ABSTRACT
The endoplasmic reticulum (ER) is a functionally and morphologically complex cellular organelle largely responsible for a variety of crucial functions, including protein folding, maturation and degradation. Furthermore, the ER plays an essential role in lipid biosynthesis, dynamic Ca2+ storage, and detoxification. Malfunctions in ER‐related processes are responsible for the genesis and progression of many diseases, such as heart failure, cancer, neurodegeneration and metabolic disorders. To fulfill many of its vital functions, the ER relies on a sufficient energy supply in the form of adenosine‐5′‐triphosphate (ATP), the main cellular energy source. Despite landmark discoveries and clarification of the functional principles of ER‐resident proteins and key ER‐related processes, the mechanism underlying ER ATP transport remains somewhat enigmatic. Here we summarize ER‐related ATP‐consuming processes and outline our knowledge about the nature and function of the ER energy supply.
Keywords: endoplasmic reticulum (ER), ATP, ATP transporter, secretory pathway, protein quality control, ER stress, unfolded protein response (UPR), ERAD
I. INTRODUCTION
In 1944, a ‘lace‐like reticulum’ appeared on electron micrographs taken by Keith Porter, Albert Claude, and Ernest Fullam, who were the first to describe the network‐resembling structure of the largest membrane‐bound organelle of the eukaryotic cell (Porter, Claude & Fullam, 1945). During the characterization of their discovery, Porter coined the name ‘endoplasmic reticulum’ (Porter & Kallman, 1952), referring to its localization in the central or ‘endoplasmic’ portions of the cytoplasm and its absence from the exterior parts of the ectoplasm. The endoplasmic reticulum (ER) can be found as flat cisternae as well as highly curved tubules comprising a single lumen enclosed by a single membrane (Friedman & Voeltz, 2011). This membrane is continuous with the outer membrane of the nucleus and thus links the two compartments – a connection which becomes evident when considering that a single set of ER proteins participates both in shaping the ER and in building the nuclear pore complex (Dawson et al., 2009). In general, contact sites between organelles are important in the coordination of cellular pathways and hence in the maintenance of cellular integrity. Whereas vesicle transport and cell signalling provide the opportunity for long‐distance conversations, the more direct interaction via membrane contact sites further promotes this interconnection. A well‐studied and remarkable example for interorganellar contact sites is provided by the so‐called mitochondria‐associated ER membranes (MAMs), which form a distinct subdomain of the smooth ER dedicated to the interaction with mitochondria. MAMs are thereby engaged in a variety of processes ranging from Ca2+ signalling and lipid metabolism to mitochondrial fission and inflammasome formation (Raturi & Simmen, 2013; Marchi, Patergnani & Pinton, 2014; Vance, 2014). The ER is divided into two major structural domains called the rough ER and the smooth ER, which can be further subdivided into functional membrane domains such as MAMs, ER quality‐control compartment (ERQC), ER exit sites (ERES) and plasma membrane‐associated membranes (PAMs) (Lynes & Simmen, 2011). The rough ER is defined by its association with ribosomes and is characterized by its involvement in the synthesis of secretory and membrane proteins and major co‐ and post‐translational modifications such as protein folding, glycosylation, secretion and degradation. The smooth ER, on the other hand, is concerned with lipid synthesis, carbohydrate metabolism and Ca2+ storage. Considering the diversity of its duties, any malfunction of the ER may have severe or even lethal effects on the cell. Defects in protein secretion or degradation, for example, lead to the accumulation of un‐ or misfolded proteins, causing so‐called ER storage diseases (Rutishauser & Spiess, 2002). Under normal circumstances ER quality control only allows correctly folded proteins to move on to the Golgi, whereas un‐ or misfolded proteins are targeted for degradation by the proteasome. Despite this efficient quality control system, unfavourable conditions can cause the accumulation of too much protein waste, which then triggers a series of signalling pathways collectively known as the unfolded protein response (UPR), leading to the activation of protective cellular mechanisms; when all of these measures fail, prolonged UPR signalling triggers apoptosis and the cells die (Rasheva & Domingos, 2009; Korennykh & Walter, 2012). Thus the ER not only provides a wide set of molecular chaperones and elaborate strategies to control protein folding but also a fallback plan (Bukau, Weissman & Horwich, 2006; Braakman & Bulleid, 2011; Gidalevitz, Stevens & Argon, 2013).
Notably, all of these processes are dependent on an energy supply in the form of ATP, and on the ATP consumed by protein phosphorylation. The first part of this review will describe the contribution of these processes to the energy demand of the ER with an attempt to identify those proteins which actually bind and hydrolyse ATP. The second part will then focus on the fueling of these processes with ATP. Remarkably, both the size and charge of the nucleotide, as well as the ER's inability to generate ATP by itself, make a specific mechanism for its transport into the lumen necessary. Yet little is known about ER ATP import (Hirschberg, Robbins & Abeijon, 1998; Csala et al., 2007) with only a few exceptions; these will be discussed critically, taking into consideration the existing knowledge of ER ATP dynamics as well as already identified ATP transporters in other organelles.
II. ATP‐CONSUMING PROCESSES IN THE ER
The fate of ATP within the ER is now explored by dissecting the functions of the ER (summarized in Fig. 1). Only those processes which take place within the ER lumen require ATP import, but for the sake of completeness ATP‐consuming events on the cytosolic side of the ER membrane are considered as well.
Figure 1.
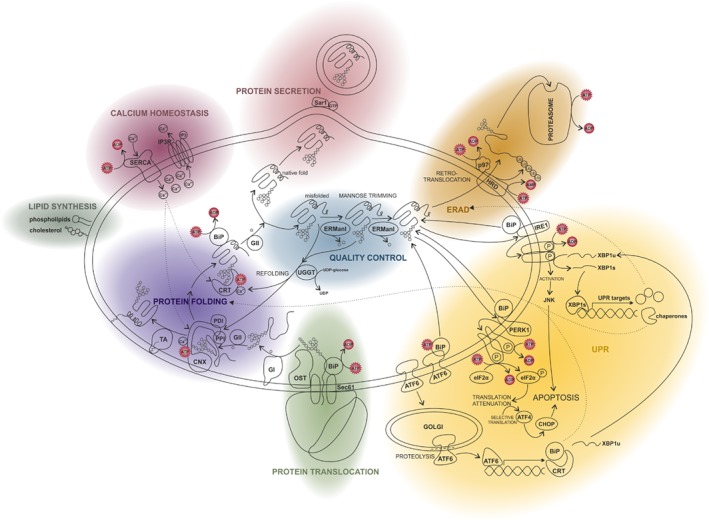
ATP‐consuming processes in the endoplasmic reticulum (ER). Schematic representation providing an overview of the most important ER‐associated processes and showing where ATP is required. Related functions are highlighted in the same colour. In green we follow protein translocation into the ER. Thereafter proteins are modified and folded (lilac). During protein quality control (blue) the folding state of the protein is monitored. Correctly folded proteins are secreted and transported to the Golgi (red). Unfolded and misfolded proteins are refolded. If all folding attempts fail, proteins undergo ER‐associated protein degradation (ERAD, orange). They are ubiquitinated and transferred into the cytosol via retrotranslocation, where they are degraded by the proteasome. The accumulation of misfolded proteins in the ER may also trigger the unfolded protein response (UPR, yellow). The UPR includes the activation of the kinases IRE1 and PERK1. IRE1 signalling results in the activation of the transcription factor XBP1 and the subsequent expression of UPR targets such as ER chaperones and ERAD factors. PERK1 causes translation inhibition by eIF2α phosphorylation leading to the successive expression of the transcription factors ATF4 and CHOP, which activates apoptosis‐promoting genes. The third important UPR mediator is the transcription factor ATF6 which induces the transcription of its target genes encoding for instance XBP1, BiP or calreticulin. Beside its central functions in protein synthesis, folding and degradation, the ER is also the most important Ca2+ store with an essential contribution to Ca2+ signalling events (magenta). Ca2+ is pumped into the ER by the ATPase SERCA and is primarily released via the IP3 receptor channel. Finally, the ER is mainly responsible for the biosynthesis of phospholipids as well as isoprenoids like cholesterol and steroid hormones (grey). ADP, adenosine diphosphate; ATF4, activating transcription factor 4; ATF6, activating transcription factor 6; ATP, adenosine triphosphate; BiP, binding immunoglobulin protein; CHOP, C/EBP homologous protein; CNX, calnexin; CRT, calreticulin; eIF2α, eukaryotic initiation factor 2α; ERManI, ER mannosidase I; GI, glucosidase I; GII, glucosidase II; GTP, guanosine triphosphate; HRD, ubiquitin‐protein ligase; IP3, inositol trisphosphate; IP3R, inositol triphosphate receptor; IRE1, inositol‐requiring enzyme 1; JNK, c‐Jun N‐terminal kinase; OST, oligosaccharyltransferase; P, indicates phosphorylation; PDI, protein disulfide isomerase; PERK1, proline‐rich receptor‐like protein kinase 1; PPI, prolyl peptidyl isomerase; Sar1, COPII‐associated small GTPase; Sec61, ER membrane protein translocator (translocon); SERCA, sarco/endoplasmic reticulum Ca2+‐ATPase; TA, GPI transamidase; UGGT, UDP‐glucose glycoprotein glucosyltransferase; UPR, unfolded protein response; XBP1, x‐box binding protein 1.
(1). Protein synthesis and the secretory pathway
Protein synthesis requires energy and the demand for ATP as an energy source is most pronounced at the transcription stage. However, the first ER‐related step of protein synthesis, which is the translation of membrane and secretory proteins by ER‐bound ribosomes, is driven by GTP hydrolysis. Therefore the ER ATP pool is not affected significantly by protein synthesis until the translocation stage.
The process of translocation involves the insertion of secretory and membrane proteins into the ER and has been reviewed previously (Rapoport, 2007; Dudek et al., 2015). Estimates suggest that about a third of the eukaryotic proteome takes the secretory pathway (Kanapin et al., 2003). To summarize, translocation starts with the binding of the signal recognition particle (SRP) to the hydrophobic N‐terminal signal sequence of the nascent peptide at the ribosome (von Heijne, 1983, 1984, 1986). In general, the SRP complex then directs the translating ribosomes to the translocon, where the peptide is inserted into a channel comprised of the heterotrimeric Sec61 complex. During translocation, the signal peptide complex cleaves the signal peptide, a reaction independent of ATP (Weihofen et al., 2002). The translocation is completed by the polypeptide chain binding protein BiP (binding immunoglobulin protein), a luminal ATP binding member of the heat shock protein 70 (HSP70) family of chaperones (Flynn et al., 1991; Lyman & Schekman, 1997; Tyedmers et al., 2003). Binding and hydrolysis of ATP by BiP is considered to provide the main driving force for protein insertion into the ER (Glick, 1995; Brodsky, 1996; Dudek et al., 2015). BiP is involved in both the gating of the channel and the completion of translocation (Dierks et al., 1996; Hamman, Hendershot & Johnson, 1998; Haigh & Johnson, 2002; Alder et al., 2005; Schäuble et al., 2012). Further chaperones like glucose‐regulated protein 94 (Grp94), which belongs to the Hsp90 family, then also assist in protein folding in an ATP‐dependent manner (see also Section II.2). Hsp70 chaperones commonly depend on two types of co‐chaperones, namely J‐domain proteins of the Hsp40 family which stimulate their ATPase activity, and nucleotide exchange factors (NEFs) which allow transition from the ADP‐ to the ATP‐bound state. In the case of BiP, Grp170 [also known as oxygen‐regulated protein 150 (Orp150) or hypoxia up‐regulated protein 1 (HYOI1)] and the BiP‐associated protein, BAP, also known as Sil1 serve as NEFs and thus control its activity via its so‐called ATPase cycle (Behnke, Feige & Hendershot, 2015; Bracher & Verghese, 2015). During this cycle, Hsp70 chaperones exist in an ATP‐bound state with a low affinity for their substrate protein and a high‐affinity ADP‐bound state (Mayer & Bukau, 2005). Thus it would seem natural that protein translocation must be somehow coupled to ATP import.
Beyond its role in translocation, BiP and other Hsp70 proteins are also involved in protein folding under normal conditions or during stress when faced with the accumulation of un‐ or misfolded proteins which have to be kept soluble for degradation (Shiber & Ravid, 2014; Clerico et al., 2015). Intriguingly, BiP and other protein‐folding factors are the most abundant ER luminal proteins, which is truly indicative of the ER's dedication to this process (Gidalevitz et al., 2013).
Protein translocation is also supported by a number of auxiliary components such as the membrane‐integrated translocating chain‐associated membrane (TRAM) protein (Görlich et al., 1992; Voigt et al., 1996; Hegde et al., 1998), the translocon‐associated protein (TRAP) which contains a transmembrane region and a luminal domain located directly below the channel (Ménétret et al., 2000, 2008; Fons, Bogert & Hegde, 2003), and the Sec62/Sec63 complex (Meyer et al., 2000; Tyedmers et al., 2003; Conti et al., 2015). The latter functions as a co‐chaperone for BiP by stimulating its ATPase activity via the J‐domain of the Hsp40 family member Sec63 (Misselwitz, Staeck & Rapoport, 1998; Rapoport, 2007). Another co‐chaperone of BiP is the Sec63‐related Hsp40 chaperone ER‐resident J‐domain protein 1 (ERj1), which again harbours a J‐domain for the interaction with BiP (Blau et al., 2005; Benedix et al., 2010). ERj1 can also associate with the ribosome and Sec61 and in this way contributes to translation regulation (Blau et al., 2005; Benedix et al., 2010). Hence ERj1 is also referred to as ribosome‐associated membrane protein (RAMP), similarly to RAMP4, also known as stress‐associated endoplasmic reticulum protein 1 (SERP1) which participates in translocation as well (Schröder et al., 1999; Yamaguchi et al., 1999). Other factors related to translocation are protein associated with the ER translocon (PAT‐10) (Meacock et al., 2002) and (palmitoylated) calnexin (Lakkaraju et al., 2012).
The membrane protein calnexin and its soluble paralogue calreticulin are both Ca2+‐dependent ER‐resident chaperones with a lectin site which enables them to bind to glycosylated proteins during and after translocation to prevent their aggregation (Molinari et al., 2004; Williams, 2006). Both proteins seem to constitute the core element of a protein‐folding complex. They interact with the ER thiol oxidoreductase ERp57, a protein disulfide isomerase (PDI) which facilitates the formation of disulfide bonds within glycoproteins, as well as with the peptidyl prolyl isomerase (PPI) cyclophilin B. Together with Hsp70 and Hsp90 chaperones, the ER‐located PDIs (Wilkinson & Gilbert, 2004; Galligan & Petersen, 2012) and PPIs (Schiene‐Fischer, 2015) are critical for the folding process. After disulfide formation, the PDI's active centre is reduced and has to be oxidized to restore its activity, which is achieved by the thiol oxidase Ero1. Returning to calnexin and calreticulin, their activity is not only regulated via Ca2+ binding but also by way of switching between an ATP‐ and an ADP‐bound state. In the case of calreticulin, a very weak ATPase activity of the chaperone itself has been demonstrated, which however might be stimulated by co‐chaperones in a similar fashion to the stimulation of the ATPase activity of Hsp70 proteins by Hsp40 co‐chaperones (Saito et al., 1999; Wijeyesakere et al., 2015). Calnexin, on the other hand, undergoes conformational changes upon ATP binding, which might also have regulatory consequences (Ou et al., 1995). Thus ATP is probably required by both calnexin and calreticulin, albeit their direct or indirect contribution to the overall ATP consumption within the ER is not well characterized.
