Padeken et al. performed a genome-wide synthetic lethality screen and show that the BRCA1/BARD1 complex is necessary for germline viability in worms lacking MET-2 but not SET-25. The synthetic sterility upon BRCA1/BARD1 and H3K9me2 loss is directly linked to the DNA damage provoked by unscheduled satellite repeat transcription.
Keywords: DNA repeats, heterochromatin, histone H3K9 methylation, BRCA1 complex, RNA:DNA hybrids, transcriptional silencing, genome instability, satellite repeats
Abstract
Caenorhabditis elegans has two histone H3 Lys9 methyltransferases, MET-2 (SETDB1 homolog) and SET-25 (G9a/SUV39H1 related). In worms, we found simple repeat sequences primarily marked by H3K9me2, while transposable elements and silent tissue-specific genes bear H3K9me3. RNA sequencing (RNA-seq) in histone methyltransferase (HMT) mutants shows that MET-2-mediated H3K9me2 is necessary for satellite repeat repression, while SET-25 silences a subset of transposable elements and tissue-specific genes through H3K9me3. A genome-wide synthetic lethality screen showed that RNA processing, nuclear RNA degradation, the BRCA1/BARD1 complex, and factors mediating replication stress survival are necessary for germline viability in worms lacking MET-2 but not SET-25. Unlike set-25 mutants, met-2-null worms accumulated satellite repeat transcripts, which form RNA:DNA hybrids on repetitive sequences, additively with the loss of BRCA1 or BARD1. BRCA1/BARD1-mediated H2A ubiquitination and MET-2 deposited H3K9me2 on satellite repeats are partially interdependent, suggesting both that the loss of silencing generates BRCA-recruiting DNA damage and that BRCA1 recruitment by damage helps silence repeats. The artificial induction of MSAT1 transcripts can itself trigger damage-induced germline lethality in a wild-type background, arguing that the synthetic sterility upon BRCA1/BARD1 and H3K9me2 loss is directly linked to the DNA damage provoked by unscheduled satellite repeat transcription.
The most broadly conserved hallmark of constitutive heterochromatin is the methylation of lysine 9 within histone H3 (H3K9me) (Saksouk et al. 2015). In humans, at least eight histone methyltransferases (HMTs) are known to contribute to H3K9 methylation (Mozzetta et al. 2015), rendering it difficult to eliminate genetically all H3K9 methylation in mammals. In contrast, H3K9me is deposited by only two HMTs in Caenorhabditis elegans: MET-2, a homolog of the mammalian ESET/SETDB1, and SET-25, bearing a catalytic domain similar to SUV39H1 and G9a (Towbin et al. 2012). In vivo, MET-2 can only catalyze H3K9me1 and H3K9me2, while SET-25 can mediate me1, me2, and me3 in somatic cells from embryonic stages to adults (Towbin et al. 2012). Surprisingly, C. elegans embryos carrying the double met-2 set-25 mutation develop to adulthood, even though the levels of H3K9 methylation are undetectable (Towbin et al. 2012; Garrigues et al. 2015; Zeller et al. 2016). While this rules out an essential role for H3K9 methylation in somatic tissue differentiation, the met-2 set-25 mutant nonetheless shows germline lethality and stochastic delays in development. This is accompanied by elevated levels of transcripts from transposable elements, satellite sequences, and simple repeats, as well as inappropriately expressed pseudogenes and tissue-specific mRNAs (Zeller et al. 2016).
All phenotypes linked to the loss of H3K9me, including the level of repeat transcription, were aggravated by growth at 25°C, as opposed to temperatures at or below 20°C. In the mutant embryos, there is a widespread accumulation of RNA:DNA hybrids (R loops), which increases at 25°C, and mutant worms accumulate genomic insertions and deletions (indels) in transcribed repeats (Zeller et al. 2016). Increased levels of indels occur in both somatic and germline tissues (Zeller et al. 2016). The ablation of C. elegans H3K9me ligands, LIN-61, HPL-2 or its cofactor LIN-13, also generated DNA damage-dependent germline sterility to varying degrees (McMurchy et al. 2017).
The importance of H3K9 methylation for organismal survival is even more pronounced in vertebrates than in nematodes, in part due to the role of H3K9me in vertebrate centromere function. Rodents that lack the HMTs, SETDB1, SUV39 isoforms, or G9a and GLP (EHMT2/1, respectively), show chromosome missegregation and either impaired early development or complete embryonic lethality (Peters et al. 2001; Tachibana et al. 2002; Dodge et al. 2004). Several reports have detected elevated levels of RNA from LINE elements and from major satellite repeats upon loss of H3K9 HMTs (Dodge et al. 2004; Matsui et al. 2010; Rowe et al. 2010; Shinkai and Tachibana 2011; Pinheiro et al. 2012; Saksouk et al. 2015). Moreover, peri-centromeric satellite repeat transcripts correlate with degenerative or pathological cell states, including a broad spectrum of mouse and human epithelial tumors (Alexiadis et al. 2007; Eymery et al. 2009; Ting et al. 2011). Elevated levels of satellite transcripts are detected in brca1-deficient tumors, as well as in normal mouse neuronal tissues depleted for BRCA1 (Zhu et al. 2011, 2018 ). Recent work confirms that G9a inhibition can generate more aggressive skin and lung tumors (Avgustinova et al. 2018; Rowbotham et al. 2018).
Nematode genomes carry all the typical types of repeat sequences found in mammals except long arrays of peri-centric satellite repeats due to their holocentric nature (for review, see Padeken et al. 2015). Nonetheless, about 3% of the worm genome is composed of simple repeats (mini- and microsatellite sequences), which are dispersed along the distal chromosome arms. The limited length of most repeats (<500 base pairs [bp]) allows them to be mapped by standard deep sequencing, which in turn has allowed an accurate mapping of the histone marks on repeats (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing [ChIP-seq]) and of RNA transcripts arising from repeats (RNA sequencing [RNA-seq]) (Zeller et al. 2016). Here, we analyze the derepression of repeat sequences in C. elegans embryos deficient for one or the other H3K9 HMT. We found that MET-2 (SETDB1 homolog) is uniquely essential for the repression of satellite sequences. SET-25, in contrast, represses a subset of DNA and RNA transposons, pseudogenes, and tissue-specific promoters, some in a manner redundant with MET-2. Along with the repeat derepression detected in met-2-deficient worms, we detect an accumulation of R loops on simple repeats and a temperature-enhanced loss of fertility. Worms lacking SET-25 do not have these phenotypes.
The viability of the met-2 set-25 double mutants at 20°C allowed us to perform a genome-wide RNAi screen for factors whose loss leads to synthetic germline lethality specifically in worms lacking H3K9 methylation. All RNAi hits that were synthetic-sterile in the met-2 set-25 mutant were also lethal in combination with met-2 alone, but not with set-25. Intriguingly, many of the genes that are synthetic-lethal with met-2 mutation are implicated in pathways that suppress RNA:DNA hybrid-induced genome instability (Huertas and Aguilera 2003; Gómez-González et al. 2009; Gan et al. 2011; Santos-Pereira and Aguilera 2015; Hamperl and Cimprich 2016). Here, we characterize the synergistic effects provoked by the loss of MET-2 together with the BRCA1 homolog, Ce BRC-1, and interacting factors. All lead to the spurious transcription of simple repeats, R-loop accumulation, DNA damage foci, and germline sterility. BRCA1/BARD1-deposited H2AK127/K129ub (Kalb et al. 2014) coincides with and appears to contribute to H3K9me2-mediated repeat repression. Thus, BRCA1/BARD1 are not only essential for germline integrity upon loss of the H3K9 dimethyltransferase, MET-2, but they also contribute to the silencing of repetitive DNA.
Results
The loss of H3K9 methylation renders C. elegans dependent on factors involved in RNA processing and replication fork stability
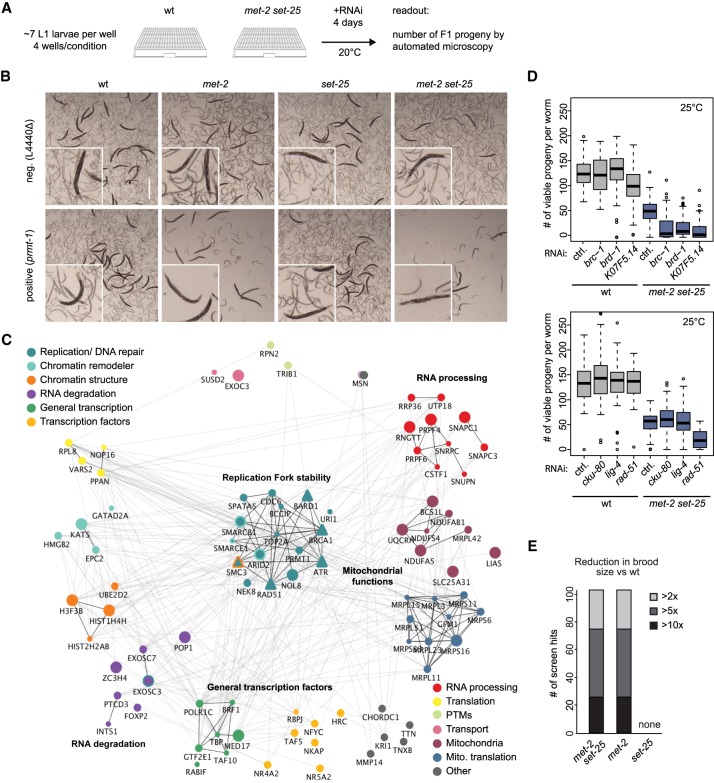
In C. elegans mutants that lack both H3K9 HMTs, 88% of the embryos reach adulthood under standard laboratory conditions despite a high degree of genomic instability that manifests as insertions and deletions in repetitive sequences (Zeller et al. 2016). Although the double mutant shows stochastic delays in development and undergoes germline apoptosis at 25°C, the mutant population survives when cultivated at or below 20°C. In order to identify the pathways that allow survival in the absence of H3K9me, we performed a synthetic lethality screen in the met-2 set-25 background at 20°C using a genome-wide RNAi library (Kamath et al. 2003). We retested all RNAi clones that significantly decreased the number of F1 progeny in the double mutant (greater than twofold) and eliminated those that also reduced fertility in a wild-type background (Fig. 1A,B). This resulted in 109 different genes whose loss impaired reproduction uniquely in met-2 set-25 worms in all four replicas (Supplemental Table S1). None of the RNAi hits was itself transcriptionally induced by the loss of H3K9me (data not shown).
