The JAV1/JUL1 system acts specifically in coordination of plant defenses without interfering with plant development or growth.
Abstract
Jasmonates regulate plant defense and development. In Arabidopsis (Arabidopsis thaliana), JASMONATE-ASSOCIATED VQ-MOTIF GENE1 (JAV1/VQ22) is a repressor of jasmonate-mediated defense responses and is degraded through the ubiquitin-26S proteasome system after herbivory. We found that JAV1-ASSOCIATED UBIQUITIN LIGASE1 (JUL1), a RING-type E3 ubiquitin ligase, interacted with JAV1. JUL1 interacted with JAV1 in the nucleus to ubiquitinate JAV1, leading to proteasomal degradation of JAV1. The transcript levels of JUL1 and JAV1 were coordinately and positively regulated by the CORONATINE INSENSITIVE1-dependent signaling pathway in the jasmonate signaling network, but in a manner that was not dependent on CORONATINE INSENSITIVE1-mediated signaling upon herbivory by Spodoptera litura. Gain or loss of function of JUL1 modulated the expression levels of the defensin gene PDF1.2 in leaves, conferring on the plants various defense properties against the generalist herbivore S. litura. Because neither the JUL1 mutant nor overexpression lines showed any obvious developmental defects, we concluded that the JAV1/JUL1 system functions as a specific coordinator of reprogramming of plant defense responses. Altogether, our findings offer insight into the mechanisms by which the JAV1/JUL1 system acts specifically to coordinate plant defense responses without interfering with plant development or growth.
Jasmonates, a lipid-derived class of plant hormones, promote gene regulation for defense responses in plants. Jasmonates, including jasmonic acid (JA) and its active derivative jasmonyl-isoleucine (JA-Ile), play a vital role in defending plants against herbivorous insects and necrotrophic pathogens (McConn et al., 1997; Browse, 2009; Pieterse et al., 2012; Okada et al., 2015; Chini et al., 2016) and mediate many developmental processes in root growth, stamen development, and senescence (Wasternack and Hause, 2013). Following JA signaling via the JA-Ile receptor CORONATINE INSENSITIVE1 (COI1), Jasmonate-ZIM domain proteins (JAZs) act as negative regulators of JA signaling (Chini et al., 2007; Thines et al., 2007). The JA-associated degradation of JAZs leads to transcriptional reprogramming of a vast array of genes controlled by transcription factors (TFs) such as MYC2, leading to the activation of defense responses and the modulation of numerous processes of plant growth and development, such as sexual maturation, trichome initiation, and root growth (Browse, 2009; Fernández-Calvo et al., 2011; Qi et al., 2011; Song et al., 2011, 2013; Wasternack and Hause, 2013; Goossens et al., 2017). Activation of JA signaling adversely affects plant growth through a tradeoff between JA-induced defense responses and gibberellin-promoted development of Arabidopsis (Arabidopsis thaliana) plants, in which angiosperm plants prioritize JA-mediated defense over growth (Yang et al., 2012; Guo et al., 2018). It appears that JA signaling controls plant growth by suppressing the GA-mediated degradation of DELLA, a GA-responsive repressor of TFs (including phytochrome-interacting factors; Yang et al., 2012). Moreover, it appears that the plant growth-defense tradeoff is promoted via the relief of transcription repression rather than by the diversion of photoassimilates from growth to defense (Campos et al., 2016).
Hu et al. (2013) identified JASMONATE-ASSOCIATED VQ-MOTIF GENE1 (JAV1, also known as VQ22) as a novel repressor of JA signaling in Arabidopsis. JAV1 is a member of a family of plant-specific proteins with the conserved short amino acid sequence motif FxxhVQxhTG, where x represents any amino acid and h represents a hydrophobic residue (Jing and Lin, 2015). This family plays various roles in plant defense responses, stress tolerance, and growth and development (Jing and Lin, 2015). JAV1 has been shown to specifically down-regulate jasmonate-mediated defense responses against a necrotrophic pathogen (Botrytis cinerea) and an herbivore (Spodoptera exigua larvae) without adversely impacting plant growth or development (Hu et al., 2013; Zhu and Zhu, 2013). Additionally, the JAV1-JAZ8-WRKY51 complex represses JA biosynthesis in healthy plants, but herbivory triggers the calmodulin-dependent phosphorylation of JAV1, which leads to JAV1 degradation and, consequently, to the release of WRKY51, thereby activating JA biosynthesis for plant defense (Yan et al., 2018).
Like JAZ degradation, JAV1 degradation requires JA signaling, which mediates JAV1 degradation through the ubiquitin-26S proteasome system (UPS) and, thereby, leads to the activation of defense genes, including PDF1.2 (Hu et al., 2013). JAV1 is able to interact with not only WRKY51 but also WRKY28, a positive regulator of PDF1.2 expression. However, the E3 ligase(s) specific to JAV1 degradation remain to be identified.
Ubiquitination is a posttranslational modification process that functions in all eukaryotes and is achieved by the consecutive action of three enzymes: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3). The Arabidopsis genome contains more than 1,400 genes encoding E3 ubiquitin ligases (Vierstra, 2009). These E3 ubiquitin ligases play critical roles in determining the substrate specificity of UPS for various target proteins, and the large number of E3 ligases implies their specific recognition of target substrates (Sadanandom et al., 2012). E3 ubiquitin ligases are classified into three families: the homology to E6-AP C-terminus E3 ligases, the really interesting new gene (RING) type of E3 ligases, and the U-box family members (Chen and Hellmann, 2013). RING-type E3 ligase family members can either ubiquitinate substrates independently or function as part of a multisubunit complex, which in plants includes Skp1-Cullin-F-box; Elongin B/C-Cul2/Cul5-SOCS-box protein; Broad‐complex, Tramtrack, and Bric‐à‐brac; UV-damaged DNA-binding protein1; and anaphase promoting complex as independent complexes (Sadanandom et al., 2012).
There are more than 460 proteins in the RING-type protein family in Arabidopsis (Stone et al., 2005; Lee and Kim, 2011). These E3 ligases act in plant biotic stress tolerance (Marino et al., 2012; Duplan and Rivas, 2014) and play essential roles in the JA signaling pathway (Zhang et al., 2012; Nagels Durand et al., 2016b; Miricescu et al., 2018 ). However, the substrate targets of only a few E3 ligases have been identified. In this study, we characterized the features of a RING-type E3 ligase that interacts with JAV1 in Arabidopsis. This E3 ligase, which we named JAV1-ASSOCIATED UBIQUITIN LIGASE1 (JUL1), was found to play a primary role in UPS for JAV1 degradation, leading to the positive regulation of defense responses.
