Abstract
Glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 (GPIHBP1), the protein that shuttles LPL to the capillary lumen, is essential for plasma triglyceride metabolism. When GPIHBP1 is absent, LPL remains stranded within the interstitial spaces and plasma triglyceride hydrolysis is impaired, resulting in severe hypertriglyceridemia. While the functions of GPIHBP1 in intravascular lipolysis are reasonably well understood, no one has yet identified DNA sequences regulating GPIHBP1 expression. In the current studies, we identified an enhancer element located ∼3.6 kb upstream from exon 1 of mouse Gpihbp1. To examine the importance of the enhancer, we used CRISPR/Cas9 genome editing to create mice lacking the enhancer (Gpihbp1Enh/Enh). Removing the enhancer reduced Gpihbp1 expression by >90% in the liver and by ∼50% in heart and brown adipose tissue. The reduced expression of GPIHBP1 was insufficient to prevent LPL from reaching the capillary lumen, and it did not lead to hypertriglyceridemia—even when mice were fed a high-fat diet. Compound heterozygotes (Gpihbp1Enh/− mice) displayed further reductions in Gpihbp1 expression and exhibited partial mislocalization of LPL (increased amounts of LPL within the interstitial spaces of the heart), but the plasma triglyceride levels were not perturbed. The enhancer element that we identified represents the first insight into DNA sequences controlling Gpihbp1 expression.
Keywords: chylomicrons, endothelial cells, lipids, lipolysis, fatty acid metabolism, triglycerides, glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1
The intravascular processing of triglyceride-rich lipoproteins (TRLs) by LPL is the central event in plasma lipid metabolism, providing nutrients for vital tissues and generating the lipoprotein remnants that play a causal role in atherogenesis (1, 2). LPL is produced by parenchymal cells (e.g., adipocytes, myocytes) and secreted into the interstitial spaces. The interstitial LPL is almost certainly bound by heparan sulfate proteoglycans near the surface of parenchymal cells, but those interactions are weak and transient, making it possible for LPL to move to glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 (GPIHBP1) on the basolateral surface of capillary endothelial cells (3, 4). After being captured by GPIHBP1, the LPL is shuttled across endothelial cells to its site of action in the capillary lumen (5). In the absence of GPIHBP1, LPL remains stranded within the interstitial spaces, affixed to heparan sulfate proteoglycans (3, 4). Aside from transporting LPL to the capillary lumen, GPIHBP1 has two other important functions in intravascular lipolysis. First, GPIHBP1 is required for the margination of TRLs within capillaries, allowing the lipolytic processing of TRLs to proceed (6). In the absence of GPIHBP1-bound LPL, TRLs do not stop along capillaries and simply “flow on by” in the bloodstream (6). Second, the GPIHBP1–LPL interaction prevents unfolding of LPL’s N-terminal catalytic domain and thereby preserves catalytic activity (7). Homozygous deficiency of GPIHBP1, either in mice or in humans, impairs TRL processing, resulting in severe hypertriglyceridemia (chylomicronemia) (2, 8). Heterozygosity for Gpihbp1 deficiency lowers tissue levels of GPIHBP1 by 50% (9–11), but half-normal amounts of GPIHBP1 appear to be quite sufficient for normal triglyceride metabolism.
GPIHBP1 is expressed by capillary endothelial cells in every peripheral tissue that has been tested, but GPIHBP1 is absent from both capillary endothelial cells of the brain and endothelial cells of large blood vessels (5). GPIHBP1 is expressed at high levels in capillaries of heart and brown adipose tissue (BAT), where TRL processing by LPL is robust, but expression is low in tissues where LPL-mediated TRL processing is less active (e.g., liver, kidney, spleen) (8). GPIHBP1 expression is absent in the brain, a tissue that relies primarily on glucose for fuel (8). However, even though GPIHBP1 has been studied for more than a decade, we have minimal insights into the regulation of GPIHBP1 expression. No one understands why GPIHBP1 expression is higher in some tissues than in others—or why GPIHBP1 expression is absent in endothelial cells of the brain and large blood vessels. Also, the DNA sequences that regulate GPIHBP1 expression have not been defined. Davies et al. (9) found that Gpihbp1 transcript levels in mice are altered by fasting/refeeding and by PPARγ agonists, but the DNA sequences controlling those responses have not been defined (9).
In the current study, we sought to gain insights into the DNA sequences that regulate Gpihbp1 expression, and in particular whether an enhancer element participates in Gpihbp1 regulation.
MATERIALS AND METHODS
Analysis of mouse and human chromatin accessibility profiles
To explore the possibility of an enhancer element upstream of Gpihbp1, several lines of experimental data were analyzed. We examined single-cell chromatin accessibility data from mouse brain, heart, kidney, liver, and lung with the assay for transposase-accessible chromatin with sequencing, or ATAC-seq (12, 13). We also utilized bulk ATAC-seq profiles from mouse liver and lung endothelial cells isolated by fluorescence activated cell sorting (FACS). We analyzed public chromatin immunoprecipitation sequencing (ChIP-seq) data for the acetylation of histone H3 on lysine 27 (H3K27ac) in adipocytes and nonadipocytes of BAT and hepatocytes and nonhepatocytes of the liver. Finally, we assessed regulatory marks in the orthologous region of the human genome with DNaseI hypersensitivity sequencing and H3K27ac ChIP-seq data (from human muscle, lung, and liver).
Single-cell ATAC-seq analyses.
Single-cell ATAC-seq data were used to define chromatin accessibility in mouse endothelial cells (13). Nine types of endothelial cells have been identified (13). Normalized chromatin accessibility data, measured in counts per million (CPM) reads, were downloaded from the Mouse sci-ATAC-seq Atlas (atlas.gs.washington.edu/mouse-atac) and plotted to the region of interest. To facilitate visualization of data, Gpihbp1 enhancer coordinates were converted to mm9 using liftOver (14). Browser tracks were generated with the Gviz package in R (15).
FACS isolation of mouse liver sinusoidal endothelial cells and lung endothelial cells.
SPRET/EiJ mice (6–10 weeks old) were purchased from Jackson Laboratories, euthanized with CO2, and nonparenchymal cell preparations were isolated (16–18). Livers were retrograde perfused for 5 min through the inferior vena cava with HBSS without Ca2+ or Mg2+ (Gibco) supplemented with 0.5 mM egtazic acid (EGTA) at a rate of 5 ml/min. Livers were digested for 5 min in a solution containing low-glucose DMEM, 0.05% Pronase E (Roche), 1 μg/ml DNaseI (Roche), and 2% FBS, followed by a 7 min digestion in DMEM containing 0.05% collagenase D (Roche), 1 μg/ml DNaseI (Roche), and 2% FBS. After passing the cell suspension through a 70 μm cell strainer, two low-speed centrifugation steps (50 g for 2 min) were used to pellet hepatocytes for removal. Next, nonparenchymal cells were collected at 700 g for 7 min, and the erythrocytes removed by osmotic lysis. To remove debris, cells were suspended in 47.5% Opti-Prep (Sigma-Aldrich) and placed beneath a 33% Opti-Prep solution, followed by centrifugation for 20 min at 932 g. Nonhepatocyte cells at the interface were collected, washed, and suspended in 33% isotonic Percoll and centrifuged at 700 g for 10 min to remove digestion debris. Finally, preparations were stained with anti-CD16/CD32 (BioLegend) to block Fc receptors and sorted by FACS on a BD FACSAria II. The staining cocktail consisted of Zombie Aqua to label dead cells, anti-CD31-PE, anti-CD45-Alexa 488, and anti-CD146-PE/Cy7; gated by LiveSingletCD45LoCD146+CD31+. All antibodies were purchased from BioLegend.
To obtain lung cells, the five lobes were dissected, minced with a razor blade, and digested in a 5 ml solution of RPMI 1640 medium, collagenase IV (1.6 mg/ml; Worthington), DNaseI (50 U/ml; Roche), and 1 μM flavopiridol (Sigma-Aldrich). The tissue was digested with gentle shaking at 37°C for 15 min, followed by erythrocyte lysis (eBioscience) on ice for 3 min. For FACS sorting, the staining cocktail was Zombie Aqua to label dead cells, anti-CD45-APC-Cy7, anti-CD31-PerCP-Cy5, and anti-CD146-FITC; gated by LiveSingletCD45−CD31+CD146+.
