The RWP-RK transcription factor Nodule Inception regulates genes required for rhizobial infection, including Rhizobium Polar Growth (RPG) and genes for nutrient (N, P, and S) uptake and assimilation.
Abstract
The symbiotic infection of root cells by nitrogen-fixing rhizobia during nodulation requires the transcription factor Nodule Inception (NIN). Our root hair transcriptomic study extends NIN’s regulon to include Rhizobium Polar Growth and genes involved in cell wall modification, gibberellin biosynthesis, and a comprehensive group of nutrient (N, P, and S) uptake and assimilation genes, suggesting that NIN’s recruitment to nodulation was based on its role as a growth module, a role shared with other NIN-Like Proteins. The expression of jasmonic acid genes in nin suggests the involvement of NIN in the resolution of growth versus defense outcomes. We find that the regulation of the growth module component Nodulation Pectate Lyase by NIN, and its function in rhizobial infection, are conserved in hologalegina legumes, highlighting its recruitment as a major event in the evolution of nodulation. We find that Nodulation Pectate Lyase is secreted to the infection chamber and the lumen of the infection thread. Gene network analysis using the transcription factor mutants for ERF Required for Nodulation1 and Nuclear Factor-Y Subunit A1 confirms hierarchical control of NIN over Nuclear Factor-Y Subunit A1 and shows that ERF Required for Nodulation1 acts independently to control infection. We conclude that while NIN shares functions with other NIN-Like Proteins, the conscription of key infection genes to NIN’s control has made it a central regulatory hub for rhizobial infection.
Most legumes can interact with nitrogen-fixing bacteria, called rhizobia, which are taken up into cells of specialized root organs called nodules. Nodulation involves two processes, rhizobial infection and nodule formation. Rhizobial infection is preceded by attachment of the bacteria to elongating root hairs, which redirects growth to entrap the rhizobia within a curl, forming an infection chamber. From this chamber, a tubular structure called the infection thread forms as an invagination of the plasma membrane and cell wall. As the infection thread grows, it becomes colonized by dividing rhizobia, directing them through a predefined route to the developing nodule (Fournier et al., 2008, 2015). Within the nodule, the rhizobia are endocytosed from the infection thread into specialized host membrane-enclosed structures called symbiosomes where nitrogen fixation takes place (Ivanov et al., 2010). Thus, nodulation depends on the successful coordination of infection and nodule formation. Legumes such as Lotus japonicus form determinate nodules, which are spherical, lack an apical meristem, and harbor some infection threads when mature (Malolepszy et al., 2018), while indeterminate-type nodules formed on legumes such as Medicago truncatula, are elongated, and feature a persistent apical meristem and an adjacent infection zone containing dense and highly branched infection threads (Guan et al., 2013; Chen et al., 2015).
Nodulation requires host perception of lipochitooligosaccharide signals called Nod factors, which are produced by rhizobia in response to plant flavonoids (Fliegmann and Bono, 2015; Liu and Murray, 2016). Several transcription factors orchestrate the downstream responses to Nod factors, leading to rhizobial infection and nodule formation, including Nodule Inception (NIN; Schauser et al., 1999; Marsh et al., 2007), Interacting Protein of DMI3 (IPD3; Horváth et al., 2011; Ovchinnikova et al., 2011) andIPD3-Like (IPD3L), ERF Required for Nodulation (ERN1/ERN2; Andriankaja et al., 2007; Middleton et al., 2007), Nuclear Factor-Y Subunit A1 (NF-YA1; Laloum et al., 2014), the GIBBERELLIC-ACID INSENSITIVE, REPRESSOR of GIBBERELLIC-ACID INSENSITIVE, and SCARECROW (GRAS) transcription factors Nodulation Signaling Pathway1 (NSP1) and NSP2 (Oldroyd and Long, 2003; Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006), and the DELLAs (Fonouni-Farde et al., 2016; Jin et al., 2016). Transcript profiling of M. truncatula root hairs on rhizobia-inoculated plants revealed several hundred genes with up-regulated expression (Breakspear et al., 2014), including many hormone-related genes and genes with roles in the cell cycle. Along with these genes, numerous transcription factors were induced in this root hair transcriptome, including NIN, NF-YA1, and ERN1, which are prime candidates for mediating a subset of the transcriptional changes that occur during rhizobial infection.
NIN is the founding member of the NIN-Like Protein (NLP) family of transcription factors (Schauser et al., 1999; Chardin et al., 2014). nin mutants are unable to form proper infection chambers and form abnormally hypercurled root hair tips. Although the nin mutant does entrap some rhizobia within curled root hairs, rhizobia cell division is limited and infection threads fail to initiate (Schauser et al., 1999; Borisov et al., 2003; Marsh et al., 2007; Fournier et al., 2015). Both nf-ya1-1 and ern1 are defective in the development of infection threads but show weaker phenotypes than nin, as they are able to form infection chambers and microcolonies (Middleton et al., 2007; Laporte et al., 2014). Similar to the nf-ya1-1 mutant, silencing of both NF-YA1 and its close homolog NF-YA2 leads to defective infection, with the strongest phenotypes showing entrapped bacteria, which form microcolonies but with no initiation of infection threads (Laloum et al., 2014; Laporte et al., 2014). NF-YA1 encodes one of the three subunits (denoted as A, B, and C) of the CCAAT-box complex, a heterotrimeric transcription factor complex named after the CCAAT-box motif, which is bound by the NF-YA subunit. The CCAAT-box complex is found across eukaryotes, and in plants each subunit is encoded by multiple genes (Zanetti et al., 2017). NF-YA1-interacting subunits B/C involved in nodulation were discovered in M. truncatula, L. japonicus, and Phaseolus vulgaris (Zanetti et al., 2010; Soyano et al., 2013; Baudin et al., 2015). ERN1 encodes an AP2/ERF family transcription factor, expressed in root hairs undergoing rhizobial infection and in developing nodules of both M. truncatula and L. japonicus (Cerri et al., 2012, 2017; Kawaharada et al., 2017; Yano et al., 2017). This expression is in part mirrored by ERN1’s close homolog, ERN2, which has relatively lower but partially overlapping expression in root tissues undergoing rhizobial infection (Cerri et al., 2012; Roux et al., 2014). Although unable to form mature nodules, M. truncatula ern1 can initiate root hair infection and nodule organogenesis (Middleton et al., 2007), which are totally abolished in the ern1 ern2 double mutant (Cerri et al., 2016). The L. japonicus Ljern1 mutant develops very few infection threads, but nodule primordia can still form (Cerri et al., 2017; Kawaharada et al., 2017; Yano et al., 2017).
In addition to their important roles during rhizobial infection, NIN, NFY-A1, and ERN1 are also required for proper nodule development in the root cortex (Marsh et al., 2007; Cerri et al., 2012, 2016; Laporte et al., 2014; Xiao et al., 2014; Vernié et al., 2015). Despite being the first nodulation gene cloned in model legumes (Schauser et al., 1999), just a few of NIN’s targets have been identified, including NODULATION PECTATE LYASE1 (NPL1), NF-YA1, NF-YB1, CLE-Root Signal1 (CLE-RS1), and CLE-RS2, by work in L. japonicus (Xie et al., 2012; Soyano et al., 2013, 2014). It was shown that NF-YA1 and NF-YA2 may act upstream of ERN1 during the early stages of rhizobial NF signaling/infection (Laloum et al., 2014; Fonouni-Farde et al., 2016). Despite these findings, the genes and the cellular processes regulated by NIN, NF-YA1, and ERN1 are still largely unknown, especially on the genomic level, in part due to the difficulty posed by the use of root samples composed of many cell types and secondary effects on gene expression that occur during later stages of nodule development.
The established relationships between NIN, NF-YA1, and ERN1 provide an opportunity to better understand how complex transcriptional networks operate. To gain insights on NIN’s role in infection and to establish the hierarchical relationships between these three regulators, we transcriptionally profiled nin, nf-ya1, and ern1 mutants using root hairs of rhizobia-inoculated plants. By using this single cell-type transcriptional profiling method, which provides greatly improved sensitivity and specificity for detecting genes involved in rhizobial infection, we have generated a more complete model for the gene regulatory network governing this process. This analysis reveals a central role for NIN in the regulation of cell growth and development in infection and identifies new symbiosis-specific genes under the control of ERN1.
RESULTS
A Gene Regulatory Network for Rhizobial Infection
To investigate the regulation of gene expression during rhizobial infection of M. truncatula, we transcriptionally profiled the ern1-1, nf-ya1-1, and nin-1 mutants using root hairs isolated from seedlings 5 d post inoculation (dpi) with Sinorhizobium meliloti (Rm1021). This time point was chosen because in our system it coincides with the onset of infection thread development and was found to be the best stage to detect gene expression changes (Breakspear et al., 2014). Three biological replicates for each treatment were used, and each replicate consisted of RNA isolated from root hairs of 100 to 200 seedlings. As a reference for comparison with the mutants, we used previously published expression data for root hairs of wild-type plants 5 dpi with rhizobia, which were obtained at the same time, using the same methodology as this study (Breakspear et al., 2014). The gene expression data presented for the wild type were described by Breakspear et al. (2014), the data for ern1, nin, and nf-ya1 have not been described elsewhere. The genome-wide expression data for root hairs of nin-1, ern1-1, and nf-ya1-1 5 dpi with S. meliloti are provided in Supplemental Data S1. All deregulated genes identified in this study are provided in Supplemental Data S2. In this updated analysis, a total of 1,353 genes (1,124 up-regulated and 229 down-regulated) had altered expression in root hairs of wild-type plants 5 dpi with Rm1021 relative to those inoculated with non-Nod factor-producing controls (SL44; Rm1021 ΔnodD1ABC) at the same time point.
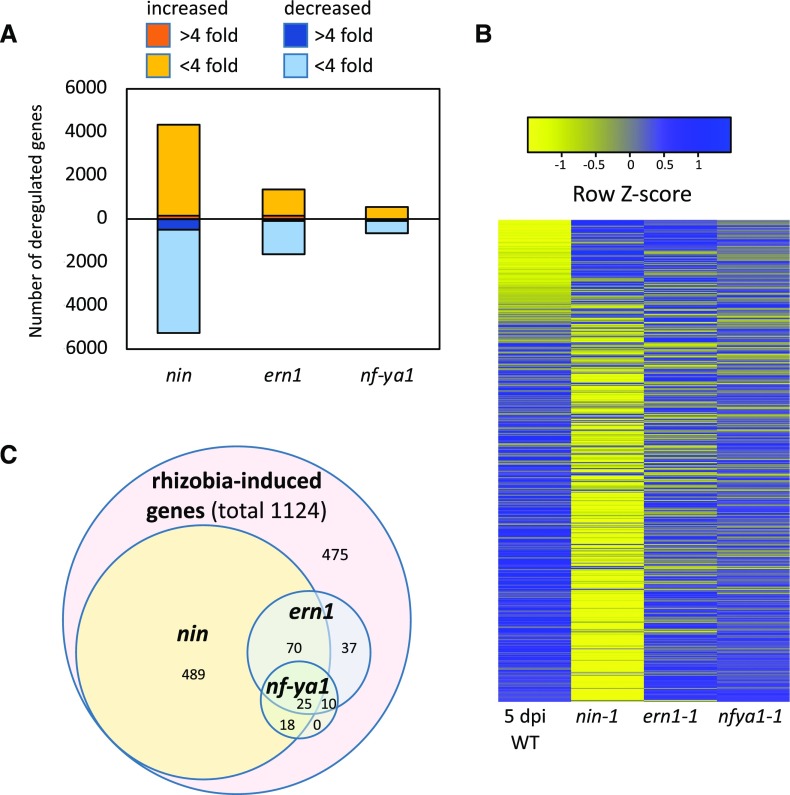
To determine the underlying cause for defective infection in the mutants, the gene expression levels were compared between each mutant and the wild type at the same time point (5 dpi). Loss of NIN had the largest impact on gene expression, with 5,178 genes down-regulated and 4,281 genes up-regulated relative to the wild type, while relatively fewer genes had changed expression in ern1 (1,612 down, 1,381 up) and nf-ya1-1 (526 up and 648 down) mutants (Fig. 1A). In the nin mutant, the down-regulated genes included 54% of infection-induced genes. In contrast, in ern1 and nf-ya1-1, only 13% and 5% of infection-induced genes failed to be expressed at wild-type levels, respectively (Fig. 1, B and C). For infection-induced genes with decreased expression in the mutants, ∼80% (489 genes) of those in nin were specific to nin, while only ∼25% (37 genes) of those in ern1 were ern1 specific (Fig. 1C). All infection genes down-regulated in nf-ya1 overlapped with nin and/or ern1 (Fig. 1C), and few of them were strongly changed. The overall transcriptional responses of nin, nf-ya1-1, and ern1 were in line with their rhizobial infection phenotypes, with loss of NIN having the largest impact on gene expression.