RESULTS
Screening of JAV1-Interacting Ubiquitin Ligases
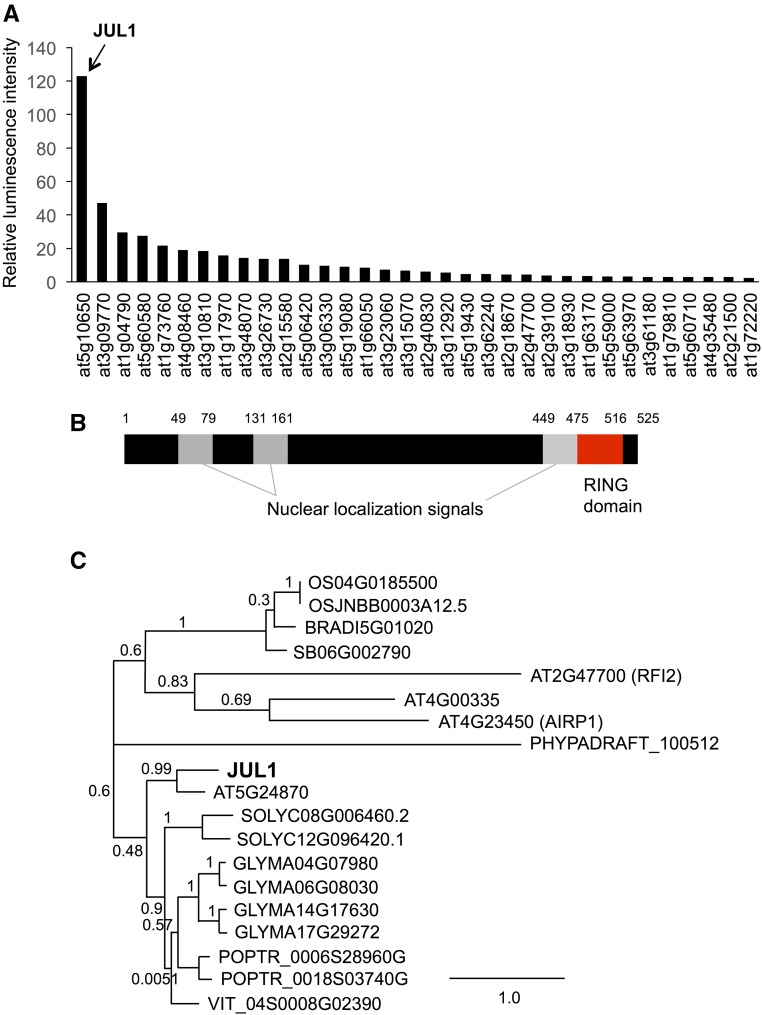
To construct a protein library of putative Arabidopsis RING-type E3 ligases, we prepared 210 cDNA clones from the RIKEN Arabidopsis full-length library (Ramadan et al., 2015), as shown in Supplemental Table S1. Transcription templates with an N-terminal FLAG-tag sequence were used to synthesize proteins using a cell-free protein synthesis system. To identify characteristic(s) of E3 ligase(s) that interact with JAV1 from the library proteins, we used the AlphaScreen-based detection system. Among the 210 synthesized proteins, JAV1 interacted most strongly with an E3 ligase (at5g10650), hereafter designated JUL1 (Fig. 1A). JUL1 has three predicted nuclear localization signals and a RING domain at the C terminus (Fig. 1B). Among the crustacean homologs exhibiting E < 10−5, none of the proteins whose functions have been predicted or characterized have sequence similarities to JUL1 (Fig. 1C). For instance, the well-characterized E3 ligases RFI2 (Chen and Ni, 2006) and AIPR1 (Ryu et al., 2010) did not have similarity to JUL1. Instead, since JUL1 showed high similarity to a predicted E3 ligase (at5g24870; 58% identity) that was not present in our RIKEN Arabidopsis full-length library, we performed the AlphaScreen assay again for interaction between JAV1 and the JUL1 homolog. However, the JAV1 and JUL1 homologs did not show interaction (Supplemental Fig. S1). Likewise, the JAV1 homolog at4g15120 did not interact with JUL1.
Figure 1.
Screening and structure of JUL1. A, Luminescence-based protein-protein interaction detection (AlphaScreen) was used for high-throughput screening of 210 FLAG-conjugated putative Arabidopsis E3 ligases for interaction with biotinylated-JAV1 (Bio-JAV1). The data for the top 34 E3 ligases that interacted strongly with Bio-JAV1 are shown (for the full set of data, see Supplemental Table S2). Luminescence intensities were normalized by those of Escherichia coli dihydrofolate reductase. Data represent means of three replicates. B, JUL1 protein is represented schematically with the nuclear localization signals and RING domain and Lys residues. C, Phylogenetic tree inferred from deduced amino acid sequences of JUL11 and other proteins from several plant taxa: Arabidopsis; BRAD, Brachypodium distachyon; GLYMA, Glycine max; OS, Oryza sativa; PHYPADRAFT, Physcomitrella patens; POPTR, Populus trichocarpa; SB, Sorghum bicolor; SOLYC, Solanum lycopersicum; VIT, Vitis vinifera.
JUL1 and JAV1 Interact in the Nucleus
JAV1 is a nuclear protein (Hu et al., 2013). Therefore, we assessed the subcellular localization of JUL1 using a JUL1-GFP fusion protein expressed in Arabidopsis mesophyll protoplast cells (Fig. 2A). In contrast to the ubiquitous localization of the native GFP protein, which served as a control, the fusion protein showed nuclear localization.
Figure 2.
Subcellular localization and interaction of JAV1 and JUL1. A, The vector containing the Cauliflower mosaic virus 35S promoter (35SP)::GFP or JUL1 fused to GFP (JUL1-GFP) was used to transform Arabidopsis leaf protoplast cells. B, BiFC analysis of JUL1-JAV1 interactions. The vector constructs of 35SP::JUL1 or the JUL1-ring domain mutant (JUL1RM) fused to the N-terminal fragment of YFP (JUL1-nYFP or JUL1RM-nYFP) and JAV1 fused to the C-terminal fragment of YFP (JAV1-cYFP), as shown at the top, were cotransformed into Arabidopsis leaf protoplast cells. The protoplasts were treated with a potent proteasome inhibitor, MG132 (+MG132), or not treated with the inhibitor (−MG132). V5, V5 epitope tag. C, The vector constructs of 35SPΩ::V5-tagged JAV1 (JAV1-V5) and FLAG-tagged JUL1 (JUL1-FLAG) or JUL1RM (JUL1RM-FLAG) were cotransformed into Arabidopsis leaf protoplast cells, and the cells then were treated with MG132 during the incubation. The extracted JAV1 and JUL1 or JUL1RM proteins (input) were immunoprecipitated with anti-FLAG antibody, subjected to SDS-PAGE, and detected using anti-V5. Three independent experiments (Expt. 1, 2, and 3) were performed. IB, immunoblotting; IP, immunoprecipitation.
Moreover, in order to assess the interaction in vivo between JUL1 and JAV1 proteins, a bimolecular fluorescence complementation (BiFC) assay was conducted. JUL1 fused to the N-terminal portion of yellow fluorescent protein (YFP; JUL1-nYFP) and JAV1 fused to the C-terminal portion of YFP (JAV1-cYFP) were transiently expressed in Arabidopsis mesophyll protoplast cells (Fig. 2B). The fluorescent signals of these recombinant YFP proteins were detected in the nuclei of Arabidopsis protoplast cells only when MG132, a proteasome inhibitor, was applied to the cells (Fig. 2B). However, fluorescence was not detected when JAV1-cYFP was coexpressed with JUL1-ring domain mutant (JUL1RM) fused to the N-terminal portion of YFP. These results indicated that the persistent access of JUL1 to JAV1 caused rapid UPS-dependent JAV1 degradation.
Moreover, immunoprecipitation assays confirmed that V5-tagged JAV1 interacted with FLAG-tagged JUL1 but not JUL1RM in the presence of MG132 (Fig. 2C).
Ubiquitin Ligase Activity of JUL1
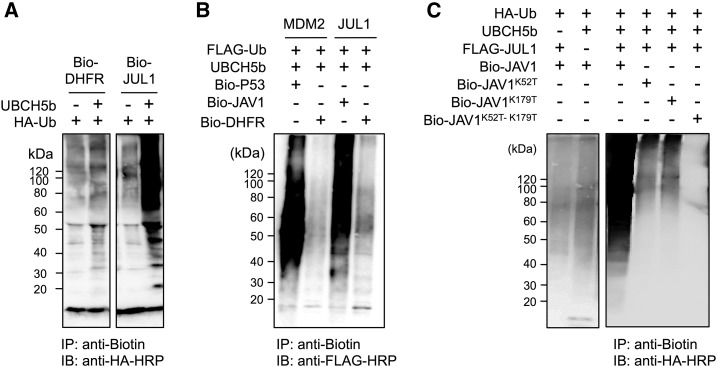
To assess the autoubiquitination activity of JUL1 in vitro, biotinylated JUL1 protein (Bio-JUL1) was incubated with HA-tagged ubiquitin (HA-Ub) in either the presence or absence of UBCH5b (Fig. 3A). UBCH5b is an E2 ubiquitin ligase that is involved in the polyubiquitination of a suite of E3 ligases, including K48 (David et al., 2011). Immunoprecipitation with anti-biotin and subsequent anti-HA antibody detection revealed that Bio-JUL1 underwent autoubiquitination in the presence of UBCH5b. Moreover, when biotinylated JAV1 protein (Bio-JAV1) was incubated with FLAG-tagged ubiquitin (FLAG-Ub), UBCH5b, and JUL1, the Bio-JAV1 protein was ubiquitinated, as was the positive control, biotinylated P53 protein (Bio-P53), when it was incubated with FLAG-Ub, UBCH5b (E2), and MDM2, a human p53-specific E3 ligase (Fig. 3B). In contrast, when biotinylated dihydrofolate reductase was used as a substrate, it was not ubiquitinated by either MDM2 or JUL1.
Figure 3.