Figure 1.
Synthetic sterility arises in met-2-deficient worms upon loss of RNA-processing/degradation and DNA damage response genes. (A) Synthetic lethality screen scheme: L1 larvae of wild-type and mutant strains were fed with RNAi-expressing bacteria at 20°C, and both adult viability and number of number of F1 progeny were assessed after 4 d. (B) Representative pictures of worms of indicated genotypes on day 4 after empty vector (L4440Δ) control or prmt-1 (a positive hit) RNAi. (C) Human homologs of RNAi-positive hits mapped onto STRING interactome (Szklarczyk et al. 2014) to illustrate general functions (color-coded) and genetic and physical interaction. Icon size reflects brood size reduction (from >2× to >10×). We include factors in the DNA replication and repair category that were tested secondarily: BRCA1, BARD1, RAD51, ATR. Visualization used visJS2jupyter (Rosenthal et al. 2018). (D) Synthetic sterility demonstrated for met-2 set-25 mutants and depletion of the indicated genes by RNAi after one generation at 25°C. N = 3; n = 25 for each condition. Box plots show median, boxes show 50%, and whiskers show 90% of all data points. (E) All 109 RNAi hits that were synthetic-sterile with met-2 set-25 mutants were tested on two met-2 single mutants (n4256 shown; ok2307 same result, not shown) and the set-25 (n5021) mutant. Number of hits and strength of synergistic effect are plotted. N = 3.
We analyzed the linkage of the hits based on available genetic and physical interaction data from multiple species (C. elegans gene names listed in Supplemental Table S1) in order to shed light on mechanisms involved. The genes fell into five functional classes grouped by either genetic or physical interaction of the gene products. These are plotted using the human homolog nomenclature and the tool visJS2jupyter (Fig. 1C; Rosenthal et al. 2018). Three of the classes include factors that are known to regulate nuclear RNA levels—that is, the exosome, RNA processing and/or export factors, and components of the basic transcription machinery (e.g., TAF3, TAF6, Mediator subunits). Collectively, these hits suggest that impaired processing or degradation of nuclear RNA, or altered control over RNA Pol II transcription, compromises survival in the absence of H3K9 methylation. A further class is labeled Replication Fork stability, as these repair and checkpoint factors all contribute to the survival of replication stress and its associated DNA damage (Fig. 1C). These include the ATR kinase, RAD51, and factors that interact with BRCA1 and BRCA2.
While many of these replication fork stability factors have additional roles, all have been identified as either interacting with factors that contribute to recovery from replication stress or else function directly to resolve aberrant structures that arise when the replication fork encounters unprocessed RNA:DNA hybrids (Table 1; Huertas and Aguilera 2003; Gómez-González et al. 2009; Gan et al. 2011; Santos-Pereira and Aguilera 2015; Hamperl and Cimprich 2016). The survival of replication-transcription interference (RTI) was shown to require the SWI/SNF chromatin remodelers SMARCAL1, ZRANB3, and HTLF (Taglialatela et al. 2017), the histone arginine methyltransferase, PRMT-1 (Yang et al. 2014), ATR kinase (Zeman and Cimprich 2014), and topoisomerase II (El Hage et al. 2010), along with the exosome (Pefanis et al. 2015), all of which were positive hits in our screen. Two uncharacterized hits, K07F5.14 and ZK1127.4, were earlier isolated as interaction partners of BRC-1, the C. elegans BRCA1 homolog (Liu et al. 2001; Li et al. 2004), or BRC-2/BRCA2, respectively (Polanowska et al. 2006; Bhatia et al. 2014). BRCA1 and BRCA2 also are thought to contribute to replication fork stability in mammals (Schlacher et al. 2012; Hatchi et al. 2015; Zhang et al. 2017; Billing et al. 2018; Przetocka et al. 2018). Finally, we recovered a class of factors that controls mitochondrial translation and/or oxidative phosphorylation. At present, it is unclear how mitochondrial misfunction relates to the other functional groups.
Table 1.
Synthetic-sterile hits involved in RNA:DNA hybrid prevention
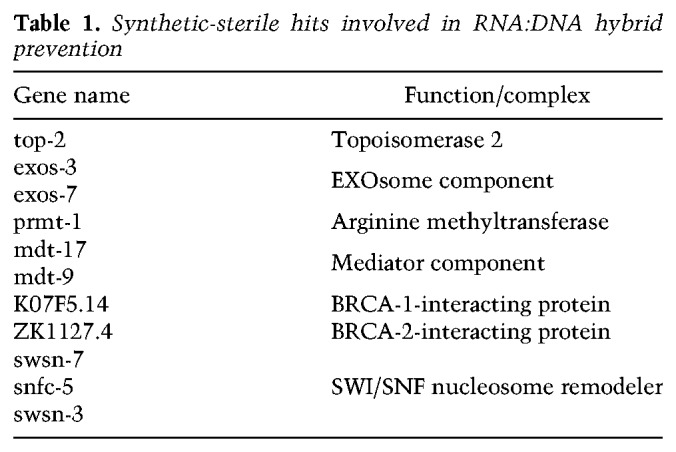
Given recent evidence that implicates BRCA1 in the survival of stalled or collapsed replication forks and the preponderance of hits involved either in elimination of RNA:DNA hybrids or in recovery from RTI, we hypothesized that the germline synthetic lethality we scored might stem from the derepression of repetitive DNA and R-loop formation (Zeller et al. 2016). Because a putative ligand of BRCA1, K07F5.14 (Li et al. 2004), was recovered in the screen, we used validated RNAi constructs to down-regulate brc-1, brd-1 (the C. elegans homolog of BARD1) (Boulton et al. 2004; Polanowska et al. 2006) , rad-51 (mammalian RAD51), and the cohesin subunit smc-3 (Terret et al. 2009), all of which are thought to work with BRCA1 in double-strand break (DSB) repair (Panier and Boulton 2014) and during replication fork arrest. In agreement with our hypothesis, all of these factors were synthetic-sterile in combination with met-2 set-25 mutations but not in wild-type worms (Fig. 1D; Supplemental Table S1). In contrast, RNAi against factors that uniquely abolish DSB repair by nonhomologous end joining (NHEJ)—i.e., cKU-80 and LIG-4—were not synthetic-lethal with the loss of H3K9me (Fig. 1D). Moreover, depletion of the C. elegans BRCA2 homolog (brc-2) did not show a clear synergistic sterility (Supplemental Fig. S1C). The sensitivity to replication fork recovery factors is consistent with previous data showing that H3K9me-deficient worms are hypersensitive to hydroxyurea (HU), which stalls replication forks by dNTP depletion, but not to γ-irradiation, which generates single- and double-strand breaks (Zeller et al. 2016).
In order to assess the relative importance of the two individual HMTs in this synthetic-sterile screen, we treated met-2 or set-25 single mutants with RNAi against all 109 confirmed hits arising from the met-2 set-25 double mutant screen at 20°C. Surprisingly, we found that all RNAi clones that led to sterility with the met-2 set-25 double mutant were synthetic-sterile in combination with the met-2 single mutant, while none was synthetically lethal with set-25 (Fig. 1B,E).
Loss of met-2 results in reduced fertility and delayed development
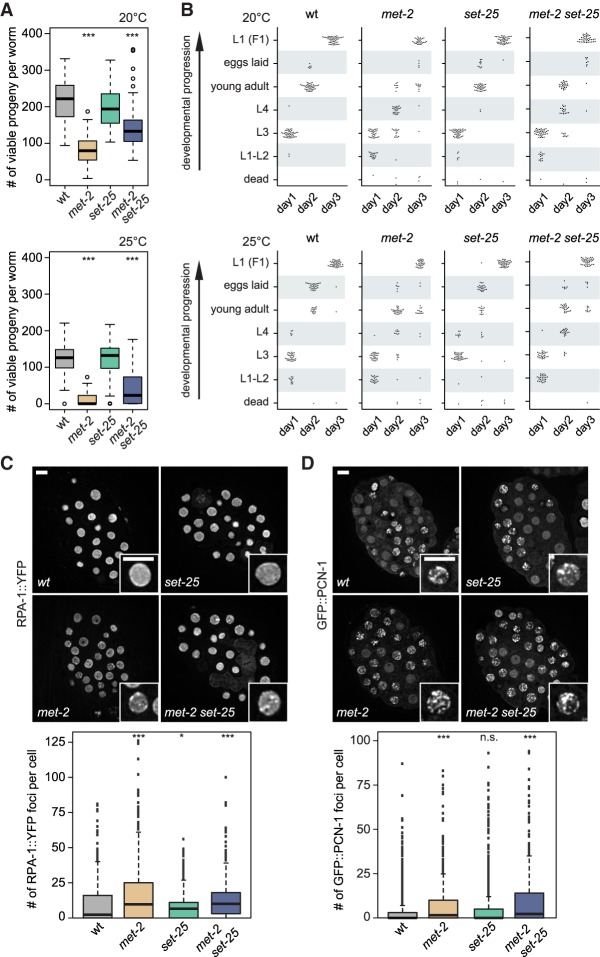
The fact that the met-2 mutant recapitulates the synthetic lethality of the met-2 set-25 double mutant, while set-25 does not, led us to examine the germline and developmental defects of the single mutants. We found that the met-2 single mutant, like the double mutant, has slightly impaired fertility at 20°C and becomes nearly sterile at 25°C (Fig. 2A). In contrast, set-25 worms had wild-type brood size (Fig. 2A). The timing of somatic development of synchronized larvae bearing the single mutations over one complete generation (3 d) showed that 87% of the met-2 larvae had stochastic delays in development at 20°C, while both wild-type and set-25 L1 stage larvae developed highly synchronously without delay (Fig. 2B). All defects associated with the met-2 mutation were accentuated at 25°C.
