||
| n | % | n | % | n | % | n | % | ||||||||||
| 2015 | 747 | 385 | 52 (A) | 248 | 57 | 33 | 46 | 97 | 75 | 7 | 6 | 98 | 25 (B) | 1 | 0.3 (E) | 0 | 0 |
| 2016 | 1,019 | 570 | 56 (A) | 319 | 52 | 61 | 53 | 168 | 86 | 22 | 12 | 207 | 36 (C) | 5 | 1 (E) | 0 | 0.2 |
| 2017 | 371 | 211 | 57 (A) | 143 | 65 | 13 | 30 | 50 | 88 | 5 | 10 | 97 | 46 (D) | 36 | 17 (F) | 5 | 2 |
| Total | 2,137 | 1,166 | 55 | 710 | 60 | 107 | 47 | 315 | 82 | 34 | 10 | 402 | 34 | 42 | 4 | 5 | 0.4 |
a Percentage was calculated by dividing the number of E. coli isolates carrying the indicated resistance genes by the total number of cefotaxime-resistant E. coli isolates obtained in each year; percentages labelled with the same letter in brackets indicated that no significant difference was observed between two variants; percentages labelled with different letters indicated that a significant difference was observed between two variants (p < 0.05); (B) vs (C): p = 0.000; (C) vs (D): p = 0.014; (B) vs (D): p = 0.000; (B) vs (C) vs (D): p = 0.000; (E) vs (F): p = 0.000.
Antimicrobial susceptibility of Escherichia coli food isolates
We determined the minimal inhibitory concentrations (MIC) of various antibiotics for all 1,166 cephalosporin-resistant E. coli isolates. These strains were found to be resistant to most of the antibiotics tested, with a resistance rate of 100% to ampicillin, cefotaxime and ceftriaxone, and resistance to the other agents as follows: 95% to sulfamethoxazole/trimethoprim, 95% to tetracycline, 86% to chloramphenicol, 82% to nalidixic acid, 64% to kanamycin and 59% to ciprofloxacin. The rate of resistance to colistin was ca 33%. On the other hand, these strains were susceptible to tigecycline (100%), amikacin (97%), ceftazidime/avibactam (96%) and meropenem (96%). For the 390 E. coli strains that were resistant to colistin, the rate of resistance to different antibiotics was slightly higher than the resistance rate of all E. coli isolates, with the of exception of tigecycline (0%), ceftazidime/avibactam (2%) and meropenem (2%) (Supplementary Table S2). Forty-two E. coli strains that were resistant to meropenem were also resistant to most of other antibiotics. Five of these 42 strains were also resistant to colistin, but they were mostly susceptible to amikacin (98%) (Supplementary Table S2). Among the 390 colistin-resistant E. coli strains, the rate of resistance to colistin increased significantly over the years, with 26% in 2015, 36% in 2016 and 46% in 2017 (p < 0.05). Among the 42 strains that were resistant to meropenem, the rate of meropenem resistance increased significantly from 0.3% in 2015 to 1% in 2016 and 17% in 2017 (p < 0.001) (Table 1). Interestingly, five meropenem-resistant strains were also resistant to colistin, accounting for 0.4% of the total number of E. coli strains isolated from food products in this study (Table 1).