In vitro ubiquitination of JUL1 on JAV1 as substrate. A, Autoubiquitination activity of JUL1. Biotinylated JUL1 proteins (Bio-JUL1) were incubated with HA-Ub in the presence or absence of UBCH5b (E2). Biotinylated dihydrofolate reductase (Bio-DHFR) served as a negative control. B, Ubiquitination activity of JUL1 on JAV1 substrate. JUL1 or MDM2 (a human p53-specific E3 ligase), which served as a positive control, was incubated in the presence or absence of FLAG-Ub, UBCH5b (E2), and biotinylated substrate proteins (Bio-p53 or Bio-JAV1). Bio-DHFR served as a substrate-negative control. For these assays, the incubated reaction mixtures were immunoprecipitated using streptavidin Alexa Fluor 647 (anti-Biotin), subjected to SDS-PAGE, and probed with anti-HA-HRP (HA-Ub) or anti-FLAG-HRP (FLAG-Ub) antibody. C, Wild type of the biotinylated JAV1 protein (Bio-JAV1) and its mutants, in which either the Lys-52 or Lys-179 residue or both Lys residues in the VQ motif domain were replaced with Thr (Bio-JAV1K52T, Bio-JAV1K179T, and Bio-JAV1K52T-K179T), were incubated with UBCH5b (E2) or FLAG-tagged JUL1 (FLAG-JUL1). Assays in the absence of either UBCH5b or Bio-JAV1 served as controls. The incubation mixtures were immunoprecipitated using streptavidin Alexa Fluor 647 (anti-Biotin), subjected to SDS-PAGE, and probed using anti-HA-HRP (HA-Ub) antibody. IB, immunoblotting; IP, immunoprecipitation.
JAV1 contains two Lys residues, Lys-52 and Lys-179. The level of ubiquitination of JAV1 by JUL1 decreased when a JAV1 mutant, in which either Lys-52 or Lys-179 was replaced by Thr, was used as the substrate in the ubiquitination assay (Fig. 3C). The ubiquitination signal was more markedly decreased when a JAV1 mutant in which both of the Lys residues were replaced with Thr was used as the substrate.
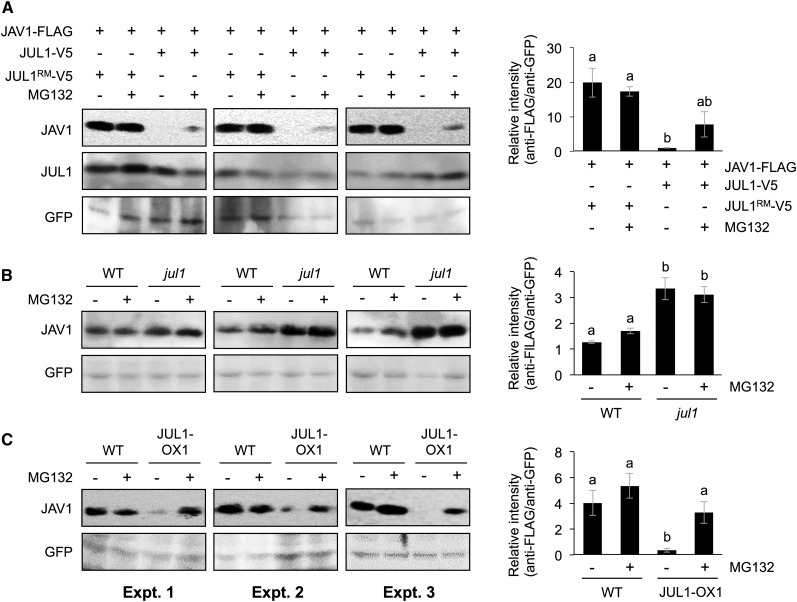
Next, in vivo degradation of JAV1 by the JUL1-UPS was evaluated. FLAG-tagged JAV1 protein (JAV1-FLAG) was expressed with V5-tagged JUL1 or JUL1RM (JUL1-V5 or JUL1RM-V5) in wild-type leaf protoplast cells (Fig. 4A). Irrespective of the presence or absence of MG132, the levels of the transiently expressed JAV1 proteins were well maintained when JUL1RM-V5 was coexpressed. In contrast, JAV1 protein levels decreased when JUL1-V5 was coexpressed in the absence of MG132. In the presence of MG132, JAV1 protein levels tended to recover slightly in comparison with those in the absence of MG132. Such JAV1 degradation in the absence of MG132 also was observed in wild-type and coi1-1 protoplast cells (Supplemental Fig. S2), indicating that COI1-dependent JA signaling is not required for JUL1 ubiquitination activity toward JAV1.
Figure 4.
In vivo protein levels of JAV1 regulated by the JUL1-26S proteasome system. A, The vector constructs of 35SPΩ::FLAG-tagged JAV1 (JAV1-FLAG) and V5-tagged JUL1 or JUL1RM (JUL1-V5 or JUL1RM-V5) were cotransformed with 35SP::GFP into Arabidopsis leaf protoplast cells, and the cells then were treated with MG132 during the incubation. B and C, 35SPΩ::JAV1-FLAG and 35SP::GFP were cotransformed into protoplast cells prepared from wild-type plants (WT) and either the jul1 mutant (B) or transgenic plants overexpressing JUL1 (JUL1-OX1; C), with (+) or without (−) MG132 treatment. Total proteins extracted from the cells were subjected to SDS-PAGE and immunoblotted with FLAG-HRP and GFP antibodies. Mean and se values from three independent experiments (Expt. 1, 2, and 3) are presented in the graphs at right. Means indicated by different letters are significantly different, based on one-way ANOVA with posthoc Tukey’s honestly significant difference (P < 0.05).
In additional assays, JAV1-FLAG was expressed in leaf protoplast cells prepared from the Arabidopsis wild type and the jul1 mutant line (see below; Fig. 4B). Irrespective of the presence or absence of MG132, the JAV1 protein levels were elevated in the jul1 cells compared with the wild-type cells (Fig. 4B). Moreover, JAV1-FLAG was expressed in leaf protoplast cells prepared from the Arabidopsis wild type and a transgenic line overexpressing JUL1 (JUL1-OX1; see below; Fig. 4C). In comparison with the level in the wild type, the JAV1 protein level was decreased in the JUL1-OX1 cells incubated in the absence of MG132 but not in those incubated in the presence of MG132. These findings confirmed that JUL1 contributes to JAV1 degradation through the UPS in leaf cells.
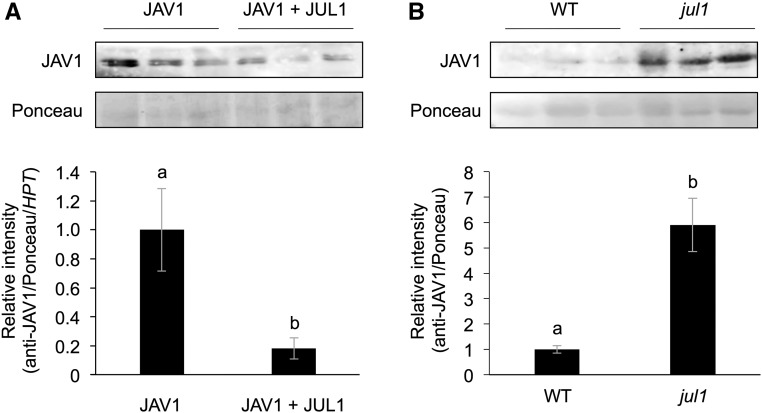
Finally, we evaluated the level of the JAV1 protein in Arabidopsis T87 cells constitutively overexpressing JAV1, with or without JUL1, using an anti-JAV1 antibody, and found a decreased level of JAV1 protein in the cells expressing both JAV1 and JUL1 in comparison with the cells expressing JAV1 without JUL1 (Fig. 5A). In contrast, the JAV1 protein level was elevated in the leaves of the jul1 mutant, in comparison with that in wild-type leaves (Fig. 5B).
Figure 5.
In vivo protein levels of JAV1 in Arabidopsis cells overexpressing JAV1 and Arabidopsis leaves. JAV1 protein levels in Arabidopsis T87 cells overexpressing JAV1 with or without JUL1 (A) and leaves of Arabidopsis wild-type (WT) and jul1 mutant plants (B) are shown. Total proteins extracted from the cells and mature leaves were subjected to SDS-PAGE and immunoblotted with an anti-JAV1 antibody. For A, data were normalized by transcript levels of the transgene of the hygromycin phosphotransferase (HPT) gene. Mean and se values from three independent experiments are presented at bottom. Means indicated by different letters are significantly different, based on Student’s t test (P < 0.05).