Figure 2.
MET-2 is essential for organismal fertility and proper developmental timing. (A) Single mutants met-2 and set-25 were tested for fertility at 20°C and 25°C (N = 3; n = 25) and compared with the met-2 set-25 double mutant. Complete viable progeny of singled worms were scored after two generations. (***) P < 0.0001, two-sided Wilcoxon signed-rank test. (B) Developmental timing was analyzed by following singled worms over 3 d of development at 20°C and 25°C of the indicated genotypes. Each dot represents a single worm. N = 3; n = 50. (C,D, top) Representative images of early embryos from adults of indicated genotypes, expressing either RPA-1::YFP (C), grown at 25°C or GFP::PCN-1, grown at 20°C (D). Bars, 5 µm. (Bottom) Quantification shows the number of foci per nucleus for the indicated genotypes. (C) N = 3; n = 25 embryos. (D) N = 2; n = 25 embryos. Box plots show median, boxes 50%, and whiskers 90%, with all other data points shown. (*) P = 0.031, (***) P < 0.0001, (n.s.) P > 0.0, two-sided Wilcoxon signed-rank test.
Given this subtle impact on development, we tested the strongest 54 hits from the screen to identify the stage of development at which the RNAi clones blocked the growth of the mutant worms (Supplemental Fig. S1B; Supplemental Table S2). Under the conditions used in the original screen, the vast majority of the RNAi clones (70%) led to arrested germ cell development, reducing both the number of oocytes produced and the number of embryos hatched in the double mutant (Supplemental Fig. S1B). The fact that somatic development from embryonic stages to adulthood was not blocked (Fig. 2B) argues that the synthetic lethality arising from RNAi against factors involved in nuclear RNA processing and degradation, and replication fork survival of R loops, likely reflects the loss of C. elegans germline integrity, rather than a failure to form differentiated tissues.
Replication-associated damage foci accumulate in met-2 embryos
In support of RTI-induced damage as the source of sterility, we examined whether the loss of met-2 led to delayed progression through S phase and/or the appearance of repair foci, which we score by RPA-1::YFP fluorescence. We found a significant increase in RPA-1 foci in met-2 mutant embryos (median approximately fourfold over wild type) and a lower increase in the set-25 mutants (median approximately twofold over wild type) (Fig. 2C). To see whether this reflects a perturbation of S phase progression, we scored the number of PCNA (GFP::PCN-1) foci in the same embryos, which marks replication factories. We found an increase in cells with PCNA foci in the met-2 mutant (median approximately sevenfold over wild type) but not in the set-25 mutant (Fig. 2D), suggesting that the loss of MET-2 perturbs replication. Indeed, the met-2 mutant is sensitive to HU, like the double mutant (Supplemental Fig. S2; Zeller et al. 2016).
Satellite repeats are MET-2 (SETDB1)-specific, while SET-25 (SUV39/G9a) targets retrotransposons and genes
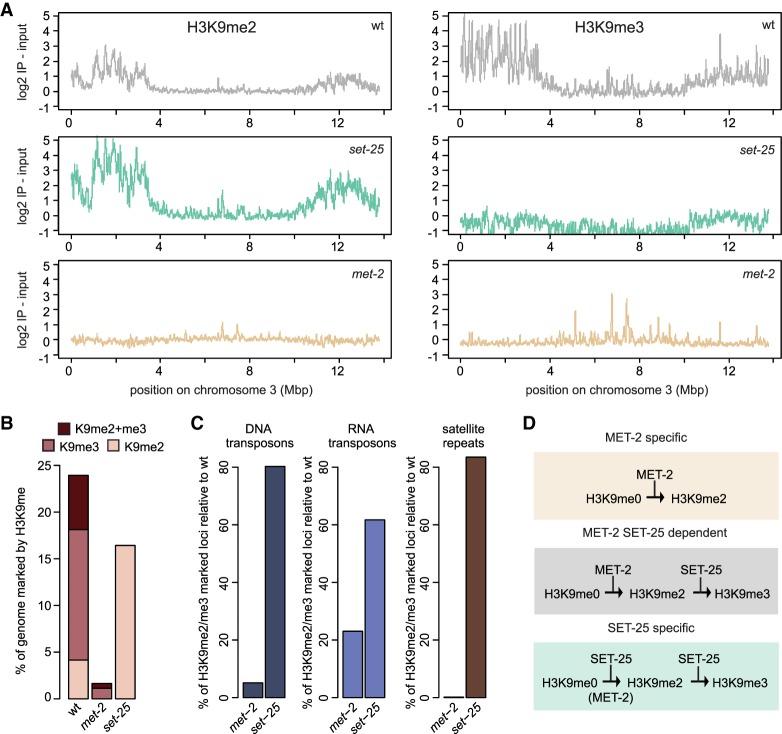
Given that all hits in our genome-wide RNAi screen were synthetic-sterile with the met-2 single mutant but did not affect fertility in a set-25-deficient background, we set out to determine the unique contributions of MET-2 vs. SET-25 HMTs to histone methylation genome-wide, with the goal of understanding the defects leading to germline sterility. We performed ChIP-seq using antibodies specific for either H3K9me2 or H3K9me3 in the two single HMT mutants (Fig. 3A) in order to determine the specificity and redundancy of the two enzymes. Earlier mass spectrometry studies of total histones had shown that set-25 ablation leads to the loss of H3K9me3 only. MET-2 is thus able to generate H3K9me2 but not me3. Intriguingly, the repeat types that were uniquely dependent on MET-2 were mini- and microsatellite repeats, as only 0.1% of these simple repeats retained H3K9me in the met-2 mutant, and almost all (∼90%) were unaffected by loss of SET-25 (Fig. 3B,C). In contrast, the mutation of met-2 alone eliminated both H3K9me2 and H3K9me3 over large regions of the genome, in particular on the repeat-rich distal arms of autosomes (shown for chromosome 3) (Fig. 3A–C; Supplemental Fig. S3A,B). The fact that the met-2 mutant loses H3K9me3 at many sites even though MET-2 cannot catalyze trimethylation argues that MET-2 activity is an essential prerequisite for SET-25 modification at these sites. In contrast, RNA transposons and the potentially active DNA transposons TC1, TC3, and TC4 retained high levels of H3K9me3 in met-2 animals, presumably because SET-25 can mediate me1, me2, and me3 on its own at these sites (Fig. 3C; Supplemental Fig S3G). This analysis defines three modes of HMT function: first, primarily at simple repeats, MET-2 alone can deposit H3K9me2; second, at other sites the two work sequentially (MET-2, then SET-25); and finally, there is a smaller set of SET-25-specific sites (H3K9me3-bearing loci) (summarized in Fig. 3D). This last class also contains sites that can be dimethylated by MET-2, in the absence of SET-25.
Figure 3.
Three pathways for MET-2 and SET-25 targeting define distinct HMT dependence. (A) H3K9me2 and H3K9me3 ChIP-seq was performed on early embryos at 20°C in wild-type (wt), set-25, and met-2 strains. The enrichment of one experiment over input along a typical autosome, chromsome III, is shown (N = 2), demonstrating retention of H3K9me2 in the set-25 mutant and the more general depletion of both marks in met-2. (B) Genome fraction in percentage bearing either H3K9me2, H3K9me3, or both H3K9me2 and me3 marks in wild-type, met-2 (SET-25 dependent), and set-25 (MET-2 dependent) embryos (H3K9me3: log2 immunoprecipitation-input >2, H3K9me2: log2 immunoprecipitation-input >1). (C) Percentage of remaining H3K9me2- or me3-positive loci in the indicated mutants relative to wild type, sorted by repeat classes: DNA-transposons, RNA-transposons, and satellite repeats. (D) Scheme of three targeting pathways for H3K9me. ChIP-seq tracks are shown and color-coded in Supplemental Figure S3G.
We conclude that the two C. elegans H3K9 HMTs do indeed have at least partially distinct targets: the set-25 mutant compromises H3K9me3 on retrotransposons and pseudogenes (Zeller et al. 2016), while MET-2 specifically labels simple or satellite repeats with H3K9me2 (Fig. 3A–D). At most DNA transposons, the two enzymes can act interchangeably or in a sequential manner. However, SET-25 could not modify nucleosomes on satellite repeats in the absence of MET-2 (Fig. 3C; Supplemental Fig. S3G). These differences argue for different mechanisms of HMT recruitment and link the synthetic sterility phenotype of met-2 with the loss of H3K9me2 on satellite repeats.
MET-2, but not SET-25, is essential to block transcription of simple, satellite repeats
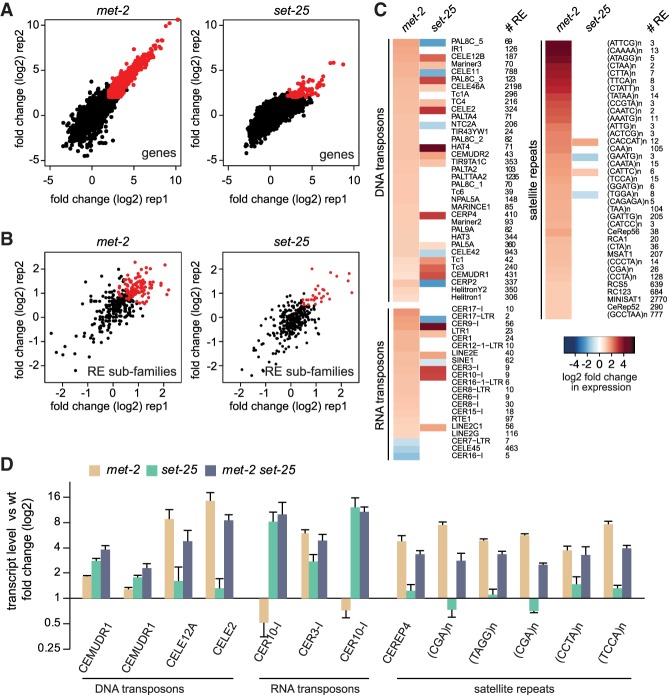
We next asked whether the differential loss of H3K9me3 or H3K9me2 correlates with different patterns of derepression. RNA-seq was performed on synchronized met-2 or set-25 embryos and revealed 562 genes for which the transcript level increased fourfold upon loss of MET-2 (fold change [log2] > 2 over wild type, false discover rate [FDR] < 0.01) (Fig. 4A). Of these, 55% were also up-regulated in met-2 set-25 double mutants (Supplemental Fig. S4B). The loss of SET-25 derepressed 180 genes (Fig. 4A, same cutoff), of which 87% were similarly affected in the met-2 set-25 embryos (Supplemental Fig. S4B). Thus, even though SET-25-dependent H3K9me3 marks many loci in embryos, MET-2-dependent H3K9me2 often compensates for the loss of SET-25 to maintain repression. Alternatively, another mechanism such as transcript degradation might be activated in the absence of SET-25 (Holoch and Moazed 2015).