Figure 1.
Impact of nin, ern1, and nf-ya1 mutations on gene expression in root hairs of S. meliloti-inoculated seedlings. A, Genes significantly deregulated in ern1-1, nf-ya1-1, and nin-1 relative to the wild type (A17) 5 dpi with S. meliloti. B, Heat map showing expression of genes induced in root hairs of the wild type (WT) 5 dpi with S. meliloti relative to the ΔnodD1ABC-inoculated control (Breakspear et al., 2014) in nin, ern1, and nf-ya1. C, Venn diagram showing the numbers of rhizobia-induced genes down-regulated in the nin, nf-ya1, and ern1 mutants.
To further investigate the relationships between these transcription factors, we carried out a gene network analysis using data from ern1, nf-ya1, nin, and the wild type. The analysis was carried out using NetProphet v2.0, which uses a weighted scoring of coexpression across samples and differential expression in mutants relative to the wild type (Haynes et al., 2013; Kang et al., 2018). This modeling approach assumes that (1) transcription factors will be coexpressed with their targets across different samples and (2) expression levels of direct targets of transcription factors will be strongly affected in their mutants. This network analysis, which considers the largest changes in gene expression, indicates that only a small set of genes show strongly ERN1-dependent expression in rhizobia-inoculated root hairs. The induction of these ERN1-dependent genes was independent of NF-YA1 and NIN. Consistent with this, the expression of ERN1 itself was not decreased in nin and nf-ya1-1, and NIN expression was not decreased in ern1, suggesting that ERN1 acts independently of the NIN and NF-YA1 regulons, which are themselves directly connected by virtue of NF-YA’s dependence on NIN (Fig. 2). This analysis highlights a large number of genes as potential targets of NIN and relatively few as targets of ERN1.
Figure 2.
Gene regulatory network for rhizobial infection. Genes induced in root hairs 5 dpi with S. meliloti were ranked using NetProphet v2.0 based on differential expression in specific mutants and regression across root hair samples. Only the top 33% scoring genes for each mutant are included for clarity of presentation. Arrows denote directionality of the regulation. Edges for genes strongly repressed (0%–15% of the wild type) for a given mutant are connected by solid lines. Yellow denotes cell wall-related genes, and green denotes genes for DNA replication/translation. Other genes of interest are denoted in red. Genes with homologs enriched for NIN binding using ChIP-seq in L. japonicus are underlined (Soyano et al., 2015). *, Infection-specific genes (identified by Breakspear et al., 2014); **, nutrient uptake genes (Table 2); ***, nodule cysteine-rich peptides.
We found that expression of NF-YA1 and NPL was dependent on NIN, consistent with the discovery of their orthologs as direct targets of NIN in L. japonicus (Xie et al., 2012; Soyano et al., 2013). Their drastically reduced expression in nin (1% or less than in the wild type) agrees with a yeast study showing that the most strongly down-regulated genes in transcription factor mutants are more likely to be directly controlled (Haynes et al., 2013). Based on this, many potential NIN targets were identified in this study; 145 genes had greatly reduced (less than 15% of the wild type) expression in nin (Fig. 1A). This includes RPG and Cystathionine-β-Synthase-like1 (CBS1; 15% and 8% of wild-type expression levels, respectively), which are important for successful rhizobial infection in M. truncatula (Arrighi et al., 2008; Sinharoy et al., 2016). We confirmed the dependence of RPG on NIN using reverse transcription-quantitative PCR (RT-qPCR) in an independent experiment (Supplemental Fig. S1), while CBS1 was shown previously to be downstream of NIN in early nodulation (Sinharoy et al., 2016). Other infection-induced genes specifically down-regulated at least 4-fold in the nin mutant were LysM receptor-like kinase 10 (LYK10), early nodulin 40 (ENOD40; Crespi et al., 1994), Annexin 1 (de Carvalho-Niebel et al., 2002), an ABI-1-LIKE1 (ABIL1) homolog encoding an actin nucleation protein (Medtr7g116710), and a SEVEN IN ABSENTIA gene (Medtr5g061290) that is Nod factor inducible (Jardinaud et al., 2016). A comparison of our findings with an earlier study by Høgslund et al. (2009), which examined gene expression changes in the root susceptible zone of the L. japonicus nin mutant after rhizobial inoculation, revealed limited overlap, possibly due to differences in sensitivity between the approaches, but found in common several of the most highly expressed and/or most strongly regulated genes, including EPR3 (ortholog of LYK10), NPL, NF-YA1, and CBS1. While many novel candidate genes were identified as potential direct targets of NIN, it should be noted that their down-regulation in nin may be secondary (i.e. mediated by downstream transcription factors) or collateral (i.e. an indirect consequence of the failure to activate specific processes that normally occur during infection).
To investigate what proportion of the genes in the NIN regulon could be direct targets, we compared our transcriptomics data for the nin mutant with a chromatin immunoprecipitation sequencing (ChIP-seq) analysis for NIN in L. japonicus (Soyano et al., 2014). The authors of the study identified the Clavata3/ESR (CLE) peptide-encoding genes CLE-RS1 and CLE-RS2 as direct targets of NIN, but a comparison of the ChIP data with the nin mutant transcriptome was not carried out. To make the comparison, we identified the M. truncatula homologs/likely orthologs of the L. japonicus genes identified by ChIP using BLAST. We found a higher degree of overlap between the genes identified using ChIP and those identified as down-regulated in nin in our study than would be expected by chance, at all fold change cutoff levels considered (Supplemental Table S1). The increase in the degree of overlap compared with the randomly sampled controls increased with increasing cutoff stringency, suggesting that the most strongly down-regulated genes are more likely to be direct targets. This analysis suggests that NIN has at least 100 direct targets. A summary of nodulins that have strongly down-regulated expression in NIN, and that had at least 15-fold enrichment in the LjNIN ChIP-seq analysis (Soyano et al., 2014), is provided in Table 1. These include the known targets CLE13 and NF-YA1 as well as RPG, symbiotic sulfate transporter 1 (SST1), and symbiotic cysteine-rich receptor-like kinase (SymCRK), which have demonstrated roles in nodulation (Krusell et al., 2005; Arrighi et al., 2008; Berrabah et al., 2014). A complete list of the genes identified by this comparison is provided in Supplemental Data S3.
Table 1. Nodulins with reduced expression in root hairs of inoculated M. truncatula nin with close L. japonicus homologs identified using ChIP-seq for LjNIN (Soyano et al., 2014).
ChIP data are from Soyano et al. (2014). CaOMT, Caffeic acid O-methytransferase; MIP, major intrinsic protein; na, no gene name yet assigned; PP2C, protein phosphatase2C; TF, transcription factor.
| M. truncatula Gene Model | L. japonicus Gene Model | Gene Name | Description | Fold Change (nin/Wild Type) | Fold Enrichment (ChIP) |
|---|---|---|---|---|---|
| Medtr1g056530 | chr5.CM0571.340.r2.m | NF-YA1 | CCAAT-box TF | 0.01 | 68.02 |
| Medtr5g011950 | chr2.CM0021.2380.r2.m | na | Lipid transfer protein | 0.02 | 25.82 |
| Medtr3g012420 | chr3.CM0005.520.r2.m | N21 | MtN21 transporter | 0.03 | 11.49 |
| Medtr6g007160 | chr2.CM0081.540.r2.d | N24 | Transmembrane protein | 0.07 | 20.17 |
| Medtr3g079850 | chr6.CM0041.530.r2.a | SymCRK | Cys-rich RK-like protein | 0.09 | 14.03 |
| Medtr6g086170 | chr2.CM0610.70.r2.m | SST1 | Sulfate transporter | 0.09 | 14.33 |
| Medtr7g114870 | LjSGA_056148.1 | na | IQ calmodulin protein | 0.10 | 17.89 |
| Medtr8g098815 | chr4.CM0004.760.r2.d | na | BEL1-related protein | 0.12 | 18.97 |
| Medtr3g074610 | chr4.CM0182.350.r2.m | na | PP2C | 0.13 | 2.87 |
| Medtr2g055940 | chr1.LjT11G20.60.r2.d | na | CaOMT | 0.16 | 11.27 |
| Medtr8g087710 | chr4.CM0046.1620.r2.m | Nod26 | MIP family transporter | 0.17 | 30.91 |
| Medtr1g090807 | chr5.CM0909.840.r2.a | RPG | Myosin-like protein | 0.17 | 8.51 |
| Medtr1g018790 | chr4.CM0536.30.r2.m | na | F-box SKIP2-like protein | 0.17 | 13.00 |
| Medtr5g026850 | chr2.CM0803.150.r2.m | IPD3L | CYCLOPS protein | 0.19 | 26.80 |
| Medtr6g081810 | LjSGA_016193.1 | na | MATE efflux family protein | 0.19 | 12.33 |
| Medtr1g085680 | chr1.CM0017.80.r2.m | na | Cytochrome P450 | 0.22 | 11.07 |
| Medtr5g014640 | chr2.CM0021.390.r2.m | na | Basic helix-loop-helix TF | 0.24 | 3.67 |
| Medtr4g079610 | CM0446.165.r2.a | CLE13 | Clavata3/ESR peptide | 0.25 | 31.35 |
