Abstract
In this work, we describe a fully automated cytokinesis-block micronucleus (CBMN) assay with a significantly shortened time to result, motivated by the need for rapid high-throughput biodosimetric estimation of radiation doses from small-volume human blood samples. The Rapid Automated Biodosimetry Tool (RABiT-II) currently consists of two commercial automated systems: a PerkinElmer cell::explorer Workstation and a GE Healthcare IN Cell Analyzer 2000 Imager. Blood samples (30 μl) from eight healthy volunteers were gamma-ray irradiated ex vivo with 0 (control), 0.5, 1.5, 2.5, 3.5 or 4.5 Gy and processed with full automation in 96-well plates on the RABiT-II system. The total cell culture time was 54 h and total assay time was 3 days. DAPI-stained fixed samples were imaged on an IN Cell Analyzer 2000 with fully-automated image analysis using the GE Healthcare IN Cell Developer Toolbox version 1.9. A CBMN dose-response calibration curve was established, after which the capability of the system to predict known doses was assessed. Various radiation doses for irradiated samples from two donors were estimated within 20% of the true dose (±0.5 Gy below 2 Gy) in 97% of the samples, with the doses in some 5 Gy irradiated samples being underestimated by up to 25%. In summary, the findings from this work demonstrate that the accelerated CBMN assay can be automated in a high-throughput format, using commercial biotech robotic systems, in 96-well plates, providing a rapid and reliable bioassay for radiation exposure.
INTRODUCTION
The cytokinesis-block micronucleus (CBMN) assay, based on the measurement of micronuclei (MN) level in binucleated (BN) cells, was suggested by Fenech and Morley (1) and is recommended by the International Atomic Energy Agency (IAEA) as one of the cytogenetic methods for radiation biodosimetry (2). In a mass casualty radiation event situation, tens, or even hundreds of thousands of people may be exposed. In such cases, automated and high-throughput tools and approaches are necessary to estimate radiation doses to individual victims (3, 4).
Different technologies have been designed and developed for at least partial automation of the cytogenetic sample preparation steps (5–7). However, such robotic systems have all been low-throughput because they were designed around 15-ml tubes and preparation of samples on individual microscope slides. As an alternative to these approaches, the Rapid Automated Biodosimetry Tool (RABiT) was originally developed as a fully-automated high-throughput system for radiation biodosimetry assays making use of the standard 96-well microplate ANSI/SLAS format (8–10) and custom-built robotics. With the recent spread of commercial high-throughput robotic systems in biotechnology and pharmaceutical industries that automate many of the routine laboratory procedures, we have previously reported the use of the standard ANSI/SLAS multi-well format as the next-generation platforms for all the steps of cytogenetic biodosimetry assays (11). Specifically we developed the RABiT-II approach to biodosimetry, where standard cytogenetic assays are implemented on commercial automated high-throughput screening platforms in a 96-well plate format (12).
The main drawback of the CBMN assay, in its classical implementation, is the long amount of time required to culture cells to division, 68–72 h (2, 13). This culture time is much longer than the 46–52 h required, for example, in the dicentric chromosome assay (2, 14). This extra day in time-to-answer represents a significant issue in the medical management of radiation injury. A shorter culture time would enhance the utility of the CBMN assay for the rapid assessment of ionizing radiation exposure after large-scale radiological events.
In classical CBMN protocols it is generally assumed that culture times less than 60 h would result in unacceptably low frequencies of BN cells in peripheral human blood lymphocytes (15). However, it was recently shown, with the use of imaging flow cytometry technology, that a radiation dose-response curve can be obtained after reducing the culture time to 48 h, and that dose reconstructions could be achieved using manually prepared blood samples of 200 and 2,000 μl with an accuracy of within ±0.5 Gy (16, 17).
Shown here are the results of the accelerated (shortened cell culturing time) CBMN assay using only 30-μl blood samples, implemented on the automated RABiT II system.