The recognition of proteins by calnexin and calreticulin depends on their glycosylation. Glycosylation is achieved by the Sec61 neighbouring oligosaccharyltransferase (OST) complex, usually in a co‐translational manner (Kelleher & Gilmore, 2006; Pfeffer et al., 2014). The glycosylation state of glycoproteins allows the surveillance of their folding progress. The OST catalyses the transfer of a preassembled glucose3‐mannose9‐N‐acetylglucosamine2 oligosaccharide from dolichol pyrophosphate onto asparagine residues of nascent proteins. During maturation, the oligosaccharide is then trimmed by glucosidase I and II starting with the two terminal glucose residues. The remaining glucose1‐mannose9‐N‐acetylglucosamine2 oligosaccharide is recognized by calnexin or calreticulin which favour proper protein folding. Next, the final glucose residue is also removed. At this point the UDP–glucose glycoprotein glucosyltransferase (UGGT) assumes control. As another protein with chaperone function, UGGT is capable of sensing the folding state of a protein. When they are not correctly folded, UGGT attaches a glucose residue to the N‐terminal glycan. This enables the protein's reassociation with calnexin or calreticulin and thus, ideally, its correct refolding. The repeated association of calnexin or calreticulin and UGGT is described as the calnexin/calreticulin cycle and contributes crucially to glycoprotein folding (Helenius & Aebi, 2004; Caramelo & Parodi, 2008). As soon as proteins reach their native state they exit this cycle and continue to pursue the secretory pathway. However, if the folding process is not successful, proteins are degraded by the ER‐associated protein degradation (ERAD) pathway (see Section II.2b ).
The oligosaccharide donor dolichol pyrophosphate provides a lipid anchor which allows the proper positioning of the oligosaccharide at the ER membrane next to the OST complex. The biosynthesis of dolichol comes within the mevalonate pathway where isopentenyl diphosphate (IPP) molecules are produced (Ferguson, Durr & Rudney, 1959). An ER‐resident isoprenyltransferase then catalyses a condensation reaction between varying numbers of IPPs with farnesyl diphosphate (FPP) to build polyprenyl diphosphate, which is further converted to dolichol in several reaction steps (Crick, Rush & Waechter, 1991; Schenk, Fernandez & Waechter, 2001a; Schenk et al., 2001b). The oligosaccharide is assembled by the sequential addition of monosaccharides (mannose, N‐acetylglucosamine or glucose) to dolichol pyrophosphate by glycosyltransferases, which takes place on the cytosolic and the luminal side of the ER membrane (Kelleher & Gilmore, 2006). While ATP is used during the mevalonate pathway, it does not seem to be required for the activity of the OST in the ER.
About 10–20% of all membrane proteins that take the secretory pathway are additionally modified within the ER by the C‐terminal attachment of a glycosylphosphatidylinositol (GPI) anchor (Orlean & Menon, 2007; Kinoshita & Fujita, 2016). In an analogous mode to the dolichol oligosaccharide, the GPI anchor is preassembled by ER‐membrane and luminal enzymes. The assembly pathway is initialized at the cytosolic face of the ER and is completed in the lumen after the seemingly ATP‐independent flipping of the anchor to the other side (Vishwakarma & Menon, 2005). After completion, the GPI anchor is finally transferred to proteins with the respective target sequence by the GPI transamidase complex.
Once the ER‐located folding machinery has successfully completed its work, proteins are packed into coat protein complex II (COPII)‐coated vesicles which are transported to the Golgi (D'Arcangelo, Stahmer & Miller, 2013; Saito & Katada, 2015). The formation of vesicles starts with the assembly of coat proteins on the cytosolic side of the ER membrane, which causes membrane curvature and formation of a vesicle bud (Barlowe et al., 1994). This process and the subsequent packing of the cargo are directed by the GTPase secretion‐associated RAS‐related protein 1(Sar1) in cooperation with the guanosine exchange factor (GEF) Sec12 and the GTPase‐activating protein (GAP) Sec23 (Barlowe & Schekman, 1993; Antonny et al., 2001). GTP binding by Sar1 initializes coat formation while GTP hydrolysis is required for effective cargo sorting and vesicle fission (Bielli et al., 2005). The cargo proteins either passively diffuse into the forming vesicle bud or are actively recruited by the adaptor protein Sec24 to be incorporated into the nascent vesicle (Miller et al., 2002, 2003).
Vesicle formation takes place at specific sites of the ER which are termed transitional ER or ERESs and can be defined by the presence of the Sec16 protein (Watson et al., 2006; Hughes et al., 2009). Targeting to and fusion with the Golgi is mediated by Rab (Ras‐related in brain) proteins and soluble N‐ethylmaleimide‐sensitive‐factor attachment receptor (SNARE) complex formation (Allan, Moyer & Balch, 2000; Moyer, Allan & Balch, 2001; Ortiz Sandoval & Simmen, 2012). The secretory pathway will not be discussed in more detail here as our focus is the ER, but more information can be found, for example, in a comprehensive review by Barlowe & Miller (2013).
(2). Protein quality control and ER stress
The accumulation of un‐ or misfolded proteins as a result of disturbing events like mutant protein expression, nutrient deprivation, viral infection or intoxication commonly causes ER stress, which is sensed by a specialized ER‐associated machinery. Misfolded proteins may be refolded or, if that fails, exported into the cytosol by a mechanism called retrotranslocation, followed by their degradation by the proteasome. This process, referred to as ERAD, together with the UPR, are considered to be the most important mechanisms of protein quality control activated upon ER stress (Bukau et al., 2006).
(a). The unfolded protein response (UPR)
Several signalling pathways are responsible for the surveillance of the ER folding machinery. The components of these pathways sense the accumulation of un‐ or misfolded proteins and induce a series of different response mechanisms which increase the ER folding capacity and are collectively described as the UPR (Schröder & Kaufman, 2005; Rasheva & Domingos, 2009; Korennykh & Walter, 2012). In mammals, the UPR is initiated by at least three ER transmembrane proteins, the kinases inositol‐requiring enzyme 1 (IRE1) (Chen & Brandizzi, 2013) and proline‐rich receptor‐like protein kinase 1 (PERK1) as well as the transcription factor activating transcription factor 6 (ATF6).
IRE1 is able to sense ER folding capacity via its luminal domain and transmits the signal to its cytosolic kinase and ribonuclease domains. ER stress induces the oligomerization of IRE1 proteins, leading to activation of the IRE1 kinase. As a consequence, the ribonuclease domain is activated and cleaves the messenger RNA (mRNA) of a basic leucine zipper (b‐Zip) transcription factor [Hac1 in yeast, X‐box binding protein 1(XBP1) in metazoans] (Yoshida et al., 2001). The cleavage allows the translation of the active form of XBP1/Hac1, which then induces the expression of UPR targets such as ER chaperones and ERAD factors. The second UPR‐connected kinase is PERK1. PERK1 harbours a luminal domain highly similar to that of IRE1, suggesting a similar sensing mechanism. However, it is not known with certainty how IRE1 and PERK1 monitor ER stress. There is evidence that BiP inactivates them by binding to their luminal domain; accumulated un‐ or misfolded proteins may then titrate BiP from this binding site, leading to the activation of IRE1 and PERK1 (Oikawa et al., 2009, 2012; Pincus et al., 2010). Although the role of BiP is not understood completely, two factors that could influence its dissociation from IRE1 and PERK1 are the availability of unfolded protein targets, and manipulation of its ATPase cycle by nucleotide levels or cofactors (Amin‐Wetzel et al., 2017). On the other hand, it is also possible that stress‐sensor activation occurs directly upon binding of un‐ or misfolded proteins to their luminal domain, which indeed contains a hydrophobic peptide binding domain (Credle et al., 2005; Gardner & Walter, 2011). Unlike that of IRE1, the cytosolic domain of PERK1 contains a eukaryotic initiation factor 2α (eIF2α) kinase, implicating a mechanism for translation regulation (Shi et al., 1998, 1999; Harding, Zhang & Ron, 1999; Harding et al., 2000; Marciniak et al., 2006): i.e. phosphorylation of eIF2α leads to its inactivation and hence inhibits translation initiation. The inhibition of translation makes sense as a consequence of accumulated misfolded proteins since the production of new proteins would only contribute to the aggravation of the situation by occupying the ER folding machinery. The third important mediator of the UPR, ATF6, is delivered to the Golgi upon ER stress (Haze et al., 1999; Wang et al., 2000). For that to happen, BiP has to dissociate from ATF6 to uncover its Golgi localization signal (Shen et al., 2002, 2005). The mechanism behind BiP dissociation from ATF6 is not clear, but one possibility is that ER stress actively dissociates BiP from ATF6 by activating the BiP ATPase cycle (Shen et al., 2005). In the Golgi, ATF6 is cleaved by resident proteases, which results in the release of the transcription factor domain from the transmembrane domain. The transcription factor domain then enters the nucleus and activates its target genes, encoding for instance XBP1, BiP or calreticulin. Since BiP activity is controlled by its ATPase cycle, its involvement in the sensing mechanism of all three UPR mediators entails the need for ATP during the initiating steps of the UPR. However, the signalling events themselves are not likely to consume notable amounts of ATP within the ER, even though IRE1 and PERK1 require ATP at the cytosolic face of the ER membrane for their kinase activity. In addition, UPR signalling ultimately results in increased production of ATP‐consuming chaperones, while translation, in general, is reduced, suggesting a reduced level of ATP hydrolysis for that purpose. Considering this, the ATP consumption rate might vary according to the phase of the UPR.
(b). ER‐associated protein degradation (ERAD)
Besides the UPR signalling events, which contribute to the restoration of regular protein folding, the removal of accumulated protein waste itself is accomplished by another set of processes involved in ER protein control, ERAD (Meusser et al., 2005; Hirsch et al., 2009). Briefly, ERAD starts with the recognition of un‐ or misfolded proteins, mainly by chaperones of the Hsp70 family. They are then targeted to membrane‐embedded ubiquitin ligases (Carvalho, Goder & Rapoport, 2006) such as the yeast ERAD‐associated E3 ubiquitin‐protein ligase (Hrd1) also known as degradation in the endoplasmic reticulum protein 1 (Der1) ligase (Bays et al., 2001; Deak & Wolf, 2001; Mehnert, Sommer & Jarosch, 2014) or its human homologues HRD1 (Kaneko et al., 2002; Nadav et al., 2003) and glycoprotein 78 (gp78) (Fang et al., 2001). After their dislocation to the cytosolic side of the ER membrane, proteins are ubiquitinated by ubiquitin ligase complexes, a reaction which requires energy in the form of ATP at the ER membrane. Ubiquitinated proteins are then released into the cytosol, which again requires ATP hydrolysis at the ER membrane for the activity of the cytosolic ATPase associated with diverse cellular activities (AAA‐ATPase) p97, referred to as cell division protein 48 (Cdc48) in yeast. The latter converts the energy released by ATP hydrolysis into the mechanical force needed for pulling proteins out of the membrane (Ye, Meyer & Rapoport, 2001). In the cytosol, proteins are degraded by the proteasome, which requires ATP hydrolysis as well. Here, ATP hydrolysis is not actually spatially linked to the ER but closely coupled with an ER process in a temporal manner (Amm, Sommer & Wolf, 2014).
An essential step during ERAD is the export of proteins into the cytosol, which requires the crossing of the ER membrane and is referred to as retrotranslocation or dislocation. To date, the identity of a retrotranslocation channel has not been determined with certainty (Hampton & Sommer, 2012). It has been suggested that the Sec61 translocation channel is directly involved in both retrotranslocation as well as translocation (Wiertz et al., 1996; Pilon, Schekman & Römisch, 1997; Plemper et al., 1997; Zhou & Schekman, 1999). On the other hand, the HRD ligase complex, which is already responsible for substrate recognition and ubiquitination, may also provide the conduit for retrotranslocation. In this context, there is also evidence to suggest that the HRD‐associated complex composed of Derlin‐1, the ATPase p97, and VCP‐interacting membrane protein (VIMP) mediates retrotranslocation (Lilley & Ploegh, 2004; Ye et al., 2004; Lilley & Ploegh, 2005). Furthermore, an alternative model was proposed by Ploegh (2007) suggesting that lipid droplet formation was involved in the removal of misfolded proteins from the ER.
The recognition of misfolded proteins relies on the N‐linked glycan which is attached to most secretory proteins in the course of translocation (Moremen & Molinari, 2006; Lederkremer, 2009; Määttänen et al., 2010; Yoshida & Tanaka, 2010). Within the ER all three glucose residues of the oligosaccharide are removed unless the protein cannot be successfully folded. Proteins that are not successfully folded repeatedly associate with the lectin‐like chaperones calnexin or calreticulin to allow refolding. However, if the folding process takes too long, a timer mechanism involving the ER mannosidase I (ERManI) and mannosidase like proteins (EDEMs) initiates ERAD (Hosokawa et al., 2001; Herscovics, Romero & Tremblay, 2002; Molinari et al., 2003; Oda et al., 2003; Olivari & Molinari, 2007). After a certain residence time in the ER, ERManI removes the terminal mannose residue of all glycosylated proteins, which serves as the mechanism for escaping further folding attempts (Frenkel et al., 2003). With no mannose left, UGGT can no longer add glucose to the oligosaccharide, which obstructs the association of misfolded proteins with calnexin or calreticulin and probably targets them for degradation. Hence proteins will be degraded if they are not properly folded within a certain time frame. Notably, ERAD processes appear to be preferentially localized to certain regions within the ER – the so‐called ER‐derived quality control compartment (ERQC) (Kamhi‐Nesher et al., 2001). In this pericentriolar compartment ERAD substrates as well as calnexin and calreticulin, PDIs, UGGT, and ERManI, but not BiP are accumulated (Frenkel et al., 2004; Avezov et al., 2008; Benyair et al., 2015). This compartmentalization may also contribute to the regulation of ERAD. Another recognition mechanism engages other ER‐resident as well as cytosolic chaperones (Nishikawa, Brodsky & Nakatsukasa, 2005; Brodsky, 2007). Among the most prominent examples is BiP, which seems to participate in all stages of protein maturation and degradation (Otero, Lizák & Hendershot, 2010), supported by other Hsp70 chaperones as well as Hsp90 chaperones such as Grp94 (Christianson et al., 2008, 2012; Eletto, Dersh & Argon, 2010) and PDIs (Feige & Hendershot, 2011; Suzuki & Schmitt, 2015).