| Medtr8g069775 | chr4.CM0161.180.r2.d | NRT2.7 | High-affinity NO3 transporter | 0.30 | 17.25 |
NIN Is Required for the Expression of Cell Wall Remodeling, Cell Cycle, and Transporter Genes
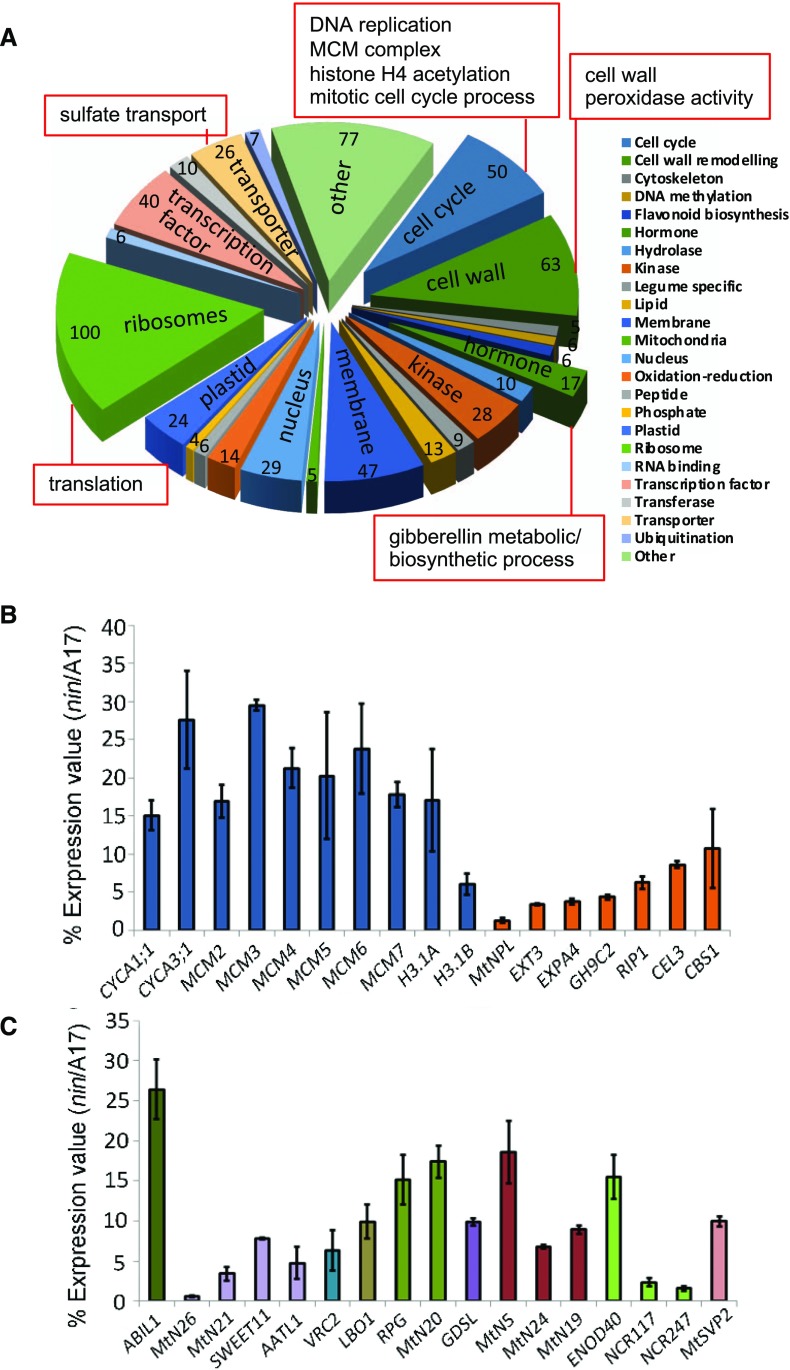
A total of 63 rhizobia-induced cell wall-related genes were down-regulated in nin root hairs after rhizobia inoculation (Fig. 3, A and B), including NPL, two cellulases, a pectinacetylesterase, a Gal oxidase, a Gal mutarotase, a galactinol synthase, an α-glucan phosphorylase, an endo-β-mannase, an expansin, and an extensin. Notably, with the exception of NPL, none of these genes had symbiosis-specific expression b (Medicago Gene Expression Atlas [MtGEA]v3). Also included in this category were the secreted peroxidases including rhizobium-induced peroxidase 1 (Wisniewski et al., 2000), which may contribute to cell wall maturation. Of 11 peroxidases previously found to be induced by S. meliloti (Breakspear et al., 2014), nine had reduced expression in nin, suggesting that NIN, either directly or indirectly, is the principal regulator of these genes during infection. All nine of these contained predicted secretion peptides. The NIN regulon also included 100 ribosomal genes and 50 genes predicted to be involved in cell cycle progression, mainly genes encoding components of the DNA replication machinery, such as all of the minichromosome maintenance (MCM) genes and many histones (Fig. 3, A and B). Genes encoding predicted membrane proteins and transporters also represent a relatively large portion of all genes under the control of NIN (Fig. 3, A and C). Many of them showed high percentages of reduced expression in nin, including several nodulins, such as the predicted membrane proteins MtN5, MtN19, and MtN24, and the transporters MtN21, MtN26, and SWEET11. The induction of SWEET11 during nodulation was previously shown to be NIN dependent (Kryvoruchko et al., 2016). The categories of cell wall, DNA replication, MCM complex, histone H4 acetylation, sulfate transport, mitotic cell cycle process, translation, and many related terms were detected as significant using GO term enrichment analysis (Fig. 3; a full list of enriched terms is provided in Supplemental Data S4). The regulation of genes involved in cell cycle-related processes (see discussion in Breakspear et al., 2014), protein translation, and cell wall remodeling by NIN suggests a major role for this transcription factor in engaging the cell cycle and inducing the genes required to support cell growth.
Figure 3.
Cellular processes regulated by NIN in root hairs during rhizobial infection. A, Pie chart showing categories of cellular processes with different numbers of infection genes (up-regulated at 5 dpi with Rm1021) that are down-regulated in nin-1. Significantly enriched Gene Ontology (GO) terms are indicated by red boxes for the different gene categories. B and C, Percentage of expression values in nin-1/the wild type of some representative genes from A. Bars indicate 95% confidence interval. CYCA, Cyclin A; MCM, MINI-CHROMOSOME MAINTENANCE; H3.1, HISTONE 3.1; NPL, NODULE PECTATE LYASE; EXT3, EXTENSIN 3; EXPA4, EXPANSIN A4; GH9C2, Glycosyl Hydrolase Family 9C; RIP1, rhizobium-induced peroxidase 1; CEL3, CELLULASE 3; CBS1, Cystathionine β Synthase-like 1; AATL1, Amino Acid Transporter-Like protein 1; VRC2, Vestitone Reductase Cluster 2; LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1; RPG, Rhizobium-directed Polar Growth; GDSL, GDSL-like lipase; NCR, Nodule Cys-Rich peptide; MtSVP2, Short Vegetative Phase protein 2.
NIN-Directed NPL Expression Is Required for Infection of M. truncatula
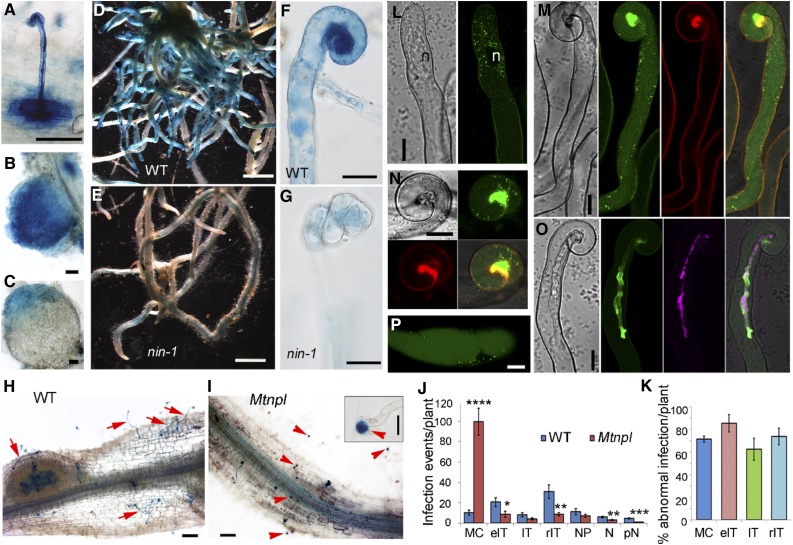
In L. japonicus, NPL encodes a pectate lyase with nodulation-enhanced expression required for infection thread formation (Xie et al., 2012). Phylogenetic analysis revealed an NPL ortholog in M. truncatula (Supplemental Fig. S2), with strongly enhanced expression in response to rhizobia (Breakspear et al., 2014). We studied its expression using promoter-GUS analysis in composite plants in both the wild type and nin mutants. In wild-type plants inoculated with S. meliloti, ProMtNPL:GUS was highly expressed at infection sites, including infected root hairs and underlying cells (Fig. 4A; Supplemental Fig. S3A). MtNPL was also expressed throughout young nodules (Fig. 4B) and in the apex of mature nodules (Fig. 4C), which contrasts with LjNPL, which is not expressed in mature nodules (Verdier et al., 2013; LjGEA). This difference is consistent with M. truncatula nodules having a persistent meristem and infection zone, which are absent in mature L. japonicus nodules. MtNPL was also found to be expressed in root tips and lateral root primordia (Supplemental Fig. S3, B and C). The wild-type plants showed strong GUS staining in roots of inoculated plants (Fig. 4D) compared with nin-1, which exhibited only residual expression (Fig. 4E). MtNPL was highly expressed in infected root hairs of wild-type plants (Fig. 4F), but its expression was greatly reduced in curled root hairs of the nin-1 mutant (Fig. 4G).
Figure 4.
MtNPL’s role, regulation, and localization during infection of M. truncatula. A to C, ProMtNPL:GUS expression in Agrobacterium rhizogenes-induced transgenic hairy roots showing expression in a root hair harboring an infection thread (A), a young nodule (B), and an elongated nodule (C). D to G, ProMtNPL:GUS expression in the wild type (WT; D and F) and Mtnin-1 (E and G) 14 dpi with S. meliloti 1021. H to K, Rhizobial infection phenotype of Mtnpl 14 dpi with S. meliloti 1021. H, A root segment from the wild type showing full elongated infection threads (red arrows) and a nodule primordium. I, Infection phenotype of Mtnpl showing a typical root segment from Mtnpl with many swelled microcolonies (red arrowheads). The inset shows higher magnification of an abnormal microcolony (red arrowhead). J, Quantification of different rhizobial infection events in the wild type and Mtnpl. K, Percentage of abnormal infection events 14 dpi in Mtnpl. MC, Microcolony; eIT, elongating infection threads; IT, full elongated infection threads in root hair; rIT, ramifying infection threads; NP, nodule primordia; N, nodules; pN, pink nodules. Error bars indicate se. Significant (Student’s t test) differences between the wild type and mutants are marked with asterisks (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; and ****, P ≤ 0.0001). L to P, Subcellular localization of MtNPL-GFP in A. rhizogenes-induced transgenic hairy roots with pMtNPL:MtNPL-GFP (L–O) or pLjUBQ1:MtNPL-GFP (P) 7 dpi with S. meliloti 1021-CFP. L, MtNPL-GFP in a root hair before any deformation occurs (left, bright field; right, GFP). M, MtNPL-GFP in a root hair-forming infection pocket (from left to right, bright field, GFP, cell wall autofluorescence, and merge). N, Another root hair showing MtNPL-GFP accumulation around and in the infection pocket (clockwise from top left: bright field, GFP, cell wall autofluorescence, and merge). O, A root hair harboring an infection thread containing rhizobia (magenta) showing MtNPL-GFP (green) in the infection thread lumen. Bars = 100 µm (A–C, H, and I), 200 mm (D and E), 20 µm (F, G, and inset in I), and 10 µm (L–P).
To determine if NPL is required in the M. truncatula-S. meliloti symbiosis, we identified a homozygous Tnt1 insertion mutant (line NF18556) that we designated as Mtnpl. The Mtnpl mutant developed more microcolonies, and most (∼70%) of them were enlarged (Fig. 4, H and I), indicating a defect in infection thread initiation leading to increased accumulation of rhizobia within the infection chamber. The mutant had significantly fewer elongating infection threads in root hairs and fewer infection threads ramified in the root cortex (Fig. 4J). The development of most infection threads forming in root hairs on the mutant was abnormal, resulting in large and misshapen infection threads that sometimes ruptured, releasing the rhizobia into the root hair cytoplasm (Fig. 4, J and K; Supplemental Fig. S3, D–F), indicating loss of integrity of the infection thread cell wall. Ruptured infection threads were not observed on the wild type in this experiment. Sometimes bacteria accumulated in intercellular spaces at sites where infections would normally bridge between cells, indicating a difficulty in initiating cell wall invagination in the Mtnpl mutant (Supplemental Fig. S3, G and H). The nodulation phenotype of Mtnpl was complemented by both a genomic fragment of MtNPL and a construct containing the MtNPL complementary DNA driven by the MtNPL native promoter (Supplemental Fig. S4). We conclude that NPL is required for infection and that it has highly specific temporal and spatial expression in rhizobia-infected cells, including in the nodule apex, in the indeterminate nodulator M. truncatula.