MATERIALS AND METHODS
Blood Sample Collection and Irradiation
Blood samples (3.5–4 ml) were collected into heparinized vacutainer tubes (BD Biosciences, Franklin Lakes, NJ) from 10 healthy volunteers (7 females and 3 males, 31–39 years old) after informed consent was given (IRB protocol no. AAAF2671). Aliquots of human blood (30 μl) were pipetted into 1-ml 2D-barcoded tubes (Matrix Storage Tubes; Thermo Fisher Scientific™ Inc., Waltham, MA), placed into an ANSI/SLAS microplate format-compatible 96-tube rack (8 × 12) and covered with the supplied top (Thermo Fisher Scientific). The covered tubes in the racks were transported to a Gammacell® 40 137cesium irradiator (Atomic Energy of Canada Ltd., Mississauga, Canada). Ninety-six tubes with blood samples from donors 1–8 were used to generate a calibration curve. The tubes were gamma-ray irradiated with 0 (control), 0.5, 1.5, 2.5, 3.5 or 4.5 Gy at a dose rate of 0.70 Gy/min. Ninety-six tubes with blood samples from donors 9 and 10 were used to test the dose prediction capabilities of the accelerated, automated assay. These tubes were gamma-ray irradiated with 0 (control), 1.0, 2.0, 3.0, 4.0 or 5.0 Gy at the same dose rate. After irradiation, 200 μl of PB-MAX™ karyotyping media (Thermo Fisher Scientific) was added to each 30-μl whole blood sample.
Automated Sample Processing
Our implementation of the fully-automated RABiT II system is based on a cell::explorer Workstation (PerkinElmer® Inc., Waltham, MA) with integrated automated humidified incubator STX500 (LiCONiC US Inc., Woburn, MA) and automated imager IN Cell Analyzer 2000 (GE Healthcare, Chicago, IL) (12). Incubation was performed in situ in the same tubes in which the blood samples were irradiated. The rack of tubes containing blood and PB-MAX karyotyping media was placed into the automated incubator STX500 at 37°C under 5% CO2 atmosphere. After 24 h of incubation, cytochalasin-B (Sigma-Aldrich® LLC, St. Louis, MO) was added to the cultures to block cytokinesis of proliferating lymphocytes at a final concentration of 6 μg/ml and cells were cultured for an additional 30 h (total incubation time: 54 h).
After completion of cell culturing, all the samples were transferred for fixation into standard height 96-well 450-μl volume microplates (Corning® Inc., Corning, NY). The cells were treated with 0.075 M potassium chloride and fixed with freshly prepared 3:1 methanol:-acetic acid. All reagents for fixation and staining were at room temperature.
Finally for imaging, each fixed cell sample was transferred to two corresponding wells of two standard height glass-bottom 96-square-well plates (630-μl well volume; Brooks Automation, Chelmsford, MA) preloaded with 300 μl of 10:1 methanol:acetic acid, centrifuged, aspirated and allowed to dry for 10 min before staining. Phosphate buffered saline (200 μl) containing 1.5 μg/ml DAPI (4′,6-diamidino-2-phenylindole; Thermo Fisher Scientific) was added for staining of nuclei and micronuclei.
Automated Imaging and Data Analysis
Glass-bottom microplates with fixed and stained samples were scanned using the IN Cell Analyzer 2000 as described elsewhere (12). TIFF images were acquired through a 20X lens with CCD camera (2,048 × 2,048 pixel resolution) using 350/50 nm excitation and 455/58 nm emission at an exposure time of 300 ms. Full well image acquisition produced 81 images per each 60-mm2 well. The IN Cell Analyzer 2000 allows hardware autofocusing using a laser module, which identifies changes in refractive index to locate the solid-liquid interface of the glass-bottom microplate. In addition to the laser module, software autofocusing was used with computer algorithms to automatically find the Z-axis position with the sharpest focus for each acquired image.