(c). ER stress‐induced apoptosis
If all attempts to eliminate ER stress fail, the continued accumulation of misfolded proteins induces cell death by apoptosis (Rasheva & Domingos, 2009). The exact mechanism by which ER stress regulates apoptosis is not well known, but it seems that apoptosis follows sustained PERK1 but not IRE1 signalling (Lin et al., 2009). PERK1‐induced phosphorylation of eIF2α prevents the synthesis of most proteins. However, translation of the transcription factor ATF4, immune to this inhibition, is selectively increased (Harding et al., 2000; Lu, Harding & Ron, 2004; Vattem & Wek, 2004). ATF4 now induces the expression of C/EBP homologous protein (CHOP), another transcription factor, which evidently promotes apoptosis in response to ER malfunction (Zinszner et al., 1998; Fawcett et al., 1999). Apoptosis induction is achieved at least in part by the CHOP‐mediated upregulation of the pro‐apoptotic factor Bim and the downregulation of the anti‐apoptotic B‐cell lymphoma 2 (Bcl2) (McCullough et al., 2001; Puthalakath et al., 2007). Furthermore, the expression of the ER thiol oxidase Ero1 (see also Section II.1) is increased; Ero1 normally promotes protein folding by disulfide formation, but also has pro‐apoptotic functions probably by producing reactive oxygen species (ROS) during this reaction (Marciniak et al., 2004). However, IRE1 signalling may also contribute to apoptosis induction via activation of the c‐Jun N‐terminal kinases (JNK) pathway (Urano et al., 2000). And finally, Ca2+ release from the ER and its transfer into mitochondria are considered to mediate apoptosis induction in response to ER stress. This process involves, for instance, the ER Ca2+‐ATPase S1T, a truncated version of the ER Ca2+ pump sarco/endoplasmic reticulum Ca2+‐ATPase (SERCA1) (see also Section II.3), which is induced via the PERK1‐eIF2α‐ATF4 pathway (Chami et al., 2008). The activity of S1T, which is localized at ER–mitochondria contact sites, leads to ER Ca2+ depletion and increases mitochondria contact sites with the overall result of increased Ca2+ transfer from the ER to mitochondria. The strong accumulation of Ca2+ in the mitochondria subsequently induces apoptosis via the opening of the mitochondrial permeability transition pore (PTP) (Pizzo & Pozzan, 2007; Pinton et al., 2008).
(3). Ca2+ homeostasis
The role of the ER as the main Ca2+ store determines its importance for all processes dependent on Ca2+ signalling, like protein folding by the Ca2+‐dependent chaperones calreticulin and calnexin or the induction of UPR and apoptosis as described above and reviewed by Krebs, Agellon & Michalak (2015). The release of Ca2+ from the stores, on the other hand, is mediated by inositol 1,4,5‐trisphosphate (InsP3) or ryanodine receptor channels. A potential regulatory connection of cellular ATP levels to ER Ca2+ homeostasis is that the InsP3 receptor channel‐opening probability is increased by high cytosolic ATP concentrations (Betzenhauser et al., 2008). The rapid mobilization of Ca2+ by InsP3 and ryanodine receptors is followed by a phase of slow entry of extracellular Ca2+ (Clapham, 2007). Depleted ER stores then have to be replenished, which is accomplished by a renewed entry of extracellular Ca2+. The second Ca2+ entry phase furthermore provides a more long‐lasting signal. Refilling of the ER is achieved by the SERCA, a P‐type ATPase, which uses ATP to pump Ca2+ against the ion concentration gradient. It should be pointed out here that owing to the role of Ca2+ as a regulator of ER‐associated processes, changing Ca2+ levels within the ER may somehow correlate with ATP concentration and transport.
(4). Lipid biosynthesis
The ER can be considered a highly productive compartment as it is crucially involved in the supply of the cell with various molecular components such as proteins, as described above. In addition, lipid synthesis also takes place largely in the (smooth) ER, including the synthesis of phospholipids (Vance, 2015) and isoprenoids such as cholesterol and steroid hormones (Holstein & Hohl, 2004).
During phospholipid synthesis, first phosphatidic acid is produced from glycerol‐3‐phosphate or dihydroxyacetone‐phosphate by the action of acyltransferases (Vance, 2015). Phosphatidic acid is then converted to diacylglycerol by phosphatidic acid phosphatase‐1 or reacts with CTP to form CDP‐diacylglycerol catalysed by CDP‐diacylglycerol synthase. The energy for the transfer reactions is provided by the use of acyl‐coenzyme A (acyl‐CoA) as the fatty acid donor. Another step in phospholipid synthesis is the formation of the headgroups and their transfer onto phosphatidic acid. Phosphatidylcholine (PC), for example, is formed via the so‐called Kennedy pathway (Kennedy & Weiss, 1956). After choline has been imported into the cell it is phosphorylated by the cytosolic choline kinase followed by the generation of CDP‐choline by the CTP:phosphocholine cytidylyltransferase which is located in the nucleus and cytosol. Finally, an ER membrane transferase catalyses the transfer of CDP‐choline to diacylglycerol to result in PC. The synthesis of phosphatidylethanolamine (PE) is analogous to PC synthesis starting with the phosphorylation of ethanolamine by a cytosolic kinase followed by the conversion into CDP‐ethanolamine by the CTP:phosphoethanolamine cytidylyltransferase and the transfer onto diacylglycerol. The second pathway for PE synthesis includes the mitochondrial phosphatidylserine (PS) decarboxylase which decarboxylates PS to PE (Borkenhagen, Kennedy & Fielding, 1961). PC and PE can now be converted into PS by replacing the choline (PS synthase‐1) or ethanolamine (PS synthase‐2) with serine. Both PS synthases are enriched in MAMs (Stone & Vance, 2000). In similar fashion the production of phosphatidylinositol (PI) and phosphoglycerol and its derivatives cardiolipin and lyso(bis)phosphatidic acid are mainly attributed to ER and secondarily to mitochondrial membranes. In summary, although the enzymes required for the production of phospholipids are primarily found within the ER membrane or activated there, the ER ATP pool is probably not affected significantly by phospholipid synthesis. On the other hand, the activation of fatty acids to acyl‐CoA as well as the phosphorylation of choline and ethanolamine require ATP outside of the lumen.
Since the ER is the main location of phospholipid synthesis, their transport to other membranes is another task for the cell. The mechanism of phospholipid transfer remains rather enigmatic. Vance (2015) discusses three possible mechanisms: transport through the cytosol via carrier proteins, transport in phospholipid vesicles, or direct exchange via membrane contact sites such as MAMs. However, since very little is known regarding this mechanism, the impact on energy consumption likewise remains obscure.
Isoprenoid synthesis first requires the formation of IPP by the mevalonate pathway (Ferguson et al., 1959), which, as already mentioned, takes place in the cytosol (see also Section II.1). Three molecules of IPP condense to form farnesyl diphosphate, which then reacts with a second molecule of farnesyl diphosphate to form squalene which is inserted into the ER membrane. This reaction is catalysed by farnesyl‐diphosphate farnesyltransferase (squalene synthase), an enzyme located in the ER membrane. The two‐step conversion of squalene to lanosterol and all further 19 reactions required to form cholesterol are also driven by ER membrane enzymes (Gaylor, 2002). Notably, cholesterol biosynthesis as well as the conversion of cholesterol into steroid hormones, which also mostly takes place in the ER, does not require ATP within the ER but rather coenzymes such as nicotinamide adenine dinucleotide (NAD), nicotinamide adenine dinucleotide phosphate (NADP) and flavin adenine dinucleotide (FAD) since most reactions fall into the category of redox reactions.
(5). ATP‐consuming ER transporters
The ER as an enclosed compartment has to provide entry and exit opportunities for molecules which cannot diffuse through the lipid bilayer. The transport of proteins into and out of the ER in the course of translocation and retrotranslocation was described in Section II.1. However, the wide range of ER‐resident enzymes necessitates the import of substrates and cofactors, such as ATP or NAD(P)(H), into the ER and the export of their products out of the ER. Thus transporters are essential for the maintenance of ER‐associated processes. An overview of ER transporters is given by Csala et al. (2007). Here we consider only ATP‐consuming transporters, while the ATP transporter itself will be discussed in Section III.1.
The presentation of peptide antigens by major histocompatibility complex class I (MHC‐I) molecules on the cell surface is a vital mechanism of the immune system (Oliveira & van Hall, 2015). The antigen‐processing pathway starts with the cleavage of proteins by the proteasome. Peptide fragments are then imported into the ER via the transporter associated with antigen processing (TAP). In the ER they are trimmed by ER aminopeptidases 1 and 2 (ERAP1/2) and then loaded onto MHC‐I molecules by the peptide‐loading complex (PLC) (Koch & Tampé, 2006). This process is assisted by chaperones including BiP, calnexin, calreticulin, the transmembrane protein tapasin as well as the oxidoreductase ERp57 (see also Section II.1). Finally, the MHC‐I antigen complex is exported from the ER and transported by vesicle transport via the trans‐Golgi to the cell surface, where the antigen is presented. The heterodimeric TAP transporter is a member of the ATP‐binding cassette (ABC) family (Ritz & Seliger, 2001; Abele & Tampé, 2011). Binding of a peptide induces ATP hydrolysis at the cytosolic ATP‐binding domains, which drives its import into the ER. Furthermore, luminal ATP is required by chaperones involved in MHC‐I loading, and hence also concerned with the regulation of this process (Wijeyesakere et al., 2015).
III. THE ELUSIVE ER ATP TRANSPORTER
All of the above‐described essential ER‐dependent cellular processes determine the energy demand of the ER and the requirement for ATP import. The magnitude of this demand, however, is subject to variations in the dynamics of these processes. Thus ATP import into the ER not only requires a means of transport but will also be subject to regulatory events. However, in mammalian cells to date, neither has the ATP transport mechanism been identified nor have ATP dynamics within the ER been accurately described. We now attempt to summarize past endeavours to identify an ER ATP transporter as well as providing possible avenues for future studies.
(1). A presumed ER ATP transport protein – characterization of ATP transport into rat liver ER
In the early 1990s, promising results regarding the transport of ATP into the ER formed the foundation for the elucidation of the underlying mechanism. Clairmont, Maio & Hirschberg (1992) demonstrated the translocation of ATP into rough ER‐derived vesicles isolated from rat liver. Because of the many ATP‐binding and ATP‐consuming proteins present in the vesicle membranes, a system based on reconstituted ER proteoliposomes was established to minimize the consumption of ATP during transport assays (see also Section III.7 and Fig. 2A). For this reason, proteins from rat liver ER membranes were reconstituted into phosphatidylcholine liposomes, which were then employed to study the import of ATP into their lumen (Guillén & Hirschberg, 1995). Transport was highly specific, temperature dependent and saturable and could be prevented by the addition of 4,4′‐diisothiocyano‐2,2′‐stilbenedisulfonic acid (DIDS), a known inhibitor of anion exchangers, but not by atractyloside, which inhibits the mitochondrial ATP/ADP translocase (ANT).
Figure 2.
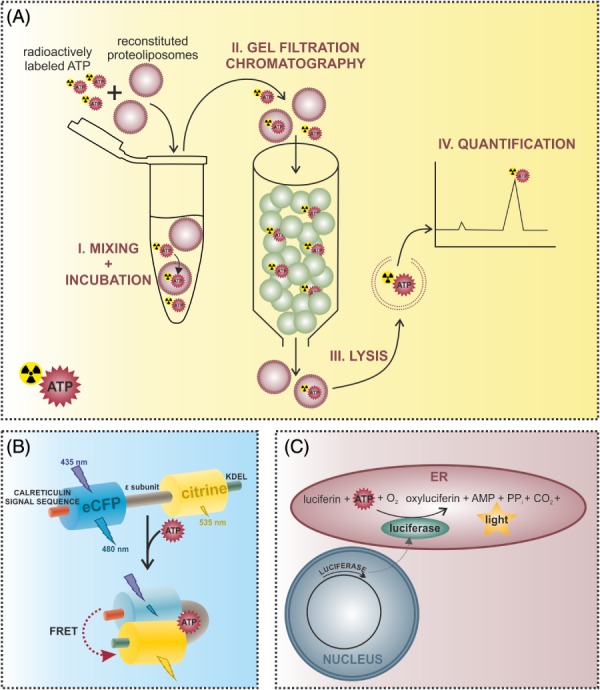
Methods for the investigation of endoplasmic reticulum (ER) ATP dynamics. (A) ATP transport into ER‐derived vesicles or reconstituted liposomes containing ER proteins can be measured using an in vitro approach. Proteoliposomes are incubated with radioactively labeled ATP and then applied to a gel filtration column. Free ATP binds to the matrix, while proteoliposomes and incorporated ATP are eluted in the void volume. Proteoliposomes are then lysed to determine the amount of imported ATP via high‐performance liquid chromatography (HPLC) or liquid scintillation counting. (B) Alterations of ER ATP levels can be measured using genetically encoded fluorescent ATP sensors targeted to the ER. The ERAT4.01 probe for example is a Förster/fluorescence resonance energy transfer (FRET)‐based sensor consisting of two fluorescent proteins – an enhanced cyan fluorescent protein (CFP) and the yellow fluorescent protein (YFP) variant citrine – flanking the ATP binding ϵ‐subunit of the FoF1‐ATP synthase from Bacillus subtilis (Imamura et al., 2009; Vishnu et al., 2014). The N‐terminal fusion of the calreticulin signal sequence and the C‐terminal addition of the ER retention signal with the amino acid sequence KDEL (lysine–aspartic acid–glutamic acid–leucine) allow the efficient targeting of the probe to the ER. Binding of ATP to the ϵ‐subunit causes a conformational change of the sensor, which increases the FRET signal intensity. (C) Alternatively, ER ATP levels can be determined using an ER‐targeted heterologously expressed firefly luciferase, which catalyses the reaction of oxyluciferin to luciferin, resulting in light emission. Since this reaction is ATP dependent, the intensity of the emitted light is proportional to the ATP concentration.