NPL Is Secreted to the Infection Chamber and the Infection Thread
To further investigate the role of NPL in infection by rhizobia, we investigated its localization in root hairs by means of a live-cell imaging system using an MtNPL-GFP (green fluorescent protein) fusion expressed under the control of the native NPL promoter (ProMtNPL:MtNPL-GFP). This construct was able to complement the mtnpl mutant, demonstrating that the MtNPL-GFP fusion protein is functional (Supplemental Fig. S4, E–G). After inoculation with S. meliloti, strong MtNPL-GFP signal was found as small puncta, reminiscent of vesicles, in the cytoplasm of the root hairs before any deformation occurred (Fig. 4L). In the curled root hairs, the punctate pattern was seen in the cytoplasm, including along the cytoplasmic strands (Fig. 4M). Also, at this stage, a large accumulation of MtNPL-GFP was observed in the infection chamber of the root hairs (Fig. 4, M and N), with vesicle-like puncta appearing to converge at the site of the initiating infection thread (Fig. 4N). In extended infection threads, MtNPL-GFP accumulated in the infection thread lumen, particularly at the tip regions (Fig. 4O). The vesicle-like punctate pattern of MtNPL-GFP was also found when driven by a strong constitutive promoter following inoculation with rhizobia (Fig. 4P). These results demonstrate the secretion of MtNPL to the infection chamber and to the infection thread, indicating its role in the cell wall remodeling required for initiation and elongation of infection threads.
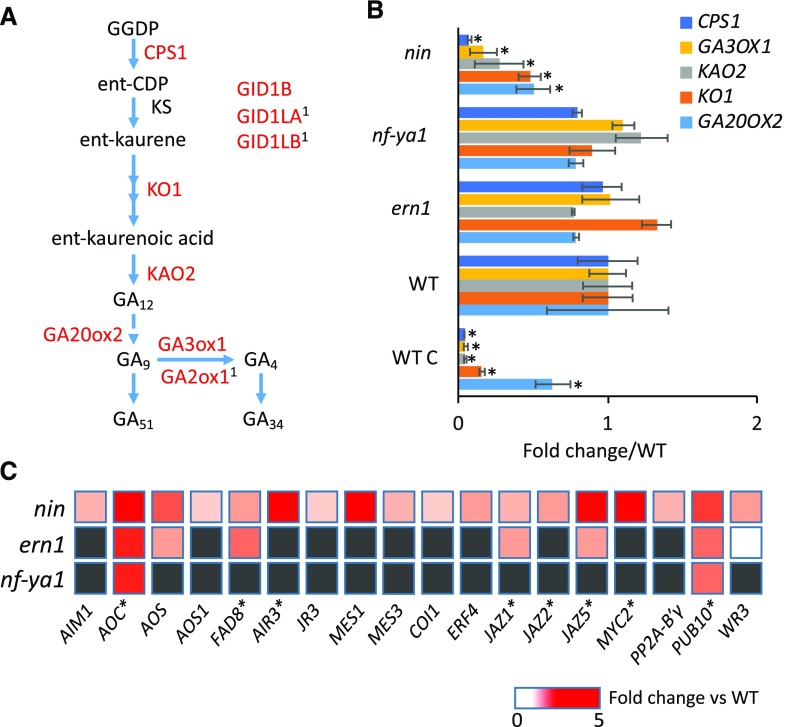
Gibberellin Biosynthesis and Jasmonate Biosynthesis/Signaling Are Differentially Regulated in nin
We found that the increased expression of several gibberellic acid (GA) biosynthesis and signaling genes following inoculation with S. meliloti is dependent on NIN. Specific to the NIN regulon were five genes for gibberellin synthesis, ENT-COPALYL DIPHOSPHATE SYNTHETASE1 (CPS1), ENT-KAURENE OXIDASE1, ENT-KAURENOIC ACID HYDROXYLASE2, GIBBERELLIN 20-OXIDASE2, and GIBBERELLIN 3-OXIDASE1, all of which were strongly reduced in nin (Fig. 5, A and B). In particular, CPS1, which controls the first committed step in GA biosynthesis, was reduced to baseline (1% of the wild type) levels. GIBBERELLIN 2-OXIDASE1 and the GA signaling components GA INSENSITIVE DWARF1B (GID1B) and GID1-LIKEA/B were down-regulated in nin and were also found to interact with NIN in L. japonicus (Fig. 5; Soyano et al., 2014). DELLA1 also had reduced expression in nin (details on these GA-related genes are provided in Supplemental Data S5). This indicates that GA biosynthesis and signaling during infection are dependent on NIN, consistent with a role in growth-related processes.
Figure 5.
Genes for GA and JA biosynthesis and signaling have decreased and increased expression, respectively, in root hairs of nin after rhizobial inoculation. A, The GA biosynthetic pathway in plants. CPS1, ENT-COPALYL DIPHOSPHATE SYNTHETASE1; KS, KAURENE SYNTHASE; KO1, ENT-KAURENE OXIDASE1; KAO2, ENT-KAURENOIC ACID HYDROXYLASE2; GA20ox2, GIBBERELLIN 20-OXIDASE2; GA3ox1, GIBBERELLIN 3-OXIDASE1; GA2ox1, GIBBERELLIN 2-OXIDASE1; GID1B, GA INSENSITIVE DWARF1B; GID1LA/B, GID1-LIKEA/B. Genes encoding the enzymes in red are significantly down-regulated in nin. Superscript 1 indicates that the gene was identified using NIN ChIP-seq by Soyano et al. (2014). B, Expression of GA biosynthesis genes in root hairs of ern1, nin, and nf-ya1 mutants relative to the wild type (WT; A17) at 5 dpi with S. meliloti Rm1021. Bars indicate 95% confidence interval. *, P ≤ 0.05, Student’s t test; mutants are compared with the wild type, and the wild type is compared with its control (WT C), which was inoculated with S. meliloti Rm1021 ΔnodD1ABC. C, Expression of JA-related genes in root hairs of nin, nf-ya1, and ern1 mutants relative to the wild type (A17) at 5 dpi with S. meliloti Rm1021. Genes reported to be induced by methyl jasmonate are indicated by asterisks (Naoumkina et al., 2007). Only genes with mean expression significantly different from that of the wild type are indicated (gray boxes indicate that expression was not significantly different).
Unexpectedly, among the 110 genes with significantly higher expression in root hairs of nin relative to the wild type after rhizobial inoculation were those with roles in jasmonic acid (JA) biosynthesis Allene Oxide Synthase and Fatty Acid Desaturase 8, metabolism (Jasmonic Acid Responsive 3, IAA-Alanine Resistant 3, and Methy Esterase 1), signaling (Jasmonate ZIM-domain Protein 5, Protein Phosphatase 2A [PP2A] β-subunit, and Ethylene Responsive Factor Binding Element 4), and other JA-related genes, including many that are methyl jasmonate inducible (Naoumkina et al., 2007; Fig. 5C; details are provided in Supplemental Data S6). Also up-regulated in nin was the gene encoding the ortholog of MYC2, a transcription factor that has been implicated in JA responses. The gene encoding the ortholog of the ubiquitin ligase, Plant U-BOX 10, which negatively regulates MYC2 (Jung et al., 2015), had increased expression in nin. In addition, the genes encoding the orthologs of the deubiquitinating enzymes Ubiquitin-Specific Protease 12 (UBP12) and UBP13, which counteract the activity of Plant U-BOX 10 to regulate the stability of MYC2, have decreased expression in nin (Jeong et al., 2017). Six of the above-mentioned JA-induced genes are also induced by pathogens (MtGEAv3; Benedito et al., 2008). None of these JA-related genes were identified using ChIP-seq for NIN (Soyano et al., 2014), suggesting that their deregulation is indirect, consistent with NIN’s role as a transcriptional activator. This indicates that the JA pathway is ectopically activated during infection in root hairs of nin plants, which could possibly contribute to the observed infection phenotype.
NIN Is Required for Expression of Nutrient Uptake and Assimilation Genes during Rhizobial Infection
NIN is a legume-specific gene belonging to the NIN-like protein family of transcription factors, which have been shown both in legumes and nonlegumes to play a role in nitrogen and general nutrient homeostasis (Castaings et al., 2009; Konishi and Yanagisawa, 2013; Marchive et al., 2013; Yu et al., 2016). No such role has been attributed to NIN, which instead has been shown to control genes with critical roles in infection, such as NF-YA1 and NPL. Recently, an NLP was shown to positively regulate the expression of CLE-RS2, which is involved in autoregulation of nodulation by nitrogen (Nishida et al., 2018), linking NIN, like other NLPs, with the regulation of nutrient homeostasis. To further investigate this possibility, we examined the expression of the M. truncatula orthologs of a set of Arabidopsis (Arabidopsis thaliana) nutrient homeostasis genes that are down-regulated in nlp7 (Castaings et al., 2009) and found that many were down-regulated in nin (Table 2). We also identified many additional genes with well-established roles in nutrient homeostasis having reduced expression in nin (Table 2). Of these 51 genes, 12 were previously identified as enriched by ChIP-seq using NIN in L. japonicus (Soyano et al., 2014). In addition, the genes with the GO terms organonitrogen compound biosynthetic process and sulfate transport were overrepresented in the NIN regulon (Supplemental Table S1). We then compared the genes down-regulated in nin root hairs with the NLP7 targets identified in Arabidopsis with ChIP-chip (Marchive et al., 2013). We found that 180 of the 850 genes identified by ChIP-chip had close homologs in M. truncatula that were also down-regulated in nin, an overlap that was higher than expected by chance (P < 1 × 10−5), including genes such as Arabidopsis LOB-Domain Containing Protein 38, CBL-interacting protein kinase 8, Nitrate Transporter 1.1 (NRT1.1), and NRT2.5, which have been implicated in nitrate uptake and signaling (Table 2). These findings suggest that NIN plays a similar role to other NLPs in the regulation of genes involved in nutrient uptake and assimilation.
Table 2. Relative expression of some nutrient uptake and assimilation genes in S. meliloti-inoculated root hairs in nin versus the wild type.
