Natural variation of 9-cis-epoxycarotenoid dioxygenase 3 (NCED3) is associated with altered stress-induced abscisic acid accumulation and NCED3 post-translational processing in Arabidopsis.
Abstract
The stress hormone abscisic acid (ABA) is critical for drought resistance; however, mechanisms controlling ABA levels are unclear. At low water potential, ABA accumulation in the Arabidopsis (Arabidopsis thaliana) accession Shahdara (Sha) was less than that in Landsberg erecta (Ler) or Columbia. Analysis of a Ler × Sha recombinant inbred line population revealed a single major-effect quantitative trait locus for ABA accumulation, which included 9-cis-epoxycarotenoid dioxygenase3 (NCED3) as a candidate gene. NCED3 encodes a rate-limiting enzyme for stress-induced ABA synthesis. Complementation experiments indicated that Sha has a reduced-function NCED3 allele. Compared with Ler, Sha did not have reduced NCED3 gene expression or protein level but did have four amino acid substitutions within NCED3. Sha differed from Ler in the apparent molecular mass of NCED3, indicative of altered NCED3 proteolytic processing in the chloroplast. Site-directed mutagenesis demonstrated that substitution at amino acid 271 was critical for the altered NCED3 molecular mass pattern, while the other Sha NCED3 polymorphisms were also involved in the reduced ABA accumulation. Sha did not have a reduced level of thylakoid-bound NCED3 but did differ from Ler in the apparent molecular mass of stromal NCED3. As Sha was not impaired in known factors critical for NCED3 function in ABA synthesis (expression, chloroplast import, and thylakoid binding), the differences between Ler and Sha NCED3 may affect NCED3 activity or other factors influencing NCED3 function. These results identify functionally important sites on NCED3 and indicate a complex pattern of NCED3 posttranslational regulation in the chloroplast.
During periods of drought stress and reduced water potential (ψw), plant endogenous abscisic acid (ABA) levels increase dramatically. Increased ABA controls rapid responses such as stomatal regulation and gene expression and also influences longer term phenotypes such as water use efficiency, shoot and root growth, and developmental changes (Finkelstein, 2013; Verslues, 2016). Analysis of ABA synthesis mutants has demonstrated that de novo ABA synthesis is required for ABA accumulation induced by low ψw and salt stress (Nambara et al., 1998; Ruggiero et al., 2004; Verslues and Bray, 2006). A key rate-limiting step in ABA synthesis is the cleavage of the carotenoids 9-cis-neoxanthin and 9-cis-violoxanthin in the chloroplast to yield xanthoxin, which is exported to the cytoplasm and metabolized to ABA (Finkelstein, 2013). This carotenoid cleavage reaction is catalyzed by the 9-cis-epoxycartenoid dioxygenases (NCEDs). Arabidopsis (Arabidopsis thaliana) has six NCEDs involved in ABA synthesis as well as three closely related carotenoid cleavage dioxygenases that metabolize other carotenoids (Tan et al., 2003). Arabidopsis NCEDs have partially overlapping functions. For example, NCED6 and NCED9 are important for ABA synthesis during seed development and maturation (Lefebvre et al., 2006), and NCED5 has some effect on both seed and vegetative ABA levels (Frey et al., 2012). However, numerous studies have shown that NCED3 gene expression is rapidly induced by drought, salt, and other stresses and have demonstrated that NCED3 has the predominant role in stress-induced ABA accumulation in vegetative tissue (Iuchi et al., 2001; Tan et al., 2003; Ruggiero et al., 2004) and also influences seed ABA levels (Ruggiero et al., 2004). Most recent studies of NCED3 have focused on understanding how its gene expression is induced by stress.
NCEDs have N-terminal stroma-targeting domains of approximately 40 to 50 amino acids to mediate plastid localization (Qin and Zeevaart, 1999; Tan et al., 2001). The maize (Zea mays) NCED Viviparous14 (VP14) has amphipathic helices and additional N-terminal sequences that mediate attachment to the thylakoid membrane (Messing et al., 2010). Binding of NCEDs to specific sites on the thylakoid membrane is thought to be essential for enzymatic activity by allowing the lipid-soluble carotenoid substrate to access the enzyme active site (Tan et al., 2001; Messing et al., 2010). Arabidopsis NCED3 has similar amphipathic helices, and it has been observed that the thylakoid-bound form accumulates during stress when ABA levels increase but decreases upon stress relief when ABA levels decline (Endo et al., 2008). In contrast, a lower molecular mass stromal form persisted during rehydration (Endo et al., 2008). This has led to the idea that release of NCED3 from the thylakoid membrane, presumably by proteolytic cleavage, is a way to regulate ABA synthesis. NCED-binding sites on the thylakoid membrane as well as the proteases involved and the sites where NCEDs are cleaved to release from the thylakoid membrane remain unknown.
Arabidopsis is naturally distributed across a range of environments that vary in environmental factors such as the amount and timing of rainfall (Lasky et al., 2012). Among Arabidopsis accessions, there is substantial natural variation in drought-related traits such as water use efficiency, growth, gene expression, Pro accumulation, ABA accumulation, and others (Des Marais et al., 2012; Lasky et al., 2014; Lovell et al., 2015; Bac-Molenaar et al., 2016; Mojica et al., 2016; Kalladan et al., 2017). The Shahdara accession (Sha; also referred to as Shakdara) differs from commonly used reference accessions such as Columbia (Col) and Landsberg erecta (Ler) in many traits, in particular, drought-related traits (Loudet et al., 2002; Bouchabke et al., 2008; Ren et al., 2010; Vallejo et al., 2010; Jasinski et al., 2012; Kesari et al., 2012; Pineau et al., 2012; Angelovici et al., 2013; Sharma et al., 2013; Szymańska et al., 2015). The type of local environment that Sha is adapted to, as well as whether Sha should be considered more or less drought tolerant than other accessions, are unclear (Trontin et al., 2011). It is clear, however, that Sha and other accessions from central Asia can be a source of divergent and informative alleles of many genes.
We found that Sha had a reduced level of low ψw-induced ABA accumulation compared with Ler or Col-0. Phenotyping of a Ler × Sha recombinant inbred line (RIL) population for ABA accumulation identified a single large-effect quantitative trait locus (QTL) containing NCED3. Sha NCED3 differed from Ler in four amino acids and had an altered molecular mass pattern of the cleaved, stromal-localized form of NCED3. These data identify new functionally important sites in NCED3 and also indicate complex posttranslational processing of NCED3.
RESULTS
A Single Major QTL Is Involved in Reduced ABA Accumulation of Sha Compared with Ler at Low ψw
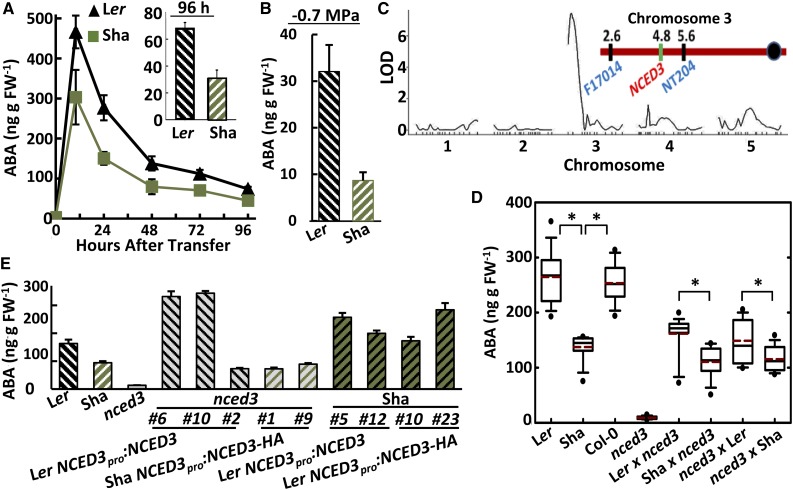
Transfer of Ler and Sha seedlings to low ψw led to a rapid increase in ABA levels, which reached a peak at approximately 10 h and then declined as the plants acclimated to the reduced ψw (Fig. 1A). By 96 h after transfer, ABA levels had reached a nearly steady value that was less than the peak ABA accumulation but still about 50-fold higher than the unstressed level. The time course of ABA accumulation in Ler was similar to that previously observed for Col-0, and the Ler ABA level at 96 h after transfer was near the median of 298 accessions previously assayed (Kalladan et al., 2017). Compared with Ler, Sha had the same temporal pattern of ABA accumulation but had 40% lower ABA content than Ler at all times after transfer (Fig. 1A). This was consistent with previous results that put Sha among the 20 accessions with lowest ABA accumulation out of 298 accessions (Kalladan et al., 2017). Sha also had significantly reduced ABA accumulation compared with Ler after transfer to −0.7 MPa, a less severe low ψw treatment (Fig. 1B). When Ler and Sha were subjected to slow soil drying, Sha also tended to have lower ABA accumulation than Ler, albeit the difference was marginally nonsignificant in individual experiments (Supplemental Fig. S1). In these experiments, Sha also maintained a slightly higher relative water content than Ler, even though both were grown in the same pot and exposed to the same severity of soil drying. Together, these data indicated that Sha had reduced ABA accumulation across a range of low-ψw severities and methods of stress imposition.
