Abstract
Liposomal spherical nucleic acids (LSNAs) are a class of nanomaterial used broadly for biomedical applications. Their intrinsic capacity to rapidly enter cells and engage cell surface and intracellular ligands stems from their unique three-dimensional architecture, which consists of densely packed and uniformly oriented oligonucleotides on the surface of a liposomal core. Such structures are promising for therapeutics because they can carry chemical cargo within the lipid core in addition to the nucleic acids that define them, in principle enabling delivery of multiple signals to a single cell. On the basis of these traits, we have designed novel dual-targeting LSNAs that deliver a nucleic acid specific for TLR9 inhibition and a small molecule (TAK-242) that inhibits TLR4. Toll-like receptors (TLRs) play a large role in pathogen recognition and disease initiation, and TLR subtypes are differentially located within the lipid membranes of the cell surface and within intracellular endosomes. Oftentimes, in acute or chronic inflammatory conditions, multiple TLRs are activated, leading to stimulation of distinct, and sometimes overlapping, downstream pathways. As such, these inflammatory conditions may respond to attenuation of more than one initiating receptor. We show that dual targeting LSNAs, comprised of unilamellar liposomal cores, the INH-18 oligonucleotide sequence, and TAK-242 robustly inhibit TLR-9 and TLR-4 respectively, in engineered TLR reporter cells and primary mouse peritoneal macrophages. Importantly, the LSNAs exhibit up to a 10- and a 1000-fold increase, respectively, in TLR inhibition compared to the linear sequence and TAK-242 alone. Moreover, the timing of delivery is shown to be a critical factor in effecting TLR-inhibition, with near-complete TLR-4 inhibition occurring when cells were pretreated with SNAs for 4 h prior to stimulation. The most pronounced effect observed from this approach is the benefit of delivering the small molecule within the SNA via the receptor-mediated internalization pathway common to SNAs.
Graphical Abstract
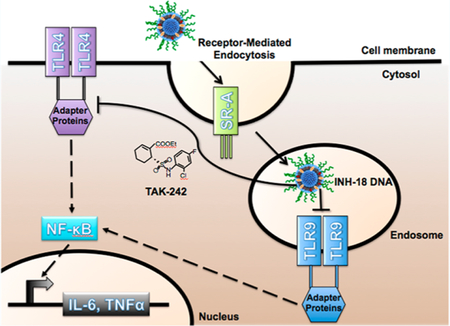
INTRODUCTION
Nanomaterials are attractive for treating human diseases because they offer advantages in terms of efficient, specific, and potent drug delivery. Specifically, increased cellular uptake, improved pharmacokinetics, biocompatibility, and biodistribution enable enhanced therapeutic efficacy and potency through high affinity binding.1 One such material at the leading edge of nanomaterial therapeutics development is the spherical nucleic acid (SNA). SNAs are a unique class of nanomaterial characterized by the dense packing of radially oriented oligonucleotides on the surface of a nanoparticle core. The spherical, multivalent architecture confers properties that distinguish SNAs from their linear DNA or RNA counterparts, such as high cellular uptake without the need for ancillary transfection reagents, increased resistance to nuclease degradation, and minimal nonspecific activation of the immune system.2–4 These properties make SNAs attractive as single entity agents for biological and medical applications particularly because the oligonucleotide shell, not the core, governs these properties.5–10 In fact, multiple SNA architectures have been designed and synthesized that were directly informed by the target disease or molecular pathway, i.e., BCL2L12-targeting siRNA-conjugated gold-based SNAs for glioblastoma,11 protein core SNAs for delivery of functional proteins,9 and liposomal SNAs (LSNAs) for the codelivery TLR9 activating DNA and tumor antigen for cancer vaccines.12
Toll-like receptors (TLRs) are attractive therapeutic targets due to their role as the molecular first-responders of innate immunity, which are found at the cell surface (TLRs 1, 2, and 4−6) or within endosomes (TLRs 3 and 7−9). Their activation relies upon specific recognition of conserved pathogenic or damage-associated motifs. Pathogen or damage-associated ligand binding to these receptors initiates a proinflammatory response resulting in the production of cytokines, chemokines, and reactive oxygen species, immune cell activation, migration, and proliferation, and eventual identification and destruction of the invading pathogen.13 While activation of TLRs contributes to the clearance of an infection, persistent overstimulation of TLRs contributes to the pathogenesis of several chronic inflammatory diseases, such as lupus, rheumatoid arthritis, sepsis, and ischemia reperfusion injury.14–17 The severity of these diseases may be, in part, due to simultaneous activation of multiple receptors leading to stimulation of downstream inflammatory pathways, such as NF-κB-mediated production of cytokines and interferons. Multireceptor activation is a common feature in many acute and chronic inflammation-mediated diseases (e.g., TLR2, TLR3, and TLR4 in sepsis; TLR3 and TLR4 in rheumatoid arthritis; TLR4 and TLR9 in liver ischemia reperfusion injury and fibrosis).35,36 Thus, the ability to simultaneously target multiple TLRs, both at the cell surface and within the endosome of the same cell, may enhance treatment of these inflammation-mediated diseases.
We have previously shown that SNAs are potent immunomodulators capable of engaging endosomal TLRs 7, 8, and 9 with their sequence-specific RNA or DNA shell.12,18 Though these endosomal TLRs are typically activated by linear oligonucleotides, TLR activation by gold- and liposome-based SNAs is orders of magnitude more potent than activation by linear nucleic acids in macrophages12 and dendritic cells.18 The liposomal version has entered Phase 1b/2 human clinical trials.19 In addition, TLR9 inhibition by LSNAs attenuates liver fibrosis in mouse models.12 It remains to be determined, however, if the incorporation of additional TLR targeting agents, such as small molecules, will synergize with the TLR-targeting nucleic acid shell of the SNA and if there is a relationship between codelivery of TLR inhibiting molecules, multireceptor inhibition, and overall impact on inhibition of downstream markers of inflammation.