Images acquired by the IN Cell Analyzer 2000 system were analyzed using the Developer Toolbox 1.9 software (GE Healthcare). Round objects of >0.5 μm2 were identified as nuclei using the nuclear segmentation module. The data for these identified nuclei objects were exported from Developer Toolbox 1.9 into Microsoft® Excel® 2010 for further data analysis. First, binucleated cells were identified with the following criteria: two nuclei with area greater than 70 μm2, similar object characteristics and with the distance between them less than three times their average radius. A micronucleus was scored if the distance from its center to the midpoint between the centers of two nuclei in the associated BN cell was less than 15 μm, and also if the area of the micronucleus was less than one third of the average area of the two nuclei in the associated BN cell. SigmaPlot™ version 12.0 for Windows (Systat® Software Inc., San Jose, CA) was used for curve fitting.
RESULTS
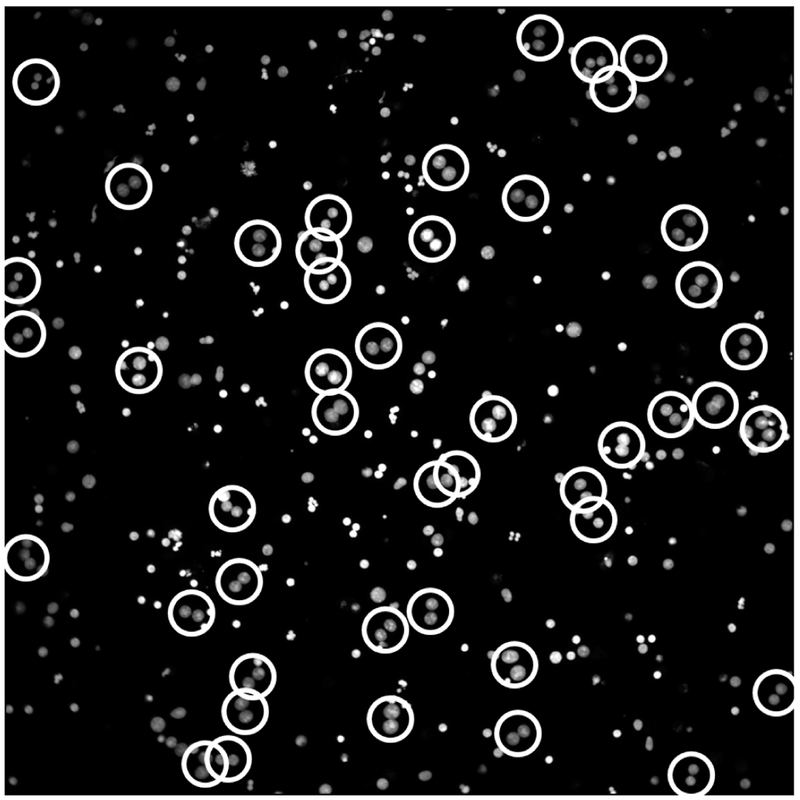
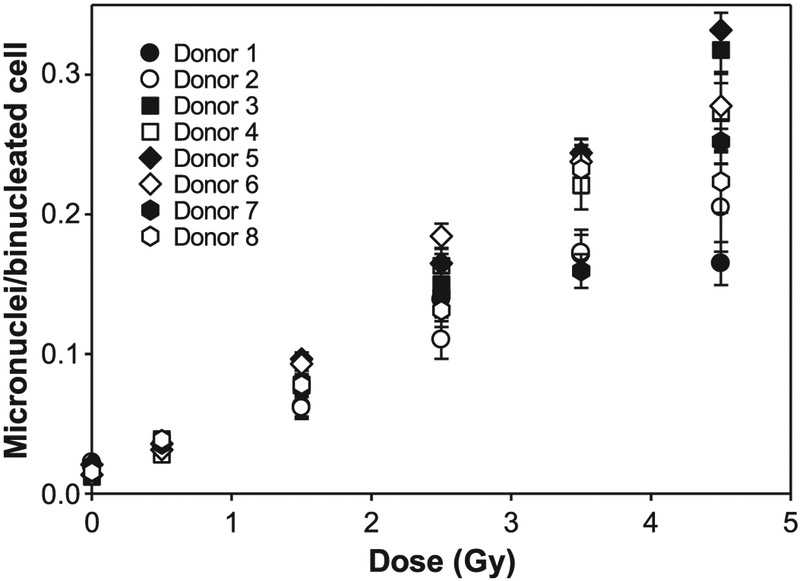
The typical image of human blood sample processed by RABiT-II for the accelerated CBMN assay with 54 h of cell incubation is shown in Fig. 1. To generate the calibration curve, yields of MN in BN cells for eight donors were obtained for 96 human blood samples irradiated ex vivo with doses in the range 0–4.5 Gy. The data are shown in Table 1 and Fig. 2. The average number of binucleated cells varies with a maximum of 3,462 BN cells for control samples and minimum of 938 BN cells for samples irradiated with the highest dose of 4.5 Gy.