Figure 1.
Reduced ABA accumulation of Sha relative to Ler is controlled by a single QTL that includes NCED3 alleles of differing function. A, Time course of ABA accumulation after transfer of 7-d-old Ler and Sha seedlings from control media (−0.25 MPa) to low-ψw stress (−1.2 MPa). Data are means ± se (n = 6–9) combined from two independent experiments. Sha ABA levels were significantly lower than those of Ler (Student’s t test, P ≤ 0.05) at each time point except time 0 (unstressed control). The inset shows the ABA contents of Ler and Sha at 96 h after transfer to low ψw. B, ABA accumulation of Ler and Sha 96 h after transfer to moderate-severity low ψw (–0.7 MPa). Data are means ± se (n = 6–12) combined from three independent experiments. ABA contents of Ler and Sha were significantly different (Student’s t test, P ≤ 0.001). C, LOD plot of QTL mapping results. A single QTL was detected at the top of chromosome 3. The red chromosome diagram shows the positions of NCED3 and flanking markers within the QTL interval. D, F1 ABA analysis from Ler, Sha, and nced3 crosses. F1 seedlings from reciprocal crosses of Ler and Sha to nced3 were transferred to –1.2 MPa, and ABA was measured 24 h later. Boxes encompass the 25th to 75th percentiles, with the solid line indicating the median and the dashed line indicating the mean. Whiskers show the 10th and 90th percentiles, and dots indicate outliers. n = 16 for all genotypes. Significant differences (Student’s t test, P ≤ 0.05) are indicated by asterisks. E, ABA contents of transgenic lines expressing the Ler or Sha NCED3 genomic fragment (promoter, 5′ UTR, and coding region) in the nced3 mutant or Sha background. NCED3 was expressed either as untagged protein or with a C-terminal hemagglutinin (HA) tag. ABA was assayed 96 h after transfer to −1.2 MPa. Data are means ± se (n = 14) combined from three independent experiments. Immunoblots verifying the NCED3 expression level in each transgenic line are shown in Supplemental Figure S1. FW, Fresh weight.
To understand the genetic basis for reduced ABA in Sha, we assayed ABA accumulation in an existing Ler × Sha RIL population (Clerkx et al., 2004; El-Lithy et al., 2004). This revealed a single large-effect QTL at the top of chromosome 3 (Fig. 1C; QTL peak at chromosome 3, 5 centimorgan, log of the odds [LOD] = 7.5). This QTL explained 26% of the variation for ABA accumulation in the RIL population and was localized in a 3-Mb region between markers F17014 and NT204 (Fig. 1C). Interestingly, this ABA QTL colocalized with previously detected QTLs for speed of seed germination (Sha faster than Ler) and ability to germinate on salt- or ABA-containing media (higher germination rate in Sha; Clerkx et al., 2004). These phenotypes are consistent with reduced ABA level in Sha. QTLs for growth-related traits were also found in this region (El-Lithy et al., 2004).
NCED3 Contributes to the Different ABA Accumulation of Ler and Sha
Interestingly, NCED3 was located in the middle of the chromosome 3 QTL interval (Fig. 1C). To test whether variation in NCED3 could be involved in the different levels of ABA accumulation observed for Ler and Sha, we crossed an nced3 T-DNA mutant (Wan and Li, 2006) to both Ler and Sha and measured ABA accumulation in F1 seedlings (Fig. 1D). In these F1 plants, the only active copy of NCED3 comes from either the Ler or Sha parent. Thus, if NCED3 variation underlies the Ler versus Sha ABA variation, it should be reflected in the F1 ABA content. Consistent with this hypothesis, the F1 of Sha × nced3 had lower ABA content than the Ler × nced3 F1, and this difference was observed consistently in reciprocal crosses (Fig. 1D). The F1 seedlings did not have ABA levels as high as the Ler or Sha parents, likely because they have only one functional copy of NCED3. The nced3 mutant is in the Col-0 background, and our assays confirmed that Col and Ler have essentially identical ABA levels; thus, the Col-0 genetic background should have minimal effect on the F1 ABA levels. This analysis also showed that while Sha had reduced ABA accumulation, it was still far higher than that of nced3 (Fig. 1D), consistent with Sha having a reduced-function NCED3 allele.
To further test the involvement of NCED3 in the reduced ABA accumulation of Sha, a 4.4-kb genomic fragment containing the NCED3-coding region, promoter, and 5′ untranslated region (UTR) sequence was cloned from both Ler and Sha and expressed in the nced3 background. Immunoblotting using NCED3-specific antisera found that two transgenic lines (#6 and #10) expressing the Ler NCED3 genomic clone had high levels of NCED3 expression (Supplemental Fig. S2) and fully complemented the reduced ABA content of nced3 (Fig. 1E). An additional transgenic line (#2) had a lower level of NCED3 protein and correspondingly less increase in ABA (Fig. 1E; Supplemental Fig. S2). In contrast, expression of the Sha NCED3 genomic fragment led to less increase in ABA levels even when the level of NCED3 expression was high as for line #9 (Fig. 1E; Supplemental Fig. S2), again consistent with Sha having a reduced-function NCED3 allele. Also consistent with this hypothesis, expression of the Ler NCED3 genomic fragment in Sha increased ABA content to a level equivalent to or greater than that of Ler (Fig. 1E). The data are consistent with NCED3 variation causing the reduced ABA accumulation of Sha; however, we do not rule out the possibility that other genes within the QTL interval also contributed to the variation in ABA accumulation.
Nonsynonymous Substitutions in Sha NCED3 Are Associated with Reduced ABA Accumulation
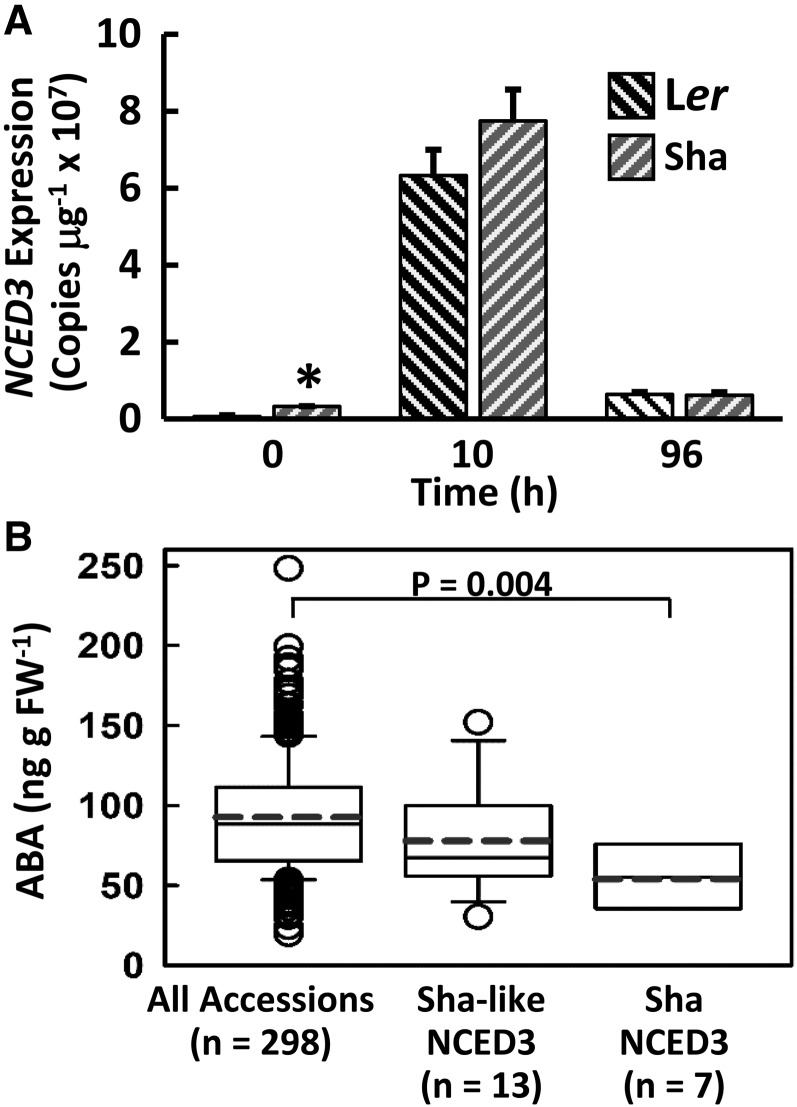
NCED3 gene expression was induced to the same, or even slightly higher, level in Sha compared with Ler within 10 h after transfer to low ψw (Fig. 2A). Such a pattern of NCED3 expression at 10 and 96 h after transfer was consistent with a previous study of Col-0 NCED3 (Sharma and Verslues, 2010) and with Ler and Sha RNA sequencing data, which showed 2-fold higher NCED3 expression in Sha than in Ler at 10 h after transfer to −1.2 MPa (Sharma et al., 2013). Thus, reduced NCED3 gene expression was not responsible for the reduced ABA accumulation of Sha.