Figure 4.
MET-2 specifically represses satellites, while SET-25 is needed for selected RNA and DNA transposon repression. (A) Scatter plot showing gene expression as log2 fold change over wild type in early embryos of indicated mutants. Two replicas shown. N = 3. (B) Scatter plot showing repeat subfamily expression as log2 fold change over wild type as in A. (C) Heat map showing transcribed repeat subfamilies in met-2 and/or set-25 worms, separated into three major repeat classes (DNA transposons, RNA transposons, simple repeats). (# RE) Copy number of annotated repeats per subfamily. (D) Quantitative PCR (qPCR) confirmation of the most highly expressed single REs demonstrating differential dependence on MET-2 or SET-25 in early embryos. N = 3. RNA5 (CELE2) overlaps with the intron of the gene C48D1.9, and (TAGG)n with the intron of mib-1; neither gene is derepressed by loss of H3K9me.
Given that most H3K9 methylation is found on repetitive elements, we mapped the total RNA-seq reads for specific repeat elements, as annotated by Repbase (http://www.girinst.org). To assign families of repeats to one or the other HMT, we collapsed all copies of a given repeat into one and analyzed 396 repeat subfamilies. For embryos lacking met-2, we found elevated transcript levels from 103 repeat subfamilies (fold change > 1.5, FDR < 0.05) (Fig. 4B; Supplemental Fig. S4A), of which 68% were also up-regulated in met-2 set-25 mutants (Supplemental Fig. S4C). In set-25 mutant embryos, we scored expression from only 24 subfamilies (Fig. 4B; Supplemental Fig. S4A), of which ∼90% were elevated in the double mutant (Supplemental Fig. S4C). Consistent with the ChIP-seq results, we found that most classes of simple repeats and satellite sequences required MET-2 for repression (shown for 37 repeat motifs in Fig. 4C), while only two were derepressed in set-25 worms. RNA and DNA transposons showed a weak response to met-2 mutation, while a distinct subset of transposons was strongly and specifically up-regulated by set-25 (Fig. 4C).
RT–PCR for individual copies of the aberrantly transcribed repeats confirmed our genome-wide observations: MET-2 is uniquely essential for the repression of simple or satellite repeats and for a subset of DNA transposons, while a significant subset of RNA transposons were sensitive specifically to the loss of SET-25 (Fig. 4D). The fact that some repeat classes (e.g., RNA transposon CER3-I) were sensitive to both set-25 and met-2 mutations parallels the ChIP-seq results (Fig. 3D). We can rule out RNAs that SET-25 silences, alone or together with MET-2, as relevant for the synthetic sterility phenotype.
MET-2 and BRCA1 mutants show temperature sensitivity and additive defects in repeat repression
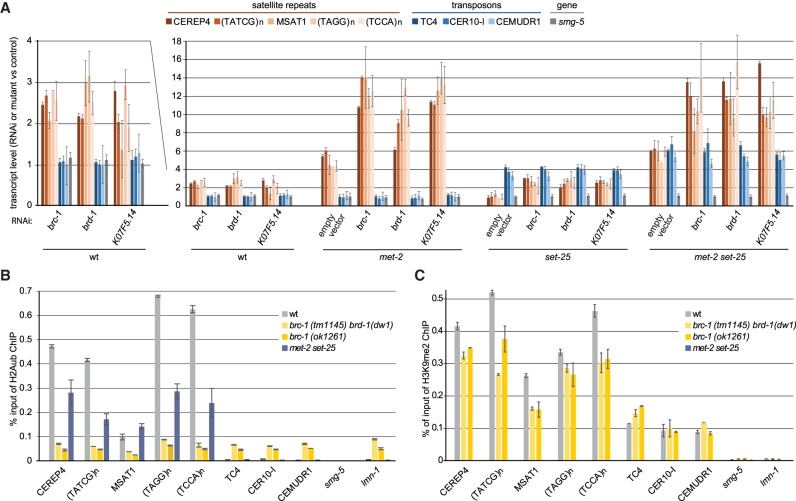
We next asked whether the derepression of satellite repeats scored in the met-2 mutant was linked to the synthetic sterility scored with the BRCA1 complex. Previous work had suggested that BRCA1 knockout mouse cells express or accumulate major and minor satellite repeat transcripts, while transposable elements like the L1 LINE element remained silent (Zhu et al. 2011). To score this in a more specific, quantitative manner, we used quantitative PCR (qPCR) to monitor transcripts from a panel of specific C. elegans repeats, including the two abundant satellite families (MSAT1 and CEREP4), two DNA transposons, and one abundant retrotransposon (TC4, CEMUDR1, and CER10-I), as well as a single-copy gene (smg-1). Total RNA was isolated from mixed embryos of either wild type, met-2 or set-25 mutants, and from the met-2 set-25 double mutant, following exposure to RNAi against the BRCA1 (brc-1), BARD1 (brd-1), KO7F5.14, or a control construct (Fig. 5A). First, as expected, we found the simple or satellite repeats specifically up-regulated upon the loss of MET-2 (brown bars), while the selected transposons were SET-25-dependent (blue bars). The constitutively expressed gene smg-1 was unaffected by either (gray). RNAi against brc-1, brd-1, and K07F5.14 in the wild-type background reproducibly showed twofold higher expression levels for specific satellite repeats, while transcripts from transposons or single copy genes were unchanged (Fig. 5A). Importantly, the combination of brc-1 complex RNAi with the met-2 single mutant showed a more than additive accumulation of repeat transcripts (Fig. 5A). None of the RNAi targets affected SET-25-dependent repeats, and there was no additivity on retrotransposon expression upon ablation of BRCA1 function (Fig. 5A). The fact that the putative cofactor of BRCA-1 (K07F5.14) had similar effects as BRCA1 itself argues that it may act together with the BRCA1/BARD1 complex in worms.
Figure 5.
Loss of BRC-1 derepresses satellite repeats, but not RNA transposons, in a strongly additive manner with loss of MET-2. (A) A qPCR analysis of the expression of MET-2-dependent satellite repeats (tan to brown), SET-25-dependent transposons (blue), and the H3K9me-independent smg-5 gene (gray) at the young adult stage of indicated genetic background (wild type, set-25, met-2, and met-2 set-25). L1 larvae were treated with RNAi against brca-1 and partners brd-1 and K07F5.14. Expression is normalized to wild type grown on empty vector-expressing bacteria. Mean and SEM. N = 3. (B) H2Aub ChIP-qPCR and (C) H3K9me2 ChIP-qPCR in early embryos of indicated genotypes on the sequences analyzed in B. Mean and SEM. N = 3. Note that brc-1 (tm1145) carries a second mutation that fully eliminates BRD-1 expression [brd-1 (dw11)] (Janisiw et al. 2018).
We next examined repeat repression following RNAi against the other replication fork restart factors that were positive hits in the synthetic-lethal screen (Fig. 1) and against factors involved in DSB repair that were not positive in the screen. The results were striking: RNAi against the BRCA1 ligand RAD51 (Schlacher et al. 2012) and the C. elegans checkpoint kinase ATR (ATL in worms [Tibbetts et al. 2000]) also resulted in increased levels of satellite transcripts in otherwise wild-type embryos and showed additive effects with met-2 mutation, but not with set-25 (Supplemental Fig. S5). Again, the ablation of replication stress factors did not affect transcript levels of retrotransposons. The knockdown of the NHEJ pathway (cku-80 and lig4), of the 9-1-1 complex (hus-1 and hpr-9) or of BRCA2 (brc-2) did not affect repeat element transcript levels, nor did they show synergy or additivity with met-2 or set-25 mutants (Supplemental Fig. S5). These results parallel precisely the pattern of synthetic lethality scored in the met-2 mutant: factors implicated in replication fork restart, rather than DSB survival, were required both for satellite repression and germline integrity.
Cross-talk between H3K9me2 and H2Aub on satellite repeats
BRCA1/BARD1 was shown to function as a heterodimeric E3 ubiquitin ligase that targets H2AK127/K129 in vitro (Hashizume et al. 2001; Kalb et al. 2014). This monoubiquitination accumulates upon DNA damage in a BRC-1/BRD-1-dependent manner (Tibbetts et al. 2000; Polanowska et al. 2006). To see if satellite sequences accumulate H2Aub-K127/K129 (hereafter H2Aub), indicating that the BRCA1 complex might associate with sites of H3K9me2-mediated repression in wild-type worms, we performed ChIP experiments. We confirmed that the mark was specific for the BRCA1 complex by performing ChIP in both the brc-1(ok1261) and brc-1(tm1145) brd-1(dw1) double mutant (Janisiw et al. 2018), followed by qPCR for a range of repeat elements. It is important to note that the repeat classes that were derepressed in the met-2 mutant were significantly enriched for H2Aub (Fig. 5B), while it was not detected on the SET-25-regulated transposable elements nor at two single copy genes (smg-5 and lmn-1) (Fig. 5B). The H2Aub signal was lost in both the single brc-1 and the double brc-1 brd-1 mutants (Fig. 5B), and we also observed a reduction of H2Aub (30%–50%) in the met-2 set-25 double mutant, specifically on satellite repeats and not on retrotransposons (Fig. 5B). The fact that even derepressed transposons, like CER10I, did not accumulate the H2Aub signal in the met-2 set-25 mutant suggests a degree of specificity for simple repeats or else for the MET-2 HMT itself. The presence of H2Aub on derepressed satellite repeats in wild-type cells, and the contribution of BRCA1 to repeat repression, argues for a feedback mechanism between damage and H3K9 methylation (Nikolov and Taddei 2016). Indeed, we monitored by ChIP a similar reduction in H3K9me2 (∼1.5-fold) selectively at satellite repeats in the brc-1 or brc-1 brd-1 mutants (Fig. 5C). The drop in H3K9me in the absence of BRCA1 suggests that damage/repair may foster MET-2 recruitment, yet the drop in H2Aub in the absence of MET-2 is counterintuitive, given that repeat transcription is elevated in the met-2 mutant. We interpret this as indicative of a potentially direct cross-regulation; for example, by methylation of BRCA1 by MET-2. In any case, the partial interdependence of BRCA1 and MET-2 may contribute to the similarity of the single mutant defects with respect to satellite derepression and to the synergistic impact that they have on germline integrity.