ATAC-seq data analysis.
ATAC-seq (12) was performed on 50,000 freshly sorted endothelial cells. After preparing sequencing libraries (19), sequencing was performed on an Illumina HiSeq 4000. Demultiplexed sequence reads were mapped to the Mus musculus genome assembly GRCm38/mm10 using Bowtie2 (20) and default parameters. Sequence alignment map (SAM) files were converted into tag directories with HOMER’s makeTagDirectory function (21). To remove normalization bias associated with different numbers of mitochondrial reads, tags mapping to the mitochondrial genome were removed, and the remaining reads were normalized across samples to 10 million reads. Data were visualized in the University of California, Santa Cruz Genome Browser (14) with the MakeMultiWigHub.pl in HOMER.
Conservation of DNA sequences.
Conserved sequences from several mammals were retrieved with Multiz (22), and sequence motifs were assessed with scanMotifGenomeWide.pl in HOMER (21).
Public datasets.
Public mouse datasets from the Gene Expression Omnibus (GEO) series GSE92590 (23) were downloaded (www.ncbi.nlm.nih.gov/geo/). Samples of SRR5121144 (BAT–adipocyte H3K27ac), SRR5121152 (BAT–non-adipocyte H3K27ac), SRR5121154 (hepatocyte H3K27ac), and SRR5121157 (non-hepatocyte H3K27ac) were retrieved for analysis (23). Raw sequence reads were aligned to the mouse genome (mm10 version) using Bowtie2 and visualized in the University of California, Santa Cruz browser. Chromatin accessibility and ChIP-seq data from human samples were downloaded from the Roadmap Epigenomics Project (24) (www.ncbi.nlm.nih.gov/geo/roadmap/epigenomics/). We retrieved raw sequence reads for GSM878637 (fetal leg muscle DNaseI), GSM595915 (fetal lung DNaseI), GSM1058767 (fetal leg muscle H3K27ac ChIP-seq), GSM1112808 (adult liver H3K27ac ChIP-seq), and GSM906395 (adult lung H3K27ac ChIP-seq). We aligned raw sequence tags to the GRCh37/hg19 human reference genome using Bowtie2 and visualized data as described earlier.
Generation of Gpihbp1Enh/Enh mice
Mice lacking the putative enhancer (Gpihbp1Enh/Enh) were created in a C57BL/6×SJL mixed genetic background by CRISPR/Cas9 genome editing (25, 26). A single guide RNA (sgRNA) target was identified upstream of the putative enhancer (G84U1: 5′-ATGGGGGACAGATATAGGCCTGG-3′) with a predicted cut site 3,627 bp upstream from the Gpihbp1 initiation codon. A sgRNA target was also identified downstream from the enhancer (G84D1: 5′-TGGGCACCAGTAGTCTACACAGG-3′) with a predicted cut site 3,306 bp upstream from the Gpihbp1 initiation codon. The sgRNA targets were cloned into plasmid pX330 (Addgene #42230) (27). The two sgRNAs were validated in mouse JM8.A3 ES cells (28) before using them for mouse zygote microinjections. The G84U1 (15 μg) and G84D1 (15 μg) sgRNA plasmids were electroporated singly or together into 8 × 106 ES cells (29). For each electroporation, 5 μg of a PGK1-puromycin resistance plasmid (30) were added, allowing for puromycin selection of ES cells 48–72 h after the electroporation. After selection, DNA was extracted from surviving cells, and a fragment of DNA was amplified with G84U1 or G84D1. A T7 endonuclease 1 assay was used to detect small deletions/insertions at the predicted Cas9 DNA cut sites (31). Both sgRNAs resulted in chromosome breaks, and when used in combination deleted the putative enhancer sequences. For pronuclear microinjection studies, circular G84D1 and G84U1 plasmids were resuspended in microinjection buffer (5 ng/μl each) (32) and then injected into C57BL/6J×SJL/J F2 zygotes (33). A total of 82 pups were born, and 12 had a deletion of the Gpihbp1 enhancer. Genotyping was performed by PCR with oligonucleotides 5′-ATAAGGTACCGCCACCACAGATCTATGTCC-3′ (forward) and 5′-CTTCCTATGAACACATGAGGGAAACCT-3′ (reverse). Gel electrophoresis was used to resolve the products (wild-type allele, 800 bp; mutant allele, 474 bp). The mice were fed a chow diet and housed in a barrier facility with a 12 h light-dark cycle. All studies were approved by the University of California, Los Angeles’ Animal Research Committee.
Quantifying mouse tissue transcripts
Mice were anesthetized with isoflurane and perfused with PBS containing 5 mM EDTA. The heart, lung, liver, mesentery, gonadal white adipose tissue, spleen, kidney, quadriceps, BAT, and brain were harvested and flash-frozen in liquid nitrogen. RNA was isolated with TRI reagent (Molecular Research), and quantitative (q)RT-PCR measurements were performed in triplicate with a 7900HT Fast real-time PCR system (Applied Biosystems) (34–36). Gene-expression was calculated with the comparative CT method and normalized to expression of cyclophilin A. Primers for Gpihbp1 were 5′-AGCAGGGACAGAGCACCTCT-3′ and 5′-AGACGAGCGTGATGCAGAAG-3′ (exons 2/3 and 3, respectively). Primers for Lpl were 5′-AGGTGGACATCGGAGAACTG-3′ and 5′-TCCCTAGCACAGAAGATGACC-3′ (exons 8 and 9, respectively). Primers for Cd31 were 5′-AACCGTATCTCCAAAGCCAGT-3′ and 5′-CCAGACGACTGGAGGAGAACT-3′ (exons 5 and 6, respectively). Primers for Cd36 were 5′-GGCCAAGCTATTGCGACAT-3′ and 5′-CAGATCCGAACACAGCGTAGA-3′ (exons 6 and 8, respectively).
Quantifying GPIHBP1 levels in plasma and tissue homogenates
Mice were anesthetized with isoflurane and perfused with PBS containing 5 mM EDTA. The heart, liver, and BAT were harvested and homogenized on ice for 12–15 s in homogenization buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 2.5 mg/ml deoxycholic acid, 0.1% SDS, and Complete EDTA-free protease inhibitor (Roche)]. Samples were centrifuged (15,000 g for 15 min) to remove cellular debris, and the supernatant fluid was collected. ELISA plates were coated overnight with a rat monoclonal antibody against the carboxyl terminus of mouse GPIHBP1 (11A12; 0.5 μg/well). On the next day, the wells were blocked for 4 h with Starting block (Thermo Fisher Scientific). Dilutions of mouse plasma (1:1, 1:2, 1:4) or tissue extracts (100, 50, 25, 12.5 μg of total protein) were added to the wells and incubated overnight at 4°C. A standard curve (from recombinant mouse GPIHBP1) was run in parallel (0–800 pg/well). After washing, the amount of GPIHBP1 captured on the wells was assessed by adding 50 ng of an HRP-labeled GPIHBP1-specific monoclonal antibody (2A8) to each well and incubating for 2 h at 4°C. After washing, wells were incubated on ice for 15–30 min with TMB substrate (100 μl/well). The reaction was stopped with 2 M sulfuric acid (100 μl/well), and the optical density was read at 450 nm.
Mouse GPIHBP1 was immunoprecipitated from heart (200 μg of protein), BAT (200 μg of protein), or liver (1 mg of protein) extracts in 500 μl of homogenization buffer by incubating the extracts with 25 μl of agarose beads coated with antibody 11A12 for 90 min at 4°C. Beads were washed with PBS/Ca/Mg containing 0.2% NP-40 and mouse GPIHBP1 was eluted by boiling the beads in 30 μl of SDS sample buffer at 90°C for 10 min. Proteins (25 μl/lane) were size-fractioned by SDS-PAGE, followed by Western blotting with an IRDye680-conjugated monoclonal antibody against mouse GPIHBP1 (11A12; 3 μg/ml) and a goat polyclonal antibody against mouse LPL (10 μg/ml) (37), followed by an IRDye800-conjugated donkey anti–goat IgG (LI-COR). Signals were visualized with an Odyssey infrared scanner (LI-COR).