Regulation of Transcript Levels of JUL1 and JAV1 via the COI1 System
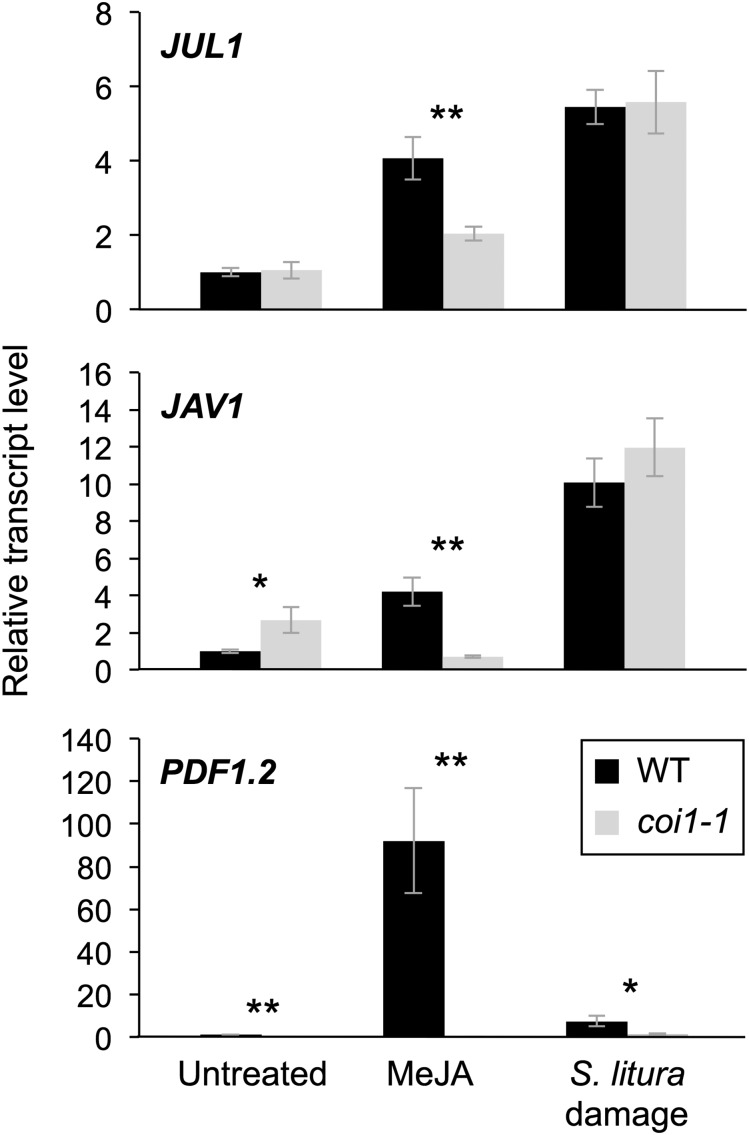
Transcript levels of JUL1, JAV1, and PLANT DEFENSIN1.2 (PDF1.2, a JA-associated defense gene) were elevated in wild-type leaves treated with methyl jasmonate (MeJA) or damaged by larvae of the generalist herbivore Spodoptera litura (Fig. 6). In contrast, the coi1-1 mutant showed defective elevation of these transcript levels in leaves after MeJA treatment. Upon herbivory, however, the transcript levels of JUL1 and JAV1 in coi1-1 leaves showed no significant defect of elevation in comparison with the levels in wild-type leaves, whereas the PDF1.2 transcript level was significantly lower in coi1-1 leaves in comparison with that in wild-type leaves.
Figure 6.
Regulation of transcript levels of JUL1, JAV1, and PDF1.2. Transcript levels are shown for JUL1, JAV1, and PDF1.2 in leaves of Arabidopsis wild-type (WT) and coi1-1 mutant plants in response to MeJA treatment for 4 h and damage with S. litura larvae for 24 h. Transcript levels of genes were measured by reverse transcription quantitative PCR and normalized by those of ACT8. Data represent means and se (n = 5–7). Asterisks indicate significant differences between wild-type and coi1-1 mutant plants, based on Student’s t test (**, P < 0.01 and *, 0.01 ≤ P < 0.05).
Intriguingly, it also should be noted that the transcript level of JAV1 was higher in unstressed coi1-1 leaves than in wild-type leaves.
Phenotypic Characterization of JUL1 Mutants and JUL1-Overexpressing Lines
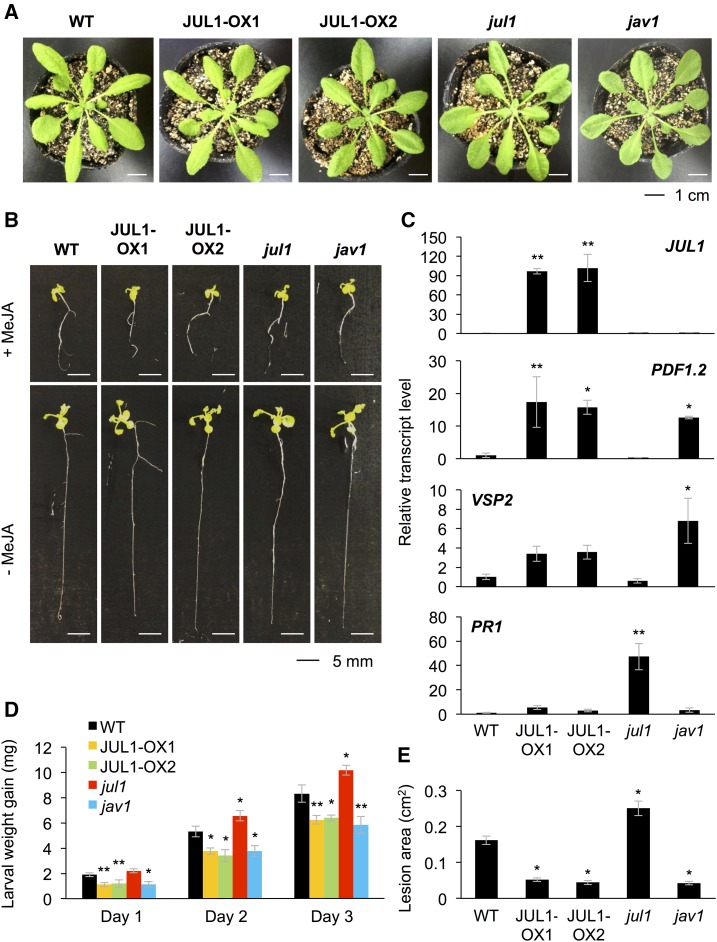
To assess the function of JUL1 in plants, we generated two JUL1-overexpressing lines (JUL1-OX1 and JUL1-OX2), exhibiting 96- and 101-fold increased levels of JUL1 expression, respectively, compared with the level in wild-type leaves (Fig. 7D). These transgenic lines, as well as the jul1 and jav1 knockdown mutants (Supplemental Fig. S3), showed no obvious differences in plant growth, development, or morphology, including root growth, vegetative stage development, or seed number, with or without MeJA treatment, in comparison with wild-type plants (Fig. 7, A and B; Supplemental Figs. S4 and S5).
Figure 7.
The JAV1/JUL1 system is responsible for defense responses but not plant growth or development properties. A and B, Phenotypes of rosette plants (A) and seedlings with (+) or without (−) MeJA treatment (B) of the wild type (WT), JUL1-overexpressing lines (JUL1-OX1 and JUL1-OX2), and jul1 and jav1 mutants. C, Transcript levels of JUL1, PDF1.2, VSP2, and PR1 in leaves of the wild type, JUL-OX lines, and mutants (n = 5–6). D, S. litura larvae were put onto potted plants. The net body weight that larvae gained during 3 d after they had been placed on the plants was determined (n = 11–13). E, Likewise, the fungus B. cinerea was inoculated onto leaves of potted plants, and areas of induced lesions were determined after 3 d (n = 10). For C to E, data represent means and se. Data marked with asterisks are significantly different from those of the wild type, based on one-way ANOVA with Holm’s sequential Bonferroni posthoc test (**, P < 0.01 and *, 0.01 ≤ P < 0.05).