Similarly to Clairmont et al. (1992), vesicles derived from rat liver rough ER were used by Kim, Shin & Park (1996) in an attempt to identify the presumed ATP transporter using photoaffinity labelling. For this purpose, a photoreactive azido derivative of ATP, 3'‐O‐(p‐azidobenzoyl)‐ATP (AB‐ATP), was synthesized. Rough ER vesicles then were incubated with [γ‐32P]AB‐ATP under varying conditions. UV radiation was applied to photolyse [γ‐32P]AB‐ATP, thereby converting it into a highly reactive form which permanently binds to and labels associated proteins. For the ER‐derived vesicles, only one protein with a molecular weight of 56 kDa was labelled, and this was declared an ATP transporter candidate. The protein was protected from photoaffinity labelling by adding non‐radioactive ATP but not GTP, demonstrating the specificity of the label for ATP binding proteins. As also reported by Guillén & Hirschberg (1995) for reconstituted ER proteoliposomes, the labelling efficiency was reduced by DIDS, but not affected by atractyloside (Kim et al., 1996), suggesting that the same possible transporter was observed in both cases. In further experiments, solubilized rough ER proteins were fractionated using DEAE chromatography and each fraction was photoaffinity labelled and reconstituted in liposomes to test transport activity. As expected the active fraction contained the photolabeled 56 kDa protein (Kim et al., 1996).
Finally, Shin et al. (2000) reported the characterization of a rat liver ER ATP transporter in reconstituted proteoliposomes. They found that ATP transport was strongly cis‐inhibited by ADP and also weakly by AMP and that preincubation of the proteoliposomes with ADP increased the ATP transport activity. Altogether their results suggest an ATP/ADP antiport mechanism in the ER ATP transporter.
(2). Is membrane trafficking involved in ATP transport? The phosphoinositide phosphatase Sac1 is connected with ER ATP import in yeast
Mayinger & Meyer (1993) described an ATP transport system of vesicles from yeast ER, which was found to be saturable and could be inhibited by DIDS and N‐ethylmaleimide. For the characterization of this system, ER membrane proteins were reconstituted into proteoliposomes to measure ATP transport (see also Section III.7 and Fig. 2A) revealing that ATP uptake was dependent on a counter substrate (ATP or ADP) in the lumen of the vesicle (Mayinger, Bankaitis & Meyer, 1995). It was therefore suggested that ATP import in vivo was linked to ADP export, implying the existence of an antiport mechanism. Protein extracts of the proteoliposome membrane were then purified with hydroxyapatite, commonly used for the purification of membrane carrier proteins. The purified fractions were again reconstituted into proteoliposomes to measure ATP transport. The fractions capable of ATP transport were repeatedly subfractionated by chromatography on hydroxyapatite and affinity chromatography with ATP agarose, resulting in a distinct enrichment of the specific ATP transport activity. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of the ultimate fraction allowed the identification of a single protein, namely the phosphatidylinositide phosphatase Sac1, which was thus considered to be involved in ATP transport. It was already known that the SAC1 gene was genetically linked with ACT1 (Novick, Osmond & Botstein, 1989) and SEC14 (Cleves, Novick & Bankaitis, 1989) suggesting its involvement in cytoskeletal organization and the secretory pathway, respectively. The transmembrane protein Sac1 localizes to the Golgi and ER and contributes to the Golgi's secretory function. Its connection with Sec14, which is a phospholipid transfer protein (Bankaitis et al., 1990), and the discovery of a phosphatidylinositol phosphate (PtdInsP) phosphatase domain within Sac1, homologous to the mammalian inositol‐5‐phosphatase synaptojanin (McPherson et al., 1996), allowed more exact placement of Sac1 in phospholipid metabolism. This domain is conserved from yeast to human and was designated the SAC1‐like domain (Guo et al., 1999). The functional association of a phosphoinositide phosphatase with the ER and Golgi complex may help us to understand the observed characteristics of Sac1 described above. Phosphoinositides are key players in signal transduction at the plasma membrane as well as the regulation of the actin cytoskeleton, membrane trafficking, and transport. Their precursor form phosphatidylinositol is synthesized in the ER and transferred into other organelles via transfer proteins or vesicular transport (Di Paolo & De Camilli, 2006). There are seven different phosphoinositides which are distributed in a characteristic way in a subset of membranes, e.g. phosphatidylinositol 4‐phosphate [PI(4)P] is enriched in the Golgi complex whereas the inositol ring is mostly not phosphorylated in the ER (Balla, 2013). The localization of phosphoinositides correlates with their phosphorylation state which creates a platform for regulatory events through alterations by phosphatases or kinases. Deletions of SAC1 lead to a massive accumulation of PI(4)P because it cannot be dephosphorylated (Guo et al., 1999). This could explain how the phosphatase activity of Sac1 may be involved in the coordination of membrane trafficking between the Golgi and ER, where the dephosphorylation of PI(4)P may occur during retrograde transport from the Golgi to the ER (Di Paolo & De Camilli, 2006; Mayinger, 2012). It might even be possible that such events play a part in ER ATP transport. Further details concerning the role of Sac1 in the ER were revealed by analysis of its contribution to ER ATP transport (Kochendörfer et al., 1999). By measuring the transport capacity of isolated microsomal membranes from a Δsac1 strain compared to wild‐type cells it was shown that Sac1 is required for ER ATP transport. When purified Sac1 protein was added to reconstituted proteoliposomes from Δsac1 cells, their ability to transport ATP was reestablished. Furthermore, overexpression of Sac1 caused increased ATP uptake. All these observations reinforced the impression that Sac1 contributes in an essential way to the transport of ATP into the ER. However, proteoliposomes containing only Sac1 were not capable of ATP transport, which means that Sac1 itself is not an ATP transporter but rather a cofactor or regulator. In further experiments Kochendörfer et al. (1999) demonstrated that Δsac1 cells exhibit an ER folding defect and constitutive activation of the UPR. It is very likely that these effects are due to reduced luminal ATP levels in Δsac1 cells, interfering dramatically with the functionality of ATP‐dependent chaperones. Sac1 has also been found to play a significant role in non‐vesicular lipid exchange between the ER and Golgi that may take place at ER/Golgi contact sites. In this case, Sac1 is speculated to help maintain a steep PI(4)P gradient from the Golgi to the ER by dephosphorylating PI(4)P in the ER. This gradient may then be used in a counter‐transport mechanism to move sterols against their gradient from the ER to the Golgi (Del Bel & Brill, 2018). However, how Sac1's roles in vesicle transport and non‐vesicular transport might be mechanistically related to ATP import into the ER remains an unsolved mystery.
Intriguingly, another yeast protein, morphogenesis checkpoint‐dependent protein 4 (Mcd4), has been connected to ATP transport and membrane trafficking (Zhong, Malhotra & Guidotti, 2003). Mcd4, which is found in intracellular membranes, is known to be required for ethanolamine phosphate transfer into the ER during GPI anchor synthesis (see Section II.1). However, Mcd4 is also able to trigger extracellular ATP release in a process mediated via the membrane trafficking pathway (Zhong et al., 2003). This process also involves the transport of ATP into the Golgi and the contribution of the vacuolar H+‐ATPase. As in the case of the Sac1 protein described above, these observations support a speculated connection between membrane trafficking and ATP transport. Moreover, the ER might also serve as an intermediate storage compartment for ATP for release into the extracellular space. A possible ATP transporter for vesicles has been identified in the form of solute carrier family 17 member 9 (SLC17A9) (Sawada et al., 2008). It is possible that this transporter also takes part in the transport of ATP via vesicles.
(3). An actual ER ATP transporter – the discovery of the Arabidopsis thaliana ER ATP/ADP‐transporter ER‐ANT1
Leroch et al. (2008) reported the discovery of an ER ATP transporter in the plant Arabidopsis thaliana. Phylogenetic analysis of the A. thaliana genome revealed that the candidate ER adenine nucleotide transporter ER‐ANT1 belongs to the family of mitochondrial carriers (MCF) forming a monophyletic group with mitochondrial ATP/ADP carriers (AACs) (Picault et al., 2004) (locus tag At5g17400). Accordingly, ER‐ANT1 features a high sequence similarity of about 60% with AAC1, AAC2, and AAC3, albeit lacking the putative N‐terminal transit peptide required for mitochondrial targeting (Murcha, Millar & Whelan, 2005; Leroch et al., 2008). Heterologous expression of ER‐ANT1 in Escherichia coli, which involved integration of the protein in the cytoplasmic membrane, was utilized to examine its ability to transport ATP (Leroch et al., 2008). The expression of ER‐ANT1 allowed the uptake of radiolabelled ATP and ADP by E. coli cells with a high specificity similar to the mitochondrial isoform AAC1 from A. thaliana. However, the transport activity of ER‐ANT1 could neither be inhibited by a number of known AAC1 inhibitors nor by inhibitors of plastidic or peroxisomal adenine nucleotide transport but was inhibited by N‐ethylmaleimide, which does not affect the mitochondrial transporter. Further experiments revealed that ER‐ANT1 also mediates the efflux of ATP and ADP, which led to a model of transport involving the exchange of the two nucleotides. In addition to the transport studies in E. coli, the A. thaliana plant itself was used to demonstrate the localization of ER‐ANT1 in the ER via cell fractionation and immunoblotting as well as electron microscopy following immunogold labelling. The expression levels of ER‐ANT1 were tested using the β‐glucuronidase (GUS) reporter system and the results showed elevated promotor activities in tissues with highly active ER metabolism, such as pollen, root tips and seeds. Leroch et al. (2008) also examined the physiological role of the ATP transporter by studying ER‐ANT1‐knockout plants. Here the knockout caused a severe growth defect and impaired root and seed development. Moreover, the expression of ER‐resident chaperones was analysed. Intriguingly, the absence of ER‐ANT1 accompanied decreased levels of the luminal binding proteins BiP1, BiP2, and BiP3 as well as calreticulin1 and calreticulin2; mRNA levels of other ATP‐dependent ER proteins were also reduced. These data emphasize the importance of ATP transport into the ER and should encourage the search for a possible mammalian counterpart, which perhaps may be one of several uncharacterized AAC‐like proteins.
(4). Characterization of ATP dynamics within the ER lumen
Independent of the mode of ATP transport into the ER, several attempts have been made to characterize this process in a variety of circumstances. Certain observations may help in the design of new strategies for the elucidation of the mechanisms of ER ATP import. For example, ER‐targeted genetically encoded ATP sensors now allow the monitoring of ATP dynamics in live cells (Vishnu et al., 2014) (see also Section III.7 and Fig. 2B). This approach has revealed that ATP levels within the ER are related to Ca2+ levels in that a reduction of Ca2+ in the ER causes a significant elevation of ATP levels. Further application of such sensors will certainly reveal other, similar connections.
(5). A general perspective on ER transport
A broader view of ER transport might contribute to a better understanding of the ER membrane's barrier function and how to overcome it. The import of nucleotide sugars, ATP and nucleotide sulfate into the ER and Golgi was reviewed by Hirschberg et al. (1998). A more recent review by Csala et al. (2007) summarizes the transport of compounds such as glucose, nucleotide sugars and oligopeptides such as glutathione across the ER membrane. As in the case of ATP, the identification of transporters for other substrates has proved difficult, and it has often only been possible to provide a functional description of transport activities. Most strategies have involved measuring transport into reconstituted proteoliposomes containing ER proteins (see also Section III.7 and Fig. 2A). Among the known transporters are those of the solute carrier (SLC37) family of sugar phosphate/phosphate exchangers (Chou & Mansfield, 2014). This family includes, among others, the SLC37A4 glucose‐6‐phosphate transporter which was found via sequence homology with a bacterial transporter for phosphate esters (Gerin et al., 1997). The search for transporters can be extended using bioinformatics. Another solute carrier family, SLC35, contains nucleotide sugar transporters resident in the ER and Golgi (Ishida & Kawakita, 2003). Nucleotide sugars are required for the glycosylation of proteins in the ER and Golgi where they are transported by an antiport mechanism in exchange for the corresponding nucleotide monophosphate, e.g. UMP for UDP‐glucose. The overall importance of SLC transporters becomes evident when considering that around 400 human transporter genes have been classified in 52 SLC families (Hediger et al., 2003). It is highly likely that an ER ATP transporter will be identified among the SLC proteins with unknown function. Indeed, as described above (Section III.3) the A. thaliana ER ATP transporter shows homology with members of the mitochondrial carrier family (AAC), which is denoted SLC25 in the human system. Alternatively, a specific ATP transport protein might not be necessary, since ER‐derived vesicles are permeable to various compounds (Le Gall, Neuhof & Rapoport, 2004). A reasonable explanation for this permeability may be found in the translocon channel (Sec61 complex) which enables the co‐ and post‐translational transport of proteins into the ER (Osborne, Rapoport & van den Berg, 2005; Gogala et al., 2014; Dudek et al., 2015; Pfeffer et al., 2015) but may also allow the rather non‐specific passage of other molecules (Csala et al., 2007). Intriguingly, a recent study in yeast, shows that glutathione can enter the ER via facilitated diffusion through Sec61, along a concentration gradient (Ponsero et al., 2017). To return to ER ATP transport, it seems plausible then that ATP could diffuse through the translocon channel during protein translocation. Furthermore, the coupling of protein and ATP import would make sense, since protein folding requires an ATP supply. Since ATP concentrations are significantly lower in the ER compared to the cytosol and mitochondria (Depaoli et al., 2018), a concentration gradient could be the driving force for ATP import.
(6). ATP transport through anion channels
Since ATP is required in all compartments of the cell, ATP transport mechanisms are also needed. The mitochondrial ATP/ADP‐translocase is described in Section III.3 and a likely vesicular ATP transporter in Section III.2. Regarding the transport of ATP into the Golgi there is some evidence that ATP import can be mediated by Golgi anion transporters, which are mainly involved with the transport of Cl− into the lumen (Thompson et al., 2006). Similarly, anion channels of the cardiac sarcoplasmic reticulum (SR) were shown to conduct ATP and other adenine nucleotides (Kawano et al., 1999). Nevertheless, the permeability or conduction of ATP through anion channels does not necessarily imply that ATP can be transported in this way in live cells.
(7). Methods for the investigation of ER ATP dynamics
Studies involving the measurement of ATP transport into reconstituted proteoliposomes have utilized an approach involving radioactively labelled ATP and size‐exclusion chromatography (Fig. 2A) (Mayinger & Meyer, 1993; Guillén & Hirschberg, 1995). First, proteoliposomes are incubated with radiolabelled ATP. The transport reaction is then stopped by loading the mixture onto a size‐exclusion column (Dovex, Sephadex). ATP dissolved in the incubation medium is incorporated into the matrix, while proteoliposomes containing imported ATP are eluted in the void volume. Thus free ATP is separated from ATP found inside the proteoliposomes. The respective ATP concentrations can be measured via high‐performance liquid chromatography (HPLC) or liquid scintillation counting and compared to determine the transport velocity.
