In this Perspective, Armstrong and Duronio discuss the findings in this issue of Genes & Developmnet by Seller et al., who developed a new technology for inhibiting maternal gene function to identify the H3K9 methyltransferase necessary for initiating constitutive heterochromatin formation during early Drosophila embryogenesis.
Keywords: cell cycle, development, embryo, heterochromatin
Abstract
Constitutive heterochromatin is a prevalent feature of eukaryotic genomes important for promoting cell differentiation and maintaining genome stability. During animal reproduction, constitutive heterochromatin is disassembled in gametes prior to formation of the zygote and then subsequently re-established as development ensues and cells differentiate. Despite progress in understanding the mechanisms that maintain heterochromatin in differentiated cell types, how constitutive heterochromatin is assembled de novo during early development remains poorly understood. In this issue of Genes & Development, Seller and colleagues (pp. 403–417) develop a new technology for inhibiting maternal gene function to identify the H3K9 methyltransferase necessary for initiating constitutive heterochromatin formation during early Drosophila embryogenesis.
Constitutive heterochromatin is a condensed form of chromatin that promotes genome stability by suppressing recombination within DNA repeats and repressing transposon mobilization in both the soma and germline. A characteristic molecular feature of constitutive heterochromatin is di- and trimethylation of K9 of histone H3 (H3K9me2/3), which binds HP1a and thereby organizes the functional features of constitutive heterochromatin in differentiated cells. Despite its name, constitutive heterochromatin is quite dynamic. Each cell cycle, it is disassembled in mitosis and reassembled in the subsequent interphase. The many layers of constitutive heterochromatin regulation, including histone tail posttranslational modifications, are erased each generation to yield a zygote with an essentially naïve genome capable of producing every differentiated cell type. As development proceeds, the genome is reorganized such that cells progressively acquire the structural and functional features of heterochromatin, but how this occurs has remained a mystery.
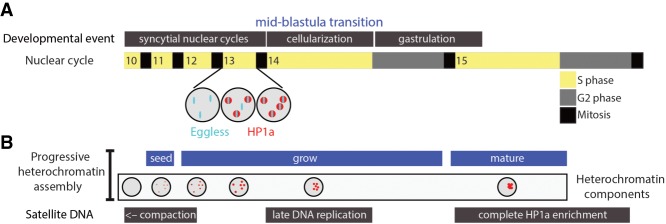
The critical features and functions of heterochromatin are evolutionarily conserved from fungi to mammals, and studies in genetically tractable organisms have begun to shed light on the mechanisms of de novo constitutive heterochromatin assembly in early development (Mutlu et al. 2018; Seller et al. 2019). A recent series of elegant studies by O'Farrell and colleagues (Seller and O'Farrell 2018; Seller et al. 2019) examined this process by combining genetics with live imaging of Drosophila embryos. Embryogenesis in Drosophila begins with 13 rapid cleavage divisions that quickly increase cell number prior to gastrulation. These nuclear divisions occur in a syncytium, lack G1 and G2 phases of the cell cycle, and rely on maternally loaded mRNAs and proteins because they occur prior to activation of zygotic transcription. Interphase length gradually increases during cycles 9–13 prior to the midblastula transition (MBT) in cycle 14, when a number of important events occur, including cellularization and activation of zygotic gene expression (Fig. 1A). H3K9me and HP1a are absent from chromatin prior to cycle 11, and mature heterochromatin with these molecular hallmarks does not appear until after the MBT, when heterochromatin acquires its particular functions (e.g., transcriptional repression) and cytology (e.g., assembly into chromocenters). The present study by Seller et al. (2019) set out to answer two outstanding questions: (1) How does heterochromatin initially assemble? (2) What controls the timing of this event?
Figure 1.
Progressive assembly of constitutive heterochromatin in Drosophila embryos. (A) Drosophila embryogenesis begins with 13 rapid syncytial nuclear cycles prior to the MBT and gastrulation. Heterochromatin is disassembled each mitosis, and, in the following interphase, Eggless/SETDB1 foci precede the appearance of HP1a foci (shown for cycle 13). (B) Eggless/SETDB1-mediated H3K9 methylation “seeds” heterochromatin assembly by recruiting HP1a. The increase in interphase duration each cycle results in increasing amounts of heterochromatin formation such that by cycle 15, heterochromatin coalesces via liquid–liquid phase transition into canonical H3K9me/HP1a/Rif1-enriched chromocenters with the functional properties of constitutive heterochromatin.