The defense ability of these lines was changed compared with that of wild-type plants. JA-inducible PDF1.2 was highly expressed in the JUL1-OX line and jav1 mutant leaves (Fig. 7C). Another JA-inducible gene, VEGETATIVE STORAGE PROTEIN2 (VSP2), was highly expressed in jav1 mutant leaves but only tended to be slightly more highly expressed in the JUL1-OX line leaves compared with wild-type leaves (Fig. 7C). Conversely, the salicylate (JA-antagonist)-inducible PATHOGENESIS-RELATED1 (Spoel et al., 2003; Beckers and Spoel, 2006) was highly expressed only in jul1 mutant leaves (Fig. 7C).
Next, to confirm the plant defense properties of those lines against larvae of the generalist herbivore S. litura, we assessed the growth of larvae on the potted plants. The larvae on JUL1-OX and jav1 plants exhibited lower weight gain for up to 3 d after the larvae were released, compared with that exhibited by the larvae on wild-type plants (Fig. 7D). In turn, the larvae on jul1 mutant plants exhibited higher weight gain after 2 and 3 d, compared with the weight gains exhibited by the larvae on wild-type plants. The feeding behavior observed at 1 d showed a similar trend to the leaf damage area when JUL1-OX and jav1 leaves were challenged with S. litura larvae for 2 h (Supplemental Fig. S6). Moreover, similar trends were observed for the leaf lesion sizes after infection with the necrotic pathogen B. cinerea for 3 d (Fig. 7E).
DISCUSSION
Arabidopsis JAV1 has been reported to be a negative regulator of JA-dependent defense responses that acts to coordinate the primary COI1-JAZ signaling pathway (Hu et al., 2013; Zhu and Zhu, 2013). JAV1 can form a complex with JAZ8 and TFs, including WRKY51, in order to suppress JA biosynthesis genes under normal conditions. However, in response to herbivory, all of these molecules are dissociated from the complex, JAV1 is then degraded via the UPS, and the free WRKY51 is able to access JA biosynthesis-related genes (Yan et al., 2018). In this study, JUL1 was found to have the characteristics of a RING-type E3 ubiquitin ligase that catalyzes ubiquitination of JAV1 as a substrate, thereby leading to the degradation of JAV1 via the UPS (Figs. 4 and 5). According to data from Hu et al. (2013) and our preliminary AlphaScreen-based assays for interactions between JAV1 and 566 TF proteins (Supplemental Table S2), it appears that the JAV1/JUL1 system is able to control a suite of WRKY family (e.g. WRKY28, WRKY48, and WRKY51) members involved in promoting the transcription of defense-related genes, including PDF1.2 (Zhu and Zhu, 2013). Such a regulatory system involving JAV1 is in accord with the basic nature of the VQ family, whose members interact specifically with the WRKY domain of groups I and IIc WRKY TFs (Cheng et al., 2012). Although Arabidopsis SIB1 (VQ23)/SIB2 (VQ16) and VQ10 have been shown to contribute to the activation of WRKY33 and WRKY8 in plant defense responses (Lai et al., 2011; Chen et al., 2018), respectively, an array of other Arabidopsis VQ proteins, including not only JAV1 but also VQ21/MKS1, VQ12, and VQ29, act as negative regulators of plant defense responses (Andreasson et al., 2005; Petersen et al., 2010; Hu et al., 2013; Wang et al., 2015).
WRKYs function as positive (WRKY28) or negative (WRKY48 and WRKY51) regulators of defense genes (Xing et al., 2008; Gao et al., 2011; Wu et al., 2011). Based on these facts, and given our finding here that both a loss of JAV1 function and a gain of JUL1 function result in enhanced expression of PDF1.2 in leaves (Fig. 7D), we conclude that the orchestration of WRKY-based transcriptional machinery, according to the level of JAV1-based defense, is ultimately related to the positive regulation of the defense response. However, this should be flexible to enable adjustment to the circumstances of the plants, for example, whether they are being stressed, in which case the transcriptional modes can be drastically reprogrammed in plant cells.
It also appeared that JUL1 and JAV1 are constitutively expressed during several stages of Arabidopsis development, based on the microarray database GENEVESTIGATOR (Zimmermann et al., 2004; Supplemental Fig. S7). Remarkably, in the leaf senescence stage, transcripts of JUL1 and JAV1 showed high and low levels, respectively, indicating that JUL1 might play a role in senescence. Moreover, both JUL1 and JAV1 transcript levels were found to be attenuated in coi1-1 leaves as compared with wild-type leaves in response to MeJA (Fig. 6), indicating that those genes are positively regulated by a COI1-dependent signaling cascade. Intriguingly, when plants were challenged with herbivores, however, neither JUL1 nor JAV1 transcripts appeared to be induced in a COI1-dependent manner. Taking all these results together, we suggest that COI1-independent factor(s) may strongly contribute to the regulation of JUL1 and JAV1 transcript levels in response to herbivory. Interestingly, the level of JAV1 transcripts was found to be negatively regulated by the COI1-dependent signaling cascade (Fig. 6). Our observation of the negative impact of COI1 on JAV1 expression is in accord with data in the microarray database GENEVESTIGATOR (Zimmermann et al., 2004; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21762) showing that JAV1 transcript levels are higher in coi1-2 leaves than in wild-type leaves under normal growth conditions (Supplemental Fig. S8). However, what this regulation implies for the JAV1/JUL1 module remains to be clarified.
RING-type E3 ligases play vital roles in the regulation of physiological and morphological processes in plants in response to various internal and external stimuli (Duplan and Rivas, 2014; Shabek and Zheng, 2014; Ding et al., 2015; Lazaro et al., 2015; Zhang et al., 2015). Nonetheless, to the best of our knowledge, RGLG3/4 and KEG are the only JA-associated RING-type E3 ligases reported thus far (Zhang et al., 2012; Pauwels et al., 2015). However, it was reported recently that RGLG3 and RGLG4 can ubiquitinate GRXS17, and thereby control the redox state of proteins, and that the RGLG system is not strongly relevant to the JA response module, in contrast to the findings in the previous report (Nagels Durand et al., 2016a). The findings of our study clearly showed that JUL1 is a novel RING-type E3 involved in JA signaling. Notably, some RING-type E3 ligases have been shown to have a diverse range of substrate targets. One of the most striking examples is CONSTITUTIVE PHOTOMORPHOGENIC1, which is involved in a wide range of plant growth and developmental processes (Kim et al., 2017). It thus remains possible that JUL1 has multiple substrate targets involved in an array of functions in plant defense, stress tolerance, and other physiological processes, and this issue should be assessed in the future.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0, its mutants (SALK_146039C [jav1] and SALK_101465C [jul1]), and transgenic plants overexpressing JUL1 (see below) were grown in plastic pots for 4 to 5 weeks in a growth chamber at 22°C ± 1°C with a photoperiod of 14 h (80 µE m–2 s–1). The coi1-1 seeds were germinated on one-half-strength Murashige and Skoog (1/2 MS) medium supplemented with 0.5% (w/v) Suc, 0.8% (w/v) agar, and 50 µm MeJA (Wako Pure Chemical Industrials) to screen the individuals showing normal root growth for 2 weeks (Xie et al., 1998). Homozygous SALK lines were confirmed according to the method described at http://signal.salk.edu/tdnaprimers.2.html.
Primers
Primers used for all the polymerase chain reactions (PCR) in this study are listed in Supplemental Table S3.
Phylogeny Analysis
The phylogenetic tree was generated using the one-click mode of the Phylogeny.fr pipeline (http://www.phylogeny.fr/; Dereeper et al., 2008).
Generation of Transgenic Arabidopsis Plants
The full-length coding region of JUL1 (at5g10650) was inserted into binary plasmid pMDC32 (2× cauliflower mosaic virus 35S promoter [35SP]::the Gateway cassette [GW] region::nopaline synthase terminator [NOST]) using the Gateway cloning system (Thermo Fisher Scientific). The resulting vector, pMDC32-JUL1, was transformed into Agrobacterium tumefaciens strain EHA105 by electroporation. Columbia-0 wild-type Arabidopsis plants that had been grown for 6 to 7 weeks were transformed via the floral dip transformation method (Clough and Bent, 1998). Transgenic T1 seeds from each transformant were tested for germination on 1/2 MS medium supplemented with 30 mg L–1 hygromycin. T2 seeds were harvested from each individual. T2 plants that showed a segregation ratio of about 3:1 were tested for hygromycin resistance again. T3 homozygous plant lines were used for further analyses.