Immunohistochemistry
Mice were anesthetized with isoflurane and perfused with PBS containing 5 mM EDTA followed by 3% paraformaldehyde. The heart, BAT, liver, and kidney were harvested and embedded in OCT medium on dry ice. In some experiments, mice were anesthetized with ketamine/xylazine and then injected with DyLight488-conjugated tomato lectin (100 μg; Vector Laboratories) via the inferior vena cava. After 30 s, the mice were perfused with PBS containing 5 mM EDTA followed by 3% paraformaldehyde. Tissue sections (7 μm for heart, liver, and kidney; 10 μm for BAT) were fixed with 3% paraformaldehyde at room temperature for 15 min, permeabilized with 0.2% Triton X-100 for 5 min, and blocked at room temperature with 5% donkey serum and 0.2% BSA in PBS/Mg/Ca. Tissues were incubated overnight at 4°C with a goat polyclonal antibody against mouse LPL (12 μg/ml) (37) and a rabbit polyclonal antibody against mouse CD31 (Abcam; 1:50), followed by a 45 min incubation at room temperature with Alexa647-conjugated antibody 11A12 (3 μg/ml), Alexa568-conjugated donkey anti–goat IgG (Thermo Fisher Scientific; 1:200), and Alexa488-conjugated donkey anti–rabbit IgG (Thermo Fisher Scientific; 1:200). For mice injected with tomato lectin, the incubations of sections with the rabbit polyclonal antibody against mouse CD31 and the Alexa488-conjugated donkey anti–rabbit IgG were omitted.
In other experiments, tissue sections were incubated overnight at 4°C with a goat polyclonal antibody against mouse LPL (12 μg/ml) (37), a rabbit polyclonal antibody against mouse β-dystroglycan (Santa Cruz Biotechnology; 1:100), and a rat monoclonal antibody against mouse CD31 (BD Pharmingen; 1:50), followed by a 45 min incubation at room temperature with Alexa488-conjugated donkey anti–rat IgG (Thermo Fisher Scientific; 1:200), Alexa568-conjugated donkey anti–rabbit IgG (Thermo Fisher Scientific; 1:200), and Alexa647-conjugated donkey anti–goat IgG (Thermo Fisher Scientific; 1:200). After washing, the tissues were fixed with 3% paraformaldehyde for 5 min and stained with DAPI to visualize DNA. Images were recorded with an Axiovert 200M microscope and processed with Zen 2010 software (all from Zeiss). The confocal microscope exposure conditions within each experiment were identical.
Measurements of plasma triglycerides
Blood was collected from 10-week-old mice by retroorbital bleeding. In some experiments, mice were given 100 μl of corn oil by gastric gavage, and blood was collected at baseline and 1, 2, 3, and 4 h postgavage by retroorbital bleeding. In other experiments, mice were fed a high-fat diet (42% calories from fat; Envigo TD.88137). Blood was collected before initiation of this diet and after 1 month on the diet. Triglycerides were measured in plasma samples with a triglyceride determination kit (Sigma-Aldrich).
Statistical analyses
All statistical analyses were performed with an unpaired two-tailed Student’s t-test.
RESULTS
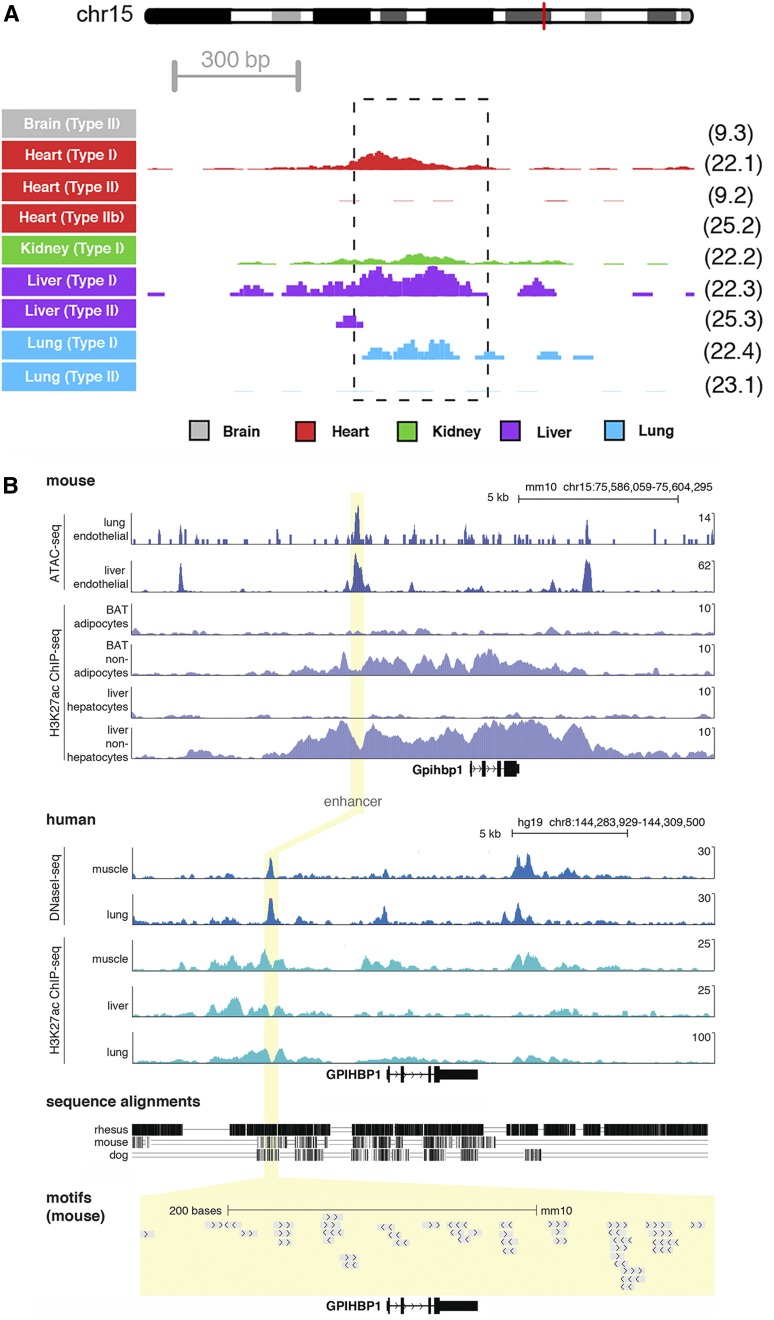
Several lines of evidence led to the identification of an ∼325 bp enhancer located ∼3.6 kb upstream of exon 1 of Gpihbp1 (Fig. 1). Single-cell ATAC-seq data on mouse endothelial cells (13) was used to define regions of accessible chromatin. A region of increased chromatin accessibility (a putative Gpihbp1 enhancer) was identified in endothelial cells from heart, liver, and lung (but not brain) (Fig. 1A). These findings were corroborated with bulk ATAC-seq profiles of mouse liver and lung endothelial cells (isolated by FACS) (Fig. 1B). We also examined public ChIP-seq data for H3K27ac (a mark of active enhancers) in adipocytes and nonadipocytes of BAT and hepatocytes and nonhepatocytes of the liver (23). We observed increased H3K27ac density adjacent to the Gpihbp1 open chromatin enhancer peak (where the most proximal nucleosomes are located). This signal was observed in nonadipocyte and nonhepatocyte cell populations (i.e., populations enriched in endothelial cells) in regions flanking the open chromatin (as identified by ATAC-seq) and was absent in adipocyte and hepatocyte populations (implying that enhancer activity was specific for endothelial cells). The sequence of the putative enhancer was conserved in humans and other mammals (supplemental Fig. S1), but not in lower vertebrates, where a gene for GPIHBP1 has never been identified and seemingly does not exist (38). We also retrieved and analyzed Roadmap Epigenomics data (24) for humans and identified marks for open chromatin and H3K27ac in muscle, liver, and lung (Fig. 1B), suggesting that the enhancer is also active in humans. DNA sequence motifs for several transcription factors, including for the endothelial cell transcription factor ERG (39), were present within the enhancer (supplemental Fig. S1).