Prevalence of mcr-1 and bla NDM in Escherichia coli isolates from food
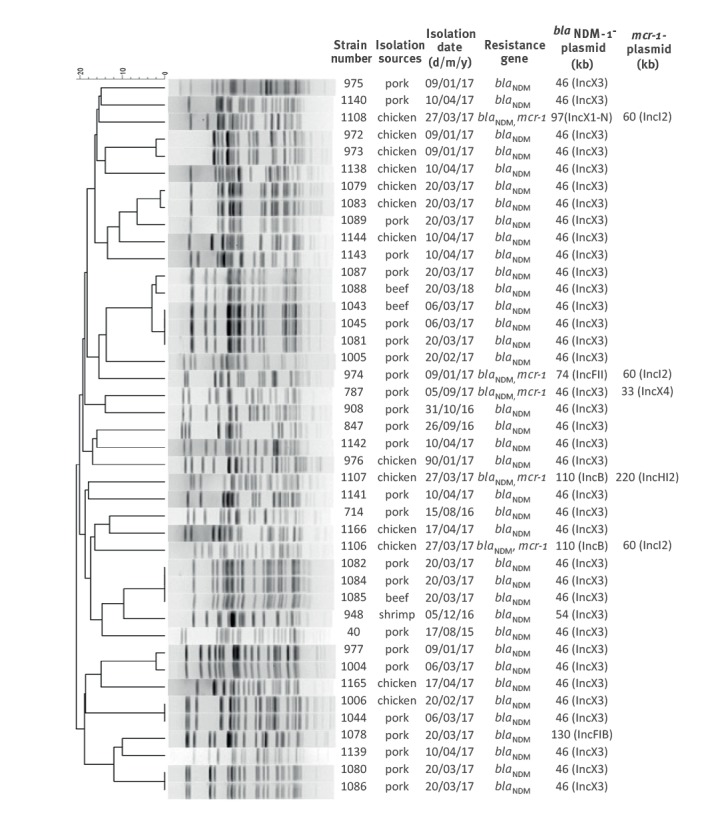
All 390 colistin-resistant E. coli strains were subjected to screening for the presence of the mcr-1, mcr-2, mcr-3 and mcr-4 genes; all of them carried the mcr-1 gene but not the other mcr genes. Screening for carbapenemase genes among the 42 meropenem-resistant E. coli strains showed that all of them harboured bla NDM-1 gene except for strains 787, 974, 977, 1079, 1106 and 1107, which were found to harbour the bla NDM-5 gene. No other carbapenemase genes, such as the bla KPC, bla VIM, bla IMP, bla OXA48-like genes, could be detected in these strains. Consistently, the five E. coli strains that were resistant to both meropenem and colistin carried both mcr-1 and bla NDM genes. Genetic characterisation by PFGE of the 42 bla NDM-1-carrying E. coli strains showed that the majority were genetically unrelated, suggesting that the spread of these strains was mainly due to acquisition of bla NDM by different E. coli strains, rather than clonal dissemination of strains that had acquired the bla NDM gene (Figure 1). Nevertheless, evidence of clonal transmission was also observed; examples include strains 1080/1086 and 972/973, which were isolated from different pork or chicken samples purchased on the same date, as well as strains 1043/1045/1081, 1082/1084/1085, 1006/1044 and 977/1004, which were isolated from different meat products on different dates.
Figure 1.
XbaI-PFGE pattern of Escherichia coli isolates resistant to meropenem, Shenzhen, 2015–2017 (n = 42)
All these plasmids were conjugative plasmids.
Genetic characterisation of bla NDM-bearing Escherichia coli strains
To investigate the genetic characteristics of E. coli strains carrying bla NDM, conjugation experiments were performed. The meropenem resistance phenotype of all 42 bla NDM-bearing strains tested was transferrable to E. coli J53 and EC600, with only data on J53 being shown here. S1-PFGE and Southern hybridisation performed on the parental and transconjugant of all 42 strains showed that bla NDM was located on plasmids of six different sizes, with the ca 46 kb IncX3-type plasmid being the most dominant, accounting for 36 of the 42 conjugative plasmids. Two strains were found to harbour an IncB plasmid of ca 110 kb, and one strain each carried a plasmid of 54 kb (IncX3), 74 kb (IncFII), 97 kb (IncX1-IncN) and 130 kb (IncFIB) in size (Figure 1).
The ca 46 kb and ca 54 kb IncX3+type bla NDM-bearing plasmids have been commonly reported in clinical isolates, while they were rarely reported in carbapenem-resistant Enterobacteriaceae strains isolated from non-clinical settings. To confirm that these plasmids, recovered from E. coli of food origin, were genetically similar to those in human clinical isolates, one ca 46 kb plasmid and one ca 54 kb plasmid were recovered from the transconjugants and subjected to complete plasmid sequencing using both Illumina and Nanopore sequencing platforms.
