Abstract
The regulation of cellular lipid storage and membrane lipid composition plays a critical role in metabolic homeostasis, and dysregulation may contribute to disorders such as obesity, fatty liver, type 2 diabetes, and cardiovascular disease. The mammalian lipin proteins (lipin 1, lipin 2, and lipin 3) are phosphatidic acid phosphatase (PAP) enzymes that modulate levels of cellular triacylglycerols and phospholipids, and also regulate lipid intermediates in cellular signaling pathways. Lipin proteins also have the ability to coactivate/corepress transcription. In humans and mice, lipin gene mutations cause severe metabolic phenotypes including rhabdomyolysis (lipin 1), autoinflammatory disease (lipin 2), and impaired intestinal lipoprotein assembly (lipin 2/lipin 3). Characterization of these diseases has revealed roles for lipin PAP activity in fundamental cellular processes such as autophagy, inflammasome activation, and lipoprotein assembly. Lipin protein activity is regulated at pre- and posttranscriptional levels, which suggests a need for their ordered response to specific physiological stimuli. Challenges for the future include better elucidation of the unique biochemical and physiological properties of individual lipin family members and determination of lipin protein structure-function relationships. Further research may propel exploration of lipin proteins as viable therapeutic targets in metabolic or inflammatory disorders.
Keywords: obesity, lipodystrophy, rhabdomyolysis, triacylglycerol, phospholipid, autophagy, inflammasome, lipoprotein, chylomicron
Graphical Abstract

A deep understanding of the mechanisms that regulate cellular lipid storage and membrane content is necessary to elucidate human metabolic disorders such as obesity and fatty liver, and related diseases including type 2 diabetes and atherosclerosis. The mammalian lipin proteins (lipin 1, lipin 2, and lipin 3) are active in the synthesis of both the major storage lipid, triacylglycerol (TAG), and major membrane phospholipids (1, 2). The lipin proteins also regulate lipid intermediates that activate signaling nodes for diverse cellular processes (3). As a result, lipin proteins are involved in many cellular processes, including the regulation of lipid storage, lipoprotein synthesis, autophagy, inflammation, and gene expression. Here, we provide an overview of the mammalian lipins and their roles in normophysiology and disease. Due to space constraints, the reader is referred to other recent reviews on mammalian lipins for topics not covered here (3–6).
LIPIN PROTEINS AND GLYCEROLIPID SYNTHESIS
In most mammalian tissues, TAG is synthesized by the glycerol 3-phosphate pathway through the sequential action of acyltransferases and lipin phosphatidic acid phosphatases (PAPs) (1, 2) (Fig. 1). An alternative pathway for TAG synthesis is active primarily in the small intestine. This pathway utilizes monoacylglycerol (MAG), an abundant product of fat digestion, as the substrate for TAG synthesis (7, 8) (Fig. 1). As a component of the glycerol 3-phosphate pathway, lipin enzymes convert phosphatidic acid (PA) to diacylglycerol (DAG), which is a precursor of TAG as well as zwitterionic phospholipids (phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine) (Fig. 1). Unlike acyltransferases, lipins do not have transmembrane domains and, instead, move freely from the cytosol to membranes containing PA, such as the ER, mitochondria, and autophagosomes/lysosomes (6).
Fig. 1.
Pathways for glycerolipid synthesis. The glycerol 3-phosphate (G3P) pathway modifies a glycerol 3-phosphate backbone by the action of acyltransferases and lipin PAP to form TAG and phospholipids. With the MAG pathway, MGAT acts on dietary-derived MAG and fatty acids to synthesize TAG. GPAT, glycerol 3-phosphate acyltransferase; AGPAT, 1-acylglycerol-3-phosphate acyltransferase; DGAT, acyl-CoA:diacylglycerol acyltransferase; LPA, lysophosphatidic acid; zwitterionic phospholipids (PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine); anionic phospholipids (CL, cardiolipin; PI, phosphatidylinositol; PG, phosphoglycerate).