Fig. 1.
Epigenetic profiles for a putative Gpihbp1 enhancer. A: Single-cell chromatin accessibility profiles for mouse tissues identified nine types of endothelial cells with distinct accessibility profiles (13). Normalized sequence tag counts are shown at the Gpihbp1 enhancer in counts per million (CPM) reads; 5 CPM is the maximum on the y axis. The dashed box indicates the nucleotides deleted in the Gpihbp1Enh allele. The cell cluster identifier, labeled in the same manner as in the original publication (13), is shown to the right of each track. The red line on the depiction of Chr15 represents the location of the Gpihbp1 locus. B: Epigenomic profiles at the Gpihbp1 locus are shown for mouse (top) and the syntenic human region (bottom), detailing open chromatin profiles (ATAC-seq and DNaseI-seq) and H3K27ac ChIP-seq in different tissue/cell types. The yellow highlight depicts the region of the enhancer deletion. Sequence alignments and transcription factor binding motifs are shown below.
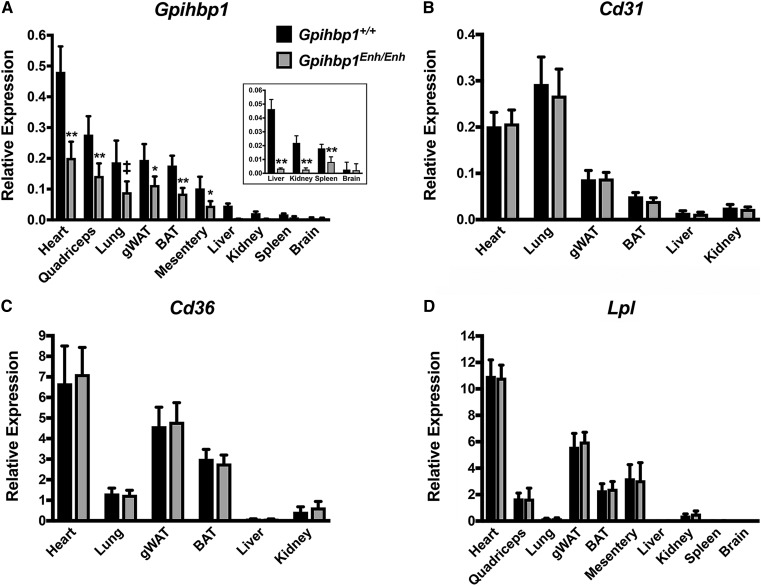
Mice lacking the putative enhancer element (Gpihbp1Enh/Enh) were created by CRISPR/Cas9 genome editing. Sequencing of the genomic DNA from Gpihbp1Enh/Enh mice defined the length of the deletion (326 bp) (supplemental Fig. S1) and excluded spurious mutations in Gpihbp1 coding sequences. Levels of Gpihbp1 transcripts in tissues were lower in Gpihbp1Enh/Enh mice than in wild-type mice, most prominently in the liver where transcript levels were reduced by >90% (Fig. 2A). In heart and BAT, Gpihbp1 transcript levels were reduced by ∼50% (Fig. 2A). Examination of a second line of Gpihbp1Enh/Enh mice (identical except that it had a deletion of 334 bp rather than 326 bp) manifested similar reductions in Gpihbp1 transcript levels (not shown). Transcript levels for Cd31 and Cd36 [expressed at high levels in endothelial cells (40, 41)] were not perturbed in Gpihbp1Enh/Enh mice (Fig. 2B, C), nor were levels of Lpl transcripts (Fig. 2D).
Fig. 2.
Gpihbp1 and Lpl transcript levels in 10-week-old Gpihbp1Enh/Enh and wild-type mice (Gpihbp1+/+). Gpihbp1 (A), Cd31 (B), Cd36 (C), and Lpl (D) transcript levels were measured by qRT-PCR (n = 10/group). Gpihbp1 expression was normalized to the expression of cyclophilin A. gWAT, gonadal white adipose tissue. Data show mean ± SD. ‡P < 0.01; *P < 0.001; **P < 0.0001.
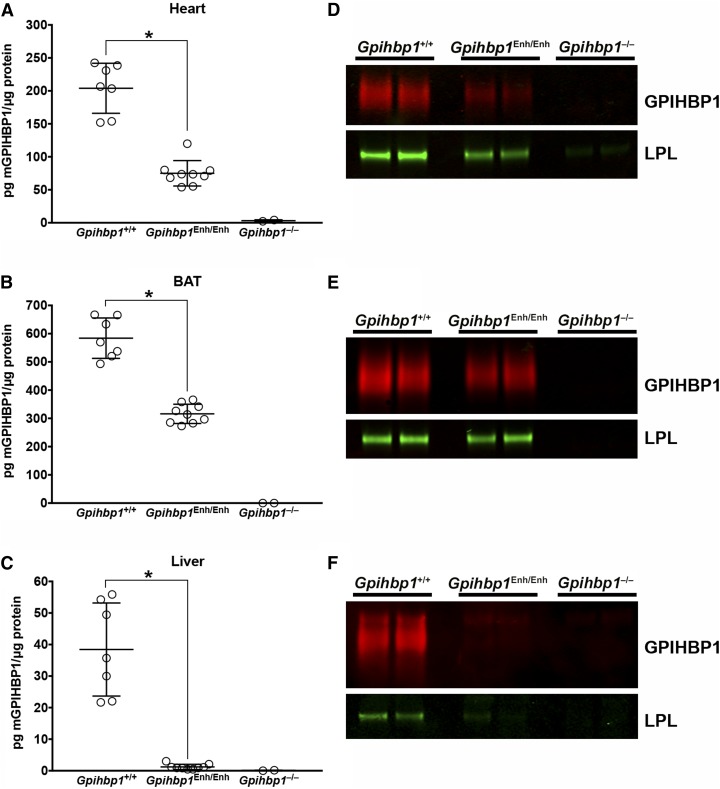
The levels of GPIHBP1 protein in tissues were assessed with a monoclonal antibody–based sandwich ELISA (Fig. 3A–C). GPIHBP1 levels in Gpihbp1Enh/Enh mice were reduced by ∼95% in the liver and by 35–50% in heart and BAT (Fig. 3A–C). GPIHBP1 was undetectable in Gpihbp1−/− mice (Fig. 3A–C). Immunoprecipitation studies on tissue extracts, performed with a GPIHBP1-specific monoclonal antibody, yielded similar findings, namely markedly reduced GPIHBP1 levels in the liver and moderately reduced GPIHBP1 levels in heart and BAT (Fig. 3D–F). The amount of LPL in BAT immunoprecipitates was roughly similar in Gpihbp1Enh/Enh and wild-type mice but appeared to be lower in heart immunoprecipitates of Gpihbp1Enh/Enh mice. LPL could be detected in liver immunoprecipitates of wild-type mice but was virtually undetectable in Gpihbp1Enh/Enh mice. The LPL:GPIHBP1 ratio in heart and BAT immunoprecipitates (as judged by infrared scanning of Western blots) was higher in Gpihbp1Enh/Enh mice than in wild-type mice (Fig. 3D–F).
Fig. 3.