Chemical and Herbivore Treatment
For chemical treatment, potted Arabidopsis plants were sprayed evenly with 1 mL of aqueous solution (0.5% [v/v] ethanol) of MeJA (0.2 mm) and incubated in a growth chamber at 22°C ± 1°C (14-h photoperiod at a light intensity of 80 µE m–2 s–1) for 4 h.
Spodoptera litura were reared on an artificial diet (Insecta LF; Nihon Nosan Nogyo) in the laboratory at 24°C ± 1°C with a photoperiod of 16 h. For herbivore treatment, four third-instar larvae were released on potted Arabidopsis plants in a growth chamber at 22°C ± 1°C (14-h photoperiod at a light intensity of 80 µE m–2 s–1) for 24 h.
Cell-Free Protein Synthesis
Cell-free protein synthesis was carried out according to the method described previously (Ramadan et al., 2015; Takahashi et al., 2016). Primers used are listed in Supplemental Table S3.
AlphaScreen System
We performed the AlphaScreen-based protein-protein interaction assay to screen the interaction of 210 Arabidopsis RING-type E3 ligases (Ramadan et al., 2015) and 566 Arabidopsis TFs (Nozawa et al., 2009) with JAV1. Assays for interaction between JUL1 and the JAV1 homolog (at4g15120) or between the JUL1 homolog (at5g24870) and JAV1 also were carried out. Assays were performed in 10 µL of 100 mm Tris-HCl (pH 8), 0.1% (v/v) Tween 20, 1 mg mL–1 BSA (AlphaScreen buffer), and 1 μL of biotinylated JAV1 (or JAV1 homolog) and FLAG-adapted E3 ligase (either JUL1 or a JUL1 homolog) or TFs per well at 25°C for 1 h on a 384-well OptiPlate (Perkin-Elmer). In accord with the AlphaScreen IgG (Protein A) detection kit (Perkin-Elmer) instruction manual, 10 μL of detection mixture containing AlphaScreen buffer, 5 μg mL–1 anti-FLAG M2 antibody (Sigma-Aldrich), 0.1 μL of streptavidin-coated donor beads, and 0.1 μL of Protein A-coated acceptor beads was added to each well of a 384-well OptiPlate, followed by an additional incubation at 25°C for 1 h. Luminescence was analyzed using the AlphaScreen detection program (Perkin-Elmer). All data represent averages of three independent experiments, and the background was controlled using a DHFR from Escherichia coli.
Mutagenesis
JAV1 point mutations (Lys-52 and Lys-179) were obtained by inverse PCR using In-Fusion HD Cloning Plus (Takara Bio), primers (JAV1-K1-fw/JAV1-K1-rv and JAV2-K1-fw/JAV1-K2-rv [Supplemental Table S3]), and JAV1 cDNA as template.
Similarly, JUL1 protein mutations were obtained at Cys-493, His-495, and His-498 of the RING domain using primers (JUL1-RING-M-F and JUL1-RING-M-R [Supplemental Table S3]) and JUL1 cDNA as template, to obtain JUL1RM based on Stone et al. (2005).
Subcellular Localization and BiFC Assays in Protoplast Cells
The full-length coding regions of JUL1 and JAV1 (except for the stop codons) were cloned into Gateway destination plasmid pGWB451 (35SP::GW::GFP; Nakagawa et al., 2007), resulting in the construction of the vector pGWB451-JUL1-GFP. This vector, plus pGWB452 (35SP::GFP::GW), which served as a control, were transiently transformed into Arabidopsis mesophyll protoplasts as described previously (Yoo et al., 2007; Wu et al., 2009). The transformed protoplasts were incubated for 18 to 20 h in the dark at 25°C, and GFP fluorescence was observed in the protoplasts with a BZ-X700 fluorescence microscope (Keyence).
The full-length coding regions of JUL1, JUL1RM (see below), and JAV1 were inserted (separately) into the Gateway destination vector 35SPΩ-GW-V5-nYFP, along with the N-terminal fragment of YFP, and 35SPΩ-GW-V5-cYFP with the C-terminal fragment of YFP, respectively (Nemoto et al., 2017). Ten micrograms of each vector was cotransformed into Arabidopsis protoplast cells. The transformed protoplasts were incubated for 14 h in the dark at 25°C, and then 50 µm MG132 (Wako) in 0.1% (v/v) dimethyl sulfoxide (DMSO) or 0.1% (v/v) DMSO solution (control) was applied. Following an additional 6-h incubation in the dark at 25°C, the protoplast cells were incubated in WI solution (5 mm MES [pH 5.7], 0.4 m mannitol, and 20 mm KCl) with 1 µg mL−1 4′,6-diamino-phenylindole (Sigma-Aldrich) for 5 min before observation. The resultant YFP fluorescence was observed in the protoplasts using a fluorescence microscope (BZ-X700).
In Vitro Ubiquitination Assay
The ubiquitination assays for JUL1 were carried out as described previously (Ramadan et al., 2015). The assays were performed in a 10-μL reaction mixture containing 1 μL of recombinant MDM2 (AF527840; mouse double minute 2 homolog) or JUL1 protein, 1 μL of recombinant substrate (P53 [AB082923; p53 tumor suppressor targeted by MDM2 E3 ubiquitin ligase], JAV1 or JAV1 mutant [see above]), 1.2 μm UbcH5b (Enzo Life Sciences), 20 mm Tris-HCl, pH 7.5, 0.2 mm DTT, 5 mm MgCl2, 10 μm zinc acetate, 3 mm ATP, 1 mg mL–1 BSA, and 400 nm FLAG-tagged or HA-tagged human ubiquitin (FLAG-Ub or HA-Ub; Boston Biochem) at 37°C for 3 h. The reactions were terminated by the addition of 5 μL of 3× SDS sample buffer and boiling for 5 min, and the products then were analyzed by 10% SDS-polyacrylamide gel electrophoresis (PAGE) followed by immunoblotting (see below) using anti-FLAG-HRP (Sigma-Aldrich) or anti-HA-HRP (clone 3F10) antibody (Roche Applied Science).
In Vivo Protein Degradation Assay
Polyethylene glycol-mediated DNA transfection of Arabidopsis mesophyll protoplasts from wild-type, jul1, and JUL1-OX1 backgrounds was performed as described previously (Yoo et al., 2007; Wu et al., 2009) with slight modifications. The full-length coding regions of JAV1 (except for the stop codons) were cloned into Gateway destination plasmid p35SΩ::GW::3×FLAG::NOST (Nemoto et al., 2017), resulting in the construction of the vector p35SΩ::JAV1 tagged with 3×FLAG at the C terminus (JAV1-FLAG)::NOST. Likewise, the full-length coding regions of JUL1 and JUL1RM (except for the stop codons) were cloned into Gateway destination plasmid p35SΩ::GW::V5::NOST (Nemoto et al., 2017), resulting in the construction of the vector p35SΩ::JUL1 tagged with V5 at the C terminus (JUL1-V5)::NOST and p35SΩ::JUL1RM tagged with V5 at the C terminus (JUL1RM-V5)::NOST, respectively. Thirty micrograms of JAV1-FLAG vector, along with 6 µg of pGWB452 (35SP::GFP::GW) vector (Nemoto et al., 2017), were transformed into 2 × 105 wild-type, jul1, or JUL1-OX1 cells. Otherwise, 30 µg of JAV1-FLAG vector and 20 µg of JUL1-V5 vector, along with 6 µg of pGWB452 (35SP::GFP::GW) vector, were transformed into 2 × 105 wild-type cells. Following incubation overnight in the dark at 22°C ± 1°C, 6 h before harvest, protoplasts were treated with 50 μm MG132 in 0.1% (v/v) DMSO or with 0.1% (v/v) DMSO (control). Total proteins were extracted from the protoplasts with 3× sample buffer. JAV1-FLAG protein was analyzed by immunoblotting using anti-FLAG HRP (see below).