PAP activity was identified more than 60 years ago (9), but due to properties of the PAP enzymes, biochemical purification eluded investigators for decades. The isolation of the lipin genes was accomplished through positional cloning of a mouse mutation that impacted TAG synthesis in adipose tissue, leading to the identification of a novel gene family that is evolutionarily conserved from humans to yeast (10). Carman and colleagues purified yeast PA phosphohydrolase activity and discovered that its sequence matched the yeast ortholog of mammalian lipins, with the DxDxT motif located in a conserved protein domain that is required for enzymatic activity (11). Finck et al. (12) demonstrated that lipin 1 also interacts with DNA-bound transcription factors through an LxxIL motif to coactivate transcription. These interactions include lipin 1 coactivation of PPARγ coactivator 1α and hepatocyte nuclear factor 4α in hepatocytes to regulate fatty acid oxidation, and corepression of nuclear factor of activated T cells c4 to regulate inflammatory gene expression in adipocytes (13–15).
TISSUE DISTRIBUTION OF THE THREE MAMMALIAN LIPIN PROTEINS
The three mammalian lipin proteins each have PAP activity, although with somewhat differing specific activities (Vmax lipin 1 >> lipin 2 > lipin 3), and exhibit distinct tissue distributions (3, 16). Lipin 1 is the exclusive lipin in human and mouse skeletal muscle and is also prominently expressed in adipose tissue, peripheral nerve, and testis, and at lower levels in liver, small intestine, and many other tissues. Lipin 2 is the most abundant lipin in liver, and also has substantial expression in small intestine, macrophages, and specific brain regions. The lipin 3 expression pattern overlaps that of lipin 1 and lipin 2, with highest levels in small intestine, adipose tissue, and liver. These expression patterns raise the possibility of cooperation among lipin proteins within cells and/or redundancy of lipin function in some cell types.
GENETIC LIPIN DEFICIENCIES HAVE REVEALED CRITICAL PHYSIOLOGICAL ROLES
The pathologies of genetic lipin deficiencies in human and mouse are consistent with their tissue distributions. Mutations in human LPIN1 (encoding lipin 1) cause episodic childhood rhabdomyolysis, which can be fatal (17–21). Lipin 1-deficient mice develop muscle pathology when they are metabolically stressed (22), and also exhibit lipodystrophy (10). The lipodystrophy phenotype led to the delineation of an important role for lipin PAP activity in adipocyte differentiation and TAG synthesis (23, 24). A mouse model in which mature adipocytes express a Lpin1 allele that lacks PAP activity but retains coactivator function exhibits reduced adipose tissue TAG synthesis and lipolysis, which indicates a requirement for PAP activity in these functions (25). Interestingly, human lipin 1 deficiency is not characterized by lipodystrophy, and it appears that human lipin 1-deficient adipocytes exhibit compensatory mechanisms that are not present in mice, including lipin-independent TAG synthesis and reduced lipolysis of stored TAG (26).
Mutations affecting human lipin 2 cause Majeed syndrome, a rare autoinflammatory disorder mainly encountered in Middle Eastern countries (27–29). Lipin 2 mutations that inactivate PAP activity while leaving coactivator function intact are sufficient to cause Majeed syndrome (30). The disease manifests as mild dyserythropoietic anemia and episodic osteomyelitis, likely related to enhanced inflammasome activation in macrophages (discussed in a later section) (27, 29, 31). Lipin 2-deficient mice recapitulate mild anemia and have reduced trabecular bone density, but do not exhibit autoinflammation, likely due to absence of a trigger for these symptoms in mice housed in pathogen-free and environmentally controlled laboratory conditions (32). Compound deficiency for lipin 1 and lipin 2 causes embryonic lethality in the mouse (32).
Human LPIN3 loss-of-function mutations have not been identified, and lipin 3-deficient mice do not have a major metabolic phenotype (33). This suggests that loss of lipin 3 can be compensated by the other two family members. Studies of mice that are deficient in lipin 3 in combination with either lipin 1 or lipin 2 have revealed collaborations among the lipin proteins in various tissues. Lipin 3 has a role alongside lipin 1 in adipocyte differentiation and adipose TAG storage, such that lipin 1/lipin3-deficient mice exhibit lipodystrophy that is even more profound than that in lipin 1-deficient mice (33). Mice with compound lipin 2/lipin 3 deficiency have impaired intestinal lipid homeostasis (discussed in a later section).