Reduced GPIHBP1 protein levels in Gpihbp1Enh/Enh mice. A–C: GPIHBP1 levels in tissue homogenates of heart (A), BAT (B), and liver (C) in 10-week-old mice were measured with a sandwich ELISA. Results are plotted as the mass of GPIHBP1 normalized to total protein (Gpihbp1+/+, n = 7; Gpihbp1Enh/Enh, n = 9; Gpihbp1−/−, n = 2). Data show mean ± SD; *P < 0.0001. D–F: GPIHBP1 and LPL levels in tissues of Gpihbp1Enh/Enh mice, as judged by Western blots. The GPIHBP1 (and any bound LPL) in 200 μg of heart (D) or BAT (E) tissue homogenates [or 1 mg of liver (F) homogenate] were immunoprecipitated with 25 μl of agarose beads coated with the GPIHBP1-specific antibody 11A12 (n = 2 mice/group). Relative amounts of GPIHBP1 and LPL in the immunoprecipitates were assessed by Western blotting with antibodies against GPIHBP1 (red) and LPL (green). GPIHBP1 signals, as judged by an infrared scanner, were quantified in Gpihbp1+/+ heart (1920000 and 1460000) and Gpihbp1Enh/Enh heart (406000 and 246000); Gpihbp1+/+ BAT (20600000 and 14900000) and Gpihbp1Enh/Enh BAT (7280000 and 9490000); and Gpihbp1+/+ liver (1110000 and 1320000) and Gpihbp1Enh/Enh liver (151000 and 163000). The LPL:GPIHBP1 ratio was calculated in Gpihbp1+/+ heart (0.427 and 0.633) and Gpihbp1Enh/Enh heart (1.138 and 1.341), Gpihbp1+/+ BAT (0.022 and 0.032) and Gpihbp1Enh/Enh BAT (0.047 and 0.045), and Gpihbp1+/+ liver (0.015 and 0.009) and Gpihbp1Enh/Enh liver (0.031 and 0.017).
We used the GPIHBP1 sandwich ELISA to measure plasma GPIHBP1 levels in Gpihbp1+/+, Gpihbp1Enh/Enh, and Gpihbp1−/− mice. In earlier studies (42), we found lower plasma GPIHBP1 levels in mutant mice that had reduced amounts of GPIHBP1 on capillary endothelial cells (42). Consistent with those studies and with the finding of reduced amounts of GPIHBP1 in tissues of Gpihbp1Enh/Enh mice (Fig. 3), the plasma GPIHBP1 levels were lower in Gpihbp1Enh/Enh mice than in wild-type mice (supplemental Fig. S2).
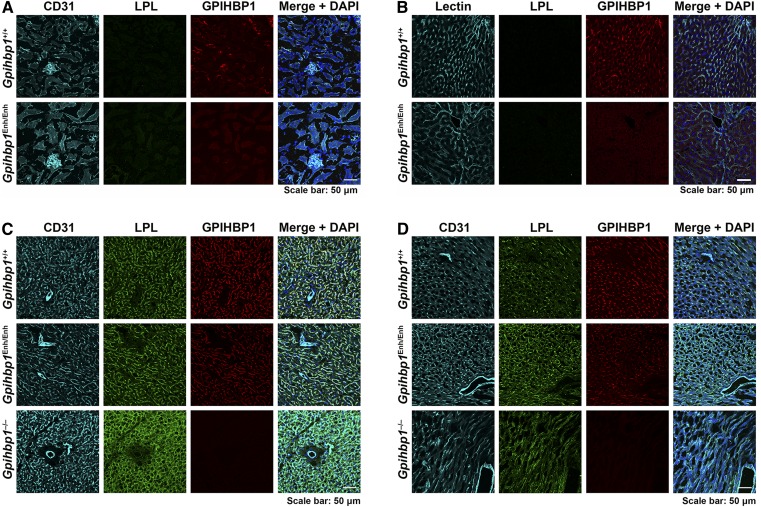
To further characterize GPIHBP1 expression in Gpihbp1Enh/Enh mice, immunohistochemistry studies were performed on tissue sections with a fluorescently labeled GPIHBP1-specific monoclonal antibody (11A12). GPIHBP1 was virtually undetectable in kidney and liver capillary endothelial cells of Gpihbp1Enh/Enh mice but was easily detectable in the kidney and liver of wild-type mice (Fig. 4A, B). In BAT and heart, GPIHBP1 staining was less intense in Gpihbp1Enh/Enh mice than in wild-type mice (Fig. 4C, D). In both Gpihbp1Enh/Enh and wild-type mice, GPIHBP1 was present in capillaries but was absent in endothelial cells of larger blood vessels (arterioles, venules) (Fig. 4C, D). GPIHBP1 expression was also absent in brain capillaries of Gpihbp1Enh/Enh mice (not shown), as reported earlier for wild-type mice (8).
Fig. 4.
Confocal immunofluorescence microscopy images of mouse tissues after staining with antibodies against CD31, GPIHBP1, and LPL. Sections of kidney (A), liver (B), BAT (C), and heart (D) were stained with antibodies against LPL (green) and GPIHBP1 (red). Kidney, BAT, and heart were stained with an antibody against CD31 (cyan); the liver was stained with tomato lectin (cyan). DNA was stained with DAPI (blue). Scale bar, 50 μm.
We also examined the binding of an LPL-specific antibody to tissues of Gpihbp1Enh/Enh mice (Fig. 4). In earlier studies, we found that nearly all of the LPL in the heart and BAT of wild-type mice is located on capillaries (5, 42), whereas it is mislocalized within the interstitial spaces in Gpihbp1−/− mice. In the current studies, we extended these experiments to include Gpihbp1Enh/Enh mice (Fig. 4). In BAT and heart of Gpihbp1Enh/Enh mice, LPL was located almost exclusively on capillaries (colocalizing with GPIHBP1), and the intensity of antibody staining appeared similar to that in wild-type mice. Also, despite lower levels of GPIHBP1 expression in Gpihbp1Enh/Enh mice, we found no evidence for partial mislocalization of LPL within tissues (i.e., we did not observe increased amounts of LPL within the interstitial spaces). Given that nearly all of the LPL in the BAT of Gpihbp1Enh/Enh mice was associated with capillaries and colocalized with GPIHBP1, we predicted that we would find LPL in the capillary lumen of Gpihbp1Enh/Enh mice. Indeed, this was the case (Fig. 5).
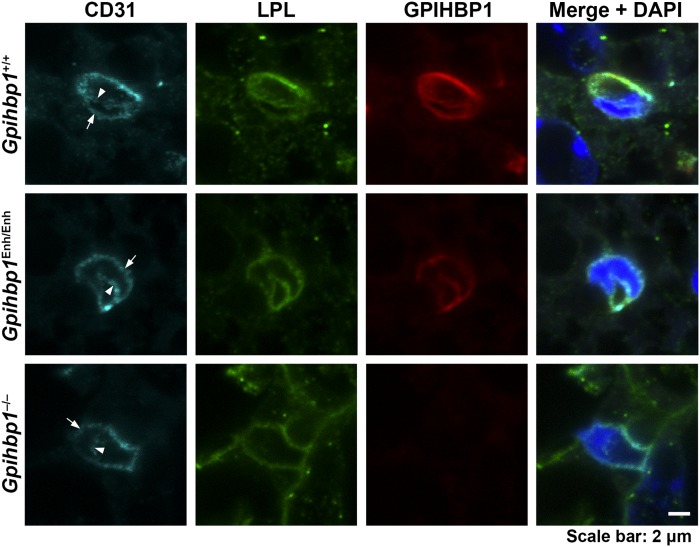
Fig. 5.
LPL reaches the capillary lumen in the BAT of Gpihbp1Enh/Enh mice despite reduced amounts of GPIHBP1 expression. Here, we examined the binding of CD31- (cyan), GPIHBP1- (red), and LPL-specific (green) antibodies to the BAT of Gpihbp1+/+, Gpihbp1Enh/Enh, and Gpihbp1−/− mice. To visualize the luminal and basolateral surfaces of capillary endothelial cells, we recorded confocal microscopy images of capillary cross-sections containing endothelial cell nuclei. In the BAT of Gpihbp1+/+ and Gpihbp1Enh/Enh mice, CD31, GPIHBP1, and LPL were visible on both the luminal (arrowheads) and basolateral surface of capillary endothelial cells (arrows). In Gpihbp1−/− mice, LPL was virtually undetectable along the luminal surface of capillary endothelial cells. DNA was stained with DAPI (blue). Scale bar, 2 μm.