FIG. 1.
Image of an accelerated (54 h of cell culturing) CBMN assay sample with DAPI-stained nuclei of human lymphocytes. Image was captured by IN Cell Analyzer 2000 using 20X objective lens with laser and software autofocusing. Binucleated cells are highlighted with white circles.
TABLE 1.
Yields of Binucleated (BN) Cells and Associated Micronuclei (MN) in 30 μl Blood Samples Detected Using the Accelerated RABiT-II CBMN Assay a
| Dose (Gy) |
Mean no. of BN cells in each sample |
Mean no. of MN associated with BN cells in sample |
|---|---|---|
| Control | 3,462 | 57.3 |
| 0.5 | 2,659 | 86.3 |
| 1.5 | 2,195 | 198.6 |
| 2.5 | 1,504 | 238.7 |
| 3.5 | 1,328 | 313.7 |
| 4.5 | 938 | 267.6 |
Average from eight donors with two replicates for each donor and dose after processing of 96 samples.
FIG. 2.
The yield of micronuclei as a function of radiation dose for eight healthy volunteers. These data were used to generate the calibration curve.
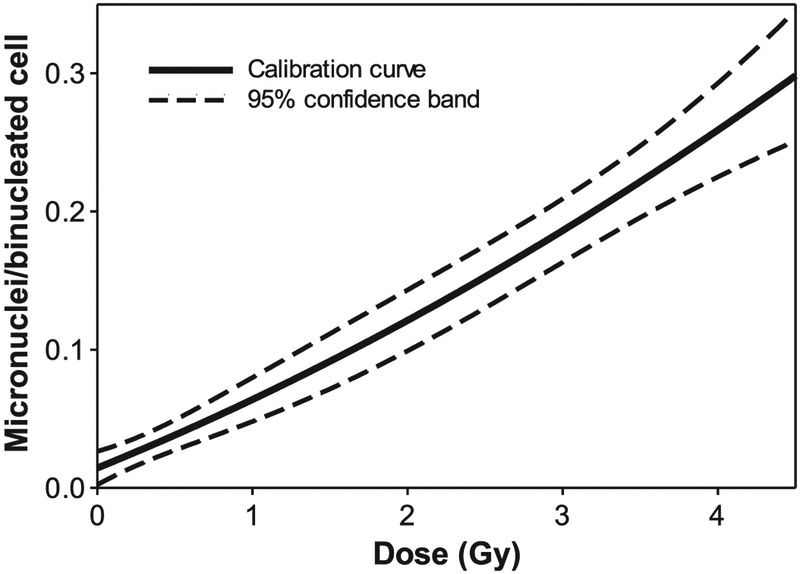
The calibration curve (Fig. 3) was fitted using data on MN yields in all samples from eight healthy volunteers with the linear-quadratic equation, Y = aD2 + bD + c, where Y is the frequency of micronuclei per BN cell and D is the radiation dose. The coefficient of determination for the regression was R2 = 0.994, and the resulting coefficients of the calibration curve were: a = 0.0039 ± 0.0021 Gy−2, b = 0.0457 ± 0.0081 Gy−1 and c = 0.0143 ± 0.0038.