A ROLE FOR LIPIN 1 IN AUTOPHAGY
As described above, lipin 1 mutations cause severe muscle dysfunction and destruction, and this has been shown to be associated with impaired PAP activity (21). Analysis of muscle ultrastructure in global or muscle-specific lipin 1-mutant mice revealed an accumulation of PA, dysfunctional mitochondria, and autophagosome accumulation, suggesting a defect in autophagy-mediated clearance of organelles (22, 34). Lipin 1 PAP activity (but not coactivator function) was required for the generation of mature autolysosomes that are responsible for the degradation of damaged organelles and proteins (22). Lipin 1 PAP activity generates DAG at the surface of autophagosomes/lysosomes, which activates a protein kinase D cascade that promotes fusion of autophagosomes with lysosomes to form functional autolysosomes (22) (see Graphical Abstract). Interestingly, statin drugs attenuate protein kinase D activity, such that the combination of lipin 1 haploinsufficiency and statin drug therapy impairs autophagic flux and leads to myopathy (17, 22). Thus, heterozygous LPIN1 mutations may be a risk factor for statin-related myopathy.
A ROLE FOR LIPIN 2 IN REGULATION OF THE INNATE IMMUNE RESPONSE
As described above, human lipin 2 deficiency causes the autoinflammatory disease, Majeed syndrome. The symptoms of this disorder are attenuated by drugs that block the cytokine interleukin (IL)-1β or the IL-1β receptor (29). This clue led Balboa and colleagues to identify a role for lipin 2 in the priming and activation of the NLRP3 inflammasome, a multi-protein complex that assembles in macrophages in response to pathogen exposure and leads to the secretion of IL-1β and IL-18 (31). NLRP3 priming in macrophages occurs in response to the activation of cell surface receptors (such as toll-like receptors) by pathogens or danger signals. Lipin 2 appears to limit the priming by affecting kinase signaling cascades and subsequent mRNA transcription of NLRP3 and IL-1β (31) (Graphical Abstract). Lipin 2 also influences the activation of the NLRP3 inflammasomes by maintaining cellular membrane cholesterol levels, which influence signaling through purinergic receptors, leading to inflammasome assembly and caspase-1 production. Caspase-1 processes the IL-1β pro-peptide to its mature active form, which is enhanced in Majeed syndrome. The role of lipin 2 in inflammasome activation is similar in macrophages isolated from mice and humans (31), suggesting that the muted inflammatory phenotype in lipin 2-deficient mice compared with humans may reflect the pathogen-free conditions that laboratory mice are raised in rather than a physiological difference between species. The role of lipin 2 in innate immune function also likely underlies the observed association of lipin 2 missense mutations with psoriasis, and suggests that lipin 2 activity should be considered in the evaluation of autoinflammatory conditions of unknown etiology (35).
Macrophages also express lipin 1. Recent studies suggest that lipin 1 may amplify inflammation by promoting macrophage production of the proinflammatory cytokine, IL-23, in response to toll-like receptor 4 activation (36). Lipin 1-deficient mice have reduced gut inflammation in experimentally induced colon cancer, which is associated with reduced infiltration of macrophages into colon tumors and reduced macrophage activation and PAP activity (37). Additionally, LPIN1 expression levels have prognostic value in the clinical course for some subtypes of colon cancer.
A ROLE FOR LIPIN 2 AND LIPIN 3 IN INTESTINAL LIPOPROTEIN SYNTHESIS
Lipin 2 and lipin 3 are expressed in a wide array of tissues, but in most cases, lipin 1 is also present and appears to be the dominant player owing to its higher PAP-specific activity. An exception is the small intestine, which expresses lipin 2 and lipin 3 without lipin 1 (16). The small intestine absorbs ∼95% of dietary TAGs, which are hydrolyzed to MAGs and free fatty acids in the intestinal lumen (7, 8). In enterocytes, these components are reassembled into TAGs, which may be transiently stored as lipid droplets or incorporated into chylomicron particles and secreted into the circulation through lymphatic ducts. It is estimated that 70–80% of intestinal TAG synthesis for chylomicron production is via the MAG acyltransferase (MGAT) pathway, and genetic ablation of Mogat2 (encoding mouse intestinal MGAT2) delays the rate of triglyceride appearance in the circulation after a meal (38). This raises a question of whether the enzymes of the glycerol 3-phosphate pathway serve a unique function in intestinal lipid metabolism or are a redundant mechanism for TAG synthesis.