The complete map of the bla NDM-5-bearing plasmid from transconjugant E. coli MTC787, designated as pMTC787, was 46,161 bp in size with a GC content of 46.7%; it was confirmed to belong to an IncX3 replicon type. BLASTN analysis of this plasmid revealed that p787-NDM displayed 100% query coverage and 99% nucleotide identity with a plasmid pCRCB-101_1 (CP024820) carried by a Citrobacter freundii strain CRCB-101 isolated from open pus of a person in South Korea, and with plasmid pNDM-EC36 (MG591703) carried by an E. coli strain EC36 isolated in Henan, China (Figure 2A). Moreover, p787-NDM showed high homology (> 98% in both identity and coverage) to other similar IncX3, bla NDM-bearing plasmids reported in GenBank, such as plasmid pNDM_MGR194 from K. pneumoniae (NC_022740.1), plasmid pP855-NDM5 (MF547508.1) and plasmid pP788A-NDM5 (MF547507.1) from clinical E. coli strains and plasmid pNDM5-SSH006 (MTKV01000083.1) from Salmonella Typhimurium strain SSH006.
Figure 2.
Schematic representation and alignment of complete sequence of mcr-1-bearing and bla NDM-bearing plasmids with homologous plasmids in GenBank, Shenzhen, 2015–2017*
Grey shading indicates homologies between the corresponding genetic loci on each plasmid. Arrows indicate coding sequences, with arrowheads indicating the direction of transcription. Red: antibiotic resistance-encoding genes; blue: mobile elements; yellow: replicon proteins; grey: maintenance/stability functioning genes or hypothetical proteins.
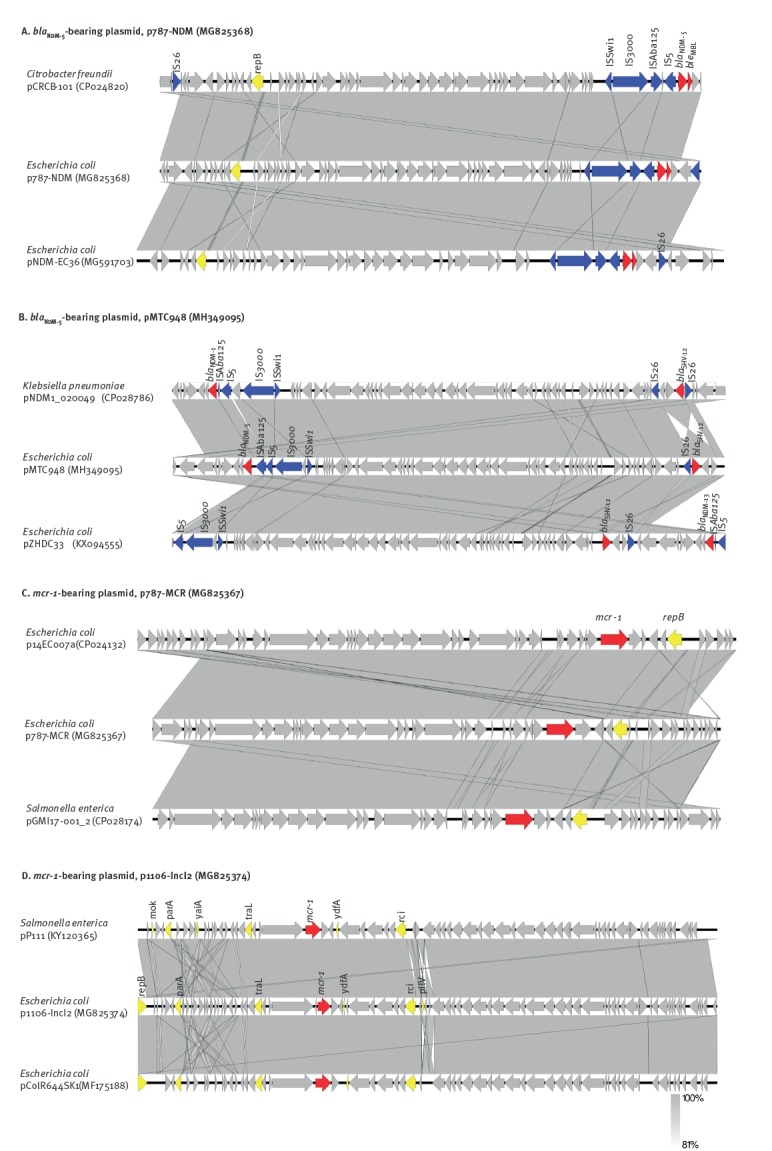
The complete sequence bla NDM-5-bearing plasmid from the transconjugant of E. coli strain 948, MTC948 was obtained and designated as pMTC948. It was confirmed as an IncX3 type with a size of 53,770 bp. Sequence analysis showed that it was highly homologous to plasmid pNDM1_020049 which is carried by a Klebsiella pneumoniae strain SCKP020049 isolated from a person in Sichuan, China and to plasmid pZHDC33 which is carried by an E. coli strain ZHDC33 isolated in Zhejiang, China. Plasmid pMTC948 aligned well with plasmid p787-NDM, with only an additional mobile element carrying IS26-bla SHV-12 (Figure 2B). This plasmid also displayed high homology (> 98% in both identity and coverage) with other IncX3, bla NDM-1-bearing plasmids reported in GenBank, such as plasmid pNDM-HN380 (NC_019162.1) from K. pneumoniae, plasmid pNDM-EcHK001 (NZ_CM008823.1) from Enterobacter cloacae strain EcHK001 and plasmid pYQ13500-NDM (KR059865.1) from En. cloacae strain EC-YQ13500.