The O'Farrell group (Yuan and O'Farrell 2016) previously developed novel technologies to visualize and describe heterochromatin emergence in living Drosophila embryos with fine-scale temporal resolution. Key among these was the use of TALE-lights, fluorescent proteins fused to TALENS that bind to specific satellite repeats present in pericentric heterochromatin (Yuan and O'Farrell 2016). A seminal observation from these studies is that not all features of constitutive heterochromatin appear simultaneously. For instance, compaction and late replication of satellite DNA sequences occur prior to the appearance of H3K9me/HP1a-enriched heterochromatin (Fig. 1B; Yuan and O'Farrell 2016; Seller and O'Farrell 2018). In the present study, Seller et al. (2019) asked which of the three Drosophila H3K9 methyltransferases—G9a, Su(var)3–9, or Eggless/SETDB1—establishes H3K9me/HP1a-enriched heterochromatin in the early Drosophila embryo. One reason for this knowledge gap is the difficulty of inhibiting the function of proteins loaded into the egg by the mother, a problem for studying chromatin organization in mammalian embryos as well (Fadloun et al. 2013). G9a and Su(var)3–9 are dispensable for Drosophila development, allowing Seller et al. (2019) to remove their maternal contribution and demonstrate that they are not required for heterochromatin assembly in the embryo. The major function of these H3K9 HMTases is likely in heterochromatin maturation and/or maintenance later in development.
In contrast to G9a and Su(var)3–9, Eggless/SETDB1 is an essential gene and is required for oogenesis, precluding the ability to generate eggs lacking this enzyme. To interrogate functions of Eggless/SETDB1 in de novo constitutive heterochromatin formation, Seller et al. (2019) implemented a novel method for functionally inactivating maternally loaded nuclear proteins called JabbaTrap. Jabba is a protein associated with cytoplasmic lipid droplets that binds to and stores maternally loaded histone H2a and H2b for use in syncytial S phases (Li et al. 2012). JabbaTrap employs an anti-GFP nanobody-Jabba fusion protein to sequester GFP-tagged proteins in the cytoplasm and exclude them from acting in the nucleus. Importantly, sequestering occurs in the syncytial embryo after oogenesis is complete, allowing the analysis of eggs in which the function of maternally deposited proteins have been inhibited. Sequestering Eggless/SETDB1 in the egg cytoplasm with JabbaTrap prevented the appearance of H3K9me2/3 and HP1a at a specific satellite sequence at the MBT. Similarly, Met-2/SETDB1 is needed for initiating heterochromatin assembly in Caenorhabditis elegans embryos (Mutlu et al. 2018). Thus, SETDB1 methyltransferases are critically important for initially establishing constitutive heterochromatin during animal development. However, what controls timing of the onset of heterochromatin formation?
In Drosophila syncytial embryos, interphase duration extends from a few minutes to ∼1 h at the MBT. Seller et al. (2019) show that Eggless/SETDB1 recruitment to pericentric satellite DNA in cycle 10 precedes the appearance and growth of small disperse foci of HP1a during syncytial cycle 11 (Fig. 1A,B). Thus, the recruitment of Eggless/SETDB1 to DNA triggers the initiation of heterochromatin formation, but how this recruitment occurs is not clear. Since heterochromatin disassembles in mitosis, the duration of interphase might limit the amount of heterochromatin that can be assembled each cycle. Indeed, as interphase duration increases each successive cycle, H3K9me/HP1a foci grow in size (Fig. 1B). Seller et al. (2019) demonstrate that changing interphase length by manipulating the activity of cell cycle regulators to extend or reduce interphase duration promotes and inhibits, respectively, Eggless/SETDB1 foci and heterochromatin formation. Thus, the developmentally programmed increase in nuclear cycle length helps initiate and promote de novo heterochromatin assembly in Drosophila. The progressive assembly and maturation of constitutive heterochromatin also occurs in C. elegans embryos, but here regulation of nuclear localization of Met-2/SETDB1 controls the onset (Mutlu et al. 2018). How de novo heterochromatin formation in early mammalian embryos is controlled is not known. Unlike Drosophila, mammalian embryos begin transcription early after zygote formation, and one possibility is that transcription of repeated sequences activates dsRNA/RNAi pathways to recruit heterochromatin factors, as occurs in fission yeast (Fadloun et al. 2013). Thus, different organisms likely have evolved distinct mechanisms to initiate constitutive heterochromatin formation early in development.
The progressive emergence of characteristics of mature heterochromatin in the early fly embryo is reminiscent of other biomolecular condensates in the nucleus, such as the nucleolus (Falahati et al. 2016). Components of biomolecular condensates often self-associate and undergo liquid–liquid phase separation (LLPS) in cells, as do both Drosophila and human HP1a (Larson et al. 2017; Strom et al. 2017). Some of these molecules can nucleate or “seed” recruitment of other factors, resulting in growth of the condensate. After small HP1a foci initially appear during syncytial cycle 11 in fly embryos, they grow in size each successive cycle and ultimately undergo a LLPS-like coalescence into large heterochromatin assemblies after the MBT (Fig. 1B; Yuan and O'Farrell 2016; Strom et al. 2017). This heterochromatin “maturation” is accompanied by a shift in HP1a behavior in the condensate from fully mobile to ∼20% of the HP1a being present in an “immobile” core associated with chromatin and potentially other cellular compartments (e.g., nuclear lamina). Seller et al. (2019) demonstrate that in the absence of Eggless/SETDB1, HP1a foci remain immature and fail to fuse, suggesting that temporal regulation of H3K9me deposition prior to and during the MBT is critical for heterochromatin maturation. However, whether H3K9me contributes to chromatin inclusion in HP1a LLPS and whether chromatin inclusion is a requirement for heterochromatin maturation remains unknown (Shin et al. 2018).