We used mouse models to investigate the role of lipins 2 and 3 in small intestinal lipid metabolism (39). The lack of either lipin 2 or lipin 3 alone leads to a reduction in intestinal PAP activity, but has little effect on intestinal lipid homeostasis. However, deletion of both lipin 2 and lipin 3 leads to 25% reduced body weight on a chow diet and lethality on a high-fat diet within a few days. Lipin 2/3-deficient enterocytes incorporate dietary fatty acids into TAGs (presumably through the MGAT pathway), but tend to store the resulting TAGs in cytosolic lipid droplets rather than incorporate them into nascent chylomicron particles in the ER lumen. Lipin 2/3 PAP activity (but not coactivator activity) regulates enterocyte phospholipid levels and membrane composition, which influences the availability of TAGs in the ER lumen to associate with the key chylomicron protein (apolipoprotein B48). Lipin PAP activity may influence enterocyte phospholipid levels in at least two ways: 1) by converting PA to DAG, a precursor of phosphatidylcholine; and 2) by modulating PA levels, which indirectly regulate the levels of the key phosphatidylcholine biosynthetic enzyme [CTP:phosphocholine cytidylyltransferase α (CCTα)] (39) (Graphical Abstract). Chylomicron assembly in lipin 2/3-deficient enterocytes can be rescued by inhibiting CCTα activity to normalize cellular phospholipid levels. Thus, lipin 2/3 PAP activity in enterocytes hydrolyzes PA, regulates phospholipid synthesis, and promotes conditions that allow TAG transport into the ER lumen for chylomicron assembly.
MULTI-LEVEL REGULATION OF LIPINS
The involvement of lipins in important physiological processes has led to interest in understanding how lipin activity is regulated. Prior to the identification of the genes encoding lipins, it was known that PAP activity is regulated by fasting, glucocorticoids, and alcohol [reviewed in (1, 4, 5)]. We now know that the lipin genes are regulated by the glucocorticoid receptor, the sterol response element-binding protein 1, estrogen-related receptor α/γ, hepatocyte nuclear factor 4α, nuclear respiratory factor 1, and PPARγ coactivator 1α [reviewed in (1, 4, 5)]. Lpin1 expression is also regulated through the action of microRNAs, which may fine-tune lipin 1 expression levels in response to stimuli such as alcohol and lipopolysaccharide (5).
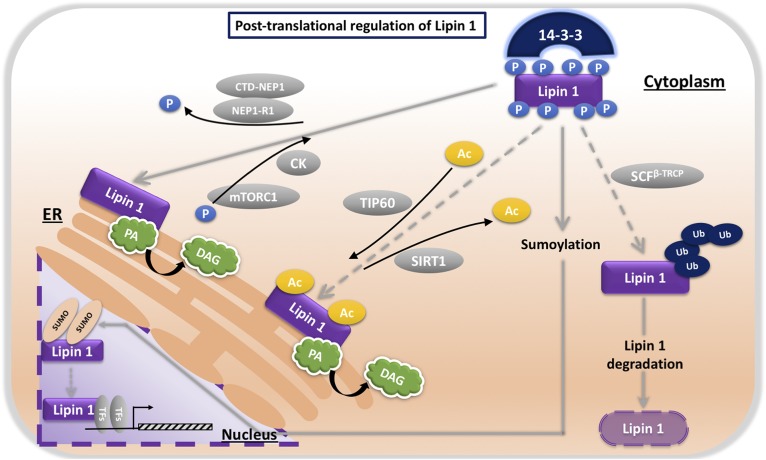
A key determinant of lipin PAP activity is posttranslational modification of lipin proteins. In mammalian lipins, posttranslational modifications include phosphorylation, sumoylation, acetylation, and ubiquitination (40–48) (Fig. 2). Modifications that promote lipin 1 translocation to the ER membrane (and membranes of other organelles) regulate PAP activity by allowing lipin protein access to its substrate, PA [reviewed in (6)]. Harris and colleagues have identified differences between lipin 1 and the other two lipin family members in posttranslational modification and subcellular localization (42, 49). While all three lipins are phosphorylated at multiple sites, only lipin 1 phosphorylation is regulated by insulin and mammalian target of rapamycin. Furthermore, whereas lipin 1 phosphorylation influences its membrane association, lipin 2 phosphorylation does not, and sequence differences in a polybasic domain underlie differences in phosphoregulation between lipin 1 and lipin 3. Understanding the properties of biochemical regulation of the three lipin family members may shed additional light on their individual physiological roles.