In this study, we synthesized LSNAs that codeliver INH-18, an oligonucleotide inhibitor of TLR9, and TAK-242,20,21 a small molecule inhibitor of TLR4 (SI Figure 1). The DNA shell of the SNA enables rapid uptake into cells and engages TLR9 within endosomes. The encapsulated TAK-242 is transported as cargo within the LSNA and is released inside the cell where it is known to associate with the intracellular domain of TLR4.22,23 To investigate their ability to attenuate inflammation, these dual TLR-targeting LSNAs were tested in both engineered reporter cells as well as primary mouse peritoneal macrophages for their ability to downregulate proinflammatory markers downstream of each receptor. These dual TLR-targeting SNAs provide two advantages: (1) they allow one to codeliver the oligonucleotide and small molecule therapeutic inhibitors to the same cells, and (2) when compared with small molecule delivery alone, they increase the rate of delivery. These experimental observations allow one to maximize the synergistic effects of small molecules with SNAs and, in principle, lay the foundation for creating combination therapies.
RESULTS AND DISCUSSION
Physical and Chemical Analysis of Liposomal Spherical Nucleic Acids.
The synthesis and characterization of liposomes with and without encapsulated TAK-242 were carried out based on reported LSNA synthesis protocols with modifications (SI Figure 1).8 Dynamic light scattering (DLS) was used to measure the average size of the bare liposomes lacking oligonucleotides, 25 nm for empty- and 36 nm for TAK-encapsulated liposomes (Figure 1A,B) with a polydispersity index of 0.15 for both, which indicates synthesis of uniform structures (Figure 1C). The concentration of lipids in bare liposomes was measured by quantifying the concentration of phosphorus in solution using inductively coupled plasma optical emission spectroscopy (ICP-OES). Because TAK-242 contains one sulfur atom, ICP-OES was used to measure the amount of sulfur within liposomes, which calculated an average of 74 TAK-242 molecules per liposome (SI Figure 2A). Finally, DLS was used to determine the size of the particles before and after attachment of DNA to their surfaces (with and without TAK-242). Consistent with SNA formation, a size shift of approximately 10 nm is observed upon oligonucleotide functionalization (a 24-mer ssDNA, Figure 1A,B). The final compositions, resultant sizes, and polydispersity indices of synthesized LSNAs are consistent with uniform structures (Figure 1C). To determine whether TAK-242 leached from the LSNAs, we employed dialysis using a 3000 MW cutoff and observed 6−8% loss of TAK-242 from LSNA and 8−17% from bare liposomes over an 8 h period (SI Figure 2B). There was minimal loss of TAK-242 from the liposomes, likely due to the hydrophobicity of the drug, and an even lesser degree of drug loss from the LSNAs, which is attributed to the increased stability of the liposomal SNA as compared with bare liposomes.12
Figure 1.
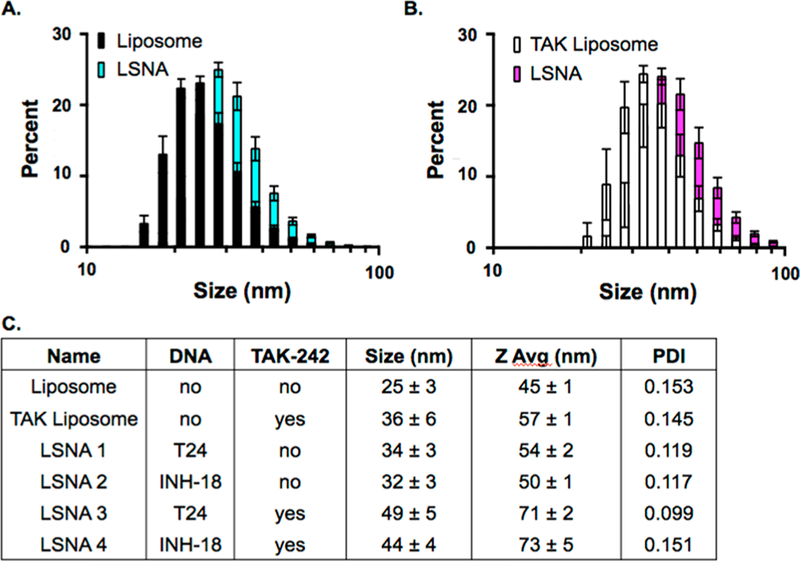
Characterization of LSNAs. A size shift of 10−15 nm was observed when empty liposomes (A) and TAK-encapsulated liposomes (B) were functionalized with a 24-mer ssDNA strand (A, black to blue shift; B, white to pink shift). The final compositions of LSNAs with and without TLR9-targeting DNA and/or TAK-242 showed uniform size shifts and polydispersity indices (PDI) (C).
In Vitro Analysis of LSNA TLR Inhibitory Potency.
To evaluate the inhibitory activity of the synthesized LSNAs, we used engineered HEK293 cell lines (HEK-Blue) that stably express either mouse TLR4 (mTLR4) or mouse TLR9 (mTLR9) and an NF-κB reporter gene. HEK-Blue cells expressing mTLR4 can only be stimulated by TLR4 ligands, such as LPS or MPLA. Likewise, HEK-Blue cells expressing mTLR9 only respond to TLR9 ligands, such as unmethylated CpG DNA. Following a 4 h coincubation of these cells with the appropriate TLR agonists and LSNAs (Figure 1C, 2A) or corresponding free inhibitors, a dose-dependent response was observed for both TLR4 and TLR9 in response to LSNA treatment. In HEK-Blue mTLR9 cells, 1 nM of LSNA2, containing the sequence INH-18 on the surface, inhibited TLR9 activation by 64% and completely inhibited TLR9 signaling at 100 nM (Figure 2B). Free INH-18 was able to inhibit TLR9 activation by 41% at 1 nM, but similar to the LSNA, completely attenuated TLR9 signaling at 100 nM and higher concentrations. In HEK-Blue mTLR4 cells, LSNA3, containing TAK-242, was able to inhibit TLR4 activation by 33% at the lowest concentration and showed strong dose dependence with 90% inhibition of TLR4 at the highest concentration (Figure 2C). By comparison, after a 4 h incubation, free TAK-242 inhibited just 20% of TLR4 signaling at the highest concentration.