Although canonically defined by H3K9me/HP1a, constitutive heterochromatin is a complex structure composed of many factors with differential dependencies on one another (Becker et al. 2017). The recruitment of Rap-interacting factor 1 (Rif1) to satellite DNA establishes late replication of heterochromatin in the syncytial Drosophila embryo (Seller and O'Farrell 2018). Notably, this occurs prior to the appearance of H3K9me or HP1a at satellite DNA, consistent with our recent work using H3K9R mutant Drosophila demonstrating that late replication is not entirely dependent on H3K9me/HP1a at postembryonic stages of development (Armstrong et al. 2018). Thus, certain heterochromatin functions (e.g., transposon repression) may require H3K9me/HP1a, whereas others (e.g., late replication) may not.
The work of Seller et al. (2019) demonstrates how understanding chromatin assembly during early development can provide a framework for addressing questions regarding how genome compartmentalization into euchromatin and heterochromatin promotes distinct transcriptional programs that drive cell differentiation. With this framework, we can perhaps better address long-standing unanswered questions, such as whether heterochromatin directly promotes differentiation of specific cell lineages (Becker et al. 2017; Nicetto et al. 2019). While the answers remain in the future, it is evident that we are beginning to unravel how multivalent regulatory networks drive heterochromatin establishment, maturation, and function.
Acknowledgments
We thank Dan McKay for helpful discussions and comments on the text. Work on chromatin in the Duronio laboratory is supported by National Institutes of Health grant GM124201.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.324731.119.
References
- Armstrong RL, Penke TJR, Strahl BD, Matera AG, McKay DJ, MacAlpine DM, Duronio RJ. 2018. Chromatin conformation and transcriptional activity are permissive regulators of DNA replication initiation in Drosophila. Genome Res 28: 1688–1700. 10.1101/gr.239913.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JS, McCarthy RL, Sidoli S, Donahue G, Kaeding KE, He Z, Lin S, Garcia BA, Zaret KS. 2017. Genomic and proteomic resolution of heterochromatin and its restriction of alternate fate genes. Mol Cell 68: 1023–1037.e1015. 10.1016/j.molcel.2017.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadloun A, Eid A, Torres-Padilla ME. 2013. Mechanisms and dynamics of heterochromatin formation during mammalian development: closed paths and open questions. Curr Top Dev Biol 104: 1–45. 10.1016/B978-0-12-416027-9.00001-2 [DOI] [PubMed] [Google Scholar]
- Falahati H, Pelham-Webb B, Blythe S, Wieschaus E. 2016. Nucleation by rRNA dictates the precision of nucleolus assembly. Curr Biol 26: 277–285. 10.1016/j.cub.2015.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. 2017. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547: 236–240. 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Thiel K, Thul PJ, Beller M, Kühnlein RP, Welte MA. 2012. Lipid droplets control the maternal histone supply of Drosophila embryos. Curr Biol 22: 2104–2113. 10.1016/j.cub.2012.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu B, Chen HM, Moresco JJ, Orelo BD, Yang B, Gaspar JM, Keppler-Ross S, Yates JR, Hall DH, Maine EM, et al. 2018. Regulated nuclear accumulation of a histone methyltransferase times the onset of heterochromatin formation in C. elegans embryos. Sci Adv 4: eaat6224 10.1126/sciadv.aat6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicetto D, Donahue G, Jain T, Peng T, Sidoli S, Sheng L, Montavon T, Becker JS, Grindheim JM, Blahnik K, et al. 2019. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 363: 294–297. 10.1126/science.aau0583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seller CA, O'Farrell PH. 2018. Rif1 prolongs the embryonic S phase at the Drosophila mid-blastula transition. PLoS Biol 16: e2005687 10.1371/journal.pbio.2005687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seller CA, Cho CY, O'Farrell PH. 2019. Rapid embryonic cell cycles defer the establishment of heterochromatin by Eggless/SetDB1 in Drosophila. Genes Dev (this issue) 10.1101/gad.321646.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M, Brangwynne CP. 2018. Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175: 1481–1491.e1413. 10.1016/j.cell.2018.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547: 241–245. 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, O'Farrell PH. 2016. TALE-light imaging reveals maternally guided, H3K9me2/3-independent emergence of functional heterochromatin in Drosophila embryos. Genes Dev 30: 579–593. 10.1101/gad.272237.115 [DOI] [PMC free article] [PubMed] [Google Scholar]