ATP transport will also be reflected in the change in ATP levels within the ER compared to other cellular compartments and the cytosol, and might thus be indirectly observed when measuring ATP concentrations in living cells. Of course, ATP levels will be affected not only by transport but also by ATP‐consuming and ATP‐producing processes. To monitor ATP dynamics, genetically encoded ATP sensors containing fluorescent proteins have been developed. A major advance in this field was the development of Förster/fluorescence resonance energy transfer (FRET)‐based indicators for ATP (‘ATeams’) consisting of a cyan and a yellow fluorescent protein linked via the ϵ‐subunit of the FoF1‐ATP synthase from Bacillus subtilis (Fig. 2B) (Imamura et al., 2009). The binding of ATP to the ϵ‐subunit causes a conformational change within the protein, resulting in increased FRET. An ER‐targeted sensor derived from an ATeam protein was constructed by Vishnu et al. (2014). For ER targeting, the signal sequence of the ER chaperone calreticulin was fused to its N‐terminus and the ER retention signal composed of the amino acid sequence – lysine (K), aspartic acid (D), glutamic acid (E), and leucine (L) – (KDEL) to its C‐terminus (Pelham, 1990; Clairmont et al., 1992). Another ATP probe was developed that used only a single fluorescent protein, circularly permuted enhanced green fluorescent protein (cpEGFP), inserted between two α‐helices of the bacterial FoF1‐ATP synthase ϵ‐subunit (Yaginuma et al., 2014). This ratiometric indicator with the descriptive name ‘quantitative evaluator of cellular energy’ (Queen) undergoes quantifiable changes in its excitation spectrum in response to ATP binding. In an analogous manner to the ATeam sensor, the ER targeting of Queen should be possible. A further fluorescent reporter was found to be suitable for monitoring ATP/ADP ratios (Berg, Hung & Yellen, 2009; Tantama et al., 2013).
Intracellular ATP content can also be measured using another phenomenon based upon photon emission: chemiluminescence. The most prominent example is firefly luciferase which uses the energy of ATP to catalyse the light‐producing reaction of luciferin with O2 to produce oxyluciferin (Fig. 2C) (DeLuca & McElroy, 1974). The intensity of emitted light is proportional to ATP concentration if the other substrates are present in excess. Purified luciferase can be added together with luciferin to permeabilized cells or isolated mitochondria to measure ATP levels (Manfredi et al., 2002). However, the heterologous expression of recombinant luciferase also allows a live‐cell approach (Ainscow & Rutter, 2002; Bell et al., 2007), and intracellular ATP levels can be described in more detail by targeting luciferase to different compartments (Dorner & Kaufman, 1994; Jouaville et al., 1999; Kennedy et al., 1999). Applications of the luciferase system were reviewed by Fan & Wood (2007) and Patergnani et al. (2014).
Measurements of the total content of ATP, ADP and AMP within the cell may also help investigators to draw conclusions about the origin of changes in ATP levels. A convenient method is HPLC, which allows the simultaneous quantification of all nucleotide species. The use of HPLC in this way is reviewed by Manfredi et al. (2002) who also outline correct sample handling, which is required for successful analysis to avoid the degradation of ATP by ATPases in the sample.
(8). The source of ER ATP
ATP is generated during glycolysis and oxidative phosphorylation. The contribution of different metabolic pathways to ATP production may vary in different cell types and under different conditions including ER stress, cell cycle, aging or carcinogenesis. In the case of cancer cells, which have an altered energy metabolism, it is tempting to speculate that there may also be abnormalities in the ER ATP supply. In general, cancer cells produce most of their ATP via high rates of aerobic glycolysis [the ‘Warburg effect’; reviewed by Liberti & Locasale, 2016], while normal cells have lower rates of glycolysis and preferentially use oxidative phosphorylation for ATP generation. Accordingly, the source of ER ATP and perhaps also the import mechanism might vary depending on the metabolic state of the cell. It seems likely that, in cells with high rates of mitochondrial respiration, ATP generated within the mitochondria is directly transferred into the ER, possibly via MAM contact sites, while glycolytic ATP might rather enter from the cytosol.
IV. CONCLUSIONS
(1) The adequate supply of the ER with ATP is important for its functions in protein synthesis and turnover, functions that underlie the role of the ER in cell growth. ATP is produced during glycolysis in the cytosol and oxidative phosphorylation in mitochondria; the ER itself is not capable of ATP generation. Thus ATP utilized by the ER must be transported there by a still‐unknown mechanism.
(2) Possible ER ATP import routes are summarized in Fig. 3. The idea that a specific ATP transporter is involved is plausible, especially given that the mitochondrial ATP/ADP‐translocase enables the transfer of ATP from its place of origin within the mitochondria to other cell compartments. However, equating the ER with mitochondria is probably not prudent, since these organelles are quite different in their architecture, function, and origin (Archibald, 2015). However, the ER is related to the Golgi: both are part of the endomembrane system (Dacks & Field, 2007). The fact that a specific ATP transporter has not been identified in either of these two compartments may indicate that no such transporter is present.
Figure 3.
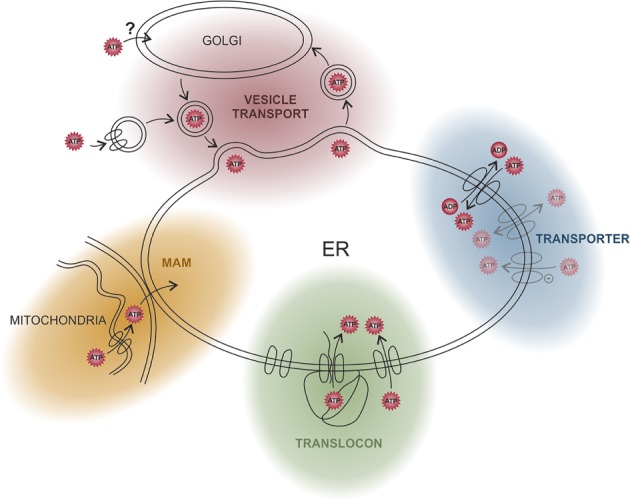
Possible ATP transport routes into the endoplasmic reticulum (ER). It is not known how ATP is transported into the ER, but several possible import pathways are shown. Experiments with rat liver microsomes provided some evidence for a specific ATP transporter and an ATP/ADP antiport mechanism. ATP may also be transported non‐specifically through anion channels. The ER membrane may also be ‘leaky’ for ATP, allowing its passive transport driven by a concentration gradient. One such source of leakiness may be translocon channels, which are widely distributed across the membrane of the rough ER; ATP could enter the ER concomitantly with the inserted peptide or by a translocon‐associated mechanism. Since ATP is mainly produced in mitochondria it seems reasonable that it is directly transported from there into the ER via membrane contact sites (mitochondria‐associated ER membranes; MAMs). Finally, the dynamics of the membrane structures evoked by membrane trafficking via vesicles might also afford the opportunity for ATP uptake.
(3) The dynamics of the ER or Golgi membrane structures during membrane trafficking events might also afford the opportunity for ATP uptake.
(4) Membrane contact sites such as mitochondria‐associated ER membranes (MAMs) could serve as a platform for ATP transfer, possibly even directly from mitochondria into the ER.
(5) ATP hydrolysis likely follows ATP import rapidly and thus an ATP concentration gradient could provide a driving force for passive diffusion through transient membrane breaks (see point 3 above), or proteinaceous channels. Indeed, ATP concentrations within the ER are considerably lower than in the cytosol and mitochondria. Passive transport could be allowed by the leakiness of the ER, caused by the great number of translocon channels spanning the membrane. However, even if the ER is not leaky for ATP, the translocon might still be associated with ATP import in a translocon‐coupled mechanism which would allow the coordination of ATP supply with the energy‐consuming processes of protein translocation and folding.
(6) Irrespective of which of these mechanisms (and perhaps several are involved) are actually responsible for ATP import into the ER, this review aims to stimulate discussion that has the potential to yield a wealth of new insights into molecular cell biology. We look forward to future developments in this field.
V. ACKNOWLEDGMENTS
The authors acknowledge the scientific advisory board of Next Generation Fluorescence Imaging (NGFI) GmbH (http://www.ngfi.eu/). The research of the authors was funded by the Ph.D. program Molecular Medicine (MOLMED) of the Medical University of Graz, by Nikon Austria within the Nikon Center of Excellence, Graz, the FWF (Austrian Science Fund: P28529‐B27 and I3716‐B27 to R.M.) and the doctoral program Metabolic and Cardiovascular Disease (DK‐W1226). The Nikon Center of Excellence, Graz, is supported by the Austrian infrastructure program 2013/2014, Nikon Austria Inc., and BioTechMed, Graz. We thank Kimberly A. Swanson for editing and proofreading.
VI. REFERENCES
- Abele, R. & Tampé, R. (2011). The TAP translocation machinery in adaptive immunity and viral escape mechanisms. Essays in Biochemistry 50, 249–264. [DOI] [PubMed] [Google Scholar]
- Ainscow, E. K. & Rutter, G. A. (2002). Glucose‐stimulated oscillations in free cytosolic ATP concentration imaged in single islet β‐cells evidence for a Ca2+‐dependent mechanism. Diabetes 51, S162–S170. [DOI] [PubMed] [Google Scholar]
- Alder, N. N. , Shen, Y. , Brodsky, J. L. , Hendershot, L. M. & Johnson, A. E. (2005). The molecular mechanisms underlying BiP‐mediated gating of the Sec61 translocon of the endoplasmic reticulum. The Journal of Cell Biology 168, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan, B. B. , Moyer, B. D. & Balch, W. E. (2000). Rab1 recruitment of p115 into a cis‐SNARE complex: programming budding COPII vesicles for fusion. Science (New York, N.Y.) 289, 444–448. [DOI] [PubMed] [Google Scholar]
- Amin‐Wetzel, N. , Saunders, R. A. , Kamphuis, M. J. , Rato, C. , Preissler, S. , Harding, H. P. & Ron, D. (2017). A J‐protein co‐chaperone recruits BiP to monomerize IRE1 and repress the unfolded protein response. Cell 171, 1625–1637.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amm, I. , Sommer, T. & Wolf, D. H. (2014). Protein quality control and elimination of protein waste: the role of the ubiquitin‐proteasome system. Biochimica et Biophysica Acta 1843, 182–196. [DOI] [PubMed] [Google Scholar]
- Antonny, B. , Madden, D. , Hamamoto, S. , Orci, L. & Schekman, R. (2001). Dynamics of the COPII coat with GTP and stable analogues. Nature Cell Biology 3, 531–537. [DOI] [PubMed] [Google Scholar]
- Archibald, J. M. (2015). Endosymbiosis and eukaryotic cell evolution. Current Biology 25, R911–R921. [DOI] [PubMed] [Google Scholar]
- Avezov, E. , Frenkel, Z. , Ehrlich, M. , Herscovics, A. & Lederkremer, G. Z. (2008). Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N‐glycan trimming to Man5‐6GlcNAc2 in glycoprotein ER‐associated degradation. Molecular Biology of the Cell 19, 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla, T. (2013). Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiological Reviews 93, 1019–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis, V. A. , Aitken, J. R. , Cleves, A. E. & Dowhan, W. (1990). An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 347, 561–562. [DOI] [PubMed] [Google Scholar]
- Barlowe, C. K. & Miller, E. A. (2013). Secretory protein biogenesis and traffic in the early secretory pathway. Genetics 193, 383–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe, C. , Orci, L. , Yeung, T. , Hosobuchi, M. , Hamamoto, S. , Salama, N. , Rexach, M. F. , Ravazzola, M. , Amherdt, M. & Schekman, R. (1994). COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895–907. [DOI] [PubMed] [Google Scholar]
- Barlowe, C. & Schekman, R. (1993). SEC12 encodes a guanine‐nucleotide‐exchange factor essential for transport vesicle budding from the ER. Nature 365, 347–349. [DOI] [PubMed] [Google Scholar]
- Bays, N. W. , Gardner, R. G. , Seelig, L. P. , Joazeiro, C. A. & Hampton, R. Y. (2001). Hrd1p/Der3p is a membrane‐anchored ubiquitin ligase required for ER‐associated degradation. Nature Cell Biology 3, 24–29. [DOI] [PubMed] [Google Scholar]
- Behnke, J. , Feige, M. J. & Hendershot, L. M. (2015). BiP and its nucleotide exchange factors Grp170 and Sil1: mechanisms of action and biological functions. Journal of Molecular Biology 427, 1589–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C. J. , Manfredi, G. , Griffiths, E. J. & Rutter, G. A. (2007). Luciferase expression for ATP imaging: application to cardiac myocytes. Methods in Cell Biology 80, 341–352. [DOI] [PubMed] [Google Scholar]
- Benedix, J. , Lajoie, P. , Jaiswal, H. , Burgard, C. , Greiner, M. , Zimmermann, R. , Rospert, S. , Snapp, E. L. & Dudek, J. (2010). BiP modulates the affinity of its co‐chaperone ERj1 for ribosomes. The Journal of Biological Chemistry 285, 36427–36433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyair, R. , Ogen‐Shtern, N. , Mazkereth, N. , Shai, B. , Ehrlich, M. & Lederkremer, G. Z. (2015). Mammalian ER mannosidase I resides in quality control vesicles, where it encounters its glycoprotein substrates. Molecular Biology of the Cell 26, 172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, J. , Hung, Y. P. & Yellen, G. (2009). A genetically encoded fluorescent reporter of ATP:ADP ratio. Nature Methods 6, 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzenhauser, M. J. , Wagner, L. E. , Iwai, M. , Michikawa, T. , Mikoshiba, K. & Yule, D. I. (2008). ATP modulation of Ca2+ release by Type‐2 and Type‐3 inositol (1, 4, 5)‐triphosphate receptors differing ATP sensitivities and molecular determinants of action. Journal of Biological Chemistry 283, 21579–21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielli, A. , Haney, C. J. , Gabreski, G. , Watkins, S. C. , Bannykh, S. I. & Aridor, M. (2005). Regulation of Sar1 NH2 terminus by GTP binding and hydrolysis promotes membrane deformation to control COPII vesicle fission. The Journal of Cell Biology 171, 919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau, M. , Mullapudi, S. , Becker, T. , Dudek, J. , Zimmermann, R. , Penczek, P. A. & Beckmann, R. (2005). ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nature Structural & Molecular Biology 12, 1015–1016. [DOI] [PubMed] [Google Scholar]
- Borkenhagen, L. F. , Kennedy, E. P. & Fielding, L. (1961). Enzymatic formation and decarboxylation of phosphatidylserine. Journal of Biological Chemistry 236, PC28–PC30. [Google Scholar]
- Braakman, I. & Bulleid, N. J. (2011). Protein folding and modification in the mammalian endoplasmic reticulum. Annual Review of Biochemistry 80, 71–99. [DOI] [PubMed] [Google Scholar]