Fig. 2.
Posttranslational lipin protein modifications regulate subcellular localization and activity. Modifications that promote lipin 1 translocation to the ER membrane (and membranes of other organelles such as autophagosomes/lysosomes) regulate PAP activity by allowing lipin protein access to its substrate, PA. Insulin, mammalian target of rapamycin (mTOR), and casein kinase activities all lead to lipin 1 phosphorylation (40, 43, 48). Hyperphosphorylated lipin 1 associates with 14-3-3 proteins and is retained in the cytoplasm (45). The CTD-NEP/NEP1-R1 phosphatase complex dephosphorylates lipin 1 and promotes membrane association (46). Lipin 1 membrane association is also promoted by acetylation via the TIP60 acetyltransferase (47), and lipin 1 sumoylation enhances nuclear localization (44). Polyubiquitination primes lipin 1 for degradation (48). P, phospho group; Ac, acetyl group, Ub, ubiquitin; CK, casein kinase; mTORC1, mammalian target of rapamycin complex 1; SIRT1, sirtuin 1; SCFb-TRCP, SCF(Skp1/Cullin1/F-box protein) E3 ubiquitin ligase complex/F-box protein b-transducin repeat-containing protein; SUMO, sumoylation sites; TFs, transcription factors.
FUTURE PERSPECTIVES
In the years since molecular cloning of the lipin genes, we have learned that Mendelian mutations in two of the lipins cause severe diseases, and have delineated critical and unforeseen roles for lipin PAP regulation of lipid intermediates in protein and organelle quality control by autophagy, regulation of NLRP3 inflammasome activation, and lipoprotein assembly. Much remains to be discovered. One key area for future attention is the determination of lipin protein structure, which will shed additional light on lipin PAP and transcriptional activities. Another area that deserves attention is the further elucidation of the biochemical and physiological division of labor between the three lipin proteins. Finally, questions arise as to whether lipin proteins are viable therapeutic targets to modulate TAG accumulation in conditions such as obesity or fatty liver. Major challenges exist on this front. As illustrated by genetic lipin deficiencies, inhibition of lipin activity may be beneficial in some tissues but detrimental in others. Furthermore, because the dual activities of lipin proteins as PAP enzymes and transcriptional cofactors may oppose one another (i.e., PAP modulation of TAG synthesis vs. transcriptional activation of fatty acid oxidation), a global inhibition of lipin function will likely not be useful. Compounds that influence lipin protein subcellular localization may be a more promising approach, but further understanding of the regulation of lipin localization and activities is required.
Footnotes
Abbreviations:
- CCTα
- CTP:phosphocholine cytidylyltransferase α
- DAG
- diacylglycerol
- IL
- interleukin
- MAG
- monoacylglycerol
- MGAT
- monoacylglycerol acyltransferase
- PA
- phosphatidic acid
- PAP
- phosphatidic acid phosphatase
- TAG
- triacylglycerol
This work was supported by National Institutes of Health Grants P01 HL028481 (K.R.) and P01 HL090553 (K.R.), and American Heart Association Grant 18POST34060200 (H.W.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no conflicts of interest.