Figure 2.
Ler and Sha have similar NCED3 expression, while the Sha and Sha-like NCED3 haplotypes are associated with lower ABA accumulation. A, NCED3 gene expression in Ler and Sha in the unstressed control (time 0) and after transfer to –1.2 MPa (10 and 96 h). Data are means ± se (n = 3). A significant difference (Student’s t test, P ≤ 0.05) of Ler versus Sha at the 0-h time point is indicated by the asterisk. The results are representative of several independent experiments. B, ABA accumulation of accessions with the Sha and Sha-like NCED3 haplotypes compared with 298 accessions phenotyped for ABA accumulation (Kalladan et al., 2017). The Sha haplotype group had significantly different ABA accumulation than the 298 accessions (Student’s t test, P ≤ 0.05). Whiskers show the 10th and 90th percentiles, and dots indicate outliers. Accessions with the Sha and Sha-like NCED3 haplotypes are listed in Supplemental Table S1. FW, Fresh weight.
The coding region of Sha NCED3 has four nonsynonymous substitutions compared with Ler. Sha has D-N-D-R at amino acids 18, 83, 184, and 271, respectively, compared with N-T-H-Q at these positions in Ler, Col-0, and more than three-fourths of the accessions in the 1,001-genome database (Alonso-Blanco et al., 2016). These substitutions are in the N-terminal half of NCED3, which includes regions that mediate chloroplast localization and attachment to the thylakoid membrane (Tan et al., 2001; Messing et al., 2010). None of the substitutions are in the C-terminal catalytic domain, and none are close to the four critical His residues that chelate iron in the NCED3 active site (His-297, His-346, His-411, and His-585 of NCED3).
To further explore NCED3 variation, all accessions not having the Ler/Col-0 haplotype at amino acids 18, 83, 184, and 271 in the 1,001-genome database were divided into groups based on their amino acid sequence at these four positions (Supplemental Table S1). The most common haplotype (other than the Ler/Col haplotype) was the D-T-D-R group (104 accessions), which differed from the Sha haplotype (D-N-D-R) in that it retains the T at amino acid 83 and is hereafter referred to as the Sha-like NCED3 haplotype. The second most abundant non-Ler/Col haplotype was the Sha haplotype (D-N-D-R), with 89 accessions. Our previous ABA phenotyping of 298 accessions included seven Sha haplotype accessions, and these all had low ABA compared with the whole group of 298 accessions (Fig. 2B). The 13 accessions in the D-T-D-R haplotype group for which we have data were more variable; however, the majority also had relatively low ABA accumulation (Fig. 2B; Supplemental Table S1) and did not significantly differ from the Sha haplotype accessions (P = 0.1, Student’s t test comparison of accession mean ABA accumulation). This observation must be treated with due caution because of the limited ABA data for accessions with the Sha or Sha-like NCED3 haplotypes. It was interesting that few accessions were intermediate between the Ler/Col haplotype and the Sha or Sha-like haplotypes. Only two accessions had one of the four amino acid changes leading to the Sha haplotype. Twenty-one accessions had two of the four amino acid changes, with 15 of these being the N-T-D-R type, where the first two polymorphic amino acids are of the Ler/Col type and the last two are of the Sha type (Supplemental Table S1).
We extended this comparative analysis by aligning NCED3 sequences from Col-0, Ler, and Sha with orthologous sequences from Arabidopsis lyrata, Brassica rapa, Brassica olereacea, and Brassica napus. We included the NCED9 sequence from Col-0 as an outgroup in this analysis. Focusing on amino acids 18, 83, 184, and 271, the A. lyrata and Brassica spp. in this sample were monomorphic: all are N-T-D-R. This configuration corresponds to the rarest observed haplotype in Arabidopsis at these four sites, comprising just 15 accessions among the 1,001-genome sample. Our result suggests that the Sha amino acids at 184 and 271 may represent the ancestral condition in Arabidopsis. Whether the other Brassicaceae species have altered NCED3 function relative to Ler and other Arabidopsis accessions remains unknown.
Our previous analysis of single-nucleotide polymorphism (SNP) associations with ABA while accounting for relatedness (Kalladan et al., 2017) did not identify NCED3 as a particularly strongly associated locus. However, single SNP associations can miss causal polymorphism in the presence of allelic heterogeneity. Using sliding and variable-width windows of SNPs across the NCED3 region to identify unique haplotypes (excluding singletons), we found significant associations with ABA, while accounting for kinship among accessions, for haplotypes in a region from 1 to 5 kb from NCED3. However, no such signal was observed in the coding region. We tested the three Sha missense mutations that were called in the imputed SNP data set using the 1,001 genomes as a reference and found that two of them (4832666 and 4833230, which correspond to NCED3 amino acids 271 and 83, respectively) had significantly lower ABA levels in ecotypes with the Sha allele (Wilcoxon test, both P = 0.04), but these differences were not significant when using a mixed model accounting for kinship. These data were consistent with an effect of the Sha allele that was not detected in our ABA genome-wide association study (GWAS), because variation at other loci in diverse ecotypes can obscure the effects of causal loci and because long-range linkage between a particular allele and many loci (due to population structure) can reduce the power to detect associations.
Sha NCED3 Has Altered Posttranslational Processing Compared with Ler/Col-0 NCED3
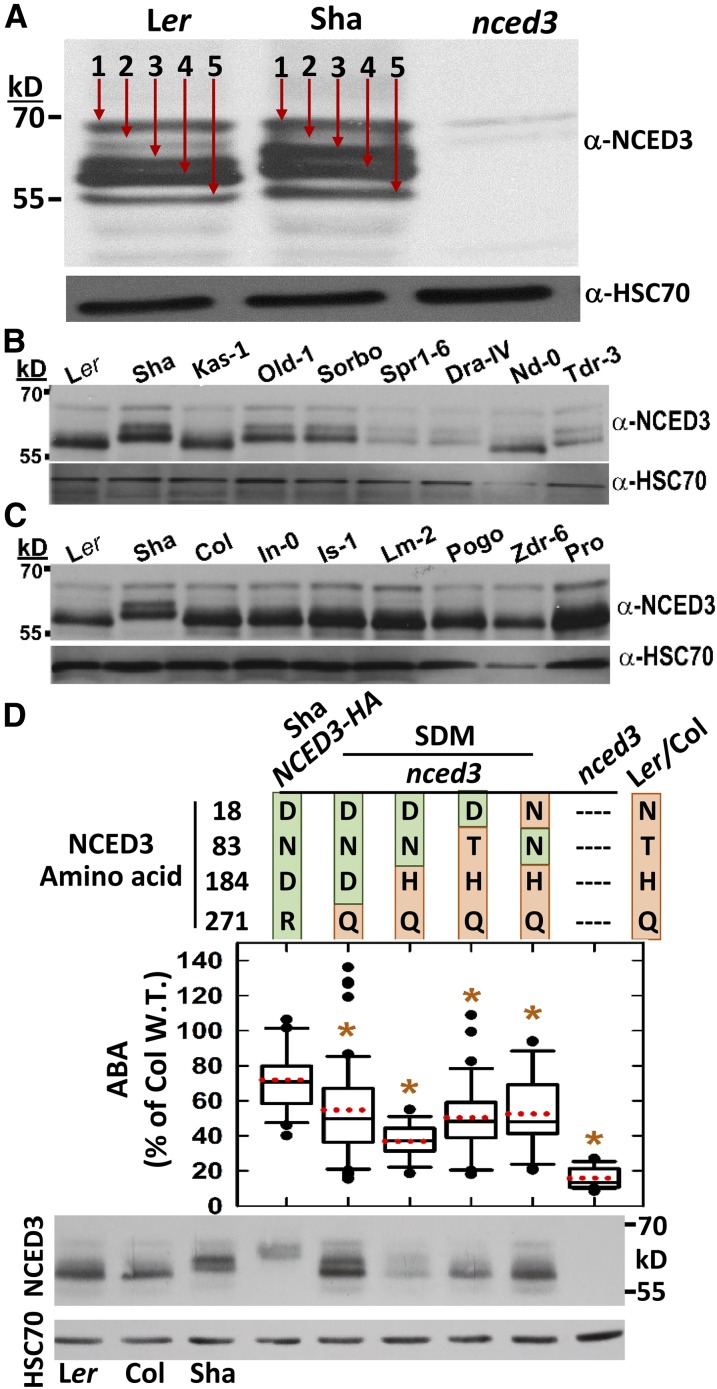
Immunoblotting of Ler and Sha after 10 h at low ψw revealed a similar level of NCED3 protein but different patterns of apparent molecular mass in Ler and Sha (Fig. 3A). We detected five distinct NCED3 bands in Ler, and immunoblot of nced3 confirmed that these bands were all derived from NCED3 (Fig. 3A). The faint signal observed in nced3 at the band 1 and 2 molecular mass values is likely from other NCEDs. The highest molecular mass band (band 1) was consistent with full-length NCED3 (66 kD) or NCED3 after cleavage of a short chloroplast-targeting domain. In addition, a series of lower molecular mass bands were observed from approximately 64 to 55 kD (Fig. 3A, bands 2–5). In Sha, NCED3 bands 3 and 4 were shifted up by approximately 4 to 5 kD compared with Ler. Endo et al. (2008) observed two NCED3 bands: a band equivalent to our band 1 corresponding to full-length (or nearly full-length) NCED3 and a lower molecular mass band corresponding to our band 3, 4, or 5 (or an unresolved combination of these bands), which they proposed to be the result of NCED3 proteolytic cleavage. Our results indicated that such NCED3 posttranslational processing is more complex than previously thought and showed that Sha had altered NCED3 posttranslational processing.