M. truncatula genes in boldface have L. japonicus homologs that are potential NIN targets (Soyano et al., 2014). Arabidopsis genes in boldface were found to be down-regulated in nlp7 (Castaings et al., 2009). Dashes indicate no data.
| N Uptake and Assimilation | S Uptake and Assimilation | P Uptake and Assimilation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M. truncatula Gene Model | nin-1 Versus the Wild Type | Arabidopsis Homolog | Gene Description (Name) | M. truncatula Gene Model | nin-1 Versus the Wild Type | Arabidopsis Homolog | Gene Description (Name) | M. truncatula Gene Model | nin-1 Versus the Wild Type | Arabidopsis Homolog | Gene Description (Name) |
| Medtr7g050870 | 0.18 | AT1G11580 | Pectin methylesterase | Medtr5g020800 | 0.38 | AT1G75280 | Isoflavone reductase | Medtr8g015960 | 0.03a | AT1G17710 | PEP carboxylase (PEPC1) |
| Medtr5g012290 | 0.11 | AT1G12110 | Nitrate transporter (NRT1.1) | Medtr5g010230 | 0.16a | AT4G23100 | Glu-Cys ligase (GSH1) | Medtr8g015950 | 0.03a | AT1G73010 | PP phosphatase (PPsPase1) |
| Medtr4g101380 | 0.56 | AT1G12110 | Nitrate transporter (NRT1.1) | Medtr4g115670 | 1.55 | AT5G34850 | PAP26 | Medtr7g067340 | 0.55 | AT5G01220 | Sulfoquinovosyltransferase |
| Medtr8g069775 | 0.30 | AT1G12940 | Nitrate transporter (NRT2.5) | Medtr3g070400 | 0.46 | AT3G55880 | S utilization efficiency (SUE4) | Medtr1g043200 | 0.29 | AT5G43360 | P transporter (PHT1;3) |
| Medtr2g030200 | 0.64 | AT1G30510 | Ferredoxin:NADP(H) oxidoreductase | Medtr4g087520 | 0.29 | AT2G43750 | Cys synthase | Medtr3g082700 | 0.07 | AT2G32830 | P transporter (PHT1;5) |
| Medtr1g078030 | 0.35 | AT1G64720 | Membrane-related protein (CP5) | Medtr6g016325 | 0.47 | AT1G19920 | ATP sulfurylase-like (APS2) | Medtr1g074930 | 0.45 | AT3G54700 | P transporter (PHT1;7) |
| contig_63842_1 | 0.50 | AT3G16560 | Protein phosphatase 2C | Medtr1g081620 | 0.43 | AT4G14880 | Cys synthase | Medtr4g083960 | 0.70 | AT1G20860 | P transporter (PHT1;9) |
| Medtr4g119830 | 0.71 | AT3G16560 | Protein phosphatase 2C | Medtr5g095470 | 0.20 | AT3G13110 | Ser acetyltransferase | Medtr7g102180 | 0.28 | AT5G03430 | PAPS reductase |
| Medtr5g026620 | 0.18 | AT3G22370 | Alternative oxidase1A (AOX1A) | Medtr3g074620 | 0.44 | AT1G34780 | APR-LIKE (APRL4) | Medtr8g010590 | 0.11 | AT3G03530 | Phospholipase C4 |
| Medtr7g071970 | 0.551 | AT5G01740 | Nuclear transport factor2 (NTF2) | Medtr1g102550 | 0.44 | AT3G22890 | ATP sulfurylase (APS1) | – | – | – | – |
| Medtr5g074500 | 0.45 | AT5G01740 | Nuclear transport factor2 (NTF2) | Medtr8g098350 | 0.59 | AT4G23100 | Glu-Cys ligase (GSH1) | – | – | – | – |
| Medtr4g077190 | 0.71 | AT5G04590 | Sulfite reductase (SIR) | Medtr2g008470 | 0.06 | AT1G22150 | S transporter (SULTR1;3) | – | – | – | – |
| Medtr5g026640 | 0.52 | AT5G45380 | Degradation of Urea3 (DUR3) | Medtr4g011970b | 0.15 | AT3G15990 | S transporter (SULTR3;4) | – | – | – | – |
| Medtr2g021255 | 0.25 | AT5G35630 | Gln synthetase2 (GS2) | Medtr2g102243 | 0.23 | AT3G15990 | S transporter (SULTR3;4) | – | – | – | – |
| Medtr5g067150 | 0.41 | AT4G24400 | CBL-interacting protein kinase (CIPK8) | Medtr6g086170 | 0.09 | AT5G19600 | S transporter (SULTR3;5) | – | – | – | – |
| Medtr1g009200 | 0.47 | AT1G52190 | Nitrate transporter (NRT1.11) | Medtr7g022870 | 0.48 | AT5G13550 | S transporter (SULTR4;1) | – | – | – | – |
| Medtr2g101650 | 0.59 | AT1G52190 | Nitrate transporter (NRT1.11) | – | – | – | – | – | – | – | – |
| Medtr1g045550 | 0.09 | AT4G13510 | Ammonium transporter1 (AMT1;1) | – | – | – | – | – | – | – | – |
| Medtr7g085410 | 0.35 | AT5G66320 | GATA transcription factor5 (GATA5) | – | – | – | – | – | – | – | – |
| Medtr4g095600 | 0.49 | AT3G49940 | LOB domain protein (LBD38) | – | – | – | – | – | – | – | – |
| Medtr2g049790 | 0.62 | AT1G30270 | CBL-interacting protein kinase (CIPK23) | – | – | – | – | – | – | – | – |
| Medtr4g133230 | 0.44 | AT5G47560 | Dicarboxylate transporter (TDT) | – | – | – | – | – | – | – | – |
| Medtr4g125810 | 0.17 | AT5G57050 | Protein phosphatase2C (ABI2) | – | – | – | – | – | – | – | – |
| Medtr4g113505 | 2.15 | AT5G47100 | Calcineurin B-like protein (CBL9) | – | – | – | – | – | – | – | – |
| Medtr5g087490 | 0.34 | AT5G20990 | Cofactor for NiR and XD1 (CNX1) | – | – | – | – | – | – | – | – |
| Medtr2g061710 | 0.43 | AT2G31955 | Cofactor for NiR and XD1 (CNX2) | – | – | – | – | – | – | – | – |
Induced in wild-type versus control root hairs 5 dpi with S. meliloti.
Medtr4g011970 = SST1.
NIN Is Required for the Expression of a Subset of the Rhizobia-Induced NF-Ys
Altogether, 40 transcription factors were found to be NIN dependent for their induction during root hair infection (Fig. 3A). In addition to NF-YA1, which was expressed at less than 3% of its wild-type level in nin, the expression of NF-YA2 and NF-YC2, whose orthologs were previously implicated in nodulation (Zanetti et al., 2010; Soyano et al., 2013; Laloum et al., 2014; Baudin et al., 2015), and NF-YB6 was also significantly dependent on NIN. The finding that NIN regulates genes encoding members of all three subunits of the CCAAT-box complex during rhizobial infection reveals that its control of this module is more extensive than previously recognized. However, the CCAAT-box subunit genes encoding NF-YC6 and NF-YB7 were still expressed at wild-type levels or higher in nin, indicating a NIN-independent expression of some subunits.
The ern1-1 Mutant Has Deregulated Expression of Receptor-Like Kinases and Reactive Oxygen Species-Related Genes and Fails to Up-Regulate a Small Set of Symbiotic Genes
Over 3,000 genes had altered expression in ern1-1 relative to the wild type in root hairs of rhizobia-inoculated seedlings. Remarkably, more than 110 receptor-like kinase (RLK) genes had increased expression in root hairs of ern1, and another 30 showed decreased expression. This included many potential TOLL-Interleukin 1-Nucleotide Binding Site-Leucine Rich Repeat resistance genes as well as RLKs involved in development, such as the ortholog of FERONIA and the symbiotic LYSM-receptor kinase NFP. Notably, in apparent contrast to findings in L. japonicus, which showed that the induction of EPR3 expression during symbiosis was dependent on ern1, we found that the M. truncatula ortholog, LYK10, is slightly (∼2 fold) increased in ern1 (adjusted P = 0.07). This widespread deregulation of RLKs in ern1 is accompanied by increased expression of nine protein phosphatase-encoding genes (PP2A and PP2B type). We also found that 44 genes involved in reactive oxygen species (ROS) generation and response were altered in ern1. This includes the ectopic up-regulation of 20 ROS-producing enzymes (respiratory burst oxidase homolog Hs, peroxidases, and glyoxal oxidases). In addition, we found evidence consistent with increased antioxidant production, in particular increased expression of both flavonoid and carotenoid biosynthesis genes, including those that are normally induced in root hairs by rhizobial inoculation (Supplemental Fig. S5; Breakspear et al., 2014).
In contrast, the gene network analysis, which considers both coexpression and strength of differential expression, indicates only a small number of genes as potential targets of ERN1 during rhizobial infection. Three of these genes are induced by rhizobia and Nod factors (Breakspear et al., 2014; Damiani et al., 2016) and are expressed in the infection zone of nodules in wild-type plants (Supplemental Table S2; Roux et al., 2014). They were strongly down-regulated (greater than 4-fold reduced) in ern1-1 and had mostly symbiotic-specific expression (MtGEAv3). Based on their homology to Arabidopsis proteins, we named these genes Cysteine-rich TM module1 (CYM1), Serine Protease Inhibitor1 (SPI1), and Symbiosis Induced Resistance gene1 (SIR1). None of these ERN1-regulated genes has been studied.
We checked the expression of SPI1, CYM1, and SIR1 using promoter-GUS fusions in transformed roots inoculated with S. meliloti and found them to be strongly expressed in infected root hairs, throughout young nodules, and at the apex of mature elongated nodules (Supplemental Fig. S6, A–C). Their ERN1-dependent up-regulation was further confirmed by RT-qPCR in another ern1 mutant allele (ern1-3; NF1390) that we isolated from the Noble Institute Tnt1 insertion mutant population (Cheng et al., 2011, 2017; R108 background). This ern1-3 mutant showed similar phenotypes to ern1-1, with strongly blocked infections and retarded nodule development (Supplemental Fig. S7; Middleton et al., 2007). Results from RT-qPCR analysis showed that SPI1, CYM1, and SIR1 were all significantly up-regulated in the wild type but not in ern1-3 after rhizobial inoculation (Supplemental Fig. S6, D–F).
Since ERN1 expression is up-regulated in root hairs responding to Nod factors (Andriankaja et al., 2007; Cerri et al., 2012), we tested whether complementation with ERN1 could rescue the Nod factor-induced expression of SPI1 and CYM1 in ern1 roots. ERN1 expressed from its own promoter in transgenic hairy roots restored Nod factor-induced expression of SPI1 and CYM1 in ern1-1 (Supplemental Fig. S6, G and H). Together, our data suggest that SPI1, CYM1, and possibly SIR1 induction by rhizobia depends on ERN1.
DISCUSSION
Taking advantage of a powerful approach using isolated root hairs, the cell type involved in the initial host-rhizobia interaction, we have transcriptionally profiled three M. truncatula infection mutants, nin, ern1, and nf-ya1, providing insight into the roles of these transcriptional regulators in accommodating root hair infection. Here, we reveal a central and specific role for NIN in the reprogramming of root hairs into infection cells, with a particular function in the regulation of genes for cell growth.
Research on NLPs in Arabidopsis has revealed a requirement for NLPs in supporting growth. NLP6/7 are required for root development (Castaings et al., 2009; Marchive et al., 2013), while loss of NLP8 inhibits nitrate-induced seed germination (Yan et al., 2016). Recent work suggests that NLP6 and NLP7 directly interact with TCP-Domain Family Protein 20 to directly control root growth (Guan et al., 2017). Our work shows that NIN similarly supports growth-related processes during infection. First, our work has shown that NIN has a similar function to other NLPs in promoting the expression of genes involved in nutrient uptake and assimilation. One of these genes, the sulfate transporter SST1, has been shown to be important for nodulation in L. japonicus (Tables 1 and 2; Krusell et al., 2005). Another, CLE13, confirms earlier reports in M. truncatula and L. japonicus for NIN (Mortier et al., 2010, 2012; Soyano et al., 2014, 2015; Nishida et al., 2016) and has recently been implicated in nitrate inhibition of nodulation (Nishida et al., 2018). Recently, it was shown that the Arabidopsis nlp7 mutant has changed expression of several CLE peptide-encoding genes (Zhao et al., 2018), suggesting that this regulation may predate the divergence of legumes. The importance of the other N/P/S-related genes in NIN’s regulon requires further study, but it seems plausible that this reflects the needs of a rapidly growing cell. The up-regulation of over 100 ribosomal genes, which is NIN dependent, suggests that infection requires a large investment in de novo protein biosynthesis. Similarly, S plays a central role in redox homeostasis via its incorporation into glutathione, and its increased uptake may support remodeling of the cell wall during growth. Finally, P uptake may be required for the apparent up-regulation of DNA synthesis, which is also heavily NIN dependent.