Figure 2.
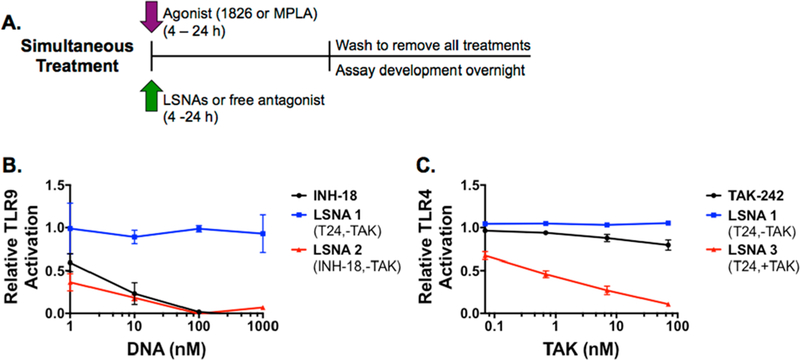
Simultaneous treatment of HEK-Blue cells with TLR agonists and antagonists. Cells were treated for 4−24 h with both TLR agonists and antagonists (A). A 4 h treatment of HEK-Blue mTLR9 cells (B) and HEK-Blue mTLR4 cells (C) with appropriate agonists ODN 1826 and MPLA, respectively, and free antagonists or LSNAs demonstrates receptor specificity and a dose-dependent inhibition of TLR activity.
These results suggest that free INH-18 and LSNAs functionalized with INH-18 may enter cells at similar rates or through the same mechanism, whereas, free TAK-242 uptake and engagement of TLR4 are likely slower than the TAK-encapsulated LSNAs. To probe the uptake mechanism of linear INH-18, TAK-242, and the LSNAs, we used 50 μg/mL fucoidan, a polysaccharide derived from seaweed and a known inhibitor of scavenger receptor A (SR-A), the receptor that mediates SNA endocytosis into cells.24,25 Indeed, when SR-A was blocked in HEK-Blue mTLR9 cells following stimulation with CpG DNA (ODN 1826), it was not possible to suppress TLR9 activation and the NF-κB pathway, suggesting that both linear INH-18 and LSNA2 rely on SR-A for entry into cells (Figure 3A). Similarly, when HEK-Blue mTLR9 cells were pretreated with fucoidan prior to stimulation with ODN 1826, these cells could not be activated (Figure 3B). However, when HEK-Blue mTLR4 cells were pretreated with fucoidan, there was minimal effect on the activation of TLR4 and the NF-κB pathway and its subsequent inhibition by free TAK-242 as demonstrated by the 44−89% inhibition over the full range of concentrations tested. However, LSNA3 with encapsulated TAK-242 was less effective at inhibiting the NF-κB pathway demonstrating 0−53% inhibition from 1 nM to 1 μM LSNA treatment (Figure 3C). This indicates that the activity of TAK-encapsulated LSNAs was diminished due to reduced uptake by SR-A mediated endocytosis.
Figure 3.
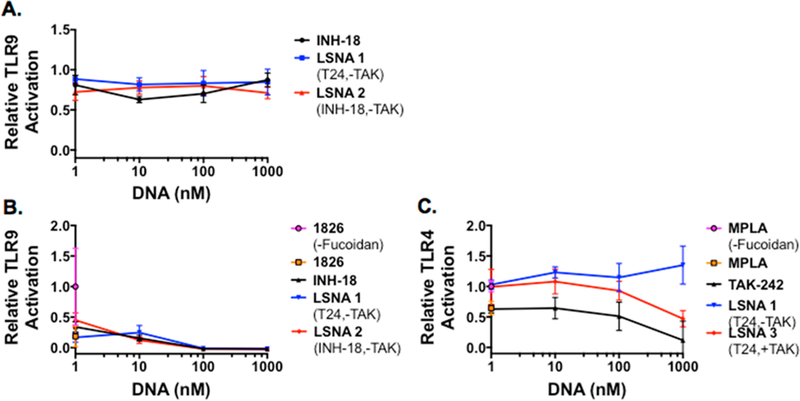
Blockage of scavenger receptor A (SR-A) using fucoidan. Following TLR9 activation using ODN 1826, SR-A blockage prevents interaction of free INH-18 and LSNAs with TLR9 (A). SR-A blockage prior to TLR activation prevents interaction of ODN 1826 CpG DNA with TLR9 as well as INH-18 and LSNAs (B) and partially inhibits LSNA delivery of TAK-242 without any effect on free TAK-242 interaction with TLR4 (C).