Genetic features of mcr-1-bearing and bla NDM-bearing plasmids in Escherichia coli
Among the 42 E. coli strains carrying bla NDM, five also carried mcr-1 (Figure 1). Conjugation experiments were performed to assess the transferability of the colistin resistance phenotype; it could be successfully transferred to E. coli J53 from all strains except strain 1107, where the mcr-1-bearing plasmid was not conjugative and could not be transferred by transformation. S1-PFGE and Southern hybridisation were performed on strain 1107 and showed that the mcr-1 gene was located on a ca 220 kb plasmid belonging to the InHI2 type. Two types of conjugative mcr-1-bearing plasmids could be recovered from strains 787, 1106 and 1108, with three strains carrying a ca 60 kb IncI2 plasmid and one harbouring a ca 33 kb, IncX4 plasmid (Figure 1). Interestingly, during conjugation, both the mcr-1-bearing and the bla NDM-1-bearing plasmid in strains 787, 974 and 1108 could be simultaneously transferred to E. coli J53 under selection of meropenem, yet only mcr-1-bearing plasmids could be transferred to J53 under selection of colistin. The data suggested that even though the mcr-1 and bla NDM-1 genes were located on different plasmids, each of these two plasmids could be transferred to other bacteria under meropenem selection pressure (Figure 3).
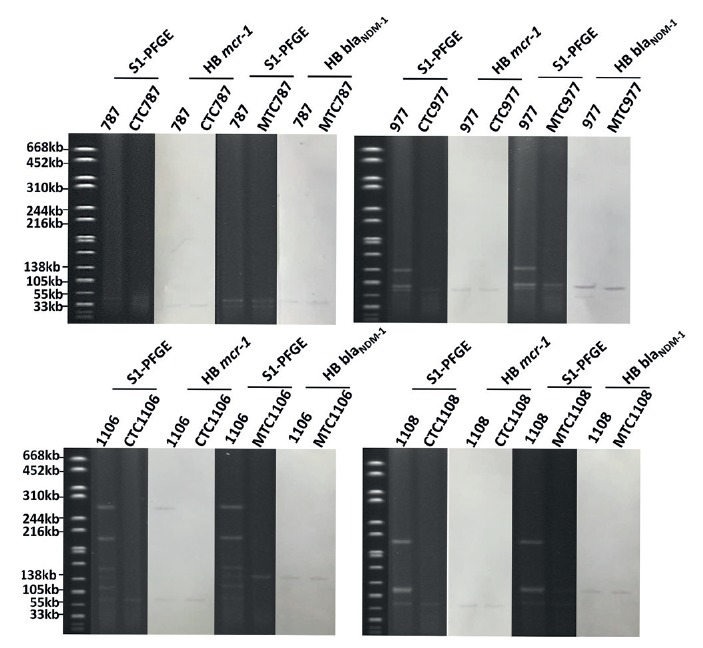
Figure 3.
S1-PFGE and Southern hybridisation of mcr-1 and bla NDM-1-bearing Escherichia coli strains and the corresponding transconjugants, Shenzhen, 2015–2017 (n = 2)
CTC: transconjugants selected with colistin; HB: Southern hybridisation; MTS: transconjugants selected with meropenem; PFGE: pulsed-field gel electrophoresis.
Both the mcr-1-bearing and the bla NDM-bearing plasmids could be transmitted to E. coli J53 under selective pressure of meropenem from strains 787, 977 and 1108.