Our second line of evidence supporting NIN’s function in growth is its regulation of GA signaling and biosynthetic genes. GA-related gene expression was reported in root hairs following Nod factor treatment and inoculation with S. meliloti (Breakspear et al., 2014; Jardinaud et al., 2016). Despite the universally accepted role for GA in cell expansion and growth, some studies indicate a negative role for GA in infection: mutations in DELLA genes, and plants treated with exogenous GA have reduced density of infection threads and nodule number, and the expression of a GA-insensitive della1-Δ18 variant increases the number of infection events (Lievens et al., 2005; Maekawa et al., 2009; Fonouni-Farde et al., 2016; Jin et al., 2016). A similar negative role was also reported for cytokinin (Murray et al., 2007; Held et al., 2014). On the other hand, GA biosynthetic mutants have decreased nodulation, suggesting a positive role for GA in nodule growth (Ferguson et al., 2005; McAdam et al., 2018). Further investigation is required to fully elucidate the role of GA at the different stages of nodulation. The concomitant activation of JA biosynthetic genes could be due to intrinsic mechanisms regulating a tradeoff between growth and defense that have been well documented for GA-JA in Arabidopsis and other plant species (Danisman et al., 2012; Yang et al., 2012; Li et al., 2015; Smakowska et al., 2016), an area that needs further study in the context of nodulation.
Finally, in addition to the acquisition of base nutrients, successful rhizobia infection requires extensive remodeling and de novo generation of cell walls and membranes. We show that an enzyme encoded by one of these genes, NPL, is secreted to the infection chamber and is required for infection thread initiation in M. truncatula. This localization is similar to that reported for ENOD11 (Fournier et al., 2015). The localization of NPL to the infection chamber suggests that loosening of the extracellular pectin matrix, which is increasingly recognized for its importance in cell wall extensibility (Saffer, 2018), is required for infection thread initiation. The expression of NIN and NPL is induced in all epidermal root hair cells when roots are treated with purified Nod factors, whereas after inoculation by rhizobia the expression of NPL is limited to infected tissues (Xie et al., 2012; Yoro et al., 2014; Vernié et al., 2015; this study). This highly localized expression could therefore reflect localized accumulation of Nod factors within the infection chamber and infection thread, resulting in the cell-specific activation of the NIN regulon. Notably, NPL localization was restricted to the infection chamber and infection thread cell wall and was not detected in the root hair cell wall, suggesting that the timing of NPL expression, as directed by NIN, determines where it is secreted, as has been shown for the transmembrane protein MtPT4 (Zhang et al., 2015). The conservation of NPL function and its regulation by NIN in M. truncatula, along with the existence of NPL orthologs in lupin (Lupinus albus) and Arachis ipaensis (Supplemental Fig. S2), suggest that this key regulatory module may be conserved across the legume family. It is unclear whether NPL’s recruitment to NIN’s regulon occurred after NIN’s recruitment to nodulation or before, as part of the broader role of NLPs in growth-related processes. Also dependent on NIN were a subset of secreted peroxidases with mostly symbiotic-specific expression (Breakspear et al., 2014; Chen et al., 2015). Peroxidases are generally considered to be involved in cell wall loosening and hardening, thus may play a role in cell wall cross-linking for formation of the cell wall and matrix of the growing infection thread (Wisniewski et al., 2000; Santos et al., 2001; Passardi et al., 2004; Chen et al., 2015).
Development of infection threads also needs extensive expansion of the plasma membrane, echoing our finding that NIN is required for induction of many membrane protein-encoding genes that are ostensibly involved in rhizobial infection, including many early nodulins. NIN also regulates the gene expression of two putative annexins, including calcium-binding Annexin 1, which localizes to the nuclear periphery in S. meliloti activated cells (de Carvalho-Niebel et al., 2002). Annexins have multiple membrane-associated functions, including the formation of calcium ion channels (Gerke et al., 2005; Mortimer et al., 2008). In addition, the infection-induced gene ABIL1 was dependent on NIN. ABIL1 has been proposed to mediate NAP1 and SCAR/WAVE binding (Basu et al., 2005) and so may act in concert with NAP1, PIR1, ARPC1, and SCARN to mediate actin nucleation, which is required for the development of infection threads (Yokota et al., 2009; Miyahara et al., 2010; Hossain et al., 2012; Qiu et al., 2015). Actin cytoskeletal dynamics play a central role in cell wall formation, particularly during polar growth (Pratap Sahi et al., 2018), further supporting NIN’s role as a growth module.
In addition to uncovering NIN’s role in growth processes, comparison of the nin transcriptome with that of ern1 and nf-ya1 allowed us to gain insights into their regulatory relationships. Generally, the impact on gene expression was much stronger in ern1 than in nf-ya1, reflecting the strength of the infection phenotypes of these mutants: in ern1, no wild-type infection threads are observed, resulting in a fix− phenotype, while in nf-ya1, many normal-looking infection threads can be found. Based on the absence of transcriptional interdependence and nonoverlap of the most strongly affected genes in the ern1 and nin mutants, we can conclude that the nin phenotype is not due to a failure to activate ERN1 expression, and vice versa. This finding appears not to be due to the redundant action of ERN2, as NIN and RPG are induced normally in response to Nod factors in the ern1 ern2 double mutant (Cerri et al., 2016). Further support for the independence of these regulons comes from L. japonicus, where ERN1 induction by rhizobia was found to be dependent on CYCLOPS and NSP2 and independent of NIN (Yano et al., 2017). Also, in accordance with results in L. japonicus, we found that the up-regulation of NF-YA1 in response to rhizobia is dependent on NIN. However, we found no evidence for positive regulation of ERN1 expression by NF-YA1 in root hairs, contrasting with results in M. truncatula showing that NF-YA1 can transcriptionally activate ERN1 (Laloum et al., 2014). However, the functional redundancy of NF-YA2 in the nf-ya1 mutant (Laloum et al., 2014) and the rhizobial induction of NIN-independent CCAAT-box subunits in the nin mutant (this study) make it impossible to conclude whether ERN1 is regulated by the CCAAT-box complex. The fact that CCAAT-box signaling is likely at least partially active in nin may have allowed us to more clearly extract information on the remaining NIN regulon. A detailed analysis of higher order CCAAT-box mutants is needed to clarify the role of this complex in nodulation. Interestingly, it was recently shown that Arabidopsis nlp7 has changed expression of several genes encoding NF-YA subunits (Zhao et al., 2018). This suggests that, as with the CLEs, these genes were part of NIN’s regulon prior to its neofunctionalization into nodulation.
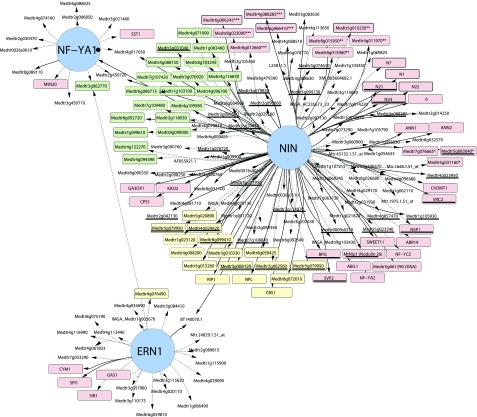
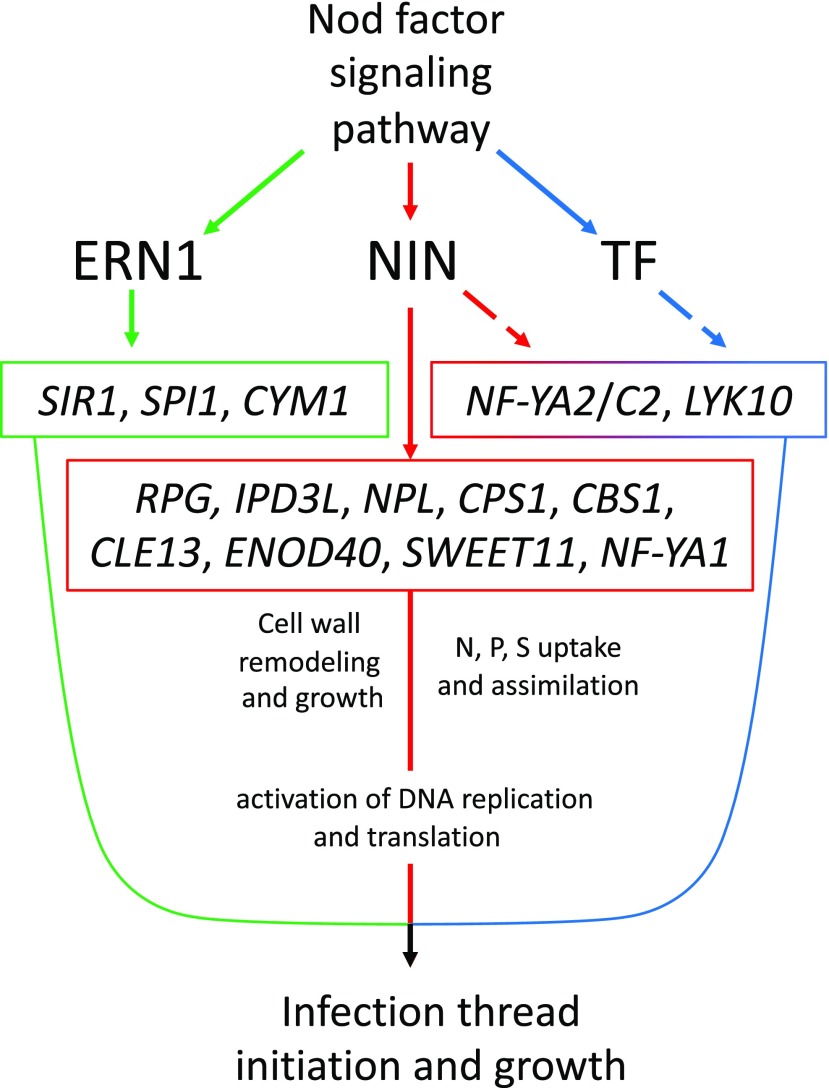
We also find evidence for the partial dependence of the induction of IPD3L and other transcription factors on NIN, suggesting further interconnectedness of the gene regulatory network directing rhizobial infection (Fig. 6).
Figure 6.
Model for the gene regulatory network underlying rhizobial infection. Nod factor signaling induces the expression of NIN (red), ERN1 (green), and other transcription factors (TF; blue). ERN1 is required for up-regulation of a few genes with symbiotic specific expression, while NIN controls a diversity of functions, including cell wall modification via NPL, GA biosynthesis by genes like CPS1, and nutrient uptake and DNA synthesis either directly or through subordinate transcription factors such as NF-YA1/A2 and NF-YC2, components of the CCAAT-box transcription factor complex. Other genes, like RPG, require NIN for their expression during infection, but their exact roles in the symbiosis are unknown. Many genes are partly dependent on NIN but require another unidentified transcription factor or factors for their full expression during infection (broken lines).