REFERENCES
- 1.Wang H., Airola M. V., and Reue K.. 2017. How lipid droplets “TAG” along: glycerolipid synthetic enzymes and lipid storage. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862: 1131–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleman R. A., and Mashek D. G.. 2011. Mammalian triacylglycerol metabolism: synthesis, lipolysis, and signaling. Chem. Rev. 111: 6359–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Csaki L. S., Dwyer J. R., Fong L. G., Tontonoz P., Young S. G., and Reue K.. 2013. Lipins, lipinopathies, and the modulation of cellular lipid storage and signaling. Prog. Lipid Res. 52: 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris T. E., and Finck B. N.. 2011. Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol. Metab. 22: 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi L., Jiang Z., and Zhou J.. 2015. The role of lipin-1 in the pathogenesis of alcoholic fatty liver. Alcohol Alcohol. 50: 146–151. [DOI] [PubMed] [Google Scholar]
- 6.Zhang P., and Reue K.. 2017. Lipin proteins and glycerolipid metabolism: roles at the ER membrane and beyond. Biochim. Biophys. Acta Biomembr. 1859: 1583–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan X., and Hussain M. M.. 2012. Gut triglyceride production. Biochim. Biophys. Acta. 1821: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yen C. L., Nelson D. W., and Yen M. I.. 2015. Intestinal triacylglycerol synthesis in fat absorption and systemic energy metabolism. J. Lipid Res. 56: 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith S. W., Weiss S. B., and Kennedy E. P.. 1957. The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 228: 915–922. [PubMed] [Google Scholar]
- 10.Péterfy M., Phan J., Xu P., and Reue K.. 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27: 121–124. [DOI] [PubMed] [Google Scholar]
- 11.Han G-S., Wu W-I., and Carman G. M.. 2006. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281: 9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finck B. N., Gropler M. C., Chen Z., Leone T. C., Croce M. A., Harris T. E., Lawrence J. C., and Kelly D. P.. 2006. Lipin 1 is an inducible amplifier of the hepatic PGC-1α/PPARα regulatory pathway. Cell Metab. 4: 199–210. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z., Gropler M. C., Norris J., Lawrence J. C. Jr., Harris T. E., and Finck B. N.. 2008. Alterations in hepatic metabolism in fld mice reveal a role for lipin 1 in regulating VLDL-triacylglyceride secretion. Arterioscler. Thromb. Vasc. Biol. 28: 1738–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z., Gropler M. C., Mitra M. S., and Finck B. N.. 2012. Complex interplay between the lipin 1 and the hepatocyte nuclear factor 4 α (HNF4α) pathways to regulate liver lipid metabolism. PLoS One. 7: e51320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H. B., Kumar A., Wang L., Liu G. H., Keller S. R., Lawrence J. C., Finck B. N., and Harris T. E.. 2010. Lipin 1 represses NFATc4 transcriptional activity in adipocytes to inhibit secretion of inflammatory factors. Mol. Cell. Biol. 30: 3126–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., and Reue K.. 2007. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282: 3450–3457. [DOI] [PubMed] [Google Scholar]
- 17.Zeharia A., Shaag A., Houtkooper R. H., Hindi T., de Lonlay P., Erez G., Hubert L., Saada A., de Keyzer Y., Eshel G., et al. 2008. Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am. J. Hum. Genet. 83: 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michot C., Hubert L., Brivet M., De Meirleir L., Valayannopoulos V., Müller-Felber W., Venkateswaran R., Ogier H., Desguerre I., Altuzarra C., et al. 2010. LPIN1 gene mutations: a major cause of severe rhabdomyolysis in early childhood. Hum. Mutat. 31: E1564–E1573. [DOI] [PubMed] [Google Scholar]
- 19.Michot C., Hubert L., Romero N. B., Gouda A., Mamoune A., Mathew S., Kirk E., Viollet L., Rahman S., Bekri S., et al. 2012. Study of LPIN1, LPIN2 and LPIN3 in rhabdomyolysis and exercise-induced myalgia. J. Inherit. Metab. Dis. 35: 1119–1128. [DOI] [PubMed] [Google Scholar]