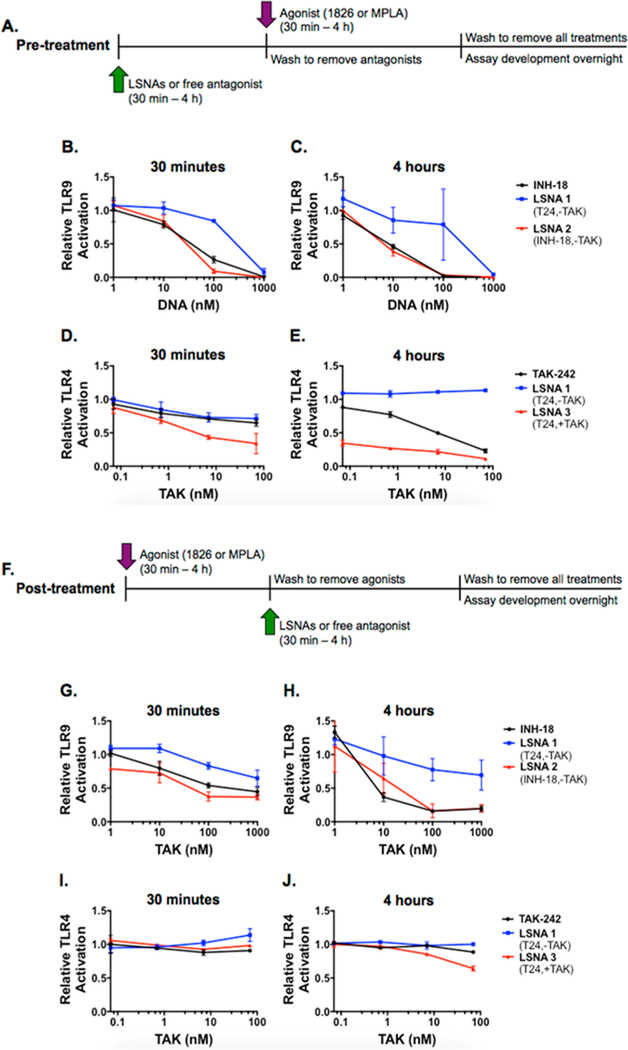
To further probe how these similarities in uptake mechanism for free INH-18 and LSNAs and differences in uptake mechanism for TAK-242 and TAK-encapsulated LSNAs might affect the overall antagonist efficacy, we varied the timing of TLR agonist and antagonist administration (Figure 4A,F). We found that increasing the pretreatment time from 30 min (Figure 4B,D) to 4 h (Figure 4C,E) increased the activity of all antagonists. The potency of LSNA2, functionalized with INH-18, at 10 nM increased from 16 to 64% inhibition when the pretreatment time was increased from 30 min to 4 h. As previously seen, the TLR9 inhibitory activity of free INH-18 in solution was very similar to that of the LSNAs bearing the same motif showing increased inhibition at 10 nM from 22 to 55% (Figure 4B,C), which supports the idea that they utilize the same uptake mechanism. On the other hand, increasing the pretreatment incubation time of TAK-encapsulated LSNAs (Figure 4D,E) made a dramatic difference. At the 4-h pretreatment time, 1 nM of TAK-242 encapsulated LSNA3 already demonstrated 66% inhibition and increased to 89% inhibition at 1 μM LSNAs, whereas TAK-242 free in solution exhibited a more dose-dependent response, demonstrating 12−78% inhibition over the same concentration range.
Figure 4.
LSNA pre- and post-treatment kinetics. Cells were treated with LSNAs or TLR specific inhibitors (blue arrow) prior to exposing them to stimulating agonists (red bar) (A). HEK-Blue mTLR9 cells (B, C) and HEK-Blue mTLR4 cells (D, E) were treated for 30 min or 4 h with free inhibitors and LSNAs prior to stimulation with ODN 1826 or MPLA, respectively. Cells were treated with stimulating agonists (red bar) for 4 h prior to exposing cells to LSNAs or TLR specific inhibitors (blue arrow) (F). HEK-Blue mTLR9 cells (G, H) and HEK-Blue mTLR4 cells (I, J) were stimulated with the ODN 1826 or MPLA, respectively, prior to a 30 min or 4 h treatment with LSNAs or free in solution TLR inhibitors. Pretreatment leads to increased potency of both TLR9 and TLR4 inhibitors as well as TLR-targeting LSNAs, whereas, post-treatment shows decreased potency. Both dose- and time-dependencies for inhibition of TLR activity were observed.
Interestingly, at the highest concentration tested, LSNA1, the nontargeting LSNA, was capable of limiting TLR9 activation. To probe the ability of the nontargeting T24 DNA strand on LSNA1 to inhibit TLR9, we compared it to the activity of its linear counterpart (SI Figure 4). Again, when used as a pretreatment, LSNA1 is capable of inhibiting TLR9 97% at 1 μM, however, linear T24 has no inhibitory effect on TLR9 (SI Figure 4B). Though some reports claim that TLR9 activity modulation is not sequence dependent,26,27 the ability of the nontargeting LSNA to engage and blunt TLR9 activation may also be due, in part, to its multivalent structure rather than its sequence.28–30
Conversely, when TLR antagonists and LSNAs were given to cells following a period of activation, the opposite trend is observed. When post-treatment time was increased from 30 min (Figure 4G,I) to 4 h (Figure 4H,J), a slight increase in antagonist activity was only observed in TLR9 stimulated cells (Figure 4G,H). At 30 min and 10 nM concentration, free INH-18 and LSNA2 exhibited 21 and 28% inhibition, respectively, but this increased to 64 and 36% inhibition, respectively, when the post-treatment time was increased to 4 h. Again, we observed that the activity of free INH-18 was similar to that of the LSNAs, with no significant differences from 1 nM to 1 μM, indicating that both molecules were capable of competing with the activating ligand for TLR9 binding with higher affinity. In TLR4 stimulated cells, there was no observed inhibitory activity of either TAK-242 or TAK-containing LSNAs at 30 min (Figure 4I), and only 36% inhibition at 4 h with the highest concentration of LSNA3 (Figure 4J). These results suggest that after MPLA activates TLR4, there may be either (1) a conformational change of the receptor’s intracellular domain that prevents TAK-242 binding22 or (2) an inability of TAK-242 to displace adapter proteins necessary for downstream NF-kB signaling once these proteins are bound to TLR4,23 unless there are high levels of the drug present in the cell.
Inhibition of Pro-Inflammatory Cytokines in Mouse Peritoneal Macrophages.