The ern1 mutant had widespread changes in the expression of ROS-related genes as well as genes encoding receptor-like kinases, a class of regulators that has many members with central roles in ROS regulation (Duan et al., 2010; Osakabe et al., 2010; Liu et al., 2013; Idänheimo et al., 2014). Interestingly, a study on the Arabidopsis ortholog of ERN1 implicated it in ROS homeostasis, suggesting potential functional conservation for this transcription factor across plants (Kim et al., 2012). The regulation of REDOX homeostasis is crucial for infection, and therefore these changes in ROS-related gene expression may contribute to the ern1 infection phenotype (Chang et al., 2009). Further genetic and biochemical studies are needed to determine the role of ERN1 and its role in symbiotic infection.
The improved sensitivity and specificity of single cell-type gene expression profiling has revealed NIN’s role in controlling genes involved in nutrient uptake, assimilation, and signaling, a role founded in the ancestral function of the NLP family. It is not yet clear whether other key infection genes like NPL and RPG were part of NIN’s original regulon or were adopted subsequent to its symbiotic recruitment. Nonetheless, the further regulation of growth-related processes, including genes for GA biosynthesis, cell cycle activation, and cell wall remodeling, points to a broader role for NIN as a growth module that serves to support rhizobial infection.
MATERIALS AND METHODS
Isolation of Root Hairs from Sinorhizobium meliloti-Inoculated Roots
Medicago truncatula root hairs were isolated as described by Breakspear et al. (2014). Essentially, seedlings were grown vertically on plates between layers of Whatman Envelope Strip filter paper (VWR), where they were inoculated with S. meliloti 1021. At 5 dpi, the seedlings were removed and immediately immersed in a liquid nitrogen bath, where the root hairs were removed using a small paint brush. Approximately 100 seedlings were used in each biological replicate, and three biological replicates were used for each mutant. RNA was then isolated, labeled, and hybridized to the Affymetrix Medicago GeneChip as previously described (Breakspear et al. (2014).
Normalization and statistical analysis were carried out using Genespring 12.0 GX. Background correction, normalization, and probe summarization were performed using Robust Multichip Averaging. Unpaired Student’s t tests were used to detect genes with significant changes in expression for each comparison. Probe sets were retained that had a value of greater than 50 for all three biological replicates in at least one of the two conditions being compared. Benjamini-Hochberg false discovery rate was used to derive asymptotically and multiple test-corrected P values. All data are available through the Medicago Gene Expression Atlas server (http://mtgea.noble.org/roothair/). GO term enrichment analysis was carried out using Phytomine.
RT-qPCR Analysis of Complemented M. truncatula Composite Roots and Seedlings
For comparison of gene expression in response to Nod factors, wild-type and ern1-1 mutant M. truncatula A17 lines carrying a stably integrated pENOD11:GUS gene fusion (Journet et al., 2001; Middleton et al., 2007) were transformed via Agrobacterium rhizogenes with respective pERN1:YFP-ERN1 or p35S:YFP binary vectors and transferred to Fahraeus-agar/paper support, as described by Boisson-Dernier et al. (2005). Root systems of composite plants were treated for 6 h with a solution of 10−9 m purified Nod factors (kindly provided by F. Maillet and J. Denarié) 3 to 5 d later. Harvested samples were kept at −80°C before RNA extraction using the Macherey-Nagel total RNA isolation kit. The DNA-free RNA samples were quantified and RNA integrity was checked by Agilent RNA Nano Chip (Agilent Technologies). First-strand complementary DNA synthesis was performed using 1 μg of total RNA with an anchored oligo(dT; 17T+V) and SuperScript II (Invitrogen) or Roche reverse transcriptases following their manufacturers’ protocols. RT-qPCR was performed on 384-well plates with the Light Cycler 480 system and the SYBR Green I Master mix (Roche) according to Cerri et al. (2012). The following primer pairs were used to amplify the respective target genes: CYM1, Medtr3g081140 (forward, 5′-ACCTCCTCCTCCTGTTGGTTAT-3′; reverse, 5′-TCCCTTCCAGAATCCATCAC-3′); SPI1, Medtr5g045470 (forward, 5′-CTCGAGTGCCAAAAATTGGT-3′; reverse, 5′-GAATGCCATGACAAGCAACA-3′); and Ubiquitin (forward, 5′-TTGTGTGTTGAATCCTAAGCAGG-3′; reverse, 5′-CAAGACCCATGCAACAAGTTCT-3′). The data shown represent transcript levels of respective genes normalized to those of endogenous Ubiquitin and are mean values obtained from two independent experiments with three technical repeats.
For experiments using seedlings, seeds of M. truncatula wild type (Jemalong A17 or R108) or mutants (nin-1, ern1-1, nin-2, and ern1-3) were sterilized, transferred onto water agar plates, and left in 4°C for 2 d. Then the seeds were transferred to 22°C for 16 h. The germinated seedlings then were transferred to plates containing 1.5% (w/v) agar and 100 nm aminoethoxyvinylglycine and inoculated as described before (Breakspear et al., 2014) with either distilled deionized water or S. meliloti (Rm1021; OD600 = 0.02). At 3 dpi, root tips of the treated seedlings were removed, and only the root zone containing root hairs was collected for extraction of RNA. This early time point prior to nodule emergence was chosen to avoid the secondary effects of gene expression caused by delayed nodulation in the mutants, which would result in reduced expression of most nodulation genes. Eight seedlings were used per replicate, and three biological replicates were used for RT-qPCR. TIP41 was used as an internal control for normalization, and the relative expression was calculated using the delta-delta Ct method.
GeneChip Experiment and Analysis
M. truncatula mutants nin-1 (Marsh et al., 2007), which has an 11-bp deletion that affects the RWP-RK DNA-binding domain, ern1-1 (Middleton et al., 2007), where the ERN1 gene is completely deleted, and nf-ya1-1, which has a nonsynonymous mutation leading to a premature stop codon at amino acid position 137 (Laporte et al., 2014), were used in this study. The methods of isolation of root hairs, RNA extraction from root hairs, and GeneChip analysis were as previously described (Breakspear et al., 2014). Briefly, analyses were performed using robust multichip averaging in Genespring 12.0 GX. Means comparisons were carried out using unpaired Student’s t tests. Multiple test corrections were made using the Benjamini-Hochberg procedure with a false discovery rate of 0.05. Normalized data were further analyzed in Microsoft Excel. For RT-qPCR confirmation of genes in the NIN regulon, the nin-2 mutant and wild-type (R108) control were sterilized and germinated at 4°C on water-agar plates for 9 d, then transferred to vermiculite:perlite substrate (1:1) for 7 d, at which point the seedlings were inoculated with Sm2011. After 1 d, the seedlings were fertilized with 0.5 mm KNO3 and then harvested for RNA isolation from the entire roots at 6 dpi. Three biological replicates were used for each genotype, and each replicate constituted three to four seedlings. Elongation Factorα (EFα) was used as a reference gene. The following primers were used: EFα, 5′-CTTTGCTTGGTGCTGTTTAGATGG-3′ and 5′-ATTCCAAAGGCGGCTGCATA-3′; CPS1, 5′-TTCTGCTTTGCTGGTCAATG-3′ and 5′-GTCCAATGCATAAGCCACCT-3′; RPG, 5′-GGCATGGAGAGTCCAAAGAG-3′ and 5′-CCTCCGTCAATTCCTTCAAA-3′; NSP1, 5′-ACGAAATGCCGAAGATGAAC-3′ and 5′-ACCGGTTATGGCTACAGCAC-3′; and Nod26, 5′-CTGGAGTTGCCACCGATACT-3′ and 5′-GGTTCATCGATGCTCCTGTT-3′.
Promoter-GUS Analysis
The promoter GUS fusion constructs of SPI1 (Medtr5g045470), CYM1 (Medtr3g081140), and SIR1 (Medtr4g016610) were made by using Gateway cloning. An upstream fragment from each gene was PCR amplified from M. truncatula A17 genomic DNA using Phusion High-Fidelity DNA Polymerase (NEB) by using primer pairs as follows: proSPI1 forward and proSPI1 reverse, proCYM1 forward and proCYM1 reverse, proSIR1 forward and proSIR1 reverse (see sequences below). The fragment was cloned into pDONR207 using Gateway BP Clonase II enzyme mix (Invitrogen). Then the fragment was introduced into a destination vector pKGWFS7 by an LR reaction to make the final promoter-GUS constructs. The ProMtNPL:GUS construct was made by Golden Gate cloning (Patron et al., 2015). A 2.5-kb MtNPL promoter, GUS gene (EC75111), and 35S terminator (EC41414) were synthesized by Life Technologies and used as level 0 modules, then cloned into level 1 vector EC47802, which was further cloned into level 2 vector EC50505 (https://www.ensa.ac.uk/). Composite plants were transferred to sand-Terra Green 4 weeks after A. rhizogenes-mediated transformation. The plants were then inoculated with Rm1021-LacZ. The roots were harvested 14 d later for GUS staining and stained for at least 4 h at 37°C in 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-glucuronic acid solution with 100 mm potassium phosphate buffer (pH 7), 1 mm potassium ferricyanide, 1 mm potassium ferrocyanide, and 10 mm EDTA. Primer sequences used were as follows: proSPI1 forward, 5′-ggggacaagtttgtacaaaaaagcaggcttcgagaagtctagtggagttatttgg-3′; proSPI1 reverse, 5′-ggggaccactttgtacaagaaagctgggttccggctagccaaggtattct-3′; proCYM1 forward, 5′-ggggacaagtttgtacaaaaaagcaggcttcttctgctgattctgcaaacata-3′; proCYM1 reverse, 5′-ggggaccactttgtacaagaaagctgggttttggaaagcaaaggaaacaa-3′; proSIR1 forward, 5′-ggggacaagtttgtacaaaaaagcaggcttccagttcatagtttgttctcacattgttt-3′; and proSIR1 reverse, 5′-ggggaccactttgtacaagaaagctgggttacgtccctacaacctttcatcatc-3′.
Mutant Screening, Genotyping, and Phenotyping
Plant roots transformed with promoter-GUS constructs were generated in M. truncatula A17 or nin-1 background by hairy root transformation mediated by A. rhizogenes Arqua1 or AR1193 (Boisson-Dernier et al., 2001). The composite plants were transferred to a mixture of sand and Terra Green (1:1) 4 weeks after transformation and inoculated with S. meliloti (Rm1021) harboring pXLGD4 (hemA:lacZ). The roots were harvested 3 weeks after inoculation for GUS staining.
Subcellular Localization
The ProMtNPL:MtNPL-GFP and ProLjUBI1:MtNPL-GFP constructs were made by using Golden Gate cloning. A 2.5-kb MtNPL promoter and MtNPL-coding DNA were synthesized by Life Technologies and assembled as described (Patron et al., 2015). Plant roots transformed with the constructs were generated by hairy root transformation mediated by A. rhizogenes AR1193. The subcellular localization experiment was done by using the imaging method described by Fournier et al. (2008). Transgenic roots were inoculated with S. meliloti (Rm1021; OD600 = 0.001) tagged with CFP, and at 7 dpi root hairs at different infection stages were imaged with a Leica SP5 confocal laser scanning microscope. CFP and GFP were excited at wavelengths 457 and 488 nm, respectively. Cell wall autofluorescence was excited at wavelength 561 nm. The emission wavelengths used were 465 to 485 nm for CFP, 500 to 530 nm for GFP, and 620 to 720 nm for cell wall autofluorescence. The confocal images were processed using Leica AF Lite software, and the bright-field images of root hairs were adjusted using Adobe Photoshop Elements 8.0.