Figure 3.
The Sha and Sha-like NCED3 haplotypes have an altered pattern of NCED3 apparent molecular mass. A, Immunoblot detection of NCED3 in Ler, Sha, and nced3 at 10 h after transfer to –1.2 MPa. In each lane, 40 μg of protein was loaded. The numbers 1 to 5 and red arrows mark the five bands of NCED3 detected. After NCED3 detection, blots were stripped and reprobed to detect HSC70 as a loading control. B, Immunoblot detection of NCED3 in accessions with the Sha NCED3 haplotype (Sha, Old-1, and Sorbo), Sha-like NCED3 haplotype (Spr1-6, Dra-iV, and Tdr3), or Ler/Col-0 NCED3 haplotype (Ler, Kas-1, and Nd-0). Samples were collected 10 h after transfer to –1.2 MPa and 40 μg of protein was loaded. C, Immunoblot detection of NCED3 in arbitrarily selected accessions with the Ler/Col-0 NCED3 haplotype. D, Complementation of nced3 with NCED3 site-directed mutants (SDMs) expressed under the control of the Ler NCED3 promoter. ABA data for the SDM lines are combined from two independent transgenic lines for each construct (n = 19–40 combined from two independent experiments) and are shown as percentages of the Col wild type (W.T.) assayed in the same experiments. Formatting of the graph is as described for Figure 1C. Asterisks indicate significant differences from Sha NCED3-HA. An expanded graph showing data for individual transgenic lines separately is shown in Supplemental Figure S3B. The Sha NCED3-HA data are replotted from Figure 1E for comparison. Immunoblotting was performed using the same conditions described for A. Note that because of the HA epitope tag, the Sha NCED3-HA lane should only be used to compare protein amount and not molecular masses.
Expression of Sha NCED3 cDNA in the nced3 mutant background produced the Sha NCED3 molecular mass pattern, while expression of Ler NCED3 cDNA produced the Ler pattern (Supplemental Fig. S3A). Thus, the NCED3 molecular mass pattern was not influenced by the genetic background. Note that since NCED3 has no introns, alternative splicing is not a factor in the NCED3 molecular mass pattern. NCED3 protein level was highest at 10 h after transfer to low ψw, consistent with the time courses of ABA accumulation (Fig. 1A) and NCED3 gene expression (Fig. 2A). As in a previous study (Endo et al., 2008), we were unable to reliably detect NCED3 in unstressed seedlings or after 96 h at low ψw.
The Sha NCED3 banding pattern was also observed in Old-1, Sorbo, Spr1-6, Dra-iV, and Tdr-3 (Fig. 3B). Sequencing of NCED3 from each of these accessions confirmed that Sha, Old-1, and Sorbo each had the Sha NCED3 haplotype (D-N-D-R), while Spr1-6, Dra-iV, and Tdr-3 had the Sha-like haplotype (D-T-D-R). This suggested that variation at amino acid 83 may not be involved in the altered NCED3 molecular mass pattern. In these experiments, we also assayed Kas-1 and found that it had the Ler/Col-0 NCED3 sequence and banding pattern (Fig. 3B). Kas-2, which is sometimes confused as Kas-1, had lower ABA accumulation and is annotated as having the Sha NCED3 sequence in the 1,001-genome database (Supplemental Table S1). Immunoblot of Col-0 and seven additional arbitrarily selected accessions from the Ler/Col NCED3 haplotype group showed that all had the same NCED3 banding pattern as Ler (Fig. 3C). Note that in these blots, NCED3 band 5 is not always detected because of its low abundance and bands 3 and 4 are sometimes not fully resolved because of their similar molecular mass. These data indicated that the N18D, H184D, and Q271R amino acid substitutions seen in the Sha and Sha-like NCED3 haplotypes were consistently associated with the altered NCED3 molecular mass pattern.
To directly test the role of the four Sha NCED3 substitutions, including combinations not seen in natural accessions, we generated SDM lines where the Sha NCED3 cDNA was partially converted to the Ler form. These new versions of NCED3 were fused to the Ler NCED3 promoter and expressed in the nced3 background. Interestingly, changing Arg-271 of Sha NCED3 to Gln (Q) was sufficient to produce the Ler NCED3 banding pattern (Fig. 3D; note that the Sha-NCED3-HA line can be used only to compare NCED3 protein level but not molecular mass pattern, because it has an HA tag fused to NCED3 while the SDM lines do not). However, the R271Q SDM lines had lower ABA levels than lines with Sha NCED3 (Fig. 3D; an expanded version of the ABA graph with data for individual transgenic lines plotted separately is shown in Supplemental Fig. S4B). Alignment of all Arabidopsis NCED sequences (from Col-0) along with maize VP14 showed that all NCEDs have His or Gln at this position (Supplemental Fig. S4). The change to Arg at amino acid 271 was sufficient to reverse the altered posttranslational processing of NCED3 observed in Sha but was not the only factor in the reduced function of Sha NCED3. Interestingly, this D-N-D-Q NCED3 haplotype was not seen in any accessions in the 1,001-genome database (Supplemental Table S1).
NCED3 SDM lines that combined the R271Q change with D184H (D-N-H-Q) also had the Ler NCED3 molecular mass pattern but low ABA accumulation (Fig. 3D). Other lines where only amino acid 18 or 83 remained in the Sha form while the other three were converted to Ler form (D-T-H-Q and N-N-H-Q) also had low ABA accumulation while having the Ler NCED3 molecular mass pattern. Of these three synthetic NCED3 haplotypes, two were not found in natural accessions and the other, D-T-H-Q, was found in only one accession (Supplemental Table S1). NCED alignment showed that these three amino acid changes are all relatively conservative (Supplemental Fig. S4). The T-to-N substitution in Sha at amino acid 83 is also found in NCED9. The H-to-D change at amino acid 184 makes Sha NCED3 more like NCED6, which has a Phe next to an Asp at this site, or NCED9, which has an Asp at position 183. The Sha substitution at amino acid 18 introduces a differently charged side chain at that position (acidic rather than basic or hydrophobic); however, the importance is difficult to judge, as this site is not conserved in all NCEDs and it is within the stromal targeting domain that may be cleaved off after import of NCED3 into the chloroplast. Together, the data indicate that none of the amino acid changes at positions 18, 83, or 184 are likely to disrupt NCED3 function. The observation that even changing three of the four polymorphic amino acids from the Sha form to the Ler form was unable to complement nced3 to the Ler level of ABA accumulation indicates that these amino acid changes together affect NCED3 function. This was consistent with the observation that only certain combinations of these amino acid polymorphisms have been found in natural accessions and was consistent with the observation that at positions 184 and 271 it is the Sha type that is similar to A. lyrata and Brassica spp. and is likely to be the ancestral NCED3 type in Arabidopsis.
Structural modeling and visualization of NCED3 structure supported the idea that none of the four polymorphic amino acids is by itself critical to NCED3 function. Amino acid 271 is in a stromal facing β-sheet, where it could affect NCED3 protein interaction (Supplemental Fig. S5). Position 184 is also on the stromal surface of NCED3 but in a loop region. Amino acid 83 is close to, but not part of, the α1 and α2 amphipathic helices involved in NCED3 binding to the thylakoid membrane (Messing et al., 2010). In all of these regions, the structure predicted for Ler/Col-0 NCED3 and Sha NCED3 were highly similar to each other and to maize VP14 (Messing et al., 2010). Thus, none of the amino acid changes in Sha were at positions rigorously conserved or structurally critical in NCEDs, further suggesting that together they have an incremental or interactive effect on NCED3 function. It was also interesting that amino acids 184 and 271, where the Sha substitutions are nearly always found together (Supplemental Table S1) and where the Sha form likely represents the ancestral form of Arabidopsis NCED3, are in close proximity to each other on the stromal surface of NCED3 (Supplemental Fig. S5). The site-directed mutagenesis and structural observations together indicated that amino acid substitutions found in Sha may lead to reduced ABA accumulation by affecting NCED3 activity, in addition to their effects on NCED3 posttranslational processing. However, it was unclear how these amino acid substitutions could alter NCED3 activity, as none had an obvious effect on the catalytic core of the enzyme.