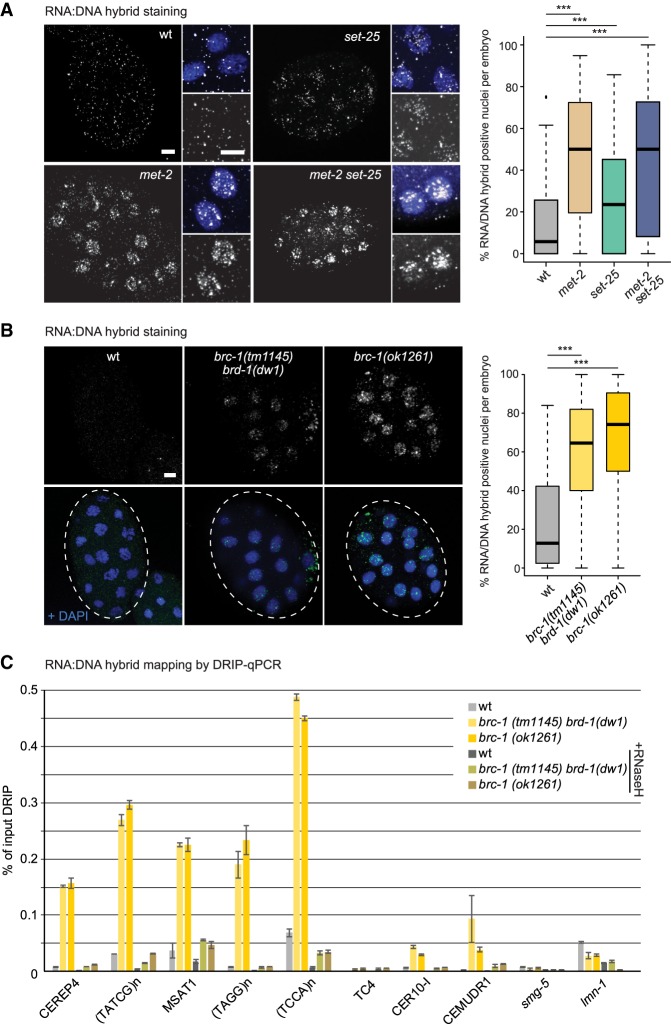
Loss of either BRC-1 or MET-2 leads to RNA:DNA hybrid accumulation on simple repeats
We previously correlated the accumulation of genomic instability (indels) and damage-dependent germline apoptosis with the accumulation of RNA:DNA hybrids in met-2 set-25 worms (Zeller et al. 2016). To extend this to the met-2 single mutant, we performed immunostaining with the S9.6 antibody in strains lacking met-2, set-25, or the double mutant (Boguslawski et al. 1986). As previously reported (Zeller et al. 2016), wild-type embryos showed very little nuclear RNA:DNA hybrid staining (6%), while the met-2 set-25 double mutant had a strong signal in 50% of the nuclei (Fig. 6A). The met-2 mutant showed a similar accumulation of nuclear RNA:DNA hybrids (50%), with a signal intensity comparable to that of the met-2 set-25 double mutant. The S9.6 staining was weaker in set-25 embryos and 24% of the nuclei were positive (Fig. 6A). We confirmed that the RNA:DNA hybrid signal is on derepressed satellite repeats in the met-2 mutant by precipitating RNA:DNA hybrids with S9.6 antibody and performing qPCR (DNA:RNA immunoprecipitation [DRIP]-qPCR) (Supplemental Fig. S6A). Interestingly, repeats that are not derepressed and single copy genes do not accumulate R loops.
Figure 6.
Loss of MET-2 and BRC-1 complex components provokes RNA:DNA hybrid accumulation on satellite repeats. (A) Representative images taken under identical conditions of early embryos of indicated genotypes grown at 20°C, stained for RNA:DNA hybrids (antibody S9.6, HB-8730, American Type Culture Collection). Enlargements at right of each quarter shows the RNA:DNA hybrid signal alone (top panel) and overlay with DAPI (bottom panel). Bar, 5 µm. At right, the percentage of cells with RNA:DNA nuclear staining per embryo is plotted for indicated genotypes. N = 3; n = 20 embryos. Box plots are as in Figure 2. (***) P < 0.0001, two-sided Wilcoxon signed-rank test. (B) RNA:DNA hybrid immunofluorescence using the S9.6 antibody in wild-type, brc-1, and brc-1 brd-1 mutant embryos, as in A. Similar quantitation for RNA:DNA hybrids is at the right. N = 3; n = 20. (C) RNA:DNA hybrid accumulation on indicated sequences scored by DRIP-qPCR with antibody S9.6. Mutants are as in A and B. qPCR monitors satellite repeats expressed in the brc-1 mutant as well as unaffected transposable elements (CEMUDR1, TC4, and CER10-I) and a single-copy gene (smg-5). RNase H-treated samples quantify unspecific DRIP signal. Mean and SEM; N = 2.
Considering the synthetic lethality of brc-1 and met-2 and evidence showing that mammalian brca1 mutant cells accumulate R loops both at highly transcribed vertebrate genes (Hatchi et al. 2015) and on aberrantly transcribed SAT1 repeats (Zhu et al. 2018), we monitored RNA:DNA hybrid accumulation in C. elegans brc-1/brd-1 mutants. Quantitation of immunostaining with S9.6 antibody of wild-type and mutant embryos showed a general increase in RNA:DNA hybrid positive nuclei in the brc-1 and brc-1 brd-1 mutants (Fig. 6B). DRIP-qPCR again mapped the RNA:DNA hybrids to up-regulated satellite repeats (Fig 6C), as in the met-2 mutant (Supplemental Fig. S6A). Indeed, at the met-2-sensitive, derepressed satellite repeats, such as MSAT-1, brc-1/brd-1 mutants showed a 10-fold higher signal for RNA:DNA hybrids than was detected in wild-type embryos (Fig. 6C). All positive signals were sensitive to treatment with RNase H, confirming the specificity of the S9.6 antibody (Fig. 6C). Given the synthetic sterility and the additive effect between met-2 and brc-1 on satellite derepression, we checked whether knockdown of brc-1 in the met-2 mutant further increases the accumulation of RNA:DNA hybrids. This is the case (Supplemental Fig. S6B).
Because RNA:DNA hybrids have been shown to perturb progression of the replication fork (Gan et al. 2011), we also quantified the number of PCNA foci in wild-type and met-2 mutant embryos that were treated with brc-1 or control RNAi. As hypothesized, we observed an additional increase in the number of cells with PCNA foci in the met-2 brc-1 RNAi, compared to the deletion of met-2 or the depletion of brc-1 alone (Supplemental Fig. S6C).
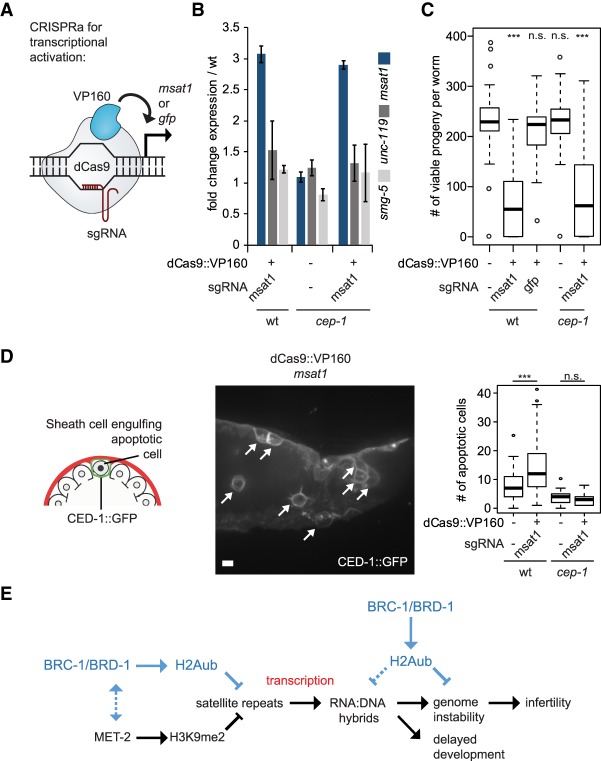
Induction of satellite repeat expression by CRISPRa is sufficient to abrogate fertility
Our results link satellite repeat transcription and DNA damage-induced germline apoptosis in a met-2 background. To prove that this link occurs in wild-type animals, we performed a gain-of-function experiment in which we induced the transcription of a single, abundant satellite repeat in otherwise wild-type C. elegans worms and monitored both brood size and germline apoptosis (Fig. 7A). We targeted the endogenous C. elegans satellite MSAT1 repeat (a 226-bp satellite repeat at ∼207 dispersed loci), using the CRISPRa system, whereby an enzymatically dead Cas9 (dCas9) fused to 10 repeats of the VP16 activator (VP160) motif is directed to an endogenous site by a guide RNA (gRNA) (Konermann et al. 2015; Long et al. 2015). As a control, we used a gRNA for an integrated gfp::h2b fusion. By qPCR, we detected a threefold increase in steady-state transcripts containing MSAT1, while there was no increase in two control genes that lack homology to MSAT1 upon introduction of the msat1 gRNA (msat1 vs. smg-1, unc-119) (Fig. 7B). Strikingly, CRISPR–cas9–VP160-induced MSAT1 transcription correlated with a significant reduction in fertility in the ensuing generation (Fig. 7C), while targeting GFP-H2B had no effect. We also scored a significant increase in an apoptotic marker, CED-1:GFP (Zhou et al. 2001) in cells with CRISPR-induced MSAT1 (Fig. 7D), although the extent of apoptosis and the sterility were both less than that scored in the met-2 set-25 double mutant (Zeller et al. 2016).