- Bracher, A. & Verghese, J. (2015). GrpE, Hsp110/Grp170, HspBP1/Sil1 and BAG domain proteins: nucleotide exchange factors for Hsp70 molecular chaperones. Sub‐Cellular Biochemistry 78, 1–33. [DOI] [PubMed] [Google Scholar]
- Brodsky, J. L. (1996). Post‐translational protein translocation: not all hsc70s are created equal. Trends in Biochemical Sciences 21, 122–126. [PubMed] [Google Scholar]
- Brodsky, J. L. (2007). The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic‐reticulum‐associated degradation). The Biochemical Journal 404, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau, B. , Weissman, J. & Horwich, A. (2006). Molecular chaperones and protein quality control. Cell 125, 443–451. [DOI] [PubMed] [Google Scholar]
- Caramelo, J. J. & Parodi, A. J. (2008). Getting in and out from calnexin/calreticulin cycles. The Journal of Biological Chemistry 283, 10221–10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, P. , Goder, V. & Rapoport, T. A. (2006). Distinct ubiquitin‐ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126, 361–373. [DOI] [PubMed] [Google Scholar]
- Chami, M. , Oulès, B. , Szabadkai, G. , Tacine, R. , Rizzuto, R. & Paterlini‐Bréchot, P. (2008). Role of SERCA1 truncated isoform in the proapoptotic calcium transfer from ER to mitochondria during ER stress. Molecular Cell 32, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. & Brandizzi, F. (2013). IRE1: ER stress sensor and cell fate executor. Trends in Cell Biology 23, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, J. Y. & Mansfield, B. C. (2014). The SLC37 family of sugar‐phosphate/phosphate exchangers. Current Topics in Membranes 73, 357–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, J. C. , Olzmann, J. A. , Shaler, T. A. , Sowa, M. E. , Bennett, E. J. , Richter, C. M. , Tyler, R. E. , Greenblatt, E. J. , Harper, J. W. & Kopito, R. R. (2012). Defining human ERAD networks through an integrative mapping strategy. Nature Cell Biology 14, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, J. C. , Shaler, T. A. , Tyler, R. E. & Kopito, R. R. (2008). OS‐9 and GRP94 deliver mutant alpha1‐antitrypsin to the Hrd1‐SEL1L ubiquitin ligase complex for ERAD. Nature Cell Biology 10, 272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clairmont, C. A. , Maio, A. D. & Hirschberg, C. B. (1992). Translocation of ATP into the lumen of rough endoplasmic reticulum‐derived vesicles and its binding to luminal proteins including BiP (GRP 78) and GRP 94. Journal of Biological Chemistry 267, 3983–3990. [PubMed] [Google Scholar]
- Clapham, D. E. (2007). Calcium signaling. Cell 131, 1047–1058. [DOI] [PubMed] [Google Scholar]
- Clerico, E. M. , Tilitsky, J. M. , Meng, W. & Gierasch, L. M. (2015). How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. Journal of Molecular Biology 427, 1575–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves, A. E. , Novick, P. J. & Bankaitis, V. A. (1989). Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. The Journal of Cell Biology 109, 2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti, B. J. , Devaraneni, P. K. , Yang, Z. , David, L. L. & Skach, W. R. (2015). Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate‐driven translocation events. Molecular Cell 58, 269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Credle, J. J. , Finer‐Moore, J. S. , Papa, F. R. , Stroud, R. M. & Walter, P. (2005). On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proceedings of the National Academy of Sciences of the United States of America 102, 18773–18784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick, D. C. , Rush, J. S. & Waechter, C. J. (1991). Characterization and localization of a long‐chain isoprenyltransferase activity in porcine brain: proposed role in the biosynthesis of dolichyl phosphate. Journal of Neurochemistry 57, 1354–1362. [DOI] [PubMed] [Google Scholar]
- Csala, M. , Marcolongo, P. , Lizák, B. , Senesi, S. , Margittai, É. , Fulceri, R. , Magyar, J. É. , Benedetti, A. & Bánhegyi, G. (2007). Transport and transporters in the endoplasmic reticulum. Biochimica et Biophysica Acta (BBA) ‐ Biomembranes 1768, 1325–1341. [DOI] [PubMed] [Google Scholar]
- Dacks, J. B. & Field, M. C. (2007). Evolution of the eukaryotic membrane‐trafficking system: origin, tempo and mode. Journal of Cell Science 120, 2977–2985. [DOI] [PubMed] [Google Scholar]
- D'Arcangelo, J. G. , Stahmer, K. R. & Miller, E. A. (2013). Vesicle‐mediated export from the ER: COPII coat function and regulation. Biochimica et Biophysica Acta 1833, 2464–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, T. R. , Lazarus, M. D. , Hetzer, M. W. & Wente, S. R. (2009). ER membrane–bending proteins are necessary for de novo nuclear pore formation. The Journal of Cell Biology 184, 659–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak, P. M. & Wolf, D. H. (2001). Membrane topology and function of Der3/Hrd1p as a ubiquitin‐protein ligase (E3) involved in endoplasmic reticulum degradation. The Journal of Biological Chemistry 276, 10663–10669. [DOI] [PubMed] [Google Scholar]
- Del Bel, L. M. & Brill, J. A. (2018). Sac1, a lipid phosphatase at the interface of vesicular and nonvesicular transport. Traffic (Copenhagen, Denmark) 19, 301–318. [DOI] [PubMed] [Google Scholar]
- DeLuca, M. & McElroy, W. D. (1974). Kinetics of the firefly luciferase catalyzed reactions. Biochemistry 13, 921–925. [DOI] [PubMed] [Google Scholar]
- Depaoli, M.R. , Karsten, F. , Madreiter-Sokolowski, C.T. , Klec, C. , Gottschalk, B. , Bischof, H. , Eroglu, E. , Waldeck-Weiermair, M. , Simmen, T. , Graier, W.F. & Malli, R. (2018). Real-Time Imaging of Mitochondrial ATP Dynamics Reveals the Metabolic Setting of Single Cells. Cell Reports 25, 501–512.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo, G. & De Camilli, P. (2006). Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657. [DOI] [PubMed] [Google Scholar]
- Dierks, T. , Volkmer, J. , Schlenstedt, G. , Jung, C. , Sandholzer, U. , Zachmann, K. , Schlotterhose, P. , Neifer, K. , Schmidt, B. & Zimmermann, R. (1996). A microsomal ATP‐binding protein involved in efficient protein transport into the mammalian endoplasmic reticulum. The EMBO Journal 15, 6931–6942. [PMC free article] [PubMed] [Google Scholar]
- Dorner, A. J. & Kaufman, R. J. (1994). The levels of endoplasmic reticulum proteins and ATP affect folding and secretion of selective proteins. Biologicals 22, 103–112. [DOI] [PubMed] [Google Scholar]
- Dudek, J. , Pfeffer, S. , Lee, P.‐H. , Jung, M. , Cavalié, A. , Helms, V. , Förster, F. & Zimmermann, R. (2015). Protein transport into the human endoplasmic reticulum. Journal of Molecular Biology 427, 1159–1175. [DOI] [PubMed] [Google Scholar]
- Eletto, D. , Dersh, D. & Argon, Y. (2010). GRP94 in ER quality control and stress responses. Seminars in Cell & Developmental Biology 21, 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, F. & Wood, K. V. (2007). Bioluminescent assays for high‐throughput screening. Assay and Drug Development Technologies 5, 127–136. [DOI] [PubMed] [Google Scholar]
- Fang, S. , Ferrone, M. , Yang, C. , Jensen, J. P. , Tiwari, S. & Weissman, A. M. (2001). The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proceedings of the National Academy of Sciences of the United States of America 98, 14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett, T. W. , Martindale, J. L. , Guyton, K. Z. , Hai, T. & Holbrook, N. J. (1999). Complexes containing activating transcription factor (ATF)/cAMP‐responsive‐element‐binding protein (CREB) interact with the CCAAT/enhancer‐binding protein (C/EBP)‐ATF composite site to regulate Gadd153 expression during the stress response. The Biochemical Journal 339 (Pt 1), 135–141. [PMC free article] [PubMed] [Google Scholar]
- Feige, M. J. & Hendershot, L. M. (2011). Disulfide bonds in ER protein folding and homeostasis. Current Opinion in Cell Biology 23, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, J. J. , Durr, I. F. & Rudney, H. (1959). The biosynthsis of mevalonic acid. Proceedings of the National Academy of Sciences of the United States of America 45, 499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn, G. C. , Pohl, J. , Flocco, M. T. & Rothman, J. E. (1991). Peptide‐binding specificity of the molecular chaperone BiP. Nature 353, 726–730. [DOI] [PubMed] [Google Scholar]
- Fons, R. D. , Bogert, B. A. & Hegde, R. S. (2003). Substrate‐specific function of the translocon‐associated protein complex during translocation across the ER membrane. The Journal of Cell Biology 160, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel, Z. , Gregory, W. , Kornfeld, S. & Lederkremer, G. Z. (2003). Endoplasmic reticulum‐associated degradation of mammalian glycoproteins involves sugar chain trimming to Man6‐5GlcNAc2. The Journal of Biological Chemistry 278, 34119–34124. [DOI] [PubMed] [Google Scholar]
- Frenkel, Z. , Shenkman, M. , Kondratyev, M. & Lederkremer, G. Z. (2004). Separate roles and different routing of calnexin and ERp57 in endoplasmic reticulum quality control revealed by interactions with asialoglycoprotein receptor chains. Molecular Biology of the Cell 15, 2133–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J. R. & Voeltz, G. K. (2011). The ER in 3D: a multifunctional dynamic membrane network. Trends in Cell Biology 21, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galligan, J. J. & Petersen, D. R. (2012). The human protein disulfide isomerase gene family. Human Genomics 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, B. M. & Walter, P. (2011). Unfolded proteins are Ire1‐activating ligands that directly induce the unfolded protein response. Science (New York, N.Y.) 333, 1891–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaylor, J. L. (2002). Membrane‐bound enzymes of cholesterol synthesis from lanosterol. Biochemical and Biophysical Research Communications 292, 1139–1146. [DOI] [PubMed] [Google Scholar]
- Gerin, I. , Veiga‐da‐Cunha, M. , Achouri, Y. , Collet, J.‐F. & Van Schaftingen, E. (1997). Sequence of a putative glucose 6‐phosphate translocase, mutated in glycogen storage disease type Ib1. FEBS Letters 419, 235–238. [DOI] [PubMed] [Google Scholar]
- Gidalevitz, T. , Stevens, F. & Argon, Y. (2013). Orchestration of secretory protein folding by ER chaperones. Biochimica et Biophysica Acta 1833, 2410–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick, B. S. (1995). Can Hsp70 proteins act as force‐generating motors? Cell 80, 11–14. [DOI] [PubMed] [Google Scholar]
- Gogala, M. , Becker, T. , Beatrix, B. , Armache, J.‐P. , Barrio‐Garcia, C. , Berninghausen, O. & Beckmann, R. (2014). Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 506, 107–110. [DOI] [PubMed] [Google Scholar]
- Görlich, D. , Hartmann, E. , Prehn, S. & Rapoport, T. A. (1992). A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature 357, 47–52. [DOI] [PubMed] [Google Scholar]
- Guillén, E. & Hirschberg, C. B. (1995). Transport of adenosine triphosphate into endoplasmic reticulum proteoliposomes. Biochemistry 34, 5472–5476. [DOI] [PubMed] [Google Scholar]
- Guo, S. , Stolz, L. E. , Lemrow, S. M. & York, J. D. (1999). SAC1‐like domains of yeast SAC1,INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. Journal of Biological Chemistry 274, 12990–12995. [DOI] [PubMed] [Google Scholar]
- Haigh, N. G. & Johnson, A. E. (2002). A new role for BiP: closing the aqueous translocon pore during protein integration into the ER membrane. The Journal of Cell Biology 156, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman, B. D. , Hendershot, L. M. & Johnson, A. E. (1998). BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell 92, 747–758. [DOI] [PubMed] [Google Scholar]
- Hampton, R. Y. & Sommer, T. (2012). Finding the will and the way of ERAD substrate retrotranslocation. Current Opinion in Cell Biology 24, 460–466. [DOI] [PubMed] [Google Scholar]
- Harding, H. P. , Zhang, Y. , Bertolotti, A. , Zeng, H. & Ron, D. (2000). Perk is essential for translational regulation and cell survival during the unfolded protein response. Molecular Cell 5, 897–904. [DOI] [PubMed] [Google Scholar]
- Harding, H. P. , Zhang, Y. & Ron, D. (1999). Protein translation and folding are coupled by an endoplasmic‐reticulum‐resident kinase. Nature 397, 271–274. [DOI] [PubMed] [Google Scholar]
- Haze, K. , Yoshida, H. , Yanagi, H. , Yura, T. & Mori, K. (1999). Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Molecular Biology of the Cell 10, 3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger, M. A. , Romero, M. F. , Peng, J.‐B. , Rolfs, A. , Takanaga, H. & Bruford, E. A. (2003). The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflügers Archiv 447, 465–468. [DOI] [PubMed] [Google Scholar]
- Hegde, R. S. , Voigt, S. , Rapoport, T. A. & Lingappa, V. R. (1998). TRAM regulates the exposure of nascent secretory proteins to the cytosol during translocation into the endoplasmic reticulum. Cell 92, 621–631. [DOI] [PubMed] [Google Scholar]
- von Heijne, G. (1983). Patterns of amino acids near signal‐sequence cleavage sites. European Journal of Biochemistry/FEBS 133, 17–21. [DOI] [PubMed] [Google Scholar]
- von Heijne, G. (1984). Analysis of the distribution of charged residues in the N‐terminal region of signal sequences: implications for protein export in prokaryotic and eukaryotic cells. The EMBO Journal 3, 2315–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne, G. (1986). A new method for predicting signal sequence cleavage sites. Nucleic Acids Research 14, 4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius, A. & Aebi, M. (2004). Roles of N‐linked glycans in the endoplasmic reticulum. Annual Review of Biochemistry 73, 1019–1049. [DOI] [PubMed] [Google Scholar]
- Herscovics, A. , Romero, P. A. & Tremblay, L. O. (2002). The specificity of the yeast and human class I ER alpha 1,2‐mannosidases involved in ER quality control is not as strict previously reported. Glycobiology 12, 14G–15G. [PubMed] [Google Scholar]
- Hirsch, C. , Gauss, R. , Horn, S. C. , Neuber, O. & Sommer, T. (2009). The ubiquitylation machinery of the endoplasmic reticulum. Nature 458, 453–460. [DOI] [PubMed] [Google Scholar]
- Hirschberg, C. B. , Robbins, P. W. & Abeijon, C. (1998). Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annual Review of Biochemistry 67, 49–69. [DOI] [PubMed] [Google Scholar]
- Holstein, S. A. & Hohl, R. J. (2004). Isoprenoids: remarkable diversity of form and function. Lipids 39, 293–309. [DOI] [PubMed] [Google Scholar]