Sha Does Not Have Reduced Thylakoid Membrane NCED3 But Differs from Ler in Stromal NCED3
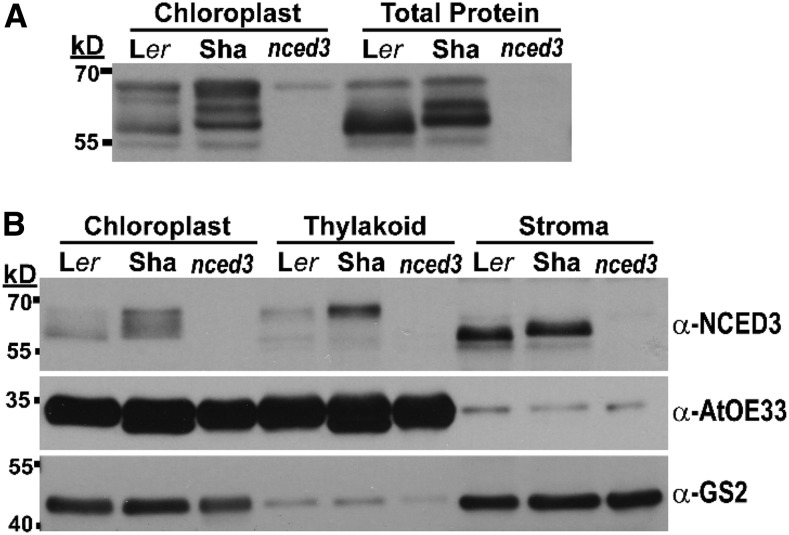
NCED3 import into chloroplasts and association with the thylakoid membrane are key factors in NCED3 activity (Tan et al., 2001; Endo et al., 2008; Messing et al., 2010) and could be a basis for reduced or altered function of Sha NCED3. Chloroplast isolation indicated that Sha chloroplasts had the same, or even slightly higher, level of NCED3 compared with Ler (Fig. 4; Supplemental Fig. S6). The high-molecular-mass thylakoid-bound NCED3 was clearly more abundant in Sha than in Ler (Fig. 4B). This greater abundance of thylakoid-bound NCED3 in Sha was reproducibly seen in repeated chloroplast fractionations using a more rapid method (Supplemental Fig. S6). Stromal NCED3 was similar in abundance but differed in molecular mass between Ler and Sha (Fig. 4B). This was also seen in repeated fractionations (Supplemental Fig. S6), although the more rapid fractionation method had some thylakoid contamination of the stromal fraction.
Figure 4.
Sha does not have reduction of thylakoid-bound NCED3 but does differ from Ler in apparent molecular mass of stromal NCED3. A, Immunoblot detection of NCED3 in total protein compared with isolated chloroplasts of Ler, Sha, and nced3. In each lane, 40 μg of protein was loaded. B, Chloroplast fractionation by Percoll gradient centrifugation and immunoblotting of thylakoid and stromal proteins. Blots were stripped and reprobed with AtOE33 as a marker of the thylakoid fraction and GS2 as a marker of the stromal fraction. In each lane, 40 μg of chloroplast protein, 30 μg of thylakoid membrane protein, or 10 μg of stromal protein was loaded. The experiment was repeated with similar results. Additional replicate assays using an alternative chloroplast fractionation procedure are shown in Supplemental Figure S6.
DISCUSSION
The wide variation in low-ψw-induced ABA accumulation between Arabidopsis accessions (Kalladan et al., 2017) is useful in uncovering molecular mechanisms affecting ABA accumulation. Several lines of evidence, including ABA contents of F1 crosses and of transgenic plants expressing Ler or Sha NCED3 in the nced3 mutant background, indicate that reduced NCED3 function is the major factor underlying reduced ABA accumulation of Sha. Further analysis indicated that NCED3 gene expression, NCED3 protein level, NCED3 import into chloroplasts, and NCED3 association with thylakoid membrane were not reduced in Sha. As these factors, known to be important for NCED3 activity, were not impaired in Sha, we are left to conclude that the lower ABA accumulation of Sha is due to other factors. A likely explanation is that Sha NCED3 has reduced enzymatic activity. However, two of the Sha amino acid substitutions are also present in closely related species and represent the ancestral form of NCED3. Along with the observation that the amino acid substitutions in Sha are not in conserved sequences critical for NCED3 enzymatic activity, this indicates that the Sha allele should not be considered as a broken or impaired form of NCED3. Instead, the bulk of Arabidopsis accessions, including Ler and Col, seem to have acquired an NCED3 allele that allows relatively high levels of ABA accumulation. As the NCED3 polymorphic amino acids we examined are not within the catalytic core of NCED3, the mechanism by which they could affect NCED3 activity is not clear. The shift in molecular mass of stromal NCED3 was one of the most striking differences in Sha. However, NCED3 SDM lines indicated that the altered molecular mass pattern of Sha NCED3 could be reverted to the Ler form without recovery of ABA accumulation to the level seen in Ler. As little is known of NCED3 posttranslational processing, the different molecular mass pattern of Ler versus Sha NCED3 is of interest, but further experiments will be required to establish its functional relevance to ABA accumulation. In either case, our data identify new amino acid sites important for NCED3 function in stress-induced ABA accumulation.
Despite the prominent role of NCED3 in stress physiology, few studies have focused on the NCED3 protein and its posttranslational regulation. Detection of multiple bands of lower molecular mass NCED3 (Fig. 3A, bands 3–5) suggests that cleavage of NCED3 to release it from the thylakoid membrane is more complex than previously thought. These multiple bands may be derived from multiple rounds of NCED3 processing or the use of more than one possible cleavage site to release NCED3 from the thylakoid membrane. Ler NCED3 bands 3 to 5 are approximately 8 to 10 kD less than band 1, consistent with cleavage just downstream of the first amphipathic helix involved in thylakoid membrane binding (this assumes that the N-terminal 30–40 amino acids are cleaved after import into the chloroplast and an additional 80–100 amino acids are removed from the N terminus to generate bands 3 and 4). Sha NCED3 bands 3 and 4 have apparent molecular mass 3 to 4 kD heavier than the corresponding Ler bands. In this case, Sha NCED3 may retain some of the first amphipathic helix or surrounding sequence, possibly affecting NCED3 function. Surprisingly, change of amino acid 271 from the Sha to the Ler form was by itself sufficient to restore the Ler banding pattern. As this amino acid is not close to the actual cleavage site(s), it is not clear how it could influence NCED3 cleavage. Perhaps a larger complex of interacting proteins is required to direct the proteolytic cleavage and release of thylakoid-bound NCED3.
The idea that NCED3 must be bound to the thylakoid membrane for activity is supported by both structural information (Messing et al., 2010) and correlation of increased thylakoid-bound NCED3 with increasing ABA content (Endo et al., 2008). Interestingly, though, both our data and those of Endo et al. (2008) indicate that there is just as much or more stromal NCED3 as thylakoid-bound NCED3 even when ABA is accumulating to very high levels. In fact, our data indicate that there is substantially more stromal than thylakoid NCED3 when one compares the intensities of band 1 versus bands 3 to 5 and considers that less stromal than thylakoid protein was loaded for the blots shown in Figure 3 and Supplemental Figure S5. This raises the question of whether stromal NCED3 is merely an inactive form awaiting degradation or whether it retains some function. Tan et al. (2003) found that NCED9 could be detected only in the stromal fraction after import into pea (Pisum sativum) chloroplasts. NCED9 is involved in seed ABA synthesis (Lefebvre et al., 2006), but it is unclear whether stromal NCED9 is active in ABA synthesis or whether NCED9 is recruited to the thylakoid under specific conditions when it is active. Further analysis of stromal NCED function is needed to understand whether the altered stromal NCED3 seen in Sha affects ABA metabolism.
The amino acid substitutions in Sha and Sha-like NCED3 are not randomly distributed among accessions but rather occur together in almost all cases. For the last two substitutions (H184D and Q271R), only seven accessions had one of these substitutions and not the other, while 209 accessions had both of these amino acid substitutions together (e.g. 97% of accessions that had one substitution also had the other one). Despite their low frequency in Arabidopsis, comparison with closely related species indicated that the Sha form at these two positions is the common ancestral form of NCED3. Interestingly, these two amino acids are also in physical proximity to each other on the stromal facing surface of NCED3. Possibly there is coordination (or compensation) such that once an accession acquires the Ler form of one of these substitutions, it will also tend to acquire the other. One other study that examined a smaller number of accessions suggested that polymorphisms at NCED3 amino acids 274 and 327 were associated with natural variation in ABA accumulation (Hao et al., 2009). Although we focused on the four amino acids differing between Ler and Sha, we do not rule out the possibility that variation at other amino acids could impact NCED3 function. Further comparison is complicated by the fact that Hao et al. (2009) measured ABA accumulation after an interval of uncontrolled soil drying. Thus, their ABA data may be confounded by differences in water use, such that not all accessions were exposed to the same severity of soil drying, whereas in our experiments, all the accessions were exposed to the same ψw.