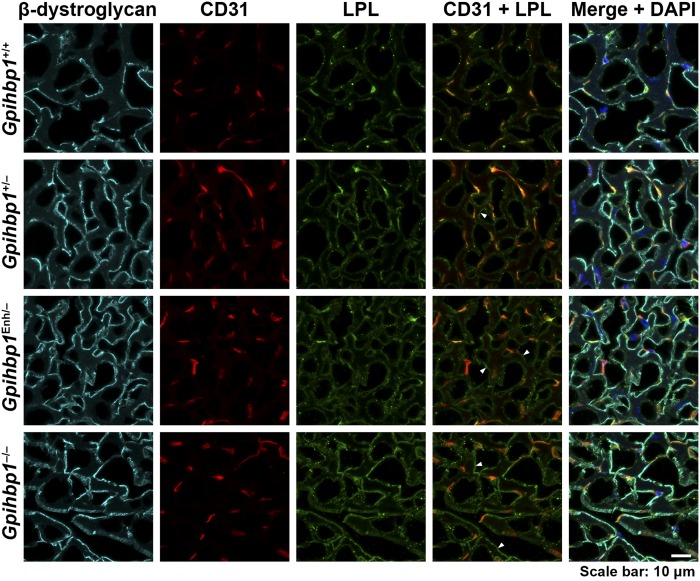
While we did not find partial mislocalization of LPL in the heart of Gpihbp1Enh/Enh mice, we suspected that we might be able to identify partial mislocalization of LPL if the levels of GPIHBP1 expression were reduced below those in Gpihbp1Enh/Enh mice. To test this suspicion, we bred Gpihbp1Enh/− mice, where Gpihbp1 transcripts were only one-half as high as in Gpihbp1Enh/Enh mice (supplemental Fig. S3). In the heart of Gpihbp1Enh/– mice, we observed partial mislocalization of LPL (Fig. 6). A substantial fraction of the LPL was located on capillaries, colocalizing with GPIHBP1, but some of the LPL was observed in the interstitial spaces near the surface of parenchymal cells (colocalizing with β-dystroglycan) (Fig. 6). Partial mislocalization of LPL to the interstitial spaces was also observed in the heart of Gpihbp1+/− mice (Fig. 6).
Fig. 6.
LPL is partially mislocalized in the heart in Gpihbp1Enh/− mice, with increased amounts of LPL in the interstitial spaces near the surface of cardiomyocytes. Confocal microscopy studies were performed on sections of heart stained with antibodies for β-dystroglycan (cyan), CD31 (red), and LPL (green). β-Dystroglycan is located along the surface of cardiomyocytes. In comparing confocal images from Gpihbp1+/+ and Gpihbp1Enh/− mice, we observed more LPL outside of capillaries in Gpihbp1Enh/− mice (colocalizing with β-dystroglycan) (arrowheads in the CD31/LPL merged image point to several such regions). An even greater amount of interstitial LPL (colocalizing with β-dystroglycan) was observed in sections from Gpihbp1−/− mice (arrowheads). A small amount of LPL was mislocalized to the interstitial spaces in Gpihbp1+/− mice (arrowhead). DNA was stained with DAPI (blue). Scale bar, 10 μm.
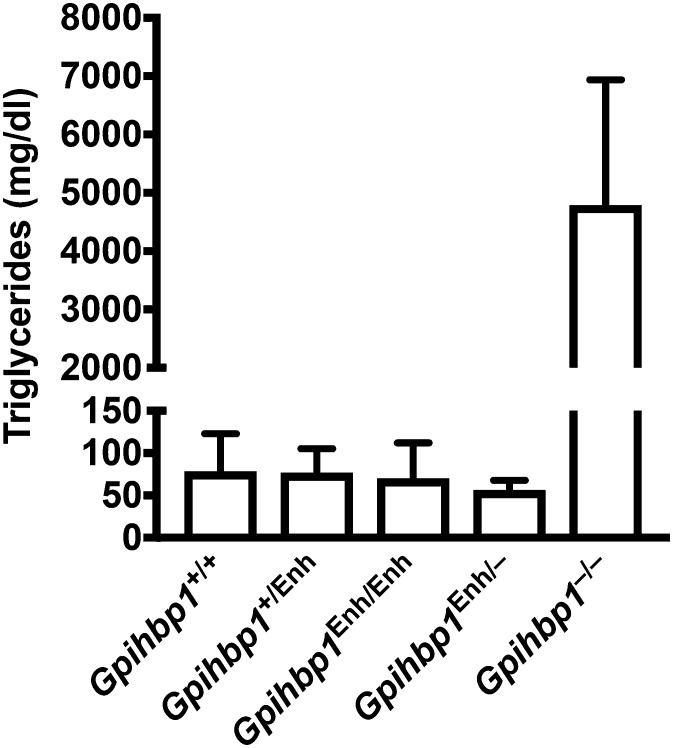
Because the intensity of the LPL staining did not appear to be reduced in the heart and BAT of Gpihbp1Enh/Enh mice (Fig. 4) and because we were able to detect LPL in the capillary lumen of Gpihbp1Enh/Enh mice (Fig. 5), we were skeptical that we would find elevated plasma triglyceride levels in Gpihbp1Enh/Enh mice. Indeed, the plasma triglyceride levels were similar in Gpihbp1Enh/Enh, Gpihbp1Enh/−, and wild-type mice (<100 mg/dl) (Fig. 7). Even after administering corn oil to Gpihbp1Enh/− mice by gastric gavage, we did not find elevated plasma triglyceride levels (supplemental Fig. S4A). Also, the plasma triglyceride levels in Gpihbp1Enh/− mice were not elevated when the mice were fed a high-fat diet (supplemental Fig. S4B).
Fig. 7.
Normal plasma triglycerides in Gpihbp1Enh/Enh and Gpihbp1Enh/− mice. Plasma triglyceride levels were measured in 10-week-old Gpihbp1+/+ (n = 16), Gpihbp1+/Enh (n = 20), Gpihbp1Enh/Enh (n = 17), Gpihbp1Enh/− (n = 8), and Gpihbp1−/− (n = 6) mice. Data show mean ± SD.
DISCUSSION
In the current study, we identified, by ATAC-seq and histone H3K27ac modifications, a Gpihbp1 enhancer in endothelial cells (an ∼325 bp element located ∼3.6 kb upstream of exon 1 of Gpihbp1). The same element was detected, by DNase1-seq and histone H3K27ac modifications, in human tissues. We suspected that the sequences that we identified could be important for regulating levels of Gpihbp1 expression and/or sites of Gpihbp1 expression. To test these possibilities, we created mice lacking the enhancer (Gpihbp1Enh/Enh). The enhancer deletion reduced levels of Gpihbp1 transcripts (but not levels of Cd31, Cd36, or Lpl transcripts) in every tissue tested (>90% in the liver, ∼50% in heart and BAT). Reductions in GPIHBP1 protein levels mirrored the decreases in transcript levels. The plasma levels of GPIHBP1 were also lower in Gpihbp1Enh/Enh mice. Reduced amounts of GPIHBP1 in Gpihbp1Enh/Enh mice were also evident by immunohistochemistry. The fact that the deletion of the enhancer did not eliminate Gpihbp1 expression was not particularly surprising. Gene expression often depends on multiple enhancers (43, 44), and deleting a single enhancer is often insufficient to abolish gene expression (45, 46).
The decrease in Gpihbp1 expression in Gpihbp1Enh/Enh mice was quite striking in the liver (where Gpihbp1 expression is normally low) but only moderate in the heart and BAT (where Gpihbp1 expression is high). We don’t understand why the impact of the deletion (as judged by the percentage decrease in Gpihbp1 expression) was greater in the liver, but it is noteworthy that the region of open chromatin (as judged by ATAC-seq) was particularly prominent in liver endothelial cells (Fig. 1).
The expression of Lpl and Gpihbp1 in the liver is normally low, but the expression of Lpl in the liver can be induced substantially by a high-cholesterol diet (47). At this time, the functional importance of GPIHBP1 and LPL in the liver is incompletely understood. Because liver capillaries are fenestrated, GPIHBP1’s role in transporting LPL across endothelial cells is likely superfluous in the liver (8, 47). However, biochemical studies have proven that GPIHBP1 preserves the structural integrity and enzymatic activity of LPL (7, 48), and this function is presumably relevant in all tissues—including the liver.