We next sought to investigate the inhibitory potential of LSNAs in a clinically relevant cell type. Peritoneal macrophages play a critical role as sentinel cells of the immune system that partake in inflammatory and reparatory processes that contribute to maintenance of tissue homeostasis. They are derived from the bone marrow (resident peritoneal macrophages) and the bloodstream (monocytes that infiltrate the peritoneal cavity in response to a stimulus and differentiate into peritoneal macrophages).31–33 Peritoneal macrophages were elicited from male and female wild type mice, and the uptake of LSNAs in cells was confirmed using dual fluorophore-labeled LSNAs with a TAMRA-labeled lipid core and Cy5-labeled oligonucleotides. Flow cytometry was used to detect cells that stained positive for both the lipid core (TAMRA) and DNA (Cy5) components of LSNAs. Results show that over 60% of isolated peritoneal cells stained double positive. We also evaluated whether stimulation of these cells affected the ability to take up LSNAs, and no change in the percent of double positive stained cells was observed (SI Figure 5). Only by the use of 50 μg/mL fucoidan to block SRA was a decrease of 7−15% seen in the percent of double positive stained cells.
Next, we evaluated the ability of free inhibitors or LSNAs to inhibit IL-6 (Figure 5A−C) and TNFα (Figure 5D-F) production in TLR4- and TLR9-stimulated peritoneal macrophages, varying concentration and treatment time similar to that which was described for the HEK-Blue studies (SI Figure 6A−F). Results show that when both receptors were activated, the highest concentration of a combination of INH-18 and TAK-242, whether free in solution or in LSNA form (LSNA4), was the most potent inhibitor of IL-6 and TNFα, capable of reducing the number of cytokine-producing cells by about 50%. Similar to what was seen in engineered cells, peritoneal macrophages pretreated with antagonists demonstrated the highest degree of inhibition of IL-6 and TNFα, with decreasing potency when antagonists were given at the time of stimulation, and very little therapeutic effect when given as a post-treatment, particularly for IL-6. Interestingly, LSNA3, which contained TAK-242 and an outer shell of nontargeting T24 DNA, was capable of suppressing IL-6 production. This suggests that in order to use these dual targeting LSNAs effectively for the remediation of a TLR-driven clinical indication, the best approach would be to use them as a prophylactic, when possible.
Figure 5.
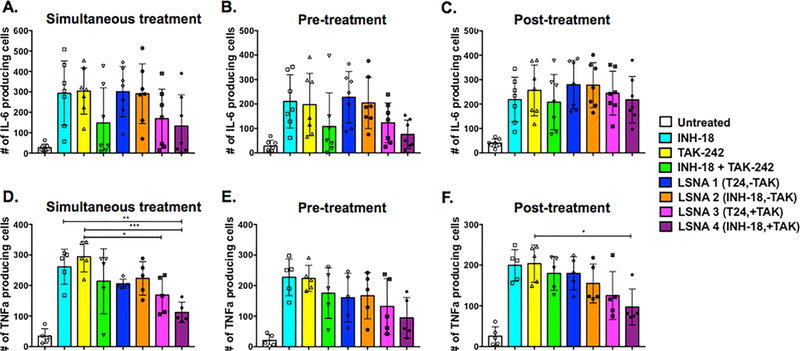
Pro-inflammatory cytokine production by peritoneal macrophages. The effects of free inhibitor or LSNA treatment on dual TLR4- and TLR9-stimulated peritoneal macrophages was measured by the number of IL-6 (A-C) and TNFα (D-F) producing cells during simultaneous treatment, pretreatment of inhibitors or LSNAs, and post-treatment of inhibitors or LSNAs. Results indicated that only a combination of TAK-242 and INH-18, whether free in solution or incorporated into an LSNA, resulted in decreased IL-6 and TNFα production. (*P < 0.05).
Of note, there appeared to be sex-specific responses both to stimulation and inhibition. IL-6 production in male mice appeared to be more sensitive to inhibitor treatment, whereas a TNFα response was only observed in female mice (Figure 5D−F and SI Figure 7). It is well-known that male and female animals, including humans, display immunological differences in their ability to mount an immune response, the type of immune response mounted, and their ability to combat certain pathogens. Though the mechanism is not well understood, it is thought that both differential gene expression and hormones play a role.34 It is possible that the kinetics of IL-6 and TNFα production by peritoneal macrophages varies with sex, thus the timing of agonist and antagonist administration in these experiments could account for the observed variability.
CONCLUSIONS
These dual TLR-targeting SNAs are important for a variety of reasons. First, in the context of TLR activation or inhibition, they are, to the best of our knowledge, the first conjugates that allow one to codeliver small molecules and nucleic acids into the same cells and requisite intracellular compartments. Second, the novel structures enhance small molecule delivery, thereby increasing potency. Third, the ability to codeliver small molecules and oligonucleotides will potentially allow one to develop therapeutic lead compounds for more complex diseases. Indeed, in diseases in which multiple TLRs have become activated, such as TLR3 and TLR4 in rheumatoid arthritis35 or TLR4 and TLR9 in liver fibrosis,36 codelivery of the key inhibitory molecules into the same cells to turn off the immune response likely will be required.37–39 Therefore, taken together, the results of this study, combined with the biocompatibility and modularity of the LSNA platform, sets the stage for the development of a variety of LSNAs with combinatorial targeting capabilities and a high potential to translate them into clinical practice.
MATERIALS AND METHODS
DNA Synthesis and Characterization.