Given the relatively low ABA accumulation of accessions with the Sha and Sha-like NCED3 haplotypes, one could ask why NCED3 was not identified as a candidate gene by GWAS (Kalladan et al., 2017). The results of biparental mapping versus GWAS may not overlap because relatively few accessions with the Sha or Sha-like NCED3 haplotypes were included in the panel of accessions used for GWAS, and genome scans are notoriously underpowered for detecting rare causal alleles. Also, the functional NCED3 polymorphisms were not included in the SNP genotyping data set used in the GWAS, and the SNPs included in the GWAS may not have been in complete linkage disequilibrium with the functional NCED3 SNPs (and were also of low frequency). While there was a single QTL around NCED3 for ABA accumulation in Ler × Sha, in other accessions the NCED3 variation may interact with other variations affecting ABA accumulation. This could explain the wide range of ABA accumulation in accessions with Sha-like NCED3.
Sha differs from other commonly used accessions in many phenotypes. Our data suggest that some of these differences may be influenced by, or even caused by, reduced ABA. The most obvious is germination on salt or osmoticum-containing medium (Clerkx et al., 2004), as this phenotype is known to be affected by NCED3 (Ruggiero et al., 2004). Likewise, increased growth of Sha at low ψw (Sharma et al., 2013) may be explained by reduced ABA levels and thus less restraint of shoot growth. More broadly, manipulation of ABA levels has been thought of as a strategy to improve water use efficiency or alter root versus shoot growth during drought. Previous attempts to construct transgenic lines with more or less ABA accumulation have not always been successful (Verslues, 2016). Our results indicate that direct alteration of the NCED sequence may be a way to change the extent of ABA accumulation while preserving its normal stress regulation and tissue specificity. Likewise, our data suggest that identification of the proteases and other proteins that control NCED processing in the chloroplast could also lead to new strategies to tune ABA accumulation to optimize plant productivity under different scenarios of drought timing and severity.
MATERIALS AND METHODS
Plant Material, Stress Treatment, and ABA Quantification
The Arabidopsis (Arabidopsis thaliana) Ler and Sha accessions as well as the Ler × Sha RIL population (Clerkx et al., 2004; El-Lithy et al., 2004) were obtained from the Arabidopsis Biological Resource Center. The nced3 T-DNA mutant (GABI_129B08) was provided by the laboratory of Wan-Hsing Cheng (Institute of Plant and Microbial Biology, Academia Sinica). Seeds were plated on agar plates containing one-half-strength Murashige and Skoog salts and 2 mM MES buffer (pH 5.7) without addition of sugar. Plates were overlaid with nylon mesh to facilitate transfer of seedlings. After stratification, plates were incubated vertically in a growth chamber (25°C, 85–100 μmol photons m−2 s−1 continuous light). For low-ψw stress treatment, 7-d-old seedlings were transferred to polyethylene glycol-8000-infused agar plates (–1.2 MPa) prepared as previously described (Verslues et al., 2006). Samples for ABA analysis were collected from 10 to 96 h after transfer as indicated in the text and figure legends.
Soil-drying experiments were conducted essentially as described by Bhaskara et al. (2015) but without partial rewatering during the drying cycle. A standard potting mix was combined with 25% (v/v) Turface (Turface MVP; Profile Products) to improve porosity and consistency of drying. Seeds of Ler and Sha were planted in sectors (four to six plants per sector) of the same 8-cm × 8-cm × 10-cm (length × width × height) plastic pots and grown in a short-day chamber (8-h light period, 25 C, light intensity of 100–120 μmol m−2 s−1). On day 18 after planting, pots were watered to saturation, allowed to drain, and weighed. Water was withheld for 24 or 28 d (leading to nearly 50% reduction in pot weight). Rosettes were then harvested for measurements of relative water content or ABA (representative rosettes were photographed just before harvest). For assay of relative water content, whole rosettes were weighed, floated on cold water for 3 to 4 h, reweighed to obtain fully hydrated weight, and then dried overnight in a 60°C oven before reweighing to obtain the dry weight.
ABA content was analyzed by gas chromatography-mass spectrometry as previously described (Verslues, 2017). In brief, 20 to 50 mg of seedlings was lyophilized, ground to a fine powder using grinding beads, and extracted in 80% (v/v) methanol with 25 pmol of [D6]ABA (Plant Biotechnology Institute) as an internal standard. Extracts were passed through a C18 solid-phase extraction cartridge (Supelco), evaporated to dryness, resuspended in diethyl ether:methanol (9:1), and derivatized by addition of trimethylsilydiazomethane. After derivatization, the remaining trimethyldiazomethane was destroyed by addition of 0.5 M acetic acid in hexane. The samples were then evaporated, resuspended in a small volume of ethyl acetate, injected onto a VF-14MS (Varian/Agilent) column, and analyzed by tandem mass spectrometry. Methanol chemical ionization was used to generate precursor ions of 261 mass-to-charge ratio (m/z) for ABA and 267 m/z for [D6]ABA. Daughter ions of 229 m/z (ABA) and 233+234 m/z ([D6]ABA) were used for quantification. The ABA amount was quantified by calculations based on a standard curve over 2 to 60 pmol of ABA using the ratio of the 229 and 233+234 peak areas. For soil-grown plants, four to six rosettes were lyophilized together and ground, and subsamples were taken for ABA analysis. Data from ABA measurements and physiological assays were statistically analyzed using Student’s t tests.
QTL and GWAS Mapping
QTL mapping used the previously described Ler × Sha RIL mapping population and a genetic map composed of 89 genetic markers (Clerkx et al., 2004). The mapping population was phenotyped for ABA content at 96 h after transfer of 7-d-old seedlings to low ψw (−1.2 MPa). The 116 RILs were randomized into eight blocks, with each RIL represented in two blocks and four to six ABA measurements conducted for each RIL (two or three samples collected in each block). The ABA measurements were conducted in sets of 72 samples each (ABA assay plate), with approximately 740 total samples assayed for the RIL population and Ler and Sha control samples. In the statistical analysis, both block and ABA assay plate were treated as random effects, and raw ABA data were log-transformed to improve the normality of residuals. We conducted QTL mapping using the R/qtl package (Broman et al., 2003). Mapping was conducted using the scanone function on a 1-centimorgan grid using a Haley-Knott regression approximation and assuming normally distributed phenotypes. A genome-wide critical threshold value for the experiment-wise type 1 error rate was set by randomly permuting the line means for ABA content among genotypes 1,000 times and using empirical false-positive rates. All mapping was conducted on RIL least-squares means after controlling for blocking and in the experimental designs using the Proc Mixed procedure in SAS. We calculated the percentage variance explained by the detected QTL using the fitqtl function. We estimated confidence intervals for the location of QTL using a standard 1.5-LOD score drop on the results from our scanone model. Scripts detailing our QTL mapping approach are available upon request.
We tested associations of haplotypes formed from alleles in the SNP array data used for GWAS (Horton et al., 2012). We formed unique haplotypes ranging from two to seven SNPs in a sliding window across the NCED3 locus. We then tested whether haplotypes were associated with different ABA levels while excluding singleton haplotypes. We used the lmekin function in the R package coxme (Therneau, 2018) and compared a model with haplotype differences and kinship-correlated random effects with a null model without haplotype differences and used the likelihood ratio test on these models to evaluate evidence for haplotype differences. To test associations between missense SNPs in ecotypes and ABA, we downloaded imputed SNP alleles from the GMI GWA Portal (https://gwas.gmi.oeaw.ac.at/#/genotype/).
Cloning, Construct Preparation, and Plant Transformation
For expression of NCED3 under the control of its native promoter, a 4.4-kb genomic fragment comprising a 2.6-kb promoter and 5′ UTR and 1.8-kb NCED3-coding region was amplified from Ler or Sha genomic DNA either with its natural stop codon intact or with it removed to generate a fusion protein with a C-terminal HA tag. The initial PCR product was cloned into pENTR/D-TOPO and transferred to pEG301 (Earley et al., 2006) destination vector by LR reaction. Constructs were transformed into Agrobacterium tumefaciens strain GV3101, and plant transformation was conducted by the floral dip method. Transgenic lines expressing 35S:NCED3 were constructed by amplifying the NCED3 cDNA sequence from genomic DNA (NCED3 has no introns), cloning into pENTR/D-TOPO (Invitrogen) Gateway entry vector, and subsequent transfer to destination vector pEG100 (Earley et al., 2006) for plant transformation. The primers used in generating NCED3 constructs are listed in Supplemental Table S2. NCED3 SDMs were generated from the Sha NCED3-coding sequence using primers listed in Supplemental Table S2. The SDMs were then fused with the Ler NCED3 promoter (3 kb) using overlap extension PCR (primers given in Supplemental Table S2). The resulting 5-kb product was cloned into pENTR/D-TOPO (Invitrogen), transferred to pEG301 by LR reaction, and transformed into the nced3 T-DNA mutant. For both the Ler and Sha NCED3 transgenic lines, it was difficult to recover plants with high NCED3 expression, presumably because increased ABA levels were detrimental to plant development. This precluded analysis of additional high-expression transgenic lines.