One ca 60 kb (IncI2) and one ca 33 kb (IncX4) mcr-1-bearing plasmid were recovered from the transconjugants and subjected to complete plasmid sequencing using both Illumina and Nanopore sequencing platforms. The complete map of the ca 30 kb mcr-1-bearing plasmid recovered from transconjugant E. coli CTC787, designated as pCTC787, was shown to belong to IncX4 type with a size of 33,301 bp. It aligned well (> 99% in both identity and coverage) with several previously reported plasmids such as p14EC007a (CP024132), which was carried by an E. coli strain 14EC007 isolated from clinical patients in China, and pGMI17–001_2 (CP028174) which was carried by a Salmonella enterica strain CFSAN064033 isolated from a turkey sample in Germany (Figure 2C). The complete sequence of a ca 60 kb mcr-1-bearing conjugative plasmids, recovered from the transconjugant E. coli CTC1106 strain was shown to belong to the IncI2 type with a size of 60,960 bp and a GC content of 42.3%. It was designated as p1106-IncI2. BLASTN analysis revealed that it exhibited the highest homology with pP111, carried by a Salmonella enterica (KY120365) strain P111 isolated in Taiwan, and with pColR644SK1, carried by an E. coli strain ColR644SK1 (MF175188) isolated in Switzerland. It also exhibited high homology (> 99% in both identity and coverage) with pHNSHP45 (KP347127) and other plasmids reported previously [25,26]. Similar to other IncI2 type plasmids, p1106-IncI2 contained genes and proteins responsible for replication (repB), plasmid stability (mok), partitioning (parA), plasmid transferability (traL), plasmid-borne site-specific recombinase (rci) and pilus-encoding loci (pilV) (Figure 2D).
Discussion
Mobile carbapenemase genes have emerged in clinical isolates but are rare in isolates from animals. In China, the first bla NDM-1 gene was reported in an E. coli strain (HK-01) in 2011 [8] and has since disseminated extensively in clinical settings [27-29]. The bla NDM-1 gene has mainly been reported on a ca 46kb and 54kb IncX3-type conjugative plasmid in China [9,29,30]. Although such a gene has since been reported in non-clinical bacterial isolates of different origins, such as animals and the environment, it was often carried by plasmids of various types mainly through insertion of the bla NDM-1-bearing mobile element into the backbone of different plasmids in bacterial strains of the same origin. These data appear to imply that bacterial strains, in particular E. coli, that commonly reside in animals and the farm environment might not be adaptable to the IncX3 type plasmids. Recently, Wang Y et al. reported the prevalence of NDM and MCR-1-producing Enterobacteriaceae strains in poultry and the farm environment, and that the prevalence of bla NDM-1 in these strains has increased sharply [19]. E. coli isolates carrying an IncX3 bla NDM-bearing plasmid were found to be prevalent in Enterobacteriaceae strains isolated from the poultry production chain [19]. Wang R et al. also reported the co-existence of mcr-1 and different bla NDM-1 variants in E. coli and K. pneumoniae isolates originating from broiler, swine and cattle samples [31]. These finding suggest that co-transmission of bla NDM and the already prevalent mcr-1 gene has occurred among Enterobacteriaceae in animals, leading to selection of strains carrying both bla NDM-1 and mcr-1 which are a risk to human health. This risk will become imminent if Enterobacteriaceae strains carrying both resistance genes become prevalent in our food products. However, isolation of E. coli carrying both the bla NDM-1 and mcr-1 gene, in particular carriage of bla NDM-1 by the IncX3-type plasmid in E. coli strains of food origin, has not been reported previously.