FIG. 3.
Fitted linear-quadratic radiation dose-response calibration curve with 95% confidence bands for accelerated RABiT-II CBMN assay in human lymphocytes for pooled data from eight donors.
In the dose-prediction study, 30-μl aliquots of human blood from two healthy volunteers were dispensed into 96 microtubes and gamma-ray irradiated with 0, 1, 2, 3, 4 or 5 Gy. The samples were processed as before with the accelerated RABiT-II CBMN protocol, and radiation doses were estimated based on the calibration curve from the first experiment (Fig. 3).
Following the standard guidelines for triage dosimetry (18–20), acceptable dose estimation ranges were defined as: within 0.5 Gy for doses up to 2 Gy; or within 20% for higher doses. The data on actual doses compared to estimated doses are shown in Table 2. The radiation doses, including those for sham-irradiated samples, were estimated acceptably in 97% of the samples; doses in the remaining samples were underestimated by between 20 and 25%.
TABLE 2.
Delivered vs. Estimated Radiation Doses Using the Accelerated RABiT-II CBMN Assaya a
| Delivered dose (Gy) |
||||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | |
| Estimated doses (Gy) | 0.19 | 0.63 | 1.89 | 2.79 | 3.31 | 4.54 |
| 0.10 | 0.82 | 1.71 | 3.11 | 3.82 | 5.16 | |
| 0.00 | 0.94 | 1.78 | 2.86 | 3.37 | 4.89 | |
| 0.00 | 0.67 | 1.84 | 2.71 | 3.85 | 4.45 | |
| 0.13 | 0.68 | 1.59 | 2.60 | 3.68 | 4.53 | |
| 0.12 | 0.87 | 1.83 | 2.68 | 4.15 | 4.56 | |
| 0.15 | 0.67 | 1.80 | 2.73 | 4.10 | 4.40 | |
| 0.00 | 0.70 | 1.80 | 2.84 | 3.86 | 4.50 | |
| 0.04 | 0.74 | 1.89 | 2.93 | 3.68 | 3.71 | |
| 0.00 | 0.74 | 1.86 | 3.09 | 4.24 | 4.26 | |
| 0.00 | 0.87 | 1.65 | 3.27 | 3.93 | 3.85 | |
| 0.00 | 0.70 | 1.73 | 3.35 | 3.64 | 4.25 | |
| 0.08 | 0.69 | 1.86 | 2.96 | 4.00 | 4.44 | |
| 0.13 | 0.69 | 1.87 | 2.66 | 3.83 | 4.36 | |
| 0.04 | 0.75 | 1.90 | 3.12 | 4.24 | 4.26 | |
| 0.03 | 0.70 | 1.74 | 2.99 | 3.71 | 3.74 | |
Data are for two donors. Numbers in bold face refer to samples with dose estimates outside the acceptable estimation range.
DISCUSSION
In this work, we have shown that the results of the CBMN assay can be obtained almost one day earlier, using a shortened cell culturing time (54 h) instead of the traditional 68–72 h, and by automation of all steps of the assay in the 96-well plate format, while preserving the capacity of the method to estimate radiation doses. Two 96-well imaging glass-bottom plates were used for 96 fixed cell samples to increase the total surface per sample, because the bottom of a well in a 96-well imaging plate has only 60 mm2, which is 30 times less than the surface of a standard microscope slide.
All steps of the accelerated CBMN assay with 54-h cell culturing were automated using the cell::explorer robot and IN Cell Analyzer 2000 and applying the RABiT-II approach (12). Since we did not use any functionality unique to this system, we expect that the same assay can be easily translated to other equivalent robotic platforms. The time that is necessary for automated preparation of 96 samples using RABiT-II after cell culturing was approximately 1 h, which is considerably less than the 2.5 h necessary for harvesting of 24 samples manually (7).