Recombinant NCED3 Production and Purification for Antibody Production
Ler NCED3 cDNA was cloned into pDONR207 and transferred to destination vector pET300 for production of fusion protein with an N-terminal His tag. Recombinant protein was produced in the Escherichia coli Rosetta strain. After induction (0.5 mM isopropylthio-β-galactoside at 37°C), cells were lysed by incubation in lysis buffer (50 mM Tri-HCl [pH 8], 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 100 μg mL−1 lysozyme, 0.1% [v/v] Triton X-100, and 1× Roche protease inhibitor) for 30 min at 37°C followed by sonication with a 10-s cycle pulse for 1 min. The suspension was centrifuged at 12,000g for 5 min at 4°C to recover the insoluble fraction, which contained most of the recombinant NCED3. The pellet was washed five times with buffer containing 50 mM Tris-HCl (pH 8), 10 mM EDTA, 200 mM NaCl, 0.25% (v/v) Triton X-100, and protease inhibitor and followed by a rinse with distilled water. Finally, the pellet was resuspended in 50 mm Tris (pH 7.4), 300 mM NaCl, 8 mm urea, and 10 mM imidazole. The solubilized protein was purified using a 6× His column (Thermo Scientific) according to the manufacturer’s instruction. Production of polyclonal antisera in rabbit was performed by LTK BioLaboratories.
To produce a more soluble protein suitable for affinity purification of the crude antisera, a truncated NCED3 clone lacking the first 330 nucleotides (amino acids 1–110, which contains the hydrophobic region involved in thylakoid membrane binding) was cloned into pDONR207 and transferred to pDEST15 for production of truncated NCED3 with N-terminal GST fusion. The recombinant protein was mainly found in the soluble fraction from the cells harvested 16 h after isopropylthio-β-galactoside induction (0.5 mM) and grown at 20°C. Cell disruption was carried out using a combination of lysozyme and sonication as in the His tag purification except that the buffer used was phosphate-buffered saline with all other components remaining same. Purification was performed using GST-Bind resin (Novagen) according to the manufacturer’s instructions. GST-NCED3 was used to generate affinity-purified NCED3 antisera (performed by LTK BioLaboratories).
Immunoblot Detection of NCED3
Protein extraction and quantification were carried out according to Kesari et al. (2012). For SDS-PAGE, 40 μg of total protein was loaded per lane. After transfer, the polyvinylidene difluoride membrane was blocked by incubating it at 4°C for 17 to 20 h with 5% (w/v) casein without shaking. Primary (affinity-purified anti-NCED3) and secondary (anti-rabbit horseradish peroxidase conjugated; Jackson Laboratories) antibodies were used at dilutions of 1:2,300 and 1:15,000, respectively. Immunoblots were developed using Pierce ECL Western blotting substrate (Thermo Scientific). Blots were stripped and reprobed with HSC70 antisera as a loading control. NCED3 gene expression was assayed as described by Sharma and Verslues (2010).
Chloroplast Isolation, Fractionation, and Western Blot
Chloroplast isolation and fractionation were carried out using both a rapid method and a more specific method involving Percoll gradient centrifugation. For both methods, tissue homogenization and chloroplast lysis were carried out in the same way and using the same buffer. Briefly, approximately 10 g of 7- to 8-d-old seedlings were collected at 10 h after transfer to low ψw. Seedlings were rinsed (to remove polyethylene glycol) and ground with 30 mL of grinding buffer (0.3 M sorbitol, 1 mM MgCl2, 50 mM HEPES, 2 mM EDTA, and freshly added 0.04% [v/v] β-mercaptoethanol and 0.1% [w/v] polyvinylpyrrolidone) using a domestic blender (three 5-s pulses). The homogenate was filtered through two layers of Miracloth placed onto a funnel in a 250-mL prechilled conical flask. For crude chloroplast fractionation, the tissue homogenate was centrifuged at 500g for 5 min at 4°C to remove debris, and the resulting supernatant was centrifuged at 1,500g for 10 min at 4°C. The pellet, containing chloroplasts, was resuspended in wash buffer (same as grinding buffer except that β-mercaptoethanol and polyvinylpyrrolidone were omitted) and centrifuged at 1,500g for 20 min at 4°C, followed by one more resuspension and centrifugation at 5,000g for 10 min at 4°C. The pellet thus obtained was resuspended in a small volume of wash buffer and used for fractionation. For rapid chloroplast fractionation, 1 volume of isolated chloroplasts was mixed with 5 volumes of lysis buffer (62.5 mM Tris-HCl [pH 7.8] and 2 mM MgCl2), incubated on ice for 30 min, and centrifuged at 15,000g for 15 min at 4°C. The supernatant and pellet thus obtained were considered as stromal and thylakoid fractions, respectively, and the pellet was further washed three times in lysis buffer. Note that the stromal and thylakoid fractions obtained using this crude method were more cross-contaminated than fractions obtained with the Percoll gradient (below), probably because of relatively low centrifugal force after the chloroplast lysis step.
Chloroplast isolation using the Percoll gradient was carried out according to Chu and Li (2011), with minor modifications. After obtaining the tissue homogenate, the filtrate was centrifuged at 3,000g for 3 min at 4°C. The pellet obtained was resuspended in a small volume of grinding buffer and loaded onto a chilled Percoll gradient, which was premade by mixing 10 mL of Percoll (50%, v/v) and 10 mL of 2× grinding buffer, followed by centrifugation at 40,000g for 30 min at 4°C. Percoll gradients loaded with chloroplast isolate were centrifuged in a swinging bucket rotor at 7,700g for 10 min at 4°C. After discarding the upper green layer containing broken chloroplasts, the lower green layer containing intact chloroplasts was retrieved and mixed with a 2- to 3-unit volume of grinding buffer and centrifuged at 15,000g for 5 min at 4°C. The pellet thus obtained contained intact chloroplasts and was redissolved in a small volume of grinding buffer for fractionation. Fractionation was carried out in the same way as for the crude preparation. The pellet obtained was washed three times with lysis buffer to obtain the thylakoid membrane fraction, while the supernatant was further centrifuged at 100,000g for 1 h at 4°C to obtain the stromal fraction (supernatant).
The stromal fraction obtained in both methods was concentrated by precipitation in 100% acetone at –20°C for 30 min. The precipitated stromal fraction and the thylakoid membrane fractions were reconstituted in protein extraction buffer, and protein was quantified by the bicinchoninic acid method. For SDS-PAGE and immunoblotting, 40, 30, and 10 μg of chloroplast, thylakoid membrane, and stromal proteins, respectively, were used. Immunoblotting was performed as described above. After detection of NCED3, the membranes were stripped and reprobed with antisera recognizing AtOE33 and GS2 (Agrisera) as markers for thylakoid membrane and stromal fractions, respectively. The AtOE33 antiserum was developed by the laboratory of Hsou-min Li (Institute of Molecular Biology, Academia Sinica). The primary antibody dilutions for NCED3, AtOE33, and GS2 were 1:2,300, 1:2,000, and 1:5,000, respectively.
NCED Sequence Analysis and Structural Modeling
Amino acid sequence alignments of Arabidopsis (Col-0) NCEDs and maize (Zea mays) VP14 were generated using T-coffee (https://www.ebi.ac.uk/Tools/msa/tcoffee/). Brassicaceae DNA sequences used to assess the ancestral sequence condition at NCED3 were downloaded from Phytozome and aligned by eye. For structural modeling, the Col-0/Ler and Sha NCED3 sequences were submitted to I-Tasser (Yang et al., 2015), and the predicted structure was visualized using Jmol (http://jmol.sourceforge.net/).
Accession Number
The accession number for NCED3 is AT3G14440.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Ler and Sha response to slow soil drying.
Supplemental Figure S2. Immunoblot analysis of NCED3 expression in transgenic lines shown in Figure 1D.
Supplemental Figure S3. NCED3 banding pattern does not depend on genetic background and ABA accumulation of NCED3 SDMs.
Supplemental Figure S4. Alignments of maize VP14 and Arabidopsis NCEDs.
Supplemental Figure S5. Structural modeling shows the proximity of amino acids 184 and 271 on the stromal facing surface of NCED3.
Supplemental Figure S6. Analysis of NCED3 chloroplast localization in Ler and Sha using a rapid chloroplast fractionation procedure.
Supplemental Table S1. Polymorphisms at NCED3 amino acids 18, 83, 184, and 271 among accessions in the 1,001-genome database.
Supplemental Table S2. Primers used in this study.