DNA oligonucleotides were synthesized on a universal, solid support (Glen Research, 26–5010) using automated phosphoramidite chemistry. Free strand TLR9 inhibitory oligonucleotide (INH-18) sequence is 5′-CCTGGATGGGAACTTACCGCTGCCA-3′; on the nanoparticle, TLR9 inhibitory sequence is 5′-CCTGGATGGGAACTTACCGCTGCCA-Spacer18-Spacer18-cholesterol-3′, where Spacer18 refers to hexaethylene glycol phosphoramidite (ChemGenes, CLP-9765) and cholesterol refers to cholesteryl-TEG phosphoramidite (Glen Research, 10–1975-90). Free in solution, nontargeting Oligonucleotide ( T24 ) sequence is 5 ′-TTTTTTTTTTTTTTTTTTTTTTTTT-3′; on the nanoparticle, the nontargetin g sequence is 5 ′-TTTTTTTTTTTTTTTTTTTTTTTT-Spacer18-Spacer18-cholesterol-3′. All oligonucleotides were synthesized with a phosphorothioate backbone. Sequences were purified by reverse-phase high-pressure liquid chromatography (Varian HPLC, Agilent) using triethylammonium acetate buffer to acetonitrile gradient and characterized using matrix assisted laser desorption ionization-time-of-flight mass spectrometry (negative mode, MALDI-TOF, Bruker Autoflex III).
Liposomal Particle Synthesis and Characterization.
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) lipid monomers were purchased from Avanti Polar Lipids, Inc. TAK-242 (CLI-095) was purchased from Invivogen. Lipids with or without TAK-242 were dissolved in chloroform and mixed. The chloroform was evaporated under nitrogen, and the lipid film was subsequently lyophilized. The lipid film was then rehydrated in a buffer containing 20 mM Tris-HCL and 150 mM NaCl. The lipid solution was probe sonicated at an amplitude of 40% for 30−40 min (QSonica). Following sonication, lipids were centrifuged at 4000 rpm for 90 min to remove larger lipid vesicles. The supernatant was removed and extruded through polycarbonate membranes of increasingly smaller pore size (80, 50, and 30 nm), until the resulting lipid particles were monodisperse, ascertained by dynamic light scattering (Malvern). The particles were passed through a column containing cross-linked Sepharose to remove any unencapsulated material. The concentration of lipids and encapsulated TAK-242 were determined by inductively coupled plasma optical emission spectroscopy (Thermo iCAP 7600). The 1.3 mM lipids were mixed with 15 μM cholesterol-terminated DNA for 3−4 h at 37 °C to allow for cholesterol intercalation into the lipid bilayer of the liposomes. Size and polydispersity were measured by DLS.
In Vitro Cell Culture of Engineered HEK Reporter Cells.
HEK-Blue mouse TLR9 (HEK-m9) cells and HEK-Blue mouse TLR4 (HEK-m4) cells were used to assess TLR9 and TLR4 activation (Invivogen). Both HEK-m9 and HEK-m4 cells were initially cultured from frozen stocks in high glucose DMEM (Gibco) containing 10% (v/v/) fetal bovine serum (Gibco), 50 U/mL penicillin, 50 μg/mL streptomycin (Thermo Fisher), and 100 μg/mL Normocin (Invivogen) for at least two passages. To select for cells expressing mouse TLR9, HEK-m9 cells were split into DMEM containing 10% (v/v) fetal bovine serum, 50 U/mL penicillin, 50 μg/mL streptomycin, 100 μg/mL Normocin, 30 μg/mL Blasticidin (Invivogen), and 100 μg/mL Zeocin (Invivogen). HEK-m4 cells were selected for in DMEM containing 10% (v/v) fetal bovine serum, 50 U/mL penicillin, 50 μg/mL streptomycin, 100 μg/mL Normocin, and 1X HEK-Blue Selection (Invivogen). When cells were ready to be tested (50−80% confluent), they were cultured in DMEM containing 10% (v/v) heat-inactivated fetal bovine serum. HEK-m9 cells were stimulated using ODN 1826 (Invivogen), and HEK-m4 cells were stimulated using MPLA (Invivogen) for set periods of time then followed by a wash step to remove agonists and antagonists from the media. The Quanti-Blue assay (Invivogen) was allowed to develop per the manufacturer’s instructions.
Primary Mouse Peritoneal Macrophage Induction and ELISPOT for Secreted Cytokines.
Peritoneal macrophages were isolated from 8 to 12 week old male and female C57Bl/6 mice as described previously.40 Animals were handled according to methods and procedures approved by the Institutional Animal Care and Use Committee at Northwestern University. Briefly, mice received an intraperitoneal (IP) injection of sterile 3.8% thioglycollate media (Sigma). Then, 3−5 days following thioglycollate injection, mice were euthanized and IP injected with cold, sterile PBS, and the abdomen was lightly massaged to dislodge macrophages. The PBS was removed from the peritoneum, and cells were centrifuged at 1600 rpm for 5 min. Red blood cell lysis was performed using ACK lysis buffer (Gibco) for 2−3 min at 37 °C. Cells were rinsed with sterile PBS and centrifuged again. The supernatant was removed, and the cells were resuspended in RPMI 1640 media (Life Technologies). Cells were counted and 15 000−20 000 cells/well were seeded onto precoated plates (Becton Dickenson, according to manufacturer protocol) and allowed to attach for 60 min. Following the attachment period, unattached cells were washed away and RPMI 1640 containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin were added. Next, TLR agonists, antagonists, and LSNAs were added. Enzyme-linked immunospot (ELISpot) assays to detect secreted cytokines were performed following the manufacturer’s protocol (Becton Dickinson). Plates were read on an ImmunoSpot analyzer (Cellular Technology Limited).
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.9b00047.