We have been conducting comprehensive surveillance of multidrug-resistant E. coli in food products in China in the past few years. E. coli strains of food origin were shown in our study to carry mcr-1 at an increasing rate, whereas the bla NDM-1 gene was rarely detected. In this study, we have shown that the bla NDM-positive isolates were increasingly detectable over time, with a prevalence rate of 0.3% and 1% in 2015 and 2016, respectively, rising sharply to 17% in 2017. This sharp increase in the prevalence of bla NDM-bearing E. coli strains in food products was mainly due to transmission of a ca 46kb IncX3 plasmid commonly harboured by clinical Enterobacteriaceae isolates. In addition, the increasing prevalence of E. coli strains carrying bla NDM implies emergence of E. coli strains that carry both bla NDM and mcr-1. In this work, all three commonly reported mcr-1-bearing plasmids were able to co-exist in a single E. coli strain. In addition to the highly prevalent IncX3 bla NDM-1-bearing plasmid, another five plasmid types were also reported in E. coli of food origin. Furthermore, all bla NDM-bearing plasmids reported in our study were conjugative, implying that they were transmissible to other E. coli strains of food origin. The increasing prevalence of Enterobacteriaceae strains carrying bla NDM and mcr-1 in food products will lead to increased colonisation of the human gastrointestinal tract with these Enterobacteriaceae strains, a phenomenon that has been associated with the high prevalence of drug-resistant infections in clinical settings [32]. Importantly, E. coli carrying bla NDM-1 or bla NDM-1 /mcr-1 could be detected in pork, beef and even shrimp, suggesting that these strains have already widely disseminated to different food products. An increasing rate of food-borne infections caused by such strains would be a serious concern. Further surveillance of bla NDM-1 and mcr-1 in other food products is warranted.
Acknowledgements
We are grateful for the help of plasmid sequence from Ruichao Li and Dong Ning from our lab. This work was supported by grant of the Thirteen’s Five-Year Plan of National key research and development (2018YFD0500300).
Supplementary Data
*Erratum
After publication, typographical errors were corrected in the title of Figure 2. These changes were made on 3 April 2019.
Conflict of interest: None declared.
Authors’ contributions: XBL, designed and performed the research and drafted the manuscript; GX participated in the supervision of the research; EWCC, participated in data analysis and manuscript writing; SC supervised the whole project, analysed the data and wrote the manuscript.
References
- 1. Jeon JH, Lee JH, Lee JJ, Park KS, Karim AM, Lee CR, et al. Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int J Mol Sci. 2015;16(5):9654-92. 10.3390/ijms16059654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. 10.3389/fmicb.2016.00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fu Y, Du X, Ji J, Chen Y, Jiang Y, Yu Y. Epidemiological characteristics and genetic structure of blaNDM-1 in non-baumannii Acinetobacter spp. in China. J Antimicrob Chemother. 2012;67(9):2114-22. 10.1093/jac/dks192 [DOI] [PubMed] [Google Scholar]
- 4. Ou W, Cui L, Li Y, Zheng B, Lv Y. Epidemiological characteristics of blaNDM-1 in Enterobacteriaceae and the Acinetobacter calcoaceticus-Acinetobacter baumannii complex in China from 2011 to 2012. PLoS One. 2014;9(12):e113852. 10.1371/journal.pone.0113852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046-54. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother. 2011;55(11):5403-7. 10.1128/AAC.00585-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pfeifer Y, Wilharm G, Zander E, Wichelhaus TA, Göttig S, Hunfeld KP, et al. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J Antimicrob Chemother. 2011;66(9):1998-2001. 10.1093/jac/dkr256 [DOI] [PubMed] [Google Scholar]
- 8. Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, et al. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One. 2011;6(3):e17989. 10.1371/journal.pone.0017989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang Q, Fang L, Fu Y, Du X, Shen Y, Yu Y. Dissemination of NDM-1-producing Enterobacteriaceae mediated by the IncX3-type plasmid. PLoS One. 2015;10(6):e0129454. 10.1371/journal.pone.0129454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Y-Y, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161-8. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 11. Quan J, Li X, Chen Y, Jiang Y, Zhou Z, Zhang H, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400-10. 10.1016/S1473-3099(16)30528-X [DOI] [PubMed] [Google Scholar]