Acknowledgments
We thank Hsou-min Li for the AtOE33 and GS2 antisera and assistance with chloroplast fractionation, Wan-Hsing Cheng for the nced3 mutant, Trent Z. Chang and Shih-Shan Huang for laboratory assistance, and Eugene Shakirov for help in sequence alignment and polymorphism characterization.
Footnotes
This work was supported by the Taiwan Ministry of Science and Technology (102-2628-B-001-003 to P.E.V.) and the U.S. National Science Foundation (IOS-0922457 to T.E.J.).
Articles can be viewed without a subscription.
References
- Alonso-Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KM, Cao J, Chae E, Dezwaan TM, Ding W, et al. (2016) 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, Buell R, Gore MA, Dellapenna D (2013) Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell 25: 4827–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bac-Molenaar JA, Granier C, Keurentjes JJB, Vreugdenhil D (2016) Genome-wide association mapping of time-dependent growth responses to moderate drought stress in Arabidopsis. Plant Cell Environ 39: 88–102 [DOI] [PubMed] [Google Scholar]
- Bhaskara GB, Yang TH, Verslues PE (2015) Dynamic proline metabolism: Importance and regulation in water limited environments. Front Plant Sci 6: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchabke O, Chang F, Simon M, Voisin R, Pelletier G, Durand-Tardif M (2008) Natural variation in Arabidopsis thaliana as a tool for highlighting differential drought responses. PLoS ONE 3: e1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Chu CC, Li HM (2011) Determining the location of an Arabidopsis chloroplast protein using in vitro import followed by fractionation and alkaline extraction. Methods Mol Biol 774: 339–350 [DOI] [PubMed] [Google Scholar]
- Clerkx EJM, El-Lithy ME, Vierling E, Ruys GJ, Blankestijn-De Vries H, Groot SPC, Vreugdenhil D, Koornneef M (2004) Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol 135: 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DL, McKay JK, Richards JH, Sen S, Wayne T, Juenger TE (2012) Physiological genomics of response to soil drying in diverse Arabidopsis accessions. Plant Cell 24: 893–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- El-Lithy ME, Clerkx EJM, Ruys GJ, Koornneef M, Vreugdenhil D (2004) Quantitative trait locus analysis of growth-related traits in a new Arabidopsis recombinant inbred population. Plant Physiol 135: 444–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A, Sawada Y, Takahashi H, Okamoto M, Ikegami K, Koiwai H, Seo M, Toyomasu T, Mitsuhashi W, Shinozaki K, et al. (2008) Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol 147: 1984–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. (2013) Abscisic acid synthesis and response. The Arabidopsis Book 11: e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A, Effroy D, Lefebvre V, Seo M, Perreau F, Berger A, Sechet J, To A, North HM, Marion-Poll A (2012) Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J 70: 501–512 [DOI] [PubMed] [Google Scholar]
- Hao GP, Zhang XH, Wang YQ, Wu ZY, Huang CL (2009) Nucleotide variation in the NCED3 region of Arabidopsis thaliana and its association study with abscisic acid content under drought stress. J Integr Plant Biol 51: 175–183 [DOI] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjálmsson BJ, et al. (2012) Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet 44: 212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Jasinski S, Lécureuil A, Miquel M, Loudet O, Raffaele S, Froissard M, Guerche P (2012) Natural variation in seed very long chain fatty acid content is controlled by a new isoform of KCS18 in Arabidopsis thaliana. PLoS ONE 7: e49261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalladan R, Lasky JR, Chang TZ, Sharma S, Juenger TE, Verslues PE (2017) Natural variation identifies genes affecting drought-induced abscisic acid accumulation in Arabidopsis thaliana. Proc Natl Acad Sci USA 114: 11536–11541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesari R, Lasky JR, Villamor JG, Des Marais DL, Chen YJC, Liu TW, Lin W, Juenger TE, Verslues PE (2012) Intron-mediated alternative splicing of Arabidopsis P5CS1 and its association with natural variation in proline and climate adaptation. Proc Natl Acad Sci USA 109: 9197–9202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky JR, Des Marais DL, McKay JK, Richards JH, Juenger TE, Keitt TH (2012) Characterizing genomic variation of Arabidopsis thaliana: The roles of geography and climate. Mol Ecol 21: 5512–5529 [DOI] [PubMed] [Google Scholar]
- Lasky JR, Des Marais DL, Lowry DB, Povolotskaya I, McKay JK, Richards JH, Keitt TH, Juenger TE (2014) Natural variation in abiotic stress responsive gene expression and local adaptation to climate in Arabidopsis thaliana. Mol Biol Evol 31: 2283–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A (2006) Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45: 309–319 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F (2002) Bay-0 × Shahdara recombinant inbred line population: A powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Lovell JT, Mullen JL, Lowry DB, Awole K, Richards JH, Sen S, Verslues PE, Juenger TE, McKay JK (2015) Exploiting differential gene expression and epistasis to discover candidate genes for drought-associated QTLs in Arabidopsis thaliana. Plant Cell 27: 969–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing SAJ, Gabelli SB, Echeverria I, Vogel JT, Guan JC, Tan BC, Klee HJ, McCarty DR, Amzel LM (2010) Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica JP, Mullen J, Lovell JT, Monroe JG, Paul JR, Oakley CG, McKay JK (2016) Genetics of water use physiology in locally adapted Arabidopsis thaliana. Plant Sci 251: 12–22 [DOI] [PubMed] [Google Scholar]
- Nambara E, Kawaide H, Kamiya Y, Naito S (1998) Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol 39: 853–858 [DOI] [PubMed] [Google Scholar]
- Pineau C, Loubet S, Lefoulon C, Chalies C, Fizames C, Lacombe B, Ferrand M, Loudet O, Berthomieu P, Richard O (2012) Natural variation at the FRD3 MATE transporter locus reveals cross-talk between Fe homeostasis and Zn tolerance in Arabidopsis thaliana. PLoS Genet 8: e1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Zeevaart JAD (1999) The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci USA 96: 15354–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Zheng Z, Chinnusamy V, Zhu J, Cui X, Iida K, Zhu JK (2010) RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc Natl Acad Sci USA 107: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero B, Koiwa H, Manabe Y, Quist TM, Inan G, Saccardo F, Joly RJ, Hasegawa PM, Bressan RA, Maggio A (2004) Uncoupling the effects of abscisic acid on plant growth and water relations: Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis. Plant Physiol 136: 3134–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Verslues PE (2010) Mechanisms independent of abscisic acid (ABA) or proline feedback have a predominant role in transcriptional regulation of proline metabolism during low water potential and stress recovery. Plant Cell Environ 33: 1838–1851 [DOI] [PubMed] [Google Scholar]
- Sharma S, Lin W, Villamor JG, Verslues PE (2013) Divergent low water potential response in Arabidopsis thaliana accessions Landsberg erecta and Shahdara. Plant Cell Environ 36: 994–1008 [DOI] [PubMed] [Google Scholar]
- Szymańska R, Nowicka B, Gabruk M, Glińska S, Michlewska S, Dłużewska J, Sawicka A, Kruk J, Laitinen R (2015) Physiological and antioxidant responses of two accessions of Arabidopsis thaliana in different light and temperature conditions. Physiol Plant 154: 194–209 [DOI] [PubMed] [Google Scholar]
- Tan BC, Cline K, McCarty DR (2001) Localization and targeting of the VP14 epoxy-carotenoid dioxygenase to chloroplast membranes. Plant J 27: 373–382 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Therneau TM. (2018) coxme: Mixed Effects Cox Models. R package version 2.2-10. https://CRAN.R-project.org/package=coxme
- Trontin C, Tisné S, Bach L, Loudet O (2011) What does Arabidopsis natural variation teach us (and does not teach us) about adaptation in plants? Curr Opin Plant Biol 14: 225–231 [DOI] [PubMed] [Google Scholar]
- Vallejo AJ, Yanovsky MJ, Botto JF (2010) Germination variation in Arabidopsis thaliana accessions under moderate osmotic and salt stresses. Ann Bot 106: 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE. (2016) ABA and cytokinins: Challenge and opportunity for plant stress research. Plant Mol Biol 91: 629–640 [DOI] [PubMed] [Google Scholar]
- Verslues PE. (2017) Rapid quantitation of abscisic acid by GC-MS/MS for studies of abiotic stress response. In Sunkar R, ed, Plant Stress Tolerance: Methods and Protocols, Vol 1631, 21. Springer, New York, NY, pp 325–336 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Bray EA (2006) Role of abscisic acid (ABA) and Arabidopsis thaliana ABA-insensitive loci in low water potential-induced ABA and proline accumulation. J Exp Bot 57: 201–212 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Wan XR, Li L (2006) Regulation of ABA level and water-stress tolerance of Arabidopsis by ectopic expression of a peanut 9-cis-epoxycarotenoid dioxygenase gene. Biochem Biophys Res Commun 347: 1030–1038 [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12: 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]