Figure 7.
Aberrant expression of a satellite repeat can cause loss of fertility. (A) Scheme showing the targeting of catalytically dead Cas9 (dCas9) to msat1 or gfp genes, and activation by 10 copies of the VP16 transactivating domain. (B) qPCR showing the selective induction of the MSAT1 satellite transcripts targeted by dCas9::VP160 as fold change over worms lacking the dCas9::VP160 fusion protein in L3–L4 larvae of wild-type and cep-1 mutant backgrounds (mean and SD.; N = 2). Expression of smg-5 and unc-119 genes serve as negative controls. (C) The total number of viable progeny of MSAT1 overexpressing wild-type and cep-1 mutant strains at 20°C. N = 3; n = 25. Strains lacking the dCas9::VP160 construct and a strain expressing dCas9::VP160 targeted to a GFP reporter transgene are negative controls. Box plots are as in Figure 1 with two-sided Wilcoxon signed-rank test (***) P < 0.0001; (n.s.) P > 0.05. (D) Representative image of a gonad showing apoptotic cells (arrows) expressing the CED-1::GFP (plasma membrane apoptotic marker) in the MSAT1 overexpression strain. Bar, 5 µm. Plotted is the quantitation of apoptosis (number of cells fully engulfed by CED-1::GFP per gonad arm) in wild type and cep-1 mutant (lacking p53 homolog) as a function of MSAT1 overexpression. N = 3; n = 60. (***) P < 0.0001; (n.s.) P > 0.05, two-sided Wilcoxon signed-rank test. (E) Schematic description of how MET-2-mediated H3K9me2 and BRC-1-mediated H2Aub work together to prevent RNA:DNA hybrid accumulation and associated phenotypes.
In met-2 set-25 mutants, sterility at 25°C correlates with p53-dependent apoptosis and increased genomic instability in germline cells (Zeller et al. 2016). While sterility is not suppressed in a p53 mutant (cep-1), the apoptotic signal arising from met-2 set-25 is lost, confirming that the loss of H3K9me leads to DNA damage in the germline (Zeller et al. 2016). This is also the case upon MSAT1 induction in the cep-1 mutant (Fig. 7D), although, in addition, we show here that p53 loss (cep-1 mutation) alone does not lead to germline lethality. Indeed, germline lethality is entirely MSAT1 expression-dependent (Fig. 7C), while the germline CED-1 fluorescence signal triggered by MSAT1 expression is entirely p53-dependent (Fig. 7D). This strongly argues that MSAT1-induced expression is the likely cause of brood size reduction in these worms.
Discussion
We present here three important conclusions. First, the loss of the BRCA1/BARD1 complex, along with other factors that regulate replication fork restart, such as Rad51 and ATR kinase, show a selective synthetic lethality with the loss of MET-2 (SETDB1), the H3K9me2 transferase in C. elegans, and not with SET-25 (G9a/SUV39). Second, we show that MET-2 is uniquely responsible for the silencing of satellite repeats, while SET-25 specifically represses pseudogenes and retrotransposons (Fig. 4; Zeller et al. 2016). Third, we are able to link repeat expression directly to a loss of germline integrity by inducing expression of the MET-2-repressed MSAT1 repeat in a wild-type background. Interestingly, we found that the BRCA1 complex is partially redundant with MET-2 in preventing the accumulation of satellite repeat transcripts, RNA:DNA hybrids, and replication defects.
Several recent studies have implicated BRCA-1 in the regulation of repeat stability. Deletion of brca-1 in mice results in the derepression of satellite sequences (Zhu et al. 2011), and human brca1-deficient tumors often accumulate satellite transcripts, as do a broad spectrum of epithelial tumors (Ting et al. 2011). In other studies, BRCA-1 was reported to be essential to prevent the spreading of Long Interspersed Element-1 (LINE-1) elements (Liu et al. 2018) and to block chromosomal structural variants arising at Alu elements (Mendez-Dorantes et al. 2018). Our synthetic sterility screen illustrates the debilitating effects of the combined loss of BRC-1 and MET-2-mediated H3K9me2. We show that H3K9me2 is highly enriched on satellite repeats, where it blocks transcription and prevents RNA:DNA hybrid formation on these elements. The more than additive appearance of satellite transcripts and R loops in met-2 and brc-1 mutants, but not in the set-25 mutant, argues that the expression of simple repeats, and not transposons, renders cells highly dependent on the BRCA1/BARD1 complex. Since different HMTs target the classes of repeats independently, this has important implications for the treatment of tumors with H3K9-HMT inhibitors (Rowbotham et al. 2018). Two recent studies have highlighted the fact that inhibitors of G9a, one of the HMTs responsible for H3K9me2 levels in mammalian cells, leads to genome instability and the delayed appearance of highly aggressive forms of skin and lung tumors (Avgustinova et al. 2018; Rowbotham et al. 2018).
Mutations that compromise the BRCA1/BARD1 complex, a multifunctional E3 ubiquitin ligase implicated in the repair of DSB, represent the highest risk factor for hereditary forms of breast and ovarian cancer (King et al. 2003). While this complex is best known to promote homologous recombination at DSB, BRCA1 is also implicated in interstrand cross-link repair and in replication fork stability, given that the complex associates both with stalled DNA and RNA polymerases (Schlacher et al. 2012; Urban et al. 2016; Stewart et al. 2018). Moreover, BRCA1 was shown to recruit the RNA/DNA helicase senataxin to sites of paused RNA polymerase II, to ensure mRNA termination and prevent ensuing damage (Hatchi et al. 2015). A recent study shows that mammalian BRCA-1 can bind RNA:DNA hybrids directly (D'Alessandro et al. 2018). BRCA1 may load Rad51 on the ssDNA generated by RNA:DNA hybridization (Schlacher et al. 2012; Santos-Pereira and Aguilera 2015). The fact that the loss of BRCA1 in worms accentuates defects arising from met-2 mutation suggests that excess satellite transcription overloads the pathways that normally deal with R loops. We do not see the same synergism with the ablation of the C. elegans homolog of BRCA-2, nor of other factors implicated in DSB repair by NHEJ. While this does not exclude that other types of DNA damage impact germline instability, in the background of promiscuous satellite repeat expression, the loss of factors that ensure fork recovery is particularly deleterious.
In C. elegans, we found that BRCA-1/BARD-1-dependent H2Aub is enriched over satellite repeat sequences. In mouse neuronal cells, the enrichment of both BRCA1 and H2Aub on satellite DNA has also been reported (Zhu et al. 2011). Upon brc-1 ablation, we monitored a drop in H3K9me2 on these elements and lost the H2Aub signal, coincident with enhanced satellite expression. There are two interpretations of this: Either H2Aub contributes to repression, or BRCA-1/BARD-1 help recruit MET-2 to modify and repress these sequences. Since the mechanism of recruiting MET-2/SETDB1 to repeats is unknown, we do not rule out either of these mechanisms.
Intriguingly, the loss of RAD51, which coats ssDNA, and ATR kinase, which signals a checkpoint activation in response to ssDNA, parallels the effects of brc-1 or brd-1 knockout, triggering satellite expression and synergistic germline lethality in the met-2. A recent study of BRCA1 in C. elegans meiosis suggests that BRCA1 not only helps recruit RAD51 to meiotic DSBs but also helps load RAD51 (Janisiw et al. 2018). This role may be taken over by BRCA2 in mammalian cells (Prakash et al. 2015). Other studies have implicated C. elegans BRC-1 in intersister recombination mediated repair in germline cells (Adamo et al. 2008). In any case, based on our synthetic-lethal screen, we identified RAD51 and ATR as well as BRCA1 and BARD1 as becoming essential for the maintenance of germline integrity in the absence of H3K9me2.
One hypothesis for synthetic sterility with loss of met-2 is that R loops form on derepressed satellite repeats in met-2 and recruit BRCA1. Loss of the BRCA1-mediated repair pathway would then lead to excessive strand breakage and damage-induced apoptosis. This damage is itself sufficient to reduce fertility, at least in the case of MSAT1 overexpression (Fig. 7). Simple repeat RNAs may be more toxic than other promiscuous transcripts, because they lack signals for processing and export. Moreover, repeat sequences may be more inclined to form fork-destabilizing fold-back structures. In this respect, it is important to underscore that the other pathways that generate germline lethality in the met-2 background are factors that regulate the processing, degradation, and export of RNA Pol II transcripts (Fig. 1). We do not know whether all synthetic-lethal hits generate R loops on repeat sequences; it is possible that impaired export and processing of RNA generates damage at unique sequence sites. The induction of DNA damage is not unique to germline cells, as we also score an increase in RPA-1 repair foci in somatic cells of C. elegans embryos lacking MET-2 (Fig. 2C). However, as C. elegans does not support apoptosis in differentiated somatic tissues, germline apoptosis is the most obvious DNA-damage induced phenotype.
Important for understanding the roles of BRCA1/BARD1 and of MET-2-dependent methylation is their parallel behavior with respect to sites affected, synergism, RPA-1 focus formation, and R-loop accumulation (this paper and Zeller et al. 2016). The cross-talk between the pathways of H3K9me and H2Aub may be conserved in other species (Zhu et al. 2011). Whether MET-2 and the BRCA1 complex interact directly is unknown, although BRCA1 is reported to be methylated at both arginine and lysine residues in breast cancer cell lines (Guendel et al. 2010). Moreover, H3K9 methylation is enhanced at DSBs in some contexts (Ayrapetov et al. 2014; Khurana et al. 2014; Clouaire et al. 2018). Exactly why H2Aub is retained on satellite repeats in wild-type embryos is unknown, but it might provide a mechanism for propagation of H3K9me at sites that are prone to damage and promiscuous transcription. This potential mechanism is illustrated in a model in Figure 7E.