The time of automated imaging using the IN Cell Analyzer was approximately 6 h for one 96-well plate. This speed per sample is comparable with that reported for the Metafer Slide Scanning System, which is used in some radiation biodosimetry laboratories (19–21). Thus, considering the time for culturing, fixing, imaging and data analysis, the results for 96 samples of the accelerated micronucleus assay can be obtained at the end of the third day after the start of cell culturing. Throughput can in principle be increased by running up to four plates per day in parallel. For comparison, the earliest report time was 4 days after sample arrival in the laboratory intercomparison work (19).
In contrast to conventional CBMN assay samples with different forms of multinucleated cells (two, three, four nuclei), it was found that practically all multinucleated cells in the accelerated CBMN samples were in their binucleated form (Fig. 1). This allows increasing of the number of cells per well due to the smaller footprint of BN cells compared to other forms of multinucleated cells. The mean yield of BN cells for the accelerated RABiT-II micronucleus assay using 30-μl blood samples (Table 1) is higher than 500 BN cells for all radiation doses used exceeding the minimum requirements of 200 BN cells for the traditional CBMN assay in triage mode scoring according to the IAEA (2).
Results of dose estimates based on the accelerated automated CBMN assay showed that only 3 of the 96 samples (3%) were underestimated. Moreover, these three samples were irradiated with 5.0 Gy, while all samples that were exposed up to 4.0 Gy were estimated correctly using the conventional uncertainty intervals described above. For comparison, in the work of Depuydt et al. (20), the success rate using a traditional CBMN assay was considerably lower: less than 90% for low-dose irradiation (<2.5 Gy) and less than 75% for high-dose irradiation (>2.5 Gy). The slightly higher number of dose estimates falling into the ±0.5 Gy interval, when employing an automatic instead of manual fixation procedure combined with semi-automated scoring on microscope slides, was found in other studies (7). Better accuracy of dose estimates using the automated accelerated CBMN assay could be explained by the automated sample preparation in which all samples in a multi-well plate are processed simultaneously and repeatedly between runs.
In summary, the results of this work demonstrate that the accelerated CBMN assay can be automated in a high-throughput format by using commercial biotech robotic systems designed for running assays in standardized multi-well plates. This approach can be used for emergency triage radiation biodosimetry with precision comparable to a standardized CBMN assay based on analysis of samples prepared on microscopic slides.
ACKNOWLEDGMENTS
This work was supported by grant no. U19 AI067773 (the Center for High-Throughput Minimally-Invasive Radiation Biodosimetry), from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIAID, NIH.
REFERENCES
- 1.Fenech M, Morley AA. Measurement of micronuclei in lymphocytes. Mutat Res 1985; 147:29–36. [DOI] [PubMed] [Google Scholar]
- 2.International Atomic Energy Agency (IAEA). Cytogenetic dosimetry: applications in preparedness for and response to radiation emergencies EPR-Biodosimetry. Vienna: IAEA; 2011. (https://bit.ly/2S7Hs0F) [Google Scholar]
- 3.Ramakumar A, Subramanian U, Prasanna PG. High-throughput sample processing and sample management; the functional evolution of classical cytogenetic assay towards automation. Mutat Res Genet Toxicol Environ Mutagen 2015; 793:132–41. [DOI] [PubMed] [Google Scholar]