In our studies, we found no evidence that the ∼90% decrease in Gpihbp1 expression in the liver and the ∼50% decrease in Gpihbp1 expression in heart and BAT influenced plasma triglyceride levels. The triglyceride levels in Gpihbp1Enh/Enh mice were similar to those in wild-type mice. In Gpihbp1Enh/− mice, where levels of Gpihbp1 expression were reduced by ∼95% in the liver and ∼75% in BAT and heart, LPL was partially mislocalized to the interstitial spaces but the plasma triglyceride levels remained normal—even when the mice were challenged with a corn oil bolus or a high-fat diet. The normal plasma triglyceride levels in Gpihbp1Enh/− mice stand in contrast to findings with hepatocyte-specific Lpl knockout mice. Eliminating LPL expression in hepatocytes resulted in modestly higher plasma triglyceride levels, both at baseline and after a bolus of corn oil (49).
Finding normal plasma triglyceride levels in Gpihbp1Enh/Enh mice was not surprising. The plasma triglyceride levels in Gpihbp1+/− mice are entirely normal (8). Also, humans heterozygous for loss-of-function GPIHBP1 mutations have normal plasma triglyceride levels (2). Again, the situation is different with LPL deficiency. Lpl+/− mice have mild–moderate increases in plasma triglyceride levels (50), and humans with one mutant LPL allele have increased plasma triglyceride levels (51, 52) along with an increased risk for coronary heart disease (53). Together, these observations imply that the limiting factor in triglyceride processing is the levels of LPL rather than the levels of GPIHBP1.
Our studies have provided the first insight into the DNA sequences regulating GPIHBP1 expression. We would hasten to admit, however, that we have only scratched the surface in understanding the regulation of GPIHBP1 expression. We have not yet identified the transcription factors that bind to the upstream Gpihbp1 enhancer, nor do we know whether GPIHBP1 expression is regulated by additional enhancer elements. We also do not understand why Gpihbp1 is expressed at high levels in the heart and BAT but is absent in the brain (5). Remarkably, the promoter for Gpihbp1 has never been defined or studied. Some of these lacunae in our understanding of GPIHBP1 expression will probably disappear with an improved understanding of endothelial cell heterogeneity and the associated heterogeneity in endothelial cell gene expression (54, 55). However, in the end, we suspect that nailing down the DNA sequences controlling GPIHBP1 expression will require additional studies with genetically modified mice, along the lines of the strategy used in the current studies.
Supplementary Material
Acknowledgments
The authors acknowledge Elizabeth Hughes for construction of genome editing reagents and Wanda Filipiak and Galina Gavrilina for mouse zygote microinjections.
Footnotes
Abbreviations:
- ATAC-seq
- assay for transposase-accessible chromatin with sequencing
- BAT
- brown adipose tissue
- ChIP-seq
- chromatin immunoprecipitation sequencing
- FACS
- fluorescence activated cell sorting
- GPIHBP1
- glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1
- H3K27ac
- acetylation of histone H3 on lysine 27
- sgRNA
- single guide RNA
- TRL
- triglyceride-rich lipoprotein
This work was supported by Fondation Leducq Transatlantic Network Grant 12CVD04 (S.G.Y.); National Heart, Lung, and Blood Institute Grants HL090553 (S.G.Y.), HL087228 (S.G.Y.), HL125335 (S.G.Y.), HL123485 (C.E.R.), and T32HL007895 (C.M.A.); and National Cancer Institute Grant T32CA009523 (T.D.T.). Fluorescence activated cell sorting studies were performed with the support of the Flow Cytometry Core at the San Diego Center for AIDS Research (P30 AI036214), the Veterans Affairs San Diego Health Care System, and the San Diego Veterans Medical Research Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors have no financial interests to declare.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Goldberg I. J. 1996. Lipoprotein lipase and lipolysis: Central roles in lipoprotein metabolism and atherogenesis. J. Lipid Res. 37: 693–707. [PubMed] [Google Scholar]
- 2.Fong L. G., Young S. G., Beigneux A. P., Bensadoun A., Oberer M., Jiang H., and Ploug M.. 2016. GPIHBP1 and plasma triglyceride metabolism. Trends Endocrinol. Metab. 27: 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allan C. M., Larsson M., Jung R. S., Ploug M., Bensadoun A., Beigneux A. P., Fong L. G., and Young S. G.. 2017. Mobility of “HSPG-bound” LPL explains how LPL is able to reach GPIHBP1 on capillaries. J. Lipid Res. 58: 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kristensen K. K., Midtgaard S. R., Mysling S., Kovrov O., Hansen L. B., Skar-Gislinge N., Beigneux A. P., Kragelund B. B., Olivecrona G., Young S. G., et al. . 2018. A disordered acidic domain in GPIHBP1 harboring a sulfated tyrosine regulates lipoprotein lipase. Proc. Natl. Acad. Sci. USA. 115: E6020–E6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies B. S., Beigneux A. P., Barnes R. H. 2nd, Tu Y., Gin P., Weinstein M. M., Nobumori C., Nyren R., Goldberg I., Olivecrona G., et al. . 2010. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 12: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goulbourne C. N., Gin P., Tatar A., Nobumori C., Hoenger A., Jiang H., Grovenor C. R., Adeyo O., Esko J. D., Goldberg I. J., et al. . 2014. The GPIHBP1-LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metab. 19: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mysling S., Kristensen K. K., Larsson M., Beigneux A. P., Gardsvoll H., Fong L. G., Bensadouen A., Jorgensen T. J., Young S. G., and Ploug M.. 2016. The acidic domain of the endothelial membrane protein GPIHBP1 stabilizes lipoprotein lipase activity by preventing unfolding of its catalytic domain. eLife. 5: e12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beigneux A. P., Davies B. S., Gin P., Weinstein M. M., Farber E., Qiao X., Peale F., Bunting S., Walzem R. L., Wong J. S., et al. . 2007. Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 5: 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies B. S., Waki H., Beigneux A. P., Farber E., Weinstein M. M., Wilpitz D. C., Tai L. J., Evans R. M., Fong L. G., Tontonoz P., et al. . 2008. The expression of GPIHBP1, an endothelial cell binding site for lipoprotein lipase and chylomicrons, is induced by peroxisome proliferator-activated receptor-gamma. Mol. Endocrinol. 22: 2496–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beigneux A. P., Miyashita K., Ploug M., Blom D. J., Ai M., Linton M. F., Khovidhunkit W., Dufour R., Garg A., McMahon M. A., et al. . 2017. Autoantibodies against GPIHBP1 as a cause of hypertriglyceridemia. N. Engl. J. Med. 376: 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plengpanich W., Young S. G., Khovidhunkit W., Bensadoun A., Karnman H., Ploug M., Gardsvoll H., Leung C. S., Adeyo O., Larsson M., et al. . 2014. Multimerization of glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) and familial chylomicronemia from a serine-to-cysteine substitution in GPIHBP1 Ly6 domain. J. Biol. Chem. 289: 19491–19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., and Greenleaf W. J.. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cusanovich D. A., Hill A. J., Aghamirzaie D., Daza R. M., Pliner H. A., Berletch J. B., Filippova G. N., Huang X., Christiansen L., DeWitt W. S., et al. . 2018. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell. 174: 1309–1324.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., and Haussler D.. 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mathé, E., and S. Davis, editors. 2016. Statistical Genomics: Methods and Protocols. Humana Press, New York, NY. [Google Scholar]
- 16.Mederacke I., Dapito D. H., Affo S., Uchinami H., and Schwabe R. F.. 2015. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat. Protoc. 10: 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muse E. D., Yu S., Edillor C. R., Tao J., Spann N. J., Troutman T. D., Seidman J. S., Henke A., Roland J. T., Ozeki K. A., et al. . 2018. Cell-specific discrimination of desmosterol and desmosterol mimetics confers selective regulation of LXR and SREBP in macrophages. Proc. Natl. Acad. Sci. USA. 115: E4680–E4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seki E., De Minicis S., Osterreicher C. H., Kluwe J., Osawa Y., Brenner D. A., and Schwabe R. F.. 2007. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat. Med. 13: 1324–1332. [DOI] [PubMed] [Google Scholar]