- Hosokawa, N. , Wada, I. , Hasegawa, K. , Yorihuzi, T. , Tremblay, L. O. , Herscovics, A. & Nagata, K. (2001). A novel ER alpha‐mannosidase‐like protein accelerates ER‐associated degradation. EMBO Reports 2, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, H. , Budnik, A. , Schmidt, K. , Palmer, K. J. , Mantell, J. , Noakes, C. , Johnson, A. , Carter, D. A. , Verkade, P. , Watson, P. & Stephens, D. J. (2009). Organisation of human ER‐exit sites: requirements for the localisation of Sec16 to transitional ER. Journal of Cell Science 122, 2924–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura, H. , Nhat, K. P. H. , Togawa, H. , Saito, K. , Iino, R. , Kato‐Yamada, Y. , Nagai, T. & Noji, H. (2009). Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer‐based genetically encoded indicators. Proceedings of the National Academy of Sciences of the United States of America 106, 15651–15656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida, N. & Kawakita, M. (2003). Molecular physiology and pathology of the nucleotide sugar transporter family (SLC35). Pflügers Archiv 447, 768–775. [DOI] [PubMed] [Google Scholar]
- Jouaville, L. S. , Pinton, P. , Bastianutto, C. , Rutter, G. A. & Rizzuto, R. (1999). Regulation of mitochondrial ATP synthesis by calcium: evidence for a long‐term metabolic priming. Proceedings of the National Academy of Sciences of the United States of America 96, 13807–13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhi‐Nesher, S. , Shenkman, M. , Tolchinsky, S. , Fromm, S. V. , Ehrlich, R. & Lederkremer, G. Z. (2001). A novel quality control compartment derived from the endoplasmic reticulum. Molecular Biology of the Cell 12, 1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanapin, A. , Batalov, S. , Davis, M. J. , Gough, J. , Grimmond, S. , Kawaji, H. , Magrane, M. , Matsuda, H. , Schönbach, C. , Teasdale, R. D. , Yuan, Z. & RIKEN GER Group & GSL Members (2003). Mouse proteome analysis. Genome Research 13, 1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, M. , Ishiguro, M. , Niinuma, Y. , Uesugi, M. & Nomura, Y. (2002). Human HRD1 protects against ER stress‐induced apoptosis through ER‐associated degradation. FEBS Letters 532, 147–152. [DOI] [PubMed] [Google Scholar]
- Kawano, S. , Kuruma, A. , Hirayama, Y. & Hiraoka, M. (1999). Anion permeability and conduction of adenine nucleotides through a chloride channel in cardiac sarcoplasmic reticulum. The Journal of Biological Chemistry 274, 2085–2092. [DOI] [PubMed] [Google Scholar]
- Kelleher, D. J. & Gilmore, R. (2006). An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R. [DOI] [PubMed] [Google Scholar]
- Kennedy, E. P. & Weiss, S. B. (1956). The function of cytidine coenzymes in the biosynthesis of phospholipides. The Journal of Biological Chemistry 222, 193–214. [PubMed] [Google Scholar]
- Kennedy, H. J. , Pouli, A. E. , Ainscow, E. K. , Jouaville, L. S. , Rizzuto, R. & Rutter, G. A. (1999). Glucose generates sub‐plasma membrane ATP microdomains in single islet β‐cells. Journal of Biological Chemistry 274, 13281–13291. [DOI] [PubMed] [Google Scholar]
- Kim, S. , Shin, S. & Park, J. (1996). Identification of the ATP transporter of rat liver rough endoplasmic reticulum via photoaffinity labeling and partial purification. Biochemistry 35, 5418–5425. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T. & Fujita, M. (2016). Biosynthesis of GPI‐anchored proteins: special emphasis on GPI lipid remodeling. Journal of Lipid Research 57, 6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, J. & Tampé, R. (2006). The macromolecular peptide‐loading complex in MHC class I‐dependent antigen presentation. Cellular and Molecular Life Sciences: CMLS 63, 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochendörfer, K. U. , Then, A. R. , Kearns, B. G. , Bankaitis, V. A. & Mayinger, P. (1999). Sac1p plays a crucial role in microsomal ATP transport, which is distinct from its function in Golgi phospholipid metabolism. The EMBO Journal 18, 1506–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh, A. & Walter, P. (2012). Structural basis of the unfolded protein response. Annual Review of Cell and Developmental Biology 28, 251–277. [DOI] [PubMed] [Google Scholar]
- Krebs, J. , Agellon, L. B. & Michalak, M. (2015). Ca(2+) homeostasis and endoplasmic reticulum (ER) stress: an integrated view of calcium signaling. Biochemical and Biophysical Research Communications 460, 114–121. [DOI] [PubMed] [Google Scholar]
- Lakkaraju, A. K. , Abrami, L. , Lemmin, T. , Blaskovic, S. , Kunz, B. , Kihara, A. , Dal Peraro, M. & van der Goot, F. G. (2012). Palmitoylated calnexin is a key component of the ribosome‐translocon complex. The EMBO Journal 31, 1823–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall, S. , Neuhof, A. & Rapoport, T. (2004). The endoplasmic reticulum membrane is permeable to small molecules. Molecular Biology of the Cell 15, 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederkremer, G. Z. (2009). Glycoprotein folding, quality control and ER‐associated degradation. Current Opinion in Structural Biology 19, 515–523. [DOI] [PubMed] [Google Scholar]
- Leroch, M. , Neuhaus, H. E. , Kirchberger, S. , Zimmermann, S. , Melzer, M. , Gerhold, J. & Tjaden, J. (2008). Identification of a novel adenine nucleotide transporter in the endoplasmic reticulum of Arabidopsis . The Plant Cell 20, 438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti, M. V. & Locasale, J. W. (2016). The Warburg effect: how does it benefit cancer cells? Trends in Biochemical Sciences 41, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley, B. N. & Ploegh, H. L. (2004). A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429, 834–840. [DOI] [PubMed] [Google Scholar]
- Lilley, B. N. & Ploegh, H. L. (2005). Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proceedings of the National Academy of Sciences of the United States of America 102, 14296–14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. H. , Li, H. , Zhang, Y. , Ron, D. & Walter, P. (2009). Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One 4, e4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, P. D. , Harding, H. P. & Ron, D. (2004). Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. The Journal of Cell Biology 167, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman, S. K. & Schekman, R. (1997). Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell 88, 85–96. [DOI] [PubMed] [Google Scholar]
- Lynes, E. M. & Simmen, T. (2011). Urban planning of the endoplasmic reticulum (ER): how diverse mechanisms segregate the many functions of the ER. Biochimica et Biophysica Acta 1813, 1893–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Määttänen, P. , Gehring, K. , Bergeron, J. J. M. & Thomas, D. Y. (2010). Protein quality control in the ER: the recognition of misfolded proteins. Seminars in Cell & Developmental Biology 21, 500–511. [DOI] [PubMed] [Google Scholar]
- Manfredi, G. , Yang, L. , Gajewski, C. D. & Mattiazzi, M. (2002). Measurements of ATP in mammalian cells. Methods 26, 317–326. [DOI] [PubMed] [Google Scholar]
- Marchi, S. , Patergnani, S. & Pinton, P. (2014). The endoplasmic reticulum–mitochondria connection: one touch, multiple functions. Biochimica et Biophysica Acta (BBA) ‐ Bioenergetics 1837, 461–469. [DOI] [PubMed] [Google Scholar]
- Marciniak, S. J. , Garcia‐Bonilla, L. , Hu, J. , Harding, H. P. & Ron, D. (2006). Activation‐dependent substrate recruitment by the eukaryotic translation initiation factor 2 kinase PERK. The Journal of Cell Biology 172, 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak, S. J. , Yun, C. Y. , Oyadomari, S. , Novoa, I. , Zhang, Y. , Jungreis, R. , Nagata, K. , Harding, H. P. & Ron, D. (2004). CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes & Development 18, 3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M. P. & Bukau, B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cellular and Molecular Life Science 62, 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayinger, P. (2012). Phosphoinositides and vesicular membrane traffic. Biochimica et Biophysica Acta (BBA) ‐ Molecular and Cell Biology of Lipids 1821, 1104–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayinger, P. , Bankaitis, V. A. & Meyer, D. I. (1995). Sac1p mediates the adenosine triphosphate transport into yeast endoplasmic reticulum that is required for protein translocation. The Journal of Cell Biology 131, 1377–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayinger, P. & Meyer, D. I. (1993). An ATP transporter is required for protein translocation into the yeast endoplasmic reticulum. The EMBO Journal 12, 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough, K. D. , Martindale, J. L. , Klotz, L. O. , Aw, T. Y. & Holbrook, N. J. (2001). Gadd153 sensitizes cells to endoplasmic reticulum stress by down‐regulating Bcl2 and perturbing the cellular redox state. Molecular and Cellular Biology 21, 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, P. S. , Garcia, E. P. , Slepnev, V. I. , David, C. , Zhang, X. , Grabs, D. , Sossini, W. S. , Bauerfeind, R. , Nemoto, Y. & De Camilli, P. (1996). A presynaptic inositol‐5‐phosphatase. Nature 379, 353–357. [DOI] [PubMed] [Google Scholar]
- Meacock, S. L. , Lecomte, F. J. L. , Crawshaw, S. G. & High, S. (2002). Different transmembrane domains associate with distinct endoplasmic reticulum components during membrane integration of a polytopic protein. Molecular Biology of the Cell 13, 4114–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert, M. , Sommer, T. & Jarosch, E. (2014). Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nature Cell Biology 16, 77–86. [DOI] [PubMed] [Google Scholar]
- Ménétret, J.‐F. , Hegde, R. S. , Aguiar, M. , Gygi, S. P. , Park, E. , Rapoport, T. A. & Akey, C. W. (2008). Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure (London, 1993) 16, 1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménétret, J. F. , Neuhof, A. , Morgan, D. G. , Plath, K. , Radermacher, M. , Rapoport, T. A. & Akey, C. W. (2000). The structure of ribosome‐channel complexes engaged in protein translocation. Molecular Cell 6, 1219–1232. [DOI] [PubMed] [Google Scholar]
- Meusser, B. , Hirsch, C. , Jarosch, E. & Sommer, T. (2005). ERAD: the long road to destruction. Nature Cell Biology 7, 766–772. [DOI] [PubMed] [Google Scholar]
- Meyer, H. A. , Grau, H. , Kraft, R. , Kostka, S. , Prehn, S. , Kalies, K. U. & Hartmann, E. (2000). Mammalian Sec61 is associated with Sec62 and Sec63. The Journal of Biological Chemistry 275, 14550–14557. [DOI] [PubMed] [Google Scholar]
- Miller, E. , Antonny, B. , Hamamoto, S. & Schekman, R. (2002). Cargo selection into COPII vesicles is driven by the Sec24p subunit. The EMBO Journal 21, 6105–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, E. A. , Beilharz, T. H. , Malkus, P. N. , Lee, M. C. S. , Hamamoto, S. , Orci, L. & Schekman, R. (2003). Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 114, 497–509. [DOI] [PubMed] [Google Scholar]
- Misselwitz, B. , Staeck, O. & Rapoport, T. A. (1998). J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Molecular Cell 2, 593–603. [DOI] [PubMed] [Google Scholar]
- Molinari, M. , Calanca, V. , Galli, C. , Lucca, P. & Paganetti, P. (2003). Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science (New York, N.Y.) 299, 1397–1400. [DOI] [PubMed] [Google Scholar]
- Molinari, M. , Eriksson, K. K. , Calanca, V. , Galli, C. , Cresswell, P. , Michalak, M. & Helenius, A. (2004). Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Molecular Cell 13, 125–135. [DOI] [PubMed] [Google Scholar]
- Moremen, K. W. & Molinari, M. (2006). N‐linked glycan recognition and processing: the molecular basis of endoplasmic reticulum quality control. Current Opinion in Structural Biology 16, 592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer, B. D. , Allan, B. B. & Balch, W. E. (2001). Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis‐‐Golgi tethering. **Traffic (Copenhagen, Denmark) 2, 268–276. [DOI] [PubMed] [Google Scholar]
- Murcha, M. W. , Millar, A. H. & Whelan, J. (2005). The N‐terminal cleavable extension of plant carrier proteins is responsible for efficient insertion into the inner mitochondrial membrane. Journal of Molecular Biology 351, 16–25. [DOI] [PubMed] [Google Scholar]
- Nadav, E. , Shmueli, A. , Barr, H. , Gonen, H. , Ciechanover, A. & Reiss, Y. (2003). A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochemical and Biophysical Research Communications 303, 91–97. [DOI] [PubMed] [Google Scholar]
- Nishikawa, S. , Brodsky, J. L. & Nakatsukasa, K. (2005). Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER‐associated degradation (ERAD). Journal of Biochemistry 137, 551–555. [DOI] [PubMed] [Google Scholar]
- Novick, P. , Osmond, B. C. & Botstein, D. (1989). Suppressors of yeast actin mutations. Genetics 121, 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda, Y. , Hosokawa, N. , Wada, I. & Nagata, K. (2003). EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science (New York, N.Y.) 299, 1394–1397. [DOI] [PubMed] [Google Scholar]
- Oikawa, D. , Kimata, Y. , Kohno, K. & Iwawaki, T. (2009). Activation of mammalian IRE1alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Experimental Cell Research 315, 2496–2504. [DOI] [PubMed] [Google Scholar]
- Oikawa, D. , Kitamura, A. , Kinjo, M. & Iwawaki, T. (2012). Direct association of unfolded proteins with mammalian ER stress sensor, IRE1β. PLoS One 7, e51290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari, S. & Molinari, M. (2007). Glycoprotein folding and the role of EDEM1, EDEM2 and EDEM3 in degradation of folding‐defective glycoproteins. FEBS Letters 581, 3658–3664. [DOI] [PubMed] [Google Scholar]
- Oliveira, C. C. & van Hall, T. (2015). Alternative antigen processing for MHC Class I: multiple roads lead to Rome. Frontiers in Immunology 6, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean, P. & Menon, A. K. (2007). Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. Journal of Lipid Research 48, 993–1011. [DOI] [PubMed] [Google Scholar]
- Ortiz Sandoval, C. & Simmen, T. (2012). Rab proteins of the endoplasmic reticulum: functions and interactors. Biochemical Society Transactions 40, 1426–1432. [DOI] [PubMed] [Google Scholar]
- Osborne, A. R. , Rapoport, T. A. & van den Berg, B. (2005). Protein translocation by the Sec61/Secy channel. Annual Review of Cell and Developmental Biology 21, 529–550. [DOI] [PubMed] [Google Scholar]