Our work and that of others (Avgustinova et al. 2018; Rowbotham et al. 2018) warn that the treatment of cancers with inhibitors of H3K9-HMTs may augment genomic instability and thus, in the long run, favor more severe forms of cancer. On the other hand, for tumors that are deficient for BRCA1, treatment with G9a or other H3K9-HMT inhibitors may enhance cell death, like the synthetic lethality we score in worms. Given that the integrity of heterochromatin is known to deteriorate with age (Tsurumi and Li 2012; Sidler et al. 2017), R-loop-induced damage on satellite repeats may be a source of age-related DNA damage and cancer proneness. We show here that MET-2-mediated H3K9me2 and SET-25-mediated H3K9me3 modifications have distinct patterns across the genome and distinct effects on repeat expression, which is likely to be true in mammalian cells as well. We can now attribute the genomic instability and germline lethality that had been previously reported for the met-2 set-25 double mutant (Garrigues et al. 2015; Zeller et al. 2016; McMurchy et al. 2017) to satellite repeat repression by MET-2. We propose that a systematic study of satellite repeat derepression and instability should be carried out before any H3K9me-HMT inhibitor is used therapeutically on patients.
Materials and methods
Experimental model and subject details
Caenorhabditis elegans strains were obtained from the Caenorhabditis Genome Center or were generated during this study (full description in Supplemental Table S3). Worms were grown on OP50 at 20°C unless stated otherwise.
CRISPR gene activation by dCas9-VP160
For the overexpression of endogenous targets, an activator protein composed of a catalytically inactive Cas9 protein (dCas9) fused to 10 tandem VP16 units (Long et al. 2015) was expressed from the ubiquitous promoter eft-3 (plasmid pIK312). In addition, we expressed an MS2-VP64 fusion protein, synthesized as a gBlock DNA fragment (Integrated DNA Technologies) from the ubiquitous promoter eft-3 (plasmid pIK303) as an attempt to potentiate activation efficiency (Konermann et al. 2015).
Extrachromosomal arrays were created containing 100 ng/µL pIK312 Peft-3::Cas9-VP160, 100 ng/µL pIK303 Peft-3::MS2-VP64, and 20 ng/µL Prab-3::mCherry coinjection marker. The injection mix targeting MINISAT1 contained two gRNAs, at 20 ng/µL each (MSAT1: [g]cggcaatttcggcaattgc) cloned into the pIK292 backbone. The injection mix targeting the minimal promoter in the control strain contained six gRNAs, at 20 ng/µL each [(G)TGCAAATTACGAGCGTTGT, (G)AAATTACGAGCGTTGTAGG, GTTGTAGGGGGCGGAGCGAT, GCGATAGGTCCTATAGGTTT, ATCATCATTCATTCATTCAT, and (G)TCCTCTTTCTGAGCTTCTC], cloned into the pIK292 backbone. Transgenic strains were established in an N2 background.
Immunofluorescence (IF), antibodies, and live microscopy
RNA:DNA hybrid IF was carried out as previously described (Zeller et al. 2016). Staining was performed in 4XSSC-T (0.1% Tween-20) and 3% BSA. Primary antibody (S9.6, HB-8730, American Type Culture Collection, gift of P. Pasero, Montpellier) was used at a dilution of 1:200 in 4XSSC-T and 3% BSA overnight at 4°C. Anti-panacetyl-H4 was included as a positive staining control (06-866, Merck Millipore, 1:500). For live cell imaging, larvae were mounted on slides coated with 2% agarose. Microscopy was carried out on a spinning disc confocal microscope (Yokogawa X1 and a Yokogawa W1 scan-head mounted on an Olympus and a Zeiss microscope, respectively, Visitron). Stacks of images were analyzed using ImageJ.
For RPA-1 and PCN-1, nuclear foci were quantified using the KNIME Analytics Platform (Dietz and Berthold 2016). In summary, nuclei were detected using a seeded watershed segmentation, and foci detected using a Laplacian-of-Gaussian (LoG) detector from TrackMate (Tinevez et al. 2017) (fmi-ij2-plugins-0.2.5 https://doi.org/10.5281/zenodo.1173536). Foci outside of a nucleus were ignored in the analysis.
Developmental timing and progeny size
Animals were adapted for two generations to the indicated temperatures, and the F3 generations were used for the experiment. For developmental timing experiments, single worms were scored by eye at 24-h intervals over 3 d of somatic development starting with stage 1 larvae (L1) synchronized by starvation. For progeny experiments, worms were singled as stage 4 larvae (L4). Adults were transferred to fresh plates to keep generations separated, and the total number of viable F1 progeny was determined.
RNAi screen
For the primary screen, worms were grown in liquid culture (NGM containing NaCl, peptone, CaCl2, MgSO4, KH2PO4 cholesterol, carb, IPTG) with RNAi-expressing bacteria of the Ahringer library in 384-well plates from Greiner (781 097) allowing for direct automated microscopy. After incubation of 4 d at 20°C in a humid chamber, worms were anesthetized by adding levamisole and imaged on a GE inCell automated microscope. The initial screen was for a reduction of between twofold to 10-fold fewer larvae in three out of four wells, after 4 d at 20°C. Rescreening was performed in 96-well format adjusting volumes appropriately using sequenced pure RNAi clones from both the Ahringer and Vidal RNAi library. Hits were quantified manually.
ChIP, DRIP, and RNA expression (RNA-seq and qPCR)
ChIP and DRIP-qPCR were performed as previously described (Zeller et al. 2016).
Libraries were prepared from chromatin IP and genomic DNA samples as described previously (Zeller et al. 2016). Read density along the genome was calculated by tiling the genome into 200-bp windows (nonoverlapping) and counting the number of sequence fragments within each window, using the qCount function of the QuasR package. To count reads on repeat subfamilies, we allowed the mapping of reads that mapped to multiple repeat copies. The sum of all reads counted to all copies of a repeat was termed repeat subfamily. To compensate for differences in the read depths, libraries were normalized to the total number of reads per library. ChIP-seq signals are displayed as average enrichment of IP—input (log2). For RNAseq, total RNA was isolated and expression levels determined as previously described (Zeller et al. 2016).
Quantification and statistical analysis
All the data were obtained from three biologically independent replicates. A two-sided Wilcoxon signed-rank test was used to analyze brood size, expression analysis by qPCR and ChIP qPCR, and quantification of microscopy data. All other data were analyzed using a Student's t-test, and P-values of <0.05 were considered significant.
P-values for differential expression analysis between different genotypes were calculated using edgeR, and P-values were adjusted for multiple testing according to the family-wise error rate (FWER) of Benjamini and Hochberg.
Accession codes
All data sets from this study have been uploaded to the Gene Expression Omnibus (GEO) under accession number GSE100726.
Supplementary Material
Acknowledgments
Strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). We thank Helge Grosshans, Yannick Hauser, and Rafal Ciosk for reagents and materials. We thank Robin van Schendel, Marcel Tijsterman, and the Friedrich Miescher Institute Genomics and Microscopy Facilities for advice and discussion. J.P. and S.P.M. are supported by long-term EMBO fellowships. S.M.G. thanks the Swiss National Science Foundation, the European Research Council, and the Novartis Research Foundation for support.
Author contributions: P.Z. and J.P. planned and executed most experiments, evaluated results, and wrote the paper; S.M.G. planned experiments, evaluated results, and wrote the paper; I.K. planned and performed the CRISPRa overexpression of MSAT1; B.T. helped to plan and execute the synthetic sterility screen; S.P.M. helped with RPA-1 and PCN-1 foci measurements; and V.K. provided invaluable technical help.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.322495.118.
Freely available online through the Genes & Development Open Access option.