- 19.Hogan N. T., Whalen M. B., Stolze L. K., Hadeli N. K., Lam M. T., Springstead J. R., Glass C. K., and Romanoski C. E.. 2017. Transcriptional networks specifying homeostatic and inflammatory programs of gene expression in human aortic endothelial cells. eLife. 6: e22536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B., and Salzberg S. L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanchette M., Kent W. J., Riemer C., Elnitski L., Smit A. F., Roskin K. M., Baertsch R., Rosenbloom K., Clawson H., Green E. D., et al. . 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14: 708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roh H. C., Tsai L. T., Lyubetskaya A., Tenen D., Kumari M., and Rosen E. D.. 2017. Simultaneous transcriptional and epigenomic profiling from specific cell types within heterogeneous tissues in vivo. Cell Reports. 18: 1048–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roadmap Epigenomics Consortium, Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. . 2015. Integrative analysis of 111 reference human epigenomes. Nature. 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., et al. . 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., and Church G. M.. 2013. RNA-guided human genome engineering via Cas9. Science. 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettitt S. J., Liang Q., Rairdan X. Y., Moran J. L., Prosser H. M., Beier D. R., Lloyd K. C., Bradley A., and Skarnes W. C.. 2009. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat. Methods. 6: 493–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes E. D., and Saunders T. L.. 2011. Gene targeting in embryonic stem cells. In Advanced Protocols for Animal Transgenesis: An ISTT Manual. S. Pease and T. L. Saunders, editors. Springer, Berlin, Heidelberg. 291–325. [Google Scholar]
- 30.McBurney M. W., Fournier S., Jardine K., and Sutherland L.. 1994. Intragenic regions of the murine Pgk-1 locus enhance integration of transfected DNAs into genomes of embryonal carcinoma cells. Somat. Cell Mol. Genet. 20: 515–528. [DOI] [PubMed] [Google Scholar]
- 31.Sakurai T., Watanabe S., Kamiyoshi A., Sato M., and Shindo T.. 2014. A single blastocyst assay optimized for detecting CRISPR/Cas9 system-induced indel mutations in mice. BMC Biotechnol. 14: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mashiko D., Fujihara Y., Satouh Y., Miyata H., Isotani A., and Ikawa M.. 2013. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 3: 3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becker K., and Jerchow B.. 2011. Generation of transgenic mice by pronuclear microinjection. In Advanced Protocols for Animal Transgenesis: An ISTT Manual. S. Pease and T. L. Saunders, editors. Springer, Berlin, Heidelberg. 99–115. [Google Scholar]
- 34.Weinstein M. M., Goulbourne C. N., Davies B. S., Tu Y., Barnes R. H. 2nd, Watkins S. M., Davis R., Reue K., Tontonoz P., Beigneux A. P., et al. . 2012. Reciprocal metabolic perturbations in the adipose tissue and liver of GPIHBP1-deficient mice. Arterioscler. Thromb. Vasc. Biol. 32: 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung H. J., Nobumori C., Goulbourne C. N., Tu Y., Lee J. M., Tatar A., Wu D., Yoshinaga Y., de Jong P. J., Coffinier C., et al. . 2013. Farnesylation of lamin B1 is important for retention of nuclear chromatin during neuronal migration. Proc. Natl. Acad. Sci. USA. 110: E1923–E1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang S. H., Chang S. Y., Yin L., Tu Y., Hu Y., Yoshinaga Y., de Jong P. J., Fong L. G., and Young S. G.. 2011. An absence of both lamin B1 and lamin B2 in keratinocytes has no effect on cell proliferation or the development of skin and hair. Hum. Mol. Genet. 20: 3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Page S., Judson A., Melford K., and Bensadoun A.. 2006. Interaction of lipoprotein lipase and receptor-associated protein. J. Biol. Chem. 281: 13931–13938. [DOI] [PubMed] [Google Scholar]
- 38.He C., Hu X., Jung R. S., Larsson M., Tu Y., Duarte-Vogel S., Kim P., Sandoval N. P., Price T. R., Allan C. M., et al. . 2017. Lipoprotein lipase reaches the capillary lumen in chickens despite an apparent absence of GPIHBP1. JCI Insight. 2: 96783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah A. V., Birdsey G. M., and Randi A. M.. 2016. Regulation of endothelial homeostasis, vascular development and angiogenesis by the transcription factor ERG. Vascul. Pharmacol. 86: 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman P. J. 1997. The biology of PECAM-1. J. Clin. Invest. 99: 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Febbraio M., Hajjar D. P., and Silverstein R. L.. 2001. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Invest. 108: 785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allan C. M., Jung C. J., Larsson M., Heizer P. J., Tu Y. P., Sandoval N. P., Dang T. L. P., Jung R. S., Beigneux A. P., de Jong P. J., et al. . 2017. Mutating a conserved cysteine in GPIHBP1 reduces amounts of GPIHBP1 in capillaries and abolishes LPL binding. J. Lipid Res. 58: 1453–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tolhuis B., Palstra R. J., Splinter E., Grosveld F., and de Laat W.. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 10: 1453–1465. [DOI] [PubMed] [Google Scholar]
- 44.Harmston N., and Lenhard B.. 2013. Chromatin and epigenetic features of long-range gene regulation. Nucleic Acids Res. 41: 7185–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moorthy S. D., Davidson S., Shchuka V. M., Singh G., Malek-Gilani N., Langroudi L., Martchenko A., So V., Macpherson N. N., and Mitchell J. A.. 2017. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 27: 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osterwalder M., Barozzi I., Tissieres V., Fukuda-Yuzawa Y., Mannion B. J., Afzal S. Y., Lee E. A., Zhu Y., Plajzer-Frick I., Pickle C. S., et al. . 2018. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 554: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weinstein M. M., Tu Y., Beigneux A. P., Davies B. S., Gin P., Voss C., Walzem R. L., Reue K., Tontonoz P., Bensadoun A., et al. . 2010. Cholesterol intake modulates plasma triglyceride levels in glycosylphosphatidylinositol HDL-binding protein 1-deficient mice. Arterioscler. Thromb. Vasc. Biol. 30: 2106–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mysling S., Kristensen K. K., Larsson M., Kovrov O., Bensadouen A., Jorgensen T. J., Olivecrona G., Young S. G., and Ploug M.. 2016. The angiopoietin-like protein ANGPTL4 catalyzes unfolding of the hydrolase domain in lipoprotein lipase and the endothelial membrane protein GPIHBP1 counteracts this unfolding. eLife. 5: e20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu G., Xu J. N., Liu D., Ding Q., Liu M. N., Chen R., Fan M., Zhang Y., Zheng C., Zou D. J., et al. . 2016. Regulation of plasma lipid homeostasis by hepatic lipoprotein lipase in adult mice. J. Lipid Res. 57: 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinstock P. H., Bisgaier C. L., Aalto-Setälä K., Radner H., Ramakrishnan R., Levak-Frank S., Essenburg A. D., Zechner R., and Breslow J. L.. 1995. Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired low density lipoprotein clearance in heterozygotes. J. Clin. Invest. 96: 2555–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miesenböck G., Hölzl B., Föger B., Brandstätter E., Paulweber B., Sandhofer F., and Patsch J. R.. 1993. Heterozygous lipoprotein lipase deficiency due to a missense mutation as the cause of impaired triglyceride tolerance with multiple lipoprotein abnormalities. J. Clin. Invest. 91: 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson D. E., Emi M., Iverius P. H., Hata A., Wu L. L., Hillas E., Williams R. R., and Lalouel J. M.. 1990. Phenotypic expression of heterozygous lipoprotein lipase deficiency in the extended pedigree of a proband homozygous for a missense mutation. J. Clin. Invest. 86: 735–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khera A. V., Won H. H., Peloso G. M., O’Dushlaine C., Liu D., Stitziel N. O., Natarajan P., Nomura A., Emdin C. A., Gupta N., et al. . 2017. Association of rare and common variation in the lipoprotein lipase gene with coronary artery disease. JAMA. 317: 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He L., Vanlandewijck M., Mae M. A., Andrae J., Ando K., Del Gaudio F., Nahar K., Lebouvier T., Lavina B., Gouveia L., et al. . 2018. Single-cell RNA sequencing of mouse brain and lung vascular and vessel-associated cell types. Sci. Data. 5: 180160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aird W. C. 2012. Endothelial cell heterogeneity. Cold Spring Harb. Perspect. Med. 2: a006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.