Schematic showing LSNA synthesis including TAK-242 encapsulation, a table showing TAK-242 loss from bare liposomes and LSNAs, a figure showing time- and dose-dependent HEK mTLR4 and HEK mTLR9 responses, a figure showing fluorescent LSNA uptake in peritoneal macrophages, and a figure showing cytokine production dose-dependent responses of peritoneal macrophages and differences between male and female mouse peritoneal macrophage cytokine production (PDF)
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Cancer Institute under Award U54CA199091 awarded to C.A.M., the National Institute of General Medical Sciences under Award F31GM119392 and Award T32GM105538 awarded to J.R.F., and the National Institute of Diabetes and Digestive and Kidney Diseases under Award K08DK101757 awarded to J.A.W. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.A.W. acknowledges funding support by the International Institute for Nanotechnology at Northwestern University and the NTU-Northwestern Institute for Nanomedicine (NNIN), as well as a grant from the Julius Frankel Foundation. This research was also supported by the Air Force Research Laboratory under Award FA8650–15-2–5518 awarded to C.A.M. The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright notation thereon. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the Air Force Research Laboratory or the U.S. Government. Research was performed using resources from the International Institute of Nanotechnology, the Simpson Querry Institute Analytical BioNanoTechnology Equipment Core, the Northwestern University Quantitative Bioelement Imaging Center, the Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Core, the Northwestern Center for Comparative Medicine, and the Feinberg School of Medicine Immune Monitoring Core.
Footnotes
Notes
The authors declare the following competing financial interest(s): C.A.M has financial interest in/relative to Exicure Inc. and Aurasense LLC, companies developing SNA therapeutic platforms, which could potentially benefit from the outcomes of this research.
REFERENCES
- (1).Anselmo AC, and Mitragotri S (2016) Nanoparticles in the clinic. Bioeng Transl Med 1, 10–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wu XA, Choi CHJ, Zhang C, Hao L, and Mirkin CA (2014) Intracellular fate of spherical nucleic acid nanoparticle conjugates. J. Am. Chem. Soc 136, 7726–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Prigodich AE, Alhasan AH, and Mirkin CA (2011) Selective enhancement of nucleases by polyvalent DNA-functionalized gold nanoparticles. J. Am. Chem. Soc 133, 2120–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Massich MD, Giljohann DA, Schmucker AL, Patel PC, and Mirkin CA (2010) Cellular response of polyvalent oligonucleotide-gold nanoparticle conjugates. ACS Nano 4, 5641–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Cutler JI, Zhang K, Zheng D, Auyeung E, Prigodich AE, and Mirkin CA (2011) Polyvalent nucleic acid nanostructures. J. Am. Chem. Soc 133, 9254–9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Young KL, Scott AW, Hao L, Mirkin SE, Liu G, and Mirkin CA (2012) Hollow Spherical Nucleic Acids for Intracellular Gene Regulation Based upon Biocompatible Silica Shells. Nano Lett 12, 3867–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Calabrese CM, Merkel TJ, Briley WE, Randeria PS, Narayan SP, Rouge JL, Walker DA, Scott AW, and Mirkin CA (2015) Biocompatible Infinite-Coordination-Polymer Nanoparticle-Nucleic-Acid Conjugates for Antisense Gene Regulation. Angew. Chem 127, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Banga RJ, Chernyak N, Narayan SP, Nguyen ST, and Mirkin CA (2014) Liposomal spherical nucleic acids. J. Am. Chem. Soc 136, 9866–9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Brodin JD, Sprangers AJ, McMillan JR, and Mirkin CA (2015) DNA-Mediated Cellular Delivery of Functional Enzymes. J. Am. Chem. Soc 137, 14838–14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Banga RJ, Meckes B, Narayan SP, Sprangers AJ, Nguyen ST, and Mirkin CA (2017) Cross-Linked Micellar Spherical Nucleic Acids from Thermoresponsive Templates. J. Am. Chem. Soc 139, 4278–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, et al. (2013) Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci. Transl. Med 5, 209ra152−209ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Radovic-Moreno AF, Chernyak N, Mader CC, Nallagatla S, Kang RS, Hao L, Walker DA, Halo TL, Merkel TJ, Rische CH, et al. (2015) Immunomodulatory spherical nucleic acids. Proc. Natl. Acad. Sci. U. S. A 112, 3892–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Akira S, and Takeda K (2004) Toll-like receptor signalling. Nat. Rev. Immunol 4, 499–511. [DOI] [PubMed] [Google Scholar]
- (14).Rahmani F, and Rezaei N (2016) Therapeutic targeting of Toll-like receptors: a review of Toll-like receptors and their signaling pathways in psoriasis. Expert Rev. Clin. Immunol 12, 1289–1298. [DOI] [PubMed] [Google Scholar]
- (15).Rakoff-Nahoum S, and Medzhitov R (2009) Toll-like receptors and cancer. Nat. Rev. Cancer 9, 57–63. [DOI] [PubMed] [Google Scholar]
- (16).van Golen RF, van Gulik TM, and Heger M (2012) The sterile immune response during hepatic ischemia/reperfusion. Cytokine Growth Factor Rev 23, 69–84. [DOI] [PubMed] [Google Scholar]
- (17).Papaioannou AI, Spathis A, Kostikas K, Karakitsos P, Papiris S, and Rossios C (2017) The role of endosomal toll-like receptors in asthma. Eur. J. Pharmacol 808, 14. [DOI] [PubMed] [Google Scholar]
- (18).Guan C, Chernyak N, Dominguez D, Cole L, Zhang B, and Mirkin CA (2018) RNA-Based Immunostimulatory Liposomal Spherical Nucleic Acids as Potent TLR7/8 Modulators. Small 14, No. 1803284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Exicure, Inc. Intratumoral AST-008 Combined With Pembrolizumab in Patients with Advanced Solid Tumors Clinical-Trials.gov Identifier: NCT03684785. https://www.clinicaltrials.gov/ct2/show/NCT03684785.