- 12. Liu X, Li R, Zheng Z, Chen K, Xie M, Chan EW, et al. Molecular characterization of Escherichia coli isolates carrying mcr-1, fosa3, and extended-spectrum-β-lactamase genes from food samples in China. Antimicrob Agents Chemother. 2017;61(6):61. 10.1128/AAC.00064-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 2016;21(9):30155. 10.2807/1560-7917.ES.2016.21.9.30155 [DOI] [PubMed] [Google Scholar]
- 14. Li R, Xie M, Zhang J, Yang Z, Liu L, Liu X, et al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother. 2017;72(2):393-401. 10.1093/jac/dkw411 [DOI] [PubMed] [Google Scholar]
- 15. Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Butaye P, Goossens H. Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet Infect Dis. 2016;16(3):283-4. 10.1016/S1473-3099(16)00012-8 [DOI] [PubMed] [Google Scholar]
- 16. Xavier BB, Lammens C, Butaye P, Goossens H, Malhotra-Kumar S. Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J Antimicrob Chemother. 2016;71(8):2342-4. 10.1093/jac/dkw191 [DOI] [PubMed] [Google Scholar]
- 17. Zhong LL, Zhang YF, Doi Y, Huang X, Zhang XF, Zeng KJ, et al. Coproduction of MCR-1 and NDM-1 by colistin-resistant Escherichia coli isolated from a healthy individual. Antimicrob Agents Chemother. 2016;61(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang RS, Feng Y, Lv XY, Duan JH, Chen J, Fang LX, et al. Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single muscovy duck (Cairina moschata). Antimicrob Agents Chemother. 2016;60(11):6899-902. 10.1128/AAC.01365-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y, Zhang R, Li J, Wu Z, Yin W, Schwarz S, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol. 2017;2(4):16260. 10.1038/nmicrobiol.2016.260 [DOI] [PubMed] [Google Scholar]
- 20. Liu BT, Song FJ, Zou M, Hao ZH, Shan H. Emergence of colistin resistance gene mcr-1 in Cronobacter sakazakii producing NDM-9 and in Escherichia coli from the same animal. Antimicrob Agents Chemother. 2017;61(2):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; 28th informational supplement. CLSI document M100-S28. Wayne, PA: CLSI 2018. Available from: http://www.iacld.ir/DL/public/CLSI-2018-M100-S28.pdf
- 22. Lin D, Xie M, Li R, Chen K, Chan EW, Chen S. IncFII conjugative plasmid-mediated transmission of blandm-1 elements among animal-borne Escherichia coli strains. Antimicrob Agents Chemother. 2016;61(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42(Database issue):D206-14. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009-10. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anjum MF, Duggett NA, AbuOun M, Randall L, Nunez-Garcia J, Ellis RJ, et al. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J Antimicrob Chemother. 2016;71(8):2306-13. 10.1093/jac/dkw149 [DOI] [PubMed] [Google Scholar]
- 26. Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother. 2016;71(8):2300-5. 10.1093/jac/dkw093 [DOI] [PubMed] [Google Scholar]
- 27. Chen Y, Zhou Z, Jiang Y, Yu Y. Emergence of NDM-1-producing Acinetobacter baumannii in China. J Antimicrob Chemother. 2011;66(6):1255-9. 10.1093/jac/dkr082 [DOI] [PubMed] [Google Scholar]
- 28. Huang Y, Yu X, Xie M, Wang X, Liao K, Xue W, et al. Widespread dissemination of carbapenem-resistant Escherichia coli sequence type 167 strains harboring blaNDM-5 in clinical settings in China. Antimicrob Agents Chemother. 2016;60(7):4364-8. 10.1128/AAC.00859-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He T, Wang Y, Sun L, Pang M, Zhang L, Wang R. Occurrence and characterization of blaNDM-5-positive Klebsiella pneumoniae isolates from dairy cows in Jiangsu, China. J Antimicrob Chemother. 2017;72(1):90-4. 10.1093/jac/dkw357 [DOI] [PubMed] [Google Scholar]
- 30. Yaici L, Haenni M, Saras E, Boudehouche W, Touati A, Madec JY. blaNDM-5-carrying IncX3 plasmid in Escherichia coli ST1284 isolated from raw milk collected in a dairy farm in Algeria. J Antimicrob Chemother. 2016;71(9):2671-2. 10.1093/jac/dkw160 [DOI] [PubMed] [Google Scholar]
- 31. Wang R, Liu Y, Zhang Q, Jin L, Wang Q, Zhang Y, et al. The prevalence of colistin resistance in Escherichia coli and Klebsiella pneumoniae isolated from food animals in China: coexistence of mcr-1 and blaNDM with low fitness cost. Int J Antimicrob Agents. 2018;51(5):739-44. 10.1016/j.ijantimicag.2018.01.023 [DOI] [PubMed] [Google Scholar]
- 32. Zhao ZC, Xu XH, Liu MB, Wu J, Lin J, Li B. Fecal carriage of carbapenem-resistant Enterobacteriaceae in a Chinese university hospital. Am J Infect Control. 2014;42(5):e61-4. 10.1016/j.ajic.2014.01.024 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