For coimmunoprecipitation assays, 30 µg of JAV1-V5 vector (p35SΩ::JAV1 tagged with V5 at the C terminus) and 20 µg of JUL1-FLAG vector (p35SΩ::JUL1 tagged with 3×FLAG at the C terminus) were transformed into 1 × 106 wild-type cells; the protoplasts were incubated overnight and treated with 50 μm MG132 for 6 h before being homogenized with immunoprecipitation lysis/wash buffer (Thermo Fisher Scientific) supplemented with 50 μm MG132 and 1% (v/v) Protease Inhibitor Cocktail (Sigma-Aldrich). Immunoprecipitation of JUL1-FLAG protein from the protoplast lysate was performed using a Pierce Crosslink Magnetic IP/Co-IP Kit (Thermo Fisher Scientific) and anti-FLAG (DYKDDDDK) monoclonal primary antibody (1E6; Wako) following the manufacturer’s protocol. After the beads were washed twice with immunoprecipitation lysis/wash buffer, the immunoprecipitated proteins were eluted with 50 μL of elution buffer. JAV1-V5 protein was analyzed by immunoblotting using anti-V5 HRP (see below). RbcL was visualized as a loading control using Ponceau S staining solution.
Generation and Analysis of Transgenic Arabidopsis T87 Cultured Cells Expressing JAV1-GFP and JUL1
The full-length coding region of JAV1 (at3g22160) was inserted into binary plasmid pGWB5 (35SP::GW:NOST) to construct pGWB5-JAV1. Additionally, the expression cassette of JUL1 in pMDC32-JUL1 was subcloned into pGWB5-JAV1 to construct pGWB5-JAV1-JUL1. pGWB5-JAV1 and pGWB5-JAV1-JUL1 were transformed into A. tumefaciens strain GV3103 by electroporation. Arabidopsis T87 cells were transformed and cultured in Jouanneau and Péaud-Lenoël medium supplemented with 10 mg L–1 hygromycin according to the method described previously (Ogawa et al., 2008). Total proteins were extracted from the transgenic cells with extraction buffer (50 mm HEPES [pH 7.4], 5 mm EDTA, 5 mm EGTA, 50 mm β-glycerophosphate, 10 mm Na3VO4, 10 mm NaF, 2 mm DTT, and 1% (v/v) Triton-X100). JAV1-GFP protein was analyzed by immunoblotting using anti-JAV1 antibody (see below). Data were normalized by transcript levels of the hygromycin phosphotransferase gene.
Immunoblot Analysis
Ten- to 15-μL samples of the boiled proteins in SDS sample buffer were subjected to 10% SDS-PAGE, and the proteins were transferred to a polyvinylidene difluoride membrane (ATTO) using the Trans-BlotTurbo Blotting System (Bio-Rad). Immunoblotting was carried out with anti-FLAG-HRP-conjugated antibody (Wako Pure Chemical Industries), anti-V5-HRP-conjugated antibody (Wako Pure Chemical Industries), or anti-GFP chicken IgY primary antibody (GeneTex) and then with anti-chicken IgY-HRP-conjugated secondary antibody (Promega). Detection was performed using Immobilon Western Chemiluminescent HRP Substrate (ECL; Millipore) according to the manufacturer’s procedure. The signals were visualized using ImageQuant LAS 4000 mini (GE Healthcare).
Immunoblotting for JAV1 was carried out with anti-JAV1 antibody obtained by immunizing rabbits with a peptide antigen (with peptide sequence CWQYNFQPHAPLQPPQR; Cosmo Bio). Unlabeled anti-rabbit IgG (#3678; Cell Signaling Technology) and HRP-linked anti-mouse IgG (#7076) antibodies were used as secondary and tertiary antibodies, respectively. The membranes were soaked with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific), and the signals were detected with an ImageQuant LAS-4000 imaging system (GE Healthcare).
RNA Extraction, cDNA Synthesis, and Reverse Transcription Quantitative PCR
Approximately 100 mg of leaf tissues or 1 mL of cell culture was homogenized in liquid nitrogen, and total RNA was isolated and purified using Sepasol-RNA I Super G (Nacalai Tesque) following the manufacturer’s protocol. First-strand cDNA synthesis and quantitative PCR were performed according to the method described previously (Uemura et al., 2018). Relative transcript abundances were determined after normalization of raw signals with the transcript abundance of the housekeeping gene ACT8 (at1g49240). We did not use samples and data when sufficient amounts of RNA were not isolated from leaves or when abnormal quantification cycle values for the actin gene were obtained. Finally, replicate analyses were conducted with five or six independent samples, except for the data in Figure 5B (three independent samples).
Root Length Measurement
Plant seedlings (14 d old) were grown on 1/2 MS medium with or without 50 µm MeJA. Root lengths were determined using ImageJ software (version 1.50i; Schneider et al. (2012)).
Herbivore Assay
We performed assays to assess the growth of S. litura larvae at 22°C ± 1°C (14-h photoperiod at 80 µE m–2 s–1). Third-instar larvae were initially weighed (1.6–2.1 mg), and each larva was released onto a potted plant for 3 d. The net body weight that S. litura larvae gained each of the following 3 d was determined. When a larva died or was lost during the assay, we excluded that sample, and final replicate analyses were conducted with 12 to 15 independent samples.
For leaf-consumption assays, five third-instar larvae, starved overnight, were released onto the shoots of potted plants for 2 h. The leaves then were scanned, and the total leaf area and the consumed leaf area were determined using ImageJ. Replicate analyses were conducted with eight to 10 independent samples.
Pathogen Infection Assay
Botrytis cinerea (strain IuRy-1) was grown on potato Suc agar under black light at 25°C for 1 week to promote the formation of conidia. For inoculation, 10 µL of conidial spore suspension (2 × 105 conidia mL−1, in 2.5% Glc) was spotted onto the left and right sides of the upper surface of a leaf of the potted plants. The inoculated plants were covered with a plastic sheet to enable the maintenance of humidity. The plants were placed in a growth chamber at 24°C ± 1°C for 3 d. The leaf lesion area was analyzed using ImageJ.
Statistical Analyses
We performed Student’s t tests for pairwise analysis and one-way ANOVA with Holm’s sequential Bonferroni posthoc test or Tukey’s honestly significant difference test using the program (http://astatsa.com/OneWay_Anova_with_TukeyHSD/) for comparing multiple samples.
Accession Numbers
The sequences and detailed information for the genes in this study can be found in the Arabidopsis Biological Resource Center database under the accession numbers listed in Supplemental Table S3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Interactions between JUL1 and JAV1 homolog (at4g15120) and between JAV1 and JUL1 homolog (at5g24870).
Supplemental Figure S2. COI1-independent protein levels of JAV1 regulated by the JUL1-26S proteasome system.
Supplemental Figure S3. Molecular analysis of T-DNA insertion mutants and gene expression profiling in the mutants.
Supplemental Figure S4. Root length of the seedlings of the wild type, JUL1-overexpressing lines, and jul1 and jav1 mutants with or without MeJA treatment.
Supplemental Figure S5. Root length of the seedlings of wild type, JUL1-overexpressing lines, and jul1 and jav1 mutants following with or without MeJA treatment.
Supplemental Figure S6. Leaf-consumption of S. litura larvae.
Supplemental Figure S7. Transition of transcript levels of JUL1 and JAV1 during the Arabidopsis life cycle, based on the GENEVESTIGATOR database.
Supplemental Figure S8. Transcript levels of JAV1 in Arabidopsis wild-type and coi1-2 mutant leaves 4 h after application of MeJA, based on the GENEVESTIGATOR database.
Supplemental Table S1. AlphaScreen-based binding intensity to JAV1.
Supplemental Table S2. AlphaScreen-based binding strength of TFs to JAV1.
Supplemental Table S3. Primers.
Acknowledgments
The plasmids pGWB451 and pGWB452 were kindly provided by Dr. Tsuyoshi Nakagawa (Shimane University).
Footnotes
This work was supported in part by a Japan Society for the Promotion of Science KAKENHI grant to G.A. (16K07407) and by Ministry of Education, Culture, Sports, Science, and Technology Grants-in-Aid for Scientific Research on Innovative Areas to G.A. (18H04630 and 18H04786).
Articles can be viewed without a subscription.