References
- Adamo A, Montemauri P, Silva N, Ward JD, Boulton SJ, La Volpe A. 2008. BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair. EMBO Rep 9: 287–292. 10.1038/sj.embor.7401167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiadis V, Ballestas ME, Sanchez C, Winokur S, Vedanarayanan V, Warren M, Ehrlich M. 2007. RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA. Biochim Biophys Acta 1769: 29–40. 10.1016/j.bbaexp.2006.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgustinova A, Symeonidi A, Castellanos A, Urdiroz-Urricelqui U, Solé-Boldo L, Martín M, Pérez-Rodríguez I, Prats N, Lehner B, Supek F, et al. 2018. Loss of G9a preserves mutation patterns but increases chromatin accessibility, genomic instability and aggressiveness in skin tumours. Nat Cell Biol 20: 1400–1409. 10.1038/s41556-018-0233-x [DOI] [PubMed] [Google Scholar]
- Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. 2014. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci 111: 9169–9174. 10.1073/pnas.1403565111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, García-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. 2014. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511: 362–365. 10.1038/nature13374 [DOI] [PubMed] [Google Scholar]
- Billing D, Horiguchi M, Wu-Baer F, Taglialatela A, Leuzzi G, Nanez SA, Jiang W, Zha S, Szabolcs M, Lin CS, et al. 2018. The BRCT domains of the BRCA1 and BARD1 tumor suppressors differentially regulate homology-directed repair and stalled fork protection. Mol Cell 72: 127–139.e8. 10.1016/j.molcel.2018.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. 1986. Characterization of monoclonal antibody to DNA · RNA and its application to immunodetection of hybrids. J Immunol Methods 89: 123–130. 10.1016/0022-1759(86)90040-2 [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Martin JS, Polanowska J, Hill DE, Gartner A, Vidal M. 2004. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr Biol 14: 33–39. 10.1016/j.cub.2003.11.029 [DOI] [PubMed] [Google Scholar]
- Clouaire T, Rocher V, Lashgari A, Arnould C, Aguirrebengoa M, Biernacka A, Skrzypczak M, Aymard F, Fongang B, Dojer N, et al. 2018. Comprehensive mapping of histone modifications at DNA double-strand breaks deciphers repair pathway chromatin signatures. Mol Cell 72: 250–262.e6. 10.1016/j.molcel.2018.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessandro G, Whelan DR, Howard SM, Vitelli V, Renaudin X, Adamowicz M, Iannelli F, Jones-Weinert CW, Lee M, Matti V, et al. 2018. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat Commun 9: 5376 10.1038/s41467-018-07799-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz C, Berthold MR. 2016. KNIME for open-source bioimage analysis: a tutorial. Adv Anat Embryol Cell Biol 219: 179–197. 10.1007/978-3-319-28549-8_7 [DOI] [PubMed] [Google Scholar]
- Dodge JE, Kang Y-K, Beppu H, Lei H, Li E. 2004. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol 24: 2478–2486. 10.1128/MCB.24.6.2478-2486.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymery A, Horard B, El Atifi-Borel M, Fourel G, Berger F, Vitte AL, Van den Broeck A, Brambilla E, Fournier A, Callanan M, et al. 2009. A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic Acids Res 37: 6340–6354. 10.1093/nar/gkp639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. 2011. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 25: 2041–2056. 10.1101/gad.17010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues JM, Sidoli S, Garcia BA, Strome S. 2015. Defining heterochromatin in C. elegans through genome-wide analysis of the heterochromatin protein 1 homolog HPL-2. Genome Res 25: 76–88. 10.1101/gr.180489.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-González B, Felipe-Abrio I, Aguilera A. 2009. The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol Cell Biol 29: 5203–5213. 10.1128/MCB.00402-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guendel I, Carpio L, Pedati C, Schwartz A, Teal C, Kashanchi F, Kehn-Hall K. 2010. Methylation of the tumor suppressor protein, BRCA1, influences its transcriptional cofactor function. PLoS One 5: e11379 10.1371/journal.pone.0011379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage AE, French SL, Beyer AL, Tollervey D. 2010. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24: 1546–1558. 10.1101/gad.573310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, Cimprich KA. 2016. Conflict resolution in the genome: how transcription and replication make it work. Cell 167: 1455–1467. 10.1016/j.cell.2016.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. 2001. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 276: 14537–14540. 10.1074/jbc.C000881200 [DOI] [PubMed] [Google Scholar]
- Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML. 2015. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell 57: 636–647. 10.1016/j.molcel.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D, Moazed D. 2015. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16: 71–84. 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Aguilera A. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721. 10.1016/j.molcel.2003.08.010 [DOI] [PubMed] [Google Scholar]
- Janisiw E, Dello Stritto MR, Jantsch V, Silva N. 2018. BRCA1-BARD1 associate with the synaptonemal complex and pro-crossover factors and influence RAD-51 dynamics during Caenorhabditis elegans meiosis. PLoS Genet 14: e1007653 10.1371/journal.pgen.1007653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb R, Mallery DL, Larkin C, Huang JT, Hiom K. 2014. BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep 8: 999–1005. 10.1016/j.celrep.2014.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, Shi L, Jailwala P, Sung MH, Hakim O, et al. 2014. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep 8: 1049–1062. 10.1016/j.celrep.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Marks JH, Mandell JB. 2003. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302: 643–646. 10.1126/science.1088759 [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H. 2015. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature 517: 583–588. 10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540–543. 10.1126/science.1091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yuan Y, Huan J, Shen Z. 2001. Inhibition of breast and brain cancer cell growth by BCCIPα, an evolutionarily conserved nuclear protein that interacts with BRCA2. Oncogene 20: 336–345. 10.1038/sj.onc.1204098 [DOI] [PubMed] [Google Scholar]
- Liu N, Lee CH, Swigut T, Grow E, Gu B, Bassik MC, Wysocka J. 2018. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 553: 228–232. 10.1038/nature25179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long L, Guo H, Yao D, Xiong K, Li Y, Liu P, Zhu Z, Liu D. 2015. Regulation of transcriptionally active genes via the catalytically inactive Cas9 in C. elegans and D. rerio. Cell Res 25: 638–641. 10.1038/cr.2015.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931. 10.1038/nature08858 [DOI] [PubMed] [Google Scholar]
- McMurchy AN, Stempor P, Gaarenstroom T, Wysolmerski B, Dong Y, Aussianikava D, Appert A, Huang N, Kolasinska-Zwierz P, Sapetschnig A, et al. 2017. A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. Elife 6: e21666 10.7554/eLife.21666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Dorantes C, Bhargava R, Stark JM. 2018. Repeat-mediated deletions can be induced by a chromosomal break far from a repeat, but multiple pathways suppress such rearrangements. Genes Dev 32: 524–536. 10.1101/gad.311084.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzetta C, Boyarchuk E, Pontis J, Ait-Si-Ali S. 2015. Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat Rev Mol Cell Biol 16: 499–513. 10.1038/nrm4029 [DOI] [PubMed] [Google Scholar]
- Nikolov I, Taddei A. 2016. Linking replication stress with heterochromatin formation. Chromosoma 125: 523–533. 10.1007/s00412-015-0545-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padeken J, Zeller P, Gasser SM. 2015. Repeat DNA in genome organization and stability. Curr Opin Genet Dev 31: 12–19. 10.1016/j.gde.2015.03.009 [DOI] [PubMed] [Google Scholar]
- Panier S, Boulton SJ. 2014. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 15: 7–18. 10.1038/nrm3719 [DOI] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu Z-P. 2015. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161: 774–789. 10.1016/j.cell.2015.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337. 10.1016/S0092-8674(01)00542-6 [DOI] [PubMed] [Google Scholar]
- Pinheiro I, Margueron R, Shukeir N, Eisold M, Fritzsch C, Richter FM, Mittler G, Genoud C, Goyama S, Kurokawa M, et al. 2012. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 150: 948–960. 10.1016/j.cell.2012.06.048 [DOI] [PubMed] [Google Scholar]
- Polanowska J, Martin JS, Garcia-Muse T, Petalcorin MI, Boulton SJ. 2006. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J 25: 2178–2188. 10.1038/sj.emboj.7601102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Zhang Y, Feng W, Jasin M. 2015. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol 7: a016600 10.1101/cshperspect.a016600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przetocka S, Porro A, Bolck HA, Walker C, Lezaja A, Trenner A, von Aesch C, Himmels SF, D'Andrea AD, Ceccaldi R, et al. 2018. CtIP-mediated fork protection synergizes with BRCA1 to suppress genomic instability upon DNA replication stress. Mol Cell 72: 568–582.e6. 10.1016/j.molcel.2018.09.014 [DOI] [PubMed] [Google Scholar]
- Rosenthal SB, Len J, Webster M, Gary A, Birmingham A, Fisch KM. 2018. Interactive network visualization in Jupyter notebooks: visJS2jupyter. Bioinformatics 34: 126–128. 10.1093/bioinformatics/btx581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbotham SP, Li F, Dost AFM, Louie SM, Marsh BP, Pessina P, Anbarasu CR, Brainson CF, Tuminello SJ, Lieberman A, et al. 2018. H3K9 methyltransferases and demethylases control lung tumor-propagating cells and lung cancer progression. Nat Commun 9: 4559 10.1038/s41467-018-07077-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240. 10.1038/nature08674 [DOI] [PubMed] [Google Scholar]
- Saksouk N, Simboeck E, Déjardin J. 2015. Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 8: 3 10.1186/1756-8935-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Pereira JM, Aguilera A. 2015. R loops: new modulators of genome dynamics and function. Nat Rev Genet 16: 583–597. 10.1038/nrg3961 [DOI] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22: 106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. 2011. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev 25: 781–788. 10.1101/gad.2027411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidler C, Kovalchuk O, Kovalchuk I. 2017. Epigenetic regulation of cellular senescence and aging. Front Genet 8: 138 10.3389/fgene.2017.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MD, Zelin E, Dhall A, Walsh T, Upadhyay E, Corn JE, Chatterjee C, King MC, Klevit RE. 2018. BARD1 is necessary for ubiquitylation of nucleosomal histone H2A and for transcriptional regulation of estrogen metabolism genes. Proc Natl Acad Sci 115: 1316–1321. 10.1073/pnas.1715467115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP. 2014. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791. 10.1101/gad.989402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela A, Alvarez S, Leuzzi G, Sannino V, Ranjha L, Huang JW, Madubata C, Anand R, Levy B, Rabadan R, et al. 2017. Restoration of replication fork stability in BRCA1- and BRCA2-deficient cells by inactivation of SNF2-family fork remodelers. Mol Cell 68: 414–430.e8. 10.1016/j.molcel.2017.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. 2009. Cohesin acetylation speeds the replication fork. Nature 462: 231–234. 10.1038/nature08550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT. 2000. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev 14: 2989–3002. 10.1101/gad.851000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW. 2017. TrackMate: an open and extensible platform for single-particle tracking. Methods (San Diego, Calif) 115: 80–90. 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S. 2011. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331: 593–596. 10.1126/science.1200801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150: 934–947. 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- Tsurumi A, Li WX. 2012. Global heterochromatin loss: a unifying theory of aging? Epigenetics 7: 680–688. 10.4161/epi.20540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban V, Dobrovolna J, Huhn D, Fryzelkova J, Bartek J, Janscak P. 2016. RECQ5 helicase promotes resolution of conflicts between replication and transcription in human cells. J Cell Biol 214: 401–415. 10.1083/jcb.201507099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, Bedford MT. 2014. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell 53: 484–497. 10.1016/j.molcel.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller P, Padeken J, van Schendel R, Kalck V, Tijsterman M, Gasser SM. 2016. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat Genet 48: 1385–1395. 10.1038/ng.3672 [DOI] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat Cell Biol 16: 2–9. 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chiang H-C, Wang Y, Zhang C, Smith S, Zhao X, Nair SJ, Michalek J, Jatoi I, Lautner M, et al. 2017. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat Commun 8: 15908 10.1038/ncomms15908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, Horvitz HR. 2001. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104: 43–56. 10.1016/S0092-8674(01)00190-8 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM. 2011. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477: 179–184. 10.1038/nature10371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Hoong N, Aslanian A, Hara T, Benner C, Heinz S, Miga KH, Ke E, Verma S, Soroczynski J, et al. 2018. Heterochromatin-encoded satellite RNAs induce breast cancer. Mol Cell 70: 842–853.e7. 10.1016/j.molcel.2018.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.