- 20.Bergounioux J., Brassier A., Rambaud C., Bustarret O., Michot C., Hubert L., Arnoux J. B., Laquerriere A., Bekri S., Galene-Gromez S., et al. 2012. Fatal rhabdomyolysis in 2 children with LPIN1 mutations. J. Pediatr. 160: 1052–1054. [DOI] [PubMed] [Google Scholar]
- 21.Schweitzer G. G., Collier S. L., Chen Z., Eaton J. M., Connolly A. M., Bucelli R. C., Pestronk A., Harris T. E., and Finck B. N.. 2015. Rhabdomyolysis-associated mutations in human LPIN1 lead to loss of phosphatidic acid phosphohydrolase activity. JIMD Rep. 23: 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang P., Verity M. A., and Reue K.. 2014. Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab. 20: 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phan J., Péterfy M., and Reue K.. 2004. Lipin expression preceding peroxisome proliferator-activated receptor-γ is critical for adipogenesis in vivo and in vitro. J. Biol. Chem. 279: 29558–29564. [DOI] [PubMed] [Google Scholar]
- 24.Phan J., and Reue K.. 2005. Lipin, a lipodystrophy and obesity gene. Cell Metab. 1: 73–83. [DOI] [PubMed] [Google Scholar]
- 25.Mitra M. S., Chen Z., Ren H., Harris T. E., Chambers K. T., Hall A. M., Nadra K., Klein S., Chrast R., Su X., et al. 2013. Mice with an adipocyte-specific lipin 1 separation-of-function allele reveal unexpected roles for phosphatidic acid in metabolic regulation. Proc. Natl. Acad. Sci. USA. 110: 642–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Temprano A., Sembongi H., Han G-S., Sebastián D., Capellades J., Moreno C., Guardiola J., Wabitsch M., Richart C., Yanes O., et al. 2016. Redundant roles of the phosphatidate phosphatase family in triacylglycerol synthesis in human adipocytes. Diabetologia. 59: 1985–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferguson P. J., Chen S., Tayeh M. K., Ochoa L., Leal S. M., Pelet A., Munnich A., Lyonnet S., Majeed H. A., and El-Shanti H.. 2005. Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome). J. Med. Genet. 42: 551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Mosawi Z. S., Al-Saad K. K., Ijadi-Maghsoodi R., El-Shanti H. I., and Ferguson P. J.. 2007. A splice site mutation confirms the role of LPIN2 in Majeed syndrome. Arthritis Rheum. 56: 960–964. [DOI] [PubMed] [Google Scholar]
- 29.Herlin T., Fiirgaard B., Bjerre M., Kerndrup G., Hasle H., Bing X., and Ferguson P. J.. 2013. Efficacy of anti-IL-1 treatment in Majeed syndrome. Ann. Rheum. Dis. 72: 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donkor J., Zhang P., Wong S., O’Loughlin L., Dewald J., Kok B. P. C., Brindley D. N., and Reue K.. 2009. A conserved serine residue is required for the phosphatidate phosphatase activity but not the transcriptional coactivator functions of lipin-1 and lipin-2. J. Biol. Chem. 284: 29968–29978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lordén G., Sanjuán-García I., de Pablo N., Meana C., Alvarez-Miguel I., Pérez-García M. T., Pelegrín P., Balsinde J., and Balboa M. A.. 2017. Lipin-2 regulates NLRP3 inflammasome by affecting P2X 7 receptor activation. J. Exp. Med. 214: 511–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dwyer J. R., Donkor J., Zhang P., Csaki L. S., Vergnes L., Lee J. M., Dewald J., Brindley D. N., Atti E., Tetradis S., et al. 2012. Mouse lipin-1 and lipin-2 cooperate to maintain glycerolipid homeostasis in liver and aging cerebellum. Proc. Natl. Acad. Sci. USA. 109: E2486–E2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Csaki L. S., Dwyer J. R., Li X., Nguyen M. H. K., Dewald J., Brindley D. N., Lusis A. J., Yoshinaga Y., de Jong P., Fong L., et al. 2013. Lipin-1 and lipin-3 together determine adiposity in vivo. Mol. Metab. 3: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schweitzer G. G., Collier S. L., Chen Z., McCommis K. S., Pittman S. K., Yoshino J., Matkovich S. J., Hsu F. F., Chrast R., Eaton J. M., et al. 2019. Loss of lipin 1-mediated phosphatidic acid phosphohydrolase activity in muscle leads to skeletal myopathy in mice. FASEB J. 33: 652–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milhavet F., Cuisset L., Hoffman H. M., Slim R., El-Shanti H., Aksentijevich I., Lesage S., Waterham H., Wise C., Sarrauste de Menthiere C., et al. 2008. The infevers autoinflammatory mutation online registry: update with new genes and functions. Hum. Mutat. 29: 803–808. [DOI] [PubMed] [Google Scholar]