References
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NH, Zhu S, Qiu JL, Micheelsen P, Rocher A, Petersen M, et al. (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 24: 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers GJ, Spoel SH (2006) Fine-tuning plant defence signalling: Salicylate versus jasmonate. Plant Biol (Stuttg) 8: 1–10 [DOI] [PubMed] [Google Scholar]
- Browse J. (2009) Jasmonate passes muster: A receptor and targets for the defense hormone. Annu Rev Plant Biol 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Campos ML, Yoshida Y, Major IT, de Oliveira Ferreira D, Weraduwage SM, Froehlich JE, Johnson BF, Kramer DM, Jander G, Sharkey TD, et al. (2016) Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun 7: 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hellmann H (2013) Plant E3 ligases: Flexible enzymes in a sessile world. Mol Plant 6: 1388–1404 [DOI] [PubMed] [Google Scholar]
- Chen M, Ni M (2006) RFI2, a RING-domain zinc finger protein, negatively regulates CONSTANS expression and photoperiodic flowering. Plant J 46: 823–833 [DOI] [PubMed] [Google Scholar]
- Chen J, Wang H, Li Y, Pan J, Hu Y, Yu D (2018) Arabidopsis VQ10 interacts with WRKY8 to modulate basal defense against Botrytis cinerea. J Integr Plant Biol 60: 956–969 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zhou Y, Yang Y, Chi YJ, Zhou J, Chen JY, Wang F, Fan B, Shi K, Zhou YH, et al. (2012) Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol 159: 810–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chini A, Gimenez-Ibanez S, Goossens A, Solano R (2016) Redundancy and specificity in jasmonate signalling. Curr Opin Plant Biol 33: 147–156 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- David Y, Ternette N, Edelmann MJ, Ziv T, Gayer B, Sertchook R, Dadon Y, Kessler BM, Navon A (2011) E3 ligases determine ubiquitination site and conjugate type by enforcing specificity on E2 enzymes. J Biol Chem 286: 44104–44115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Zhang B, Qin F (2015) Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 27: 3228–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplan V, Rivas S (2014) E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front Plant Sci 5: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao QM, Venugopal S, Navarre D, Kachroo A (2011) Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol 155: 464–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens J, Mertens J, Goossens A (2017) Role and functioning of bHLH transcription factors in jasmonate signalling. J Exp Bot 68: 1333–1347 [DOI] [PubMed] [Google Scholar]
- Guo Q, Major IT, Howe GA (2018) Resolution of growth-defense conflict: Mechanistic insights from jasmonate signaling. Curr Opin Plant Biol 44: 72–81 [DOI] [PubMed] [Google Scholar]
- Hu P, Zhou W, Cheng Z, Fan M, Wang L, Xie D (2013) JAV1 controls jasmonate-regulated plant defense. Mol Cell 50: 504–515 [DOI] [PubMed] [Google Scholar]
- Jing Y, Lin R (2015) The VQ motif-containing protein family of plant-specific transcriptional regulators. Plant Physiol 169: 371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Song JT, Seo HS (2017) COP1 regulates plant growth and development in response to light at the post-translational level. J Exp Bot 68: 4737–4748 [DOI] [PubMed] [Google Scholar]
- Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z (2011) Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23: 3824–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Mouriz A, Piñeiro M, Jarillo JA (2015) Red light-mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis. Plant Cell 27: 2437–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim WT (2011) Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol Cells 31: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D, Peeters N, Rivas S (2012) Ubiquitination during plant immune signaling. Plant Physiol 160: 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J (1997) Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miricescu A, Goslin K, Graciet E (2018) Ubiquitylation in plants: Signaling hub for the integration of environmental signals. J Exp Bot 69: 4511–4527 [DOI] [PubMed] [Google Scholar]
- Nagels Durand A, Iñigo S, Ritter A, Iniesto E, De Clercq R, Staes A, Van Leene J, Rubio V, Gevaert K, De Jaeger G, et al. (2016a) The Arabidopsis iron-sulfur protein GRXS17 is a target of the ubiquitin E3 ligases RGLG3 and RGLG4. Plant Cell Physiol 57: 1801–1813 [DOI] [PubMed] [Google Scholar]
- Nagels Durand A, Pauwels L, Goossens A (2016b) The ubiquitin system and jasmonate signaling. Plants (Basel) 5: E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nemoto K, Ramadan A, Arimura GI, Imai K, Tomii K, Shinozaki K, Sawasaki T (2017) Tyrosine phosphorylation of the GARU E3 ubiquitin ligase promotes gibberellin signalling by preventing GID1 degradation. Nat Commun 8: 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa A, Matsubara Y, Tanaka Y, Takahashi H, Akagi T, Seki M, Shinozaki K, Endo Y, Sawasaki T (2009) Construction of a protein library of Arabidopsis transcription factors using a wheat cell-free protein production system and its application for DNA binding analysis. Biosci Biotechnol Biochem 73: 1661–1664 [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Dansako T, Yano K, Sakurai N, Suzuki H, Aoki K, Noji M, Saito K, Shibata D (2008) Efficient and high-throughput vector construction and Agrobacterium-mediated transformation of Arabidopsis thaliana suspension-cultured cells for functional genomics. Plant Cell Physiol 49: 242–250 [DOI] [PubMed] [Google Scholar]
- Okada K, Abe H, Arimura G (2015) Jasmonates induce both defense responses and communication in monocotyledonous and dicotyledonous plants. Plant Cell Physiol 56: 16–27 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Ritter A, Goossens J, Durand AN, Liu H, Gu Y, Geerinck J, Boter M, Vanden Bossche R, De Clercq R, et al. (2015) The RING E3 ligase KEEP ON GOING modulates JASMONATE ZIM-DOMAIN12 stability. Plant Physiol 169: 1405–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K, Qiu JL, Lütje J, Fiil BK, Hansen S, Mundy J, Petersen M (2010) Arabidopsis MKS1 is involved in basal immunity and requires an intact N-terminal domain for proper function. PLoS ONE 5: e14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The Jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan A, Nemoto K, Seki M, Shinozaki K, Takeda H, Takahashi H, Sawasaki T (2015) Wheat germ-based protein libraries for the functional characterisation of the Arabidopsis E2 ubiquitin conjugating enzymes and the RING-type E3 ubiquitin ligase enzymes. BMC Plant Biol 15: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MY, Cho SK, Kim WT (2010) The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol 154: 1983–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S (2012) The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol 196: 13–28 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N, Zheng N (2014) Plant ubiquitin ligases as signaling hubs. Nat Struct Mol Biol 21: 293–296 [DOI] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D (2011) The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Xie D (2013) Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol Plant 6: 1065–1073 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux JP, Brown R, Kazan K, et al. (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Uematsu A, Yamanaka S, Imamura M, Nakajima T, Doi K, Yasuoka S, Takahashi C, Takeda H, Sawasaki T (2016) Establishment of a wheat cell-free synthesized protein array containing 250 human and mouse E3 ubiquitin ligases to identify novel interaction between E3 ligases and substrate proteins. PLoS ONE 11: e0156718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Uemura T, Yashiro T, Oda R, Shioya N, Nakajima T, Hachisu M, Kobayashi S, Nishiyama C, Arimura GI (2018) Intestinal anti-inflammatory activity of perillaldehyde. J Agric Food Chem 66: 3443–3448 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wang H, Hu Y, Pan J, Yu D (2015) Arabidopsis VQ motif-containing proteins VQ12 and VQ29 negatively modulate basal defense against Botrytis cinerea. Sci Rep 5: 14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B (2013) Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111: 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis Sandwich: A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LT, Zhong GM, Wang JM, Li XF, Song X, Yang Y (2011) Arabidopsis WRKY28 transcription factor is required for resistance to necrotrophic pathogen, Botrytis cinerea. Afr J Microbiol Res 5: 5481–5488 [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xing DH, Lai ZB, Zheng ZY, Vinod KM, Fan BF, Chen ZX (2008) Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol Plant 1: 459–470 [DOI] [PubMed] [Google Scholar]
- Yan C, Fan M, Yang M, Zhao J, Zhang W, Su Y, Xiao L, Deng H, Xie D (2018) Injury activates Ca2+/calmodulin-dependent phosphorylation of JAV1-JAZ8-WRKY51 complex for jasmonate biosynthesis. Mol Cell 70: 136–149.e7 [DOI] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang H, Cui F, Wu Y, Lou L, Liu L, Tian M, Ning Y, Shu K, Tang S, Xie Q (2015) The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27: 214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wu Q, Ren J, Qian W, He S, Huang K, Yu X, Gao Y, Huang P, An C (2012) Two novel RING-type ubiquitin ligases, RGLG3 and RGLG4, are essential for jasmonate-mediated responses in Arabidopsis. Plant Physiol 160: 808–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhu JK (2013) Double repression in jasmonate-mediated plant defense. Mol Cell 50: 459–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]