- (20).Yamada M, Ichikawa T, Ii M, Sunamoto M, Itoh K, Tamura N, and Kitazaki T. (2005) Discovery of novel and potent small-molecule inhibitors of NO and cytokine production as antisepsis agents: synthesis and biological activity of alkyl 6-(N-substituted sulfamoyl)cyclohex-1-ene-1-carboxylate. J. Med. Chem 48, 7457–7467. [DOI] [PubMed] [Google Scholar]
- (21).Ii M, Matsunaga N, Hazeki K, Nakamura K, Takashima K, Seya T, Hazeki O, Kitazaki T, and Iizawa Y (2006) A novel cyclohexene derivative, ethyl (6R)-6-[N-(2-Chloro-4-fluorophenyl)-sulfamoyl]cyclohex-1-ene-1-carboxylate (TAK-242), selectively inhibits toll-like receptor 4-mediated cytokine production through suppression of intracellular signaling. Mol. Pharmacol 69, 1288–1295. [DOI] [PubMed] [Google Scholar]
- (22).Kawamoto T, Ii M, Kitazaki T, Iizawa Y, and Kimura H. (2008) TAK-242 selectively suppresses Toll-like receptor 4-signaling mediated by the intracellular domain. Eur. J. Pharmacol 584, 40–48. [DOI] [PubMed] [Google Scholar]
- (23).Matsunaga N, Tsuchimori N, Matsumoto T, and Ii M (2011) TAK-242 (resatorvid), a small-molecule inhibitor of Toll-like receptor (TLR) 4 signaling, binds selectively to TLR4 and interferes with interactions between TLR4 and its adaptor molecules. Mol. Pharmacol 79, 34–41. [DOI] [PubMed] [Google Scholar]
- (24).Patel PC, Giljohann DA, Daniel WL, Zheng D, Prigodich AE, and Mirkin CA (2010) Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjugate Chem 21, 2250–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Choi CHJ, Hao L, Narayan SP, Auyeung E, and Mirkin CA (2013) Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. U. S. A 110, 7625–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Li Y, Berke IC, and Modis Y (2012) DNA binding to proteolytically activated TLR9 is sequence-independent and enhanced by DNA curvature. EMBO J 31, 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Yasuda K, Rutz M, Schlatter B, Metzger J, Luppa PB, Schmitz F, Haas T, Heit A, Bauer S, and Wagner H (2006) CpG motif-independent activation of TLR9 upon endosomal translocation of “natural” phosphodiester DNA. Eur. J. Immunol 36, 431–436. [DOI] [PubMed] [Google Scholar]
- (28).Wu CCN, Lee J, Raz E, Corr M, and Carson DA (2004) Necessity of oligonucleotide aggregation for toll-like receptor 9 activation. J. Biol. Chem 279, 33071–33078. [DOI] [PubMed] [Google Scholar]
- (29).Avalos AM, and Ploegh HL (2011) Competition by inhibitory oligonucleotides prevents binding of CpG to C-terminal TLR9. Eur. J. Immunol 41, 2820–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Rutz M, Metzger J, Gellert T, Luppa P, Lipford GB, Wagner H, and Bauer S (2004) Toll-like receptor 9 binds single-stranded CpG-DNA in a sequence- and pH-dependent manner. Eur. J. Immunol 34, 2541–2550. [DOI] [PubMed] [Google Scholar]
- (31).Ajuebor MN, Das AM, Viraǵ L, Flower RJ, Szabó C, and Perretti M. (1999) Role of resident peritoneal macrophages and mast cells in chemokine production and neutrophil migration in acute inflammation: evidence for an inhibitory loop involving endogenous IL-10. J. Immunol 162, 1685–1691. [PubMed] [Google Scholar]
- (32).Ghosn EEB, Cassado AA, Govoni GR, Fukuhara T, Yang Y, Monack DM, Bortoluci KR, Almeida SR, Herzenberg LA, and Herzenberg LA (2010) Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc. Natl. Acad. Sci. U. S. A 107, 2568–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Cassado ADA, D’Impeŕio Lima MR, and Bortoluci KR. (2015) Revisiting mouse peritoneal macrophages: heterogeneity, development, and function. Front. Immunol 6, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Klein SL, and Flanagan KL (2016) Sex differences in immune responses. Nat. Rev. Immunol 16, 626–638. [DOI] [PubMed] [Google Scholar]
- (35).Gao W, Xiong Y, Li Q, and Yang H (2017) Inhibition of Toll-Like Receptor Signaling as a Promising Therapy for Inflammatory Diseases: A Journey from Molecular to Nano Therapeutics. Front. Physiol 8, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Hong-Geller E, Chaudhary A, and Lauer S (2008) Targeting toll-like receptor signaling pathways for design of novel immune therapeutics. Curr. Drug Discovery Technol 5, 29–38. [DOI] [PubMed] [Google Scholar]
- (37).Lenert PS (2010) Classification, mechanisms of action, and therapeutic applications of inhibitory oligonucleotides for Toll-like receptors (TLR) 7 and 9. Mediators Inflammation 2010, 986596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Peng S, Li C, Wang X, Liu X, Han C, Jin T, Liu S, Zhang X, Zhang H, He X, et al. (2016) Increased Toll-Like Receptors Activity and TLR Ligands in Patients with Autoimmune Thyroid Diseases. Front. Immunol 7, 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Clancy RM, Markham AJ, and Buyon JP (2016) Endosomal Toll-like receptors in clinically overt and silent auto-immunity. Immunol. Rev 269, 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ray A, and Dittel BN (2010) Isolation of mouse peritoneal cavity cells. J. Visualized Exp, e1488. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.9b00047.
Schematic showing LSNA synthesis including TAK-242 encapsulation, a table showing TAK-242 loss from bare liposomes and LSNAs, a figure showing time- and dose-dependent HEK mTLR4 and HEK mTLR9 responses, a figure showing fluorescent LSNA uptake in peritoneal macrophages, and a figure showing cytokine production dose-dependent responses of peritoneal macrophages and differences between male and female mouse peritoneal macrophage cytokine production (PDF)