- Otero, J. H. , Lizák, B. & Hendershot, L. M. (2010). Life and death of a BiP substrate. Seminars in Cell & Developmental Biology 21, 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, W. J. , Bergeron, J. J. , Li, Y. , Kang, C. Y. & Thomas, D. Y. (1995). Conformational changes induced in the endoplasmic reticulum luminal domain of calnexin by Mg‐ATP and Ca2+. The Journal of Biological Chemistry 270, 18051–18059. [DOI] [PubMed] [Google Scholar]
- Patergnani, S. , Baldassari, F. , De Marchi, E. , Karkucinska‐Wieckowska, A. , Wieckowski, M. R. & Pinton, P. (2014). Methods to monitor and compare mitochondrial and glycolytic ATP production. Methods in Enzymology 542, 313–332. [DOI] [PubMed] [Google Scholar]
- Pelham, H. R. B. (1990). The retention signal for soluble proteins of the endoplasmic reticulum. Trends in Biochemical Sciences 15, 483–486. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S. , Burbaum, L. , Unverdorben, P. , Pech, M. , Chen, Y. , Zimmermann, R. , Beckmann, R. & Förster, F. (2015). Structure of the native Sec61 protein‐conducting channel. Nature Communications 6, 8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer, S. , Dudek, J. , Gogala, M. , Schorr, S. , Linxweiler, J. , Lang, S. , Becker, T. , Beckmann, R. , Zimmermann, R. & Förster, F. (2014). Structure of the mammalian oligosaccharyl‐transferase complex in the native ER protein translocon. Nature Communications 5, 3072. [DOI] [PubMed] [Google Scholar]
- Picault, N. , Hodges, M. , Palmieri, L. & Palmieri, F. (2004). The growing family of mitochondrial carriers in Arabidopsis. Trends in Plant Science 9, 138–146. [DOI] [PubMed] [Google Scholar]
- Pilon, M. , Schekman, R. & Römisch, K. (1997). Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. The EMBO Journal 16, 4540–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus, D. , Chevalier, M. W. , Aragón, T. , van Anken, E. , Vidal, S. E. , El‐Samad, H. & Walter, P. (2010). BiP binding to the ER‐stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biology 8, e1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinton, P. , Giorgi, C. , Siviero, R. , Zecchini, E. & Rizzuto, R. (2008). Calcium and apoptosis: ER‐mitochondria Ca2+ transfer in the control of apoptosis. Oncogene 27, 6407–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzo, P. & Pozzan, T. (2007). Mitochondria‐endoplasmic reticulum choreography: structure and signaling dynamics. Trends in Cell Biology 17, 511–517. [DOI] [PubMed] [Google Scholar]
- Plemper, R. K. , Böhmler, S. , Bordallo, J. , Sommer, T. & Wolf, D. H. (1997). Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388, 891–895. [DOI] [PubMed] [Google Scholar]
- Ploegh, H. L. (2007). A lipid‐based model for the creation of an escape hatch from the endoplasmic reticulum. Nature 448, 435–438. [DOI] [PubMed] [Google Scholar]
- Ponsero, A. J. , Igbaria, A. , Darch, M. A. , Miled, S. , Outten, C. E. , Winther, J. R. , Palais, G. , D'Autréaux, B. , Delaunay‐Moisan, A. & Toledano, M. B. (2017). Endoplasmic reticulum transport of glutathione by Sec61 is regulated by Ero1 and Bip. Molecular Cell 67, 962–973.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, K. R. , Claude, A. & Fullam, E. F. (1945). A study of tissue culture cells by electron microscopy. The Journal of Experimental Medicine 81, 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, K. R. & Kallman, F. L. (1952). Significance of cell particulates as seen by electron microscopy. Annals of the New York Academy of Sciences 54, 882–891. [DOI] [PubMed] [Google Scholar]
- Puthalakath, H. , O'Reilly, L. A. , Gunn, P. , Lee, L. , Kelly, P. N. , Huntington, N. D. , Hughes, P. D. , Michalak, E. M. , McKimm‐Breschkin, J. , Motoyama, N. , Gotoh, T. , Akira, S. , Bouillet, P. & Strasser, A. (2007). ER stress triggers apoptosis by activating BH3‐only protein Bim. Cell 129, 1337–1349. [DOI] [PubMed] [Google Scholar]
- Rapoport, T. A. (2007). Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669. [DOI] [PubMed] [Google Scholar]
- Rasheva, V. I. & Domingos, P. M. (2009). Cellular responses to endoplasmic reticulum stress and apoptosis. Apoptosis 14, 996–1007. [DOI] [PubMed] [Google Scholar]
- Raturi, A. & Simmen, T. (2013). Where the endoplasmic reticulum and the mitochondrion tie the knot: the mitochondria‐associated membrane (MAM). Biochimica et Biophysica Acta (BBA) ‐ Molecular Cell Research 1833, 213–224. [DOI] [PubMed] [Google Scholar]
- Ritz, U. & Seliger, B. (2001). The transporter associated with antigen processing (TAP): structural integrity, expression, function, and its clinical relevance. Molecular Medicine (Cambridge, Mass.) 7, 149–158. [PMC free article] [PubMed] [Google Scholar]
- Rutishauser, J. & Spiess, M. (2002). Endoplasmic reticulum storage diseases. Swiss Medical Weekly 132, 211–222. [DOI] [PubMed] [Google Scholar]
- Saito, K. & Katada, T. (2015). Mechanisms for exporting large‐sized cargoes from the endoplasmic reticulum. Cellular and Molecular Life Sciences 72, 3709–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, Y. , Ihara, Y. , Leach, M. R. , Cohen‐Doyle, M. F. & Williams, D. B. (1999). Calreticulin functions in vitro as a molecular chaperone for both glycosylated and non‐glycosylated proteins. The EMBO Journal 18, 6718–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada, K. , Echigo, N. , Juge, N. , Miyaji, T. , Otsuka, M. , Omote, H. , Yamamoto, A. & Moriyama, Y. (2008). Identification of a vesicular nucleotide transporter. Proceedings of the National Academy of Sciences of the United States of America 105, 5683–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäuble, N. , Lang, S. , Jung, M. , Cappel, S. , Schorr, S. , Ulucan, Ö. , Linxweiler, J. , Dudek, J. , Blum, R. , Helms, V. , Paton, A. W. , Paton, J. C. , Cavalié, A. & Zimmermann, R. (2012). BiP‐mediated closing of the Sec61 channel limits Ca2+ leakage from the ER. The EMBO Journal 31, 3282–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, B. , Fernandez, F. & Waechter, C. J. (2001a). The ins(ide) and out(side) of dolichyl phosphate biosynthesis and recycling in the endoplasmic reticulum. Glycobiology 11, 61R–70R. [DOI] [PubMed] [Google Scholar]
- Schenk, B. , Rush, J. S. , Waechter, C. J. & Aebi, M. (2001b). An alternative cis‐isoprenyltransferase activity in yeast that produces polyisoprenols with chain lengths similar to mammalian dolichols. Glycobiology 11, 89–98. [DOI] [PubMed] [Google Scholar]
- Schiene‐Fischer, C. (2015). Multidomain peptidyl prolyl cis/trans isomerases. Biochimica et Biophysica Acta 1850, 2005–2016. [DOI] [PubMed] [Google Scholar]
- Schröder, K. , Martoglio, B. , Hofmann, M. , Hölscher, C. , Hartmann, E. , Prehn, S. , Rapoport, T. A. & Dobberstein, B. (1999). Control of glycosylation of MHC class II‐associated invariant chain by translocon‐associated RAMP4. The EMBO Journal 18, 4804–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder, M. & Kaufman, R. J. (2005). The mammalian unfolded protein response. Annual Review of Biochemistry 74, 739–789. [DOI] [PubMed] [Google Scholar]
- Shen, J. , Chen, X. , Hendershot, L. & Prywes, R. (2002). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Developmental Cell 3, 99–111. [DOI] [PubMed] [Google Scholar]
- Shen, J. , Snapp, E. L. , Lippincott‐Schwartz, J. & Prywes, R. (2005). Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Molecular and Cellular Biology 25, 921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , An, J. , Liang, J. , Hayes, S. E. , Sandusky, G. E. , Stramm, L. E. & Yang, N. N. (1999). Characterization of a mutant pancreatic eIF‐2alpha kinase, PEK, and co‐localization with somatostatin in islet delta cells. The Journal of Biological Chemistry 274, 5723–5730. [DOI] [PubMed] [Google Scholar]
- Shi, Y. , Vattem, K. M. , Sood, R. , An, J. , Liang, J. , Stramm, L. & Wek, R. C. (1998). Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha‐subunit kinase, PEK, involved in translational control. Molecular and Cellular Biology 18, 7499–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiber, A. & Ravid, T. (2014). Chaperoning proteins for destruction: diverse roles of Hsp70 chaperones and their co‐chaperones in targeting misfolded proteins to the proteasome. Biomolecules 4, 704–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, S. J. , Lee, W. K. , Lim, H. W. & Park, J.‐S. (2000). Characterization of the ATP transporter in the reconstituted rough endoplasmic reticulum proteoliposomes. Biochimica et Biophysica Acta (BBA) ‐ Biomembranes 1468, 55–62. [DOI] [PubMed] [Google Scholar]
- Stone, S. J. & Vance, J. E. (2000). Phosphatidylserine synthase‐1 and ‐2 are localized to mitochondria‐associated membranes. The Journal of Biological Chemistry 275, 34534–34540. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y. & Schmitt, M. J. (2015). Redox diversity in ERAD‐mediated protein retrotranslocation from the endoplasmic reticulum: a complex puzzle. Biological Chemistry 396, 539–554. [DOI] [PubMed] [Google Scholar]
- Tantama, M. , Martínez‐François, J. R. , Mongeon, R. & Yellen, G. (2013). Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP‐to‐ADP ratio. Nature Communications 4, 2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, R. J. , Akana, H. C. S. R. , Finnigan, C. , Howell, K. E. & Caldwell, J. H. (2006). Anion channels transport ATP into the Golgi lumen. American Journal of Physiology. Cell Physiology 290, C499–C514. [DOI] [PubMed] [Google Scholar]
- Tyedmers, J. , Lerner, M. , Wiedmann, M. , Volkmer, J. & Zimmermann, R. (2003). Polypeptide‐binding proteins mediate completion of co‐translational protein translocation into the mammalian endoplasmic reticulum. EMBO Reports 4, 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano, F. , Wang, X. , Bertolotti, A. , Zhang, Y. , Chung, P. , Harding, H. P. & Ron, D. (2000). Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science (New York, N.Y.) 287, 664–666. [DOI] [PubMed] [Google Scholar]
- Vance, J. E. (2014). MAM (mitochondria‐associated membranes) in mammalian cells: lipids and beyond. Biochimica et Biophysica Acta (BBA) ‐ Molecular and Cell Biology of Lipids 1841, 595–609. [DOI] [PubMed] [Google Scholar]
- Vance, J. E. (2015). Phospholipid synthesis and transport in mammalian cells. Traffic (Copenhagen, Denmark) 16, 1–18. [DOI] [PubMed] [Google Scholar]
- Vattem, K. M. & Wek, R. C. (2004). Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proceedings of the National Academy of Sciences 101, 11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnu, N. , Jadoon Khan, M. , Karsten, F. , Groschner, L. N. , Waldeck‐Weiermair, M. , Rost, R. , Hallström, S. , Imamura, H. , Graier, W. F. & Malli, R. (2014). ATP increases within the lumen of the endoplasmic reticulum upon intracellular Ca2+ release. Molecular Biology of the Cell 25, 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma, R. A. & Menon, A. K. (2005). Flip‐flop of glycosylphosphatidylinositols (GPI's) across the ER. Chemical Communications (Cambridge, England) 4, 453–455. [DOI] [PubMed] [Google Scholar]
- Voigt, S. , Jungnickel, B. , Hartmann, E. & Rapoport, T. A. (1996). Signal sequence‐dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. The Journal of Cell Biology 134, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Shen, J. , Arenzana, N. , Tirasophon, W. , Kaufman, R. J. & Prywes, R. (2000). Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. The Journal of Biological Chemistry 275, 27013–27020. [DOI] [PubMed] [Google Scholar]
- Watson, P. , Townley, A. K. , Koka, P. , Palmer, K. J. & Stephens, D. J. (2006). Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic (Copenhagen, Denmark) 7, 1678–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen, A. , Binns, K. , Lemberg, M. K. , Ashman, K. & Martoglio, B. (2002). Identification of signal peptide peptidase, a presenilin‐type aspartic protease. Science (New York, N.Y.) 296, 2215–2218. [DOI] [PubMed] [Google Scholar]
- Wiertz, E. J. , Tortorella, D. , Bogyo, M. , Yu, J. , Mothes, W. , Jones, T. R. , Rapoport, T. A. & Ploegh, H. L. (1996). Sec61‐mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384, 432–438. [DOI] [PubMed] [Google Scholar]
- Wijeyesakere, S. J. , Gagnon, J. K. , Arora, K. , Brooks, C. L. & Raghavan, M. (2015). Regulation of calreticulin‐major histocompatibility complex (MHC) class I interactions by ATP. Proceedings of the National Academy of Sciences of the United States of America 112, E5608–E5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, B. & Gilbert, H. F. (2004). Protein disulfide isomerase. Biochimica et Biophysica Acta 1699, 35–44. [DOI] [PubMed] [Google Scholar]
- Williams, D. B. (2006). Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. Journal of Cell Science 119, 615–623. [DOI] [PubMed] [Google Scholar]
- Yaginuma, H. , Kawai, S. , Tabata, K. V. , Tomiyama, K. , Kakizuka, A. , Komatsuzaki, T. , Noji, H. & Imamura, H. (2014). Diversity in ATP concentrations in a single bacterial cell population revealed by quantitative single‐cell imaging. Scientific Reports 4, 6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, A. , Hori, O. , Stern, D. M. , Hartmann, E. , Ogawa, S. & Tohyama, M. (1999). Stress‐associated endoplasmic reticulum protein 1 (SERP1)/Ribosome‐associated membrane protein 4 (RAMP4) stabilizes membrane proteins during stress and facilitates subsequent glycosylation. The Journal of Cell Biology 147, 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Y. , Meyer, H. H. & Rapoport, T. A. (2001). The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652–656. [DOI] [PubMed] [Google Scholar]
- Ye, Y. , Shibata, Y. , Yun, C. , Ron, D. & Rapoport, T. A. (2004). A membrane protein complex mediates retro‐translocation from the ER lumen into the cytosol. Nature 429, 841–847. [DOI] [PubMed] [Google Scholar]
- Yoshida, H. , Matsui, T. , Yamamoto, A. , Okada, T. & Mori, K. (2001). XBP1 mRNA is induced by ATF6 and Spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891. [DOI] [PubMed] [Google Scholar]
- Yoshida, Y. & Tanaka, K. (2010). Lectin‐like ERAD players in ER and cytosol. Biochimica et Biophysica Acta 1800, 172–180. [DOI] [PubMed] [Google Scholar]
- Zhong, X. , Malhotra, R. & Guidotti, G. (2003). ATP uptake in the Golgi and extracellular release require Mcd4 protein and the vacuolar H+‐ATPase. The Journal of Biological Chemistry 278, 33436–33444. [DOI] [PubMed] [Google Scholar]
- Zhou, M. & Schekman, R. (1999). The engagement of Sec61p in the ER dislocation process. Molecular Cell 4, 925–934. [DOI] [PubMed] [Google Scholar]
- Zinszner, H. , Kuroda, M. , Wang, X. , Batchvarova, N. , Lightfoot, R. T. , Remotti, H. , Stevens, J. L. & Ron, D. (1998). CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes & Development 12, 982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]