- 36.Meana C., Peña L., Lordén G., Esquinas E., Guijas C., Valdearcos M., Balsinde J., and Balboa M. A.. 2014. Lipin-1 integrates lipid synthesis with proinflammatory responses during TLR activation in macrophages. J. Immunol. 193: 4614–4622. [DOI] [PubMed] [Google Scholar]
- 37.Meana C., García-Rostán G., Peña L., Lordén G., Cubero Á., Orduña A., Győrffy B., Balsinde J., and Balboa M. A.. 2018. The phosphatidic acid phosphatase lipin-1 facilitates inflammation-driven colon carcinogenesis. JCI Insight. 3: 97506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen C-L. E., Cheong M-L., Grueter C., Zhou P., Moriwaki J., Wong J. S., Hubbard B., Marmor S., and Farese R. V.. 2009. Deficiency of the intestinal enzyme acyl CoA:monoacylglycerol acyltransferase-2 protects mice from metabolic disorders induced by high-fat feeding. Nat. Med. 15: 442–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang P., Csaki L. S., Ronquillo E., Baufeld L. J., Lin J. Y., Gutierrez A., Dwyer J. R., Brindley D. N., Fong L. G., Tontonoz P., et al. 2019. Lipin 2/3 phosphatidic acid phosphatases maintain phospholipid homeostasis to regulate chylomicron synthesis. J. Clin. Invest. 129: 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., and Lawrence J. C.. 2007. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282: 277–286. [DOI] [PubMed] [Google Scholar]
- 41.Eaton J. M., Mullins G. R., Brindley D. N., and Harris T. E.. 2013. Phosphorylation of lipin 1 and charge on the phosphatidic acid head group control its phosphatidic acid phosphatase activity and membrane association. J. Biol. Chem. 288: 9933–9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eaton J. M., Takkellapati S., Lawrence R. T., McQueeney K. E., Boroda S., Mullins G. R., Sherwood S. G., Finck B. N., Villen J., and Harris T. E.. 2014. Lipin 2 binds phosphatidic acid by the electrostatic hydrogen bond switch mechanism independent of phosphorylation. J. Biol. Chem. 289: 18055–18066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson T. R., Sengupta S. S., Harris T. E., Carmack A. E., Kang S. A., Balderas E., Guertin D. A., Madden K. L., Carpenter A. E., Finck B. N., et al. 2011. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 146: 408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu G-H., and Gerace L.. 2009. Sumoylation regulates nuclear localization of lipin-1α in neuronal cells. PLoS One. 4: e7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Péterfy M., Harris T. E., Fujita N., and Reue K.. 2010. Insulin-stimulated interaction with 14-3-3 promotes cytoplasmic localization of lipin-1 in adipocytes. J. Biol. Chem. 285: 3857–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han S., Bahmanyar S., Zhang P., Grishin N., Oegema K., Crooke R., Graham M., Reue K., Dixon J. E., and Goodman J. M.. 2012. Nuclear envelope phosphatase 1-regulatory subunit 1 (formerly TMEM188) is the metazoan Spo7p ortholog and functions in the lipin activation pathway. J. Biol. Chem. 287: 3123–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li T. Y., Song L., Sun Y., Li J., Yi C., Lam S. M., Xu D., Zhou L., Li X., Yang Y., et al. 2018. Tip60-mediated lipin 1 acetylation and ER translocation determine triacylglycerol synthesis rate. Nat. Commun. 9: 1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu K., Fukushima H., Ogura K., Lien E. C., Nihira N. T., Zhang J., North B. J., Guo A., Nagashima K., Nakagawa T., et al. 2017. The SCFβ-TRCP E3 ubiquitin ligase complex targets Lipin1 for ubiquitination and degradation to promote hepatic lipogenesis. Sci. Signal. 10: eaah4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boroda S., Takkellapati S., Lawrence R. T., Entwisle S. W., Pearson J. M., Granade M. E., Mullins G. R., Eaton J. M., Villén J., and Harris T. E.. 2017. The phosphatidic acid–binding, polybasic domain is responsible for the differences in the phosphoregulation of lipins 1 and 3. J. Biol. Chem. 292: 20481–20493. [DOI] [PMC free article] [PubMed] [Google Scholar]