- 4.Garty G, Chen Y, Salerno A, Turner H, Zhang J, Lyulko O, et al. The RABIT: a rapid automated biodosimetry tool for radiological triage. Health Phys 2010; 98:209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vrolijk J, Korthof G, Vletter G, van der Geest CR, Gerrese GW, Pearson PL. The automation of culturing and harvesting of human chromosome specimens. Histochemistry 1986; 84:586–93. [DOI] [PubMed] [Google Scholar]
- 6.Hayata I, Tabuchi H, Furukawa A, Okabe N, Yamamoto M, Sato K. Robot system for preparing lymphocyte chromosome. J Radiat Res (Tokyo) 1992; 33 Suppl:231–41. [DOI] [PubMed] [Google Scholar]
- 7.Beinke C, Port M, Abend M. Automatic versus manual lymphocyte fixation: impact on dose estimation using the cytokinesis-block micronucleus assay. Radiat Environ Biophys 2015; 54:81–90. [DOI] [PubMed] [Google Scholar]
- 8.Garty G, Turner HC, Salerno A, Bertucci A, Zhang J, Chen Y, et al. The decade of the RABiT (2005–15). Radiat Prot Dosimetry 2016; 172:201–06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner HC, Sharma P, Perrier JR, Bertucci A, Smilenov L, Johnson G, et al. The RABiT: high-throughput technology for assessing global DSB repair. Radiat Environ Biophys 2014; 53:265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertucci A, Smilenov LB, Turner HC, Amundson SA, Brenner DJ. In vitro RABiT measurement of dose rate effects on radiation induction of micronuclei in human peripheral blood lymphocytes. Radiat Environ Biophys 2016; 55:53–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Repin M, Turner HC, Garty G, Brenner DJ. Next generation platforms for high-throughput biodosimetry. Radiat Prot Dosimetry 2014; 159:105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Repin M, Pampou S, Karan C, Brenner DJ, Garty G. RABiT-II: implementation of a high-throughput micronucleus biodosimetry assay on commercial biotech robotic systems. Radiat Res 2017; 187:492–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenech M, Cytokinesis-block micronucleus cytome assay. Nat Protoc 2007; 2:1084–104. [DOI] [PubMed] [Google Scholar]
- 14.Beinke C, Barnard S, Boulay-Greene H, De Amicis A, De Sanctis S, Herodin F, et al. Laboratory intercomparison of the dicentric chromosome analysis assay. Radiat Res 2013; 180:129–37. [DOI] [PubMed] [Google Scholar]
- 15.Lee TK, Johnson J, Wiley AL Jr, Means JA Assessment of two protocols for the human lymphocyte cytokinesis-blocked micronucleus assay. Mutagenesis 1995; 10:375–7. [DOI] [PubMed] [Google Scholar]
- 16.Rodrigues MA, Probst CE, Beaton-Green LA, Wilkins RC. The effect of an optimized imaging flow cytometry analysis template on sample throughput in the reduced culture cytokinesis-block micronucleus assay. Radiat Prot Dosimetry 2016; 172:223–9. [DOI] [PubMed] [Google Scholar]
- 17.Rodrigues MA, Beaton-Green LA, Wilkins RC. Validation of the cytokinesis-block micronucleus assay using imaging flow cytometry for high throughput radiation biodosimetry. Health Phys 2016; 110:29–36. [DOI] [PubMed] [Google Scholar]
- 18.Lloyd DC, Edwards AA, Moquet JE, Guerrero-Carbajal YC. The role of cytogenetics in early triage of radiation casualties. Appl Radiat Isot 2000; 52:1107–12. [DOI] [PubMed] [Google Scholar]
- 19.Romm H, Barnard S, Boulay-Greene H, De Amicis A, De Sanctis S, Franco M, et al. Laboratory intercomparison of the cytokinesis-block micronucleus assay. Radiat Res 2013; 180:120–8. [DOI] [PubMed] [Google Scholar]
- 20.Depuydt J, Baeyens A, Barnard S, Beinke C, Benedek A, Beukes P, et al. RENEB intercomparison exercises analyzing micronuclei (cytokinesis-block micronucleus assay). Int J Radiat Biol 2017; 93:36–47. [DOI] [PubMed] [Google Scholar]
- 21.Vral A, Fenech M, Thierens H. The micronucleus assay as a biological dosimeter of in vivo ionising radiation exposure. Mutagenesis 2011; 26:11–7. [DOI] [PubMed] [Google Scholar]