Gene Network Analysis
Gene network analysis was carried out using NetProphet v2.0 (Haynes et al., 2013; Kang et al., 2018). Before the gene network analysis, the data from this work were normalized with previously published data (Breakspear et al., 2014) using LIMMA. LIMMA uses preprocessing steps prior to statistical analysis that preserve information (Ritchie et al., 2015) and provides a superior analysis to what was originally reported by Breakspear et al. (2014), so we provide these results in Supplemental Table S2. For NetProphet analysis, we included probe sets that were significantly up-regulated greater than 2-fold relative to their respective controls (SL44; S. meliloti ΔnodD1ABC) at 1, 3, or 5 dpi by S. meliloti inoculation. For differential expression, data for each mutant inoculated with Rm1021 (nin-1, ern1-1, and nf-ya1-1) were compared with the Rm1021-inoculated control (5 dpi). For coexpression analysis, the following samples were used (nin-1, ern1-1, nf-ya1-1, wild type 1, 3, and 5 dpi with Rm1021, and wild type 1 d post treatment with Nod factors). Although nin-1 and nf-ya1-1 are null alleles (Marsh et al., 2007; Laporte et al., 2014), they still produce stable transcripts that are induced upon infection. It was therefore necessary to set the expression values for NIN and NF-YA1 probe sets in the mutants to baseline expression levels for the analysis. NetProphet uses a weighted scoring of coexpression and differential expression values in a gene perturbation analysis to rank transcription factor potential targets. All data used in this analysis are available through http://mtgea.noble.org/roothair/ and the Gene Expression Omnibus.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers provided in Tables 1 and 2 and Supplemental Data S2. The gene models corresponding to the mutants analyzed in this article were NIN (Medtr5g099060), NF-YA1 (Medtr1g056530), ERN1 (Medtr7g085810), and NPL (Medtr3g086320).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Relative expression of selected genes identified using GeneChip analysis for the wild type (R108) versus nin-2 using qPCR.
Supplemental Figure S2. Phylogenetic tree of pectate lyases.
Supplemental Figure S3. Promoter-GUS analysis of the MtNPL gene and rhizobial infection phenotype of Mtnpl.
Supplemental Figure S4. Complementation of Mtnpl mutant.
Supplemental Figure S5. Genes involved in production of antioxidant compounds are more highly expressed in ern1-1 following rhizobial inoculation.
Supplemental Figure S6. The expression of SPI1, CYM1, and SIR1 is symbiosis specific and is dependent on ERN1.
Supplemental Figure S7. Infection phenotype of the ern1-3 mutant.
Supplemental Table S1. The number of genes present in the M. truncatula NIN regulon for which homologs were identified in L. japonicus NIN using ChIP-seq (Soyano et al., 2014).
Supplemental Table S2. Expression of ERN1, ERN2, NF-YA1, NIN, and genes downstream of ERN1 across nodule zones.
Supplemental Data S1. Gene expression levels in root hairs of nin, ern1, and nf-ya1 versus the wild type after inoculation with S. meliloti.
Supplemental Data S2. Comparison of relative gene expression levels in root hairs of nin, ern1, and nf-ya1 versus the wild type after inoculation with S. meliloti.
Supplemental Data S3. Genes that are down-regulated in M. truncatula nin that have close homologs that were identified in an independent ChIP-seq analysis for LjNIN by Soyano et al. (2014).
Supplemental Data S4. GO term enrichment analysis of genes down-regulated in nin mutant root hairs 5 dpi with rhizobia.
Supplemental Data S5. Most GA biosynthesis and signaling genes are down-regulated in nin.
Supplemental Data S6. Expression of JA-related genes is increased in nin.
Acknowledgments
We thank Fang Xie for providing the MtNPL complementation construct, Grant Calder and Eva Wegel for help with microscopy imaging, and Allan Downie and Ping Xu for helpful comments on the article.
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (grant nos. BB/L010305/1 and BB/L010305/1) and the John Innes Foundation (to D.G. and S.R.). F.d.C.-N. and A.N. were supported by the TULIP (ANR-10-LABX-41), NODCCAAT (ANR-15-CE20-0012-01; A.N.) and COME-IN (ANR-14-CE35-0007-01; F.d.C.-N.) grants.
Articles can be viewed without a subscription.
References
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de Carvalho-Niebel F (2007) AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Godfroy O, de Billy F, Saurat O, Jauneau A, Gough C (2008) The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc Natl Acad Sci USA 105: 9817–9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Le J, El-Din El-Essal S, Huang S, Zhang C, Mallery EL, Koliantz G, Staiger CJ, Szymanski DB (2005) DISTORTED3/SCAR2 is a putative Arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis. Plant Cell 17: 502–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin M, Laloum T, Lepage A, Rípodas C, Ariel F, Frances L, Crespi M, Gamas P, Blanco FA, Zanetti ME, et al. (2015) A phylogenetically conserved group of nuclear factor-Y transcription factors interact to control nodulation in legumes. Plant Physiol 169: 2761–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Berrabah F, Bourcy M, Eschstruth A, Cayrel A, Guefrachi I, Mergaert P, Wen J, Jean V, Mysore KS, Gourion B, et al. (2014) A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol 203: 1305–1314 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Andriankaja A, Chabaud M, Niebel A, Journet EP, Barker DG, de Carvalho-Niebel F (2005) MtENOD11 gene activation during rhizobial infection and mycorrhizal arbuscule development requires a common AT-rich-containing regulatory sequence. Mol Plant Microbe Interact 18: 1269–1276 [DOI] [PubMed] [Google Scholar]
- Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N, et al. (2003) The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol 131: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear A, Liu C, Roy S, Stacey N, Rogers C, Trick M, Morieri G, Mysore KS, Wen J, Oldroyd GED, et al. (2014) The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26: 4680–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou JP, Daniel-Vedele F, Fernandez E, et al. (2009) The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J 57: 426–435 [DOI] [PubMed] [Google Scholar]
- Cerri MR, Frances L, Laloum T, Auriac MC, Niebel A, Oldroyd GE, Barker DG, Fournier J, de Carvalho-Niebel F (2012) Medicago truncatula ERN transcription factors: Regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol 160: 2155–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri MR, Frances L, Kelner A, Fournier J, Middleton PH, Auriac MC, Mysore KS, Wen J, Erard M, Barker DG, et al. (2016) The symbiosis-related ERN transcription factors act in concert to coordinate rhizobial host root infection. Plant Physiol 171: 1037–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri MR, Wang Q, Stolz P, Folgmann J, Frances L, Katzer K, Li X, Heckmann AB, Wang TL, Downie JA, et al. (2017) The ERN1 transcription factor gene is a target of the CCaMK/CYCLOPS complex and controls rhizobial infection in Lotus japonicus. New Phytol 215: 323–337 [DOI] [PubMed] [Google Scholar]
- Chang C, Damiani I, Puppo A, Frendo P (2009) Redox changes during the legume-rhizobium symbiosis. Mol Plant 2: 370–377 [DOI] [PubMed] [Google Scholar]
- Chardin C, Girin T, Roudier F, Meyer C, Krapp A (2014) The plant RWP-RK transcription factors: Key regulators of nitrogen responses and of gametophyte development. J Exp Bot 65: 5577–5587 [DOI] [PubMed] [Google Scholar]
- Chen DS, Liu CW, Roy S, Cousins D, Stacey N, Murray JD (2015) Identification of a core set of rhizobial infection genes using data from single cell-types. Front Plant Sci 6: 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Wen J, Tadege M, Ratet P, Mysore KS (2011) Reverse genetics in Medicago truncatula using Tnt1 insertion mutants. Methods Mol Biol 678: 179–190 [DOI] [PubMed] [Google Scholar]
- Cheng X, Krom N, Zhang S, Mysore KS, Udvardi M, Wen J (2017) Enabling reverse genetics in Medicago truncatula using high-throughput sequencing for Tnt1 flanking sequence recovery. Methods Mol Biol 1610: 25–37 [DOI] [PubMed] [Google Scholar]
- Crespi MD, Jurkevitch E, Poiret M, d’Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A (1994) enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J 13: 5099–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani I, Drain A, Guichard M, Balzergue S, Boscari A, Boyer JC, Brunaud V, Cottaz S, Rancurel C, Da Rocha M, et al. (2016) Nod factor effects on root hair-specific transcriptome of Medicago truncatula: Focus on plasma membrane transport systems and reactive oxygen species networks. Front Plant Sci 7: 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S, van der Wal F, Dhondt S, Waites R, de Folter S, Bimbo A, van Dijk AD, Muino JM, Cutri L, Dornelas MC, et al. (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol 159: 1511–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho-Niebel F, Timmers AC, Chabaud M, Defaux-Petras A, Barker DG (2002) The Nod factor-elicited annexin MtAnn1 is preferentially localised at the nuclear periphery in symbiotically activated root tissues of Medicago truncatula. Plant J 32: 343–352 [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM (2010) FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BJ, Ross JJ, Reid JB (2005) Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol 138: 2396–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegmann J, Bono JJ (2015) Lipo-chitooligosaccharidic nodulation factors and their perception by plant receptors. Glycoconj J 32: 455–464 [DOI] [PubMed] [Google Scholar]
- Fonouni-Farde C, Tan S, Baudin M, Brault M, Wen J, Mysore KS, Niebel A, Frugier F, Diet A (2016) DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat Commun 7: 12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier J, Timmers AC, Sieberer BJ, Jauneau A, Chabaud M, Barker DG (2008) Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol 148: 1985–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier J, Teillet A, Chabaud M, Ivanov S, Genre A, Limpens E, de Carvalho-Niebel F, Barker DG (2015) Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol 167: 1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V, Creutz CE, Moss SE (2005) Annexins: Linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol 6: 449–461 [DOI] [PubMed] [Google Scholar]
- Guan D, Stacey N, Liu C, Wen J, Mysore KS, Torres-Jerez I, Vernié T, Tadege M, Zhou C, Wang ZY, et al. (2013) Rhizobial infection is associated with the development of peripheral vasculature in nodules of Medicago truncatula. Plant Physiol 162: 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P, Ripoll JJ, Wang R, Vuong L, Bailey-Steinitz LJ, Ye D, Crawford NM (2017) Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc Natl Acad Sci USA 114: 2419–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BC, Maier EJ, Kramer MH, Wang PI, Brown H, Brent MR (2013) Mapping functional transcription factor networks from gene expression data. Genome Res 23: 1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142: 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held M, Hou H, Miri M, Huynh C, Ross L, Hossain MS, Sato S, Tabata S, Perry J, Wang TL, et al. (2014) Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. Plant Cell 26: 678–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høgslund N, Radutoiu S, Krusell L, Voroshilova V, Hannah MA, Goffard N, Sanchez DH, Lippold F, Ott T, Sato S, et al. (2009) Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. PLoS ONE 4: e6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth B, Yeun LH, Domonkos A, Halász G, Gobbato E, Ayaydin F, Miró K, Hirsch S, Sun J, Tadege M, et al. (2011) Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol Plant Microbe Interact 24: 1345–1358 [DOI] [PubMed] [Google Scholar]
- Hossain MS, Liao J, James EK, Sato S, Tabata S, Jurkiewicz A, Madsen LH, Stougaard J, Ross L, Szczyglowski K (2012) Lotus japonicus ARPC1 is required for rhizobial infection. Plant Physiol 160: 917–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idänheimo N, Gauthier A, Salojärvi J, Siligato R, Brosché M, Kollist H, Mähönen AP, Kangasjärvi J, Wrzaczek M (2014) The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem Biophys Res Commun 445: 457–462 [DOI] [PubMed] [Google Scholar]
- Ivanov S, Fedorova E, Bisseling T (2010) Intracellular plant microbe associations: Secretory pathways and the formation of perimicrobial compartments. Curr Opin Plant Biol 13: 372–377 [DOI] [PubMed] [Google Scholar]
- Jardinaud MF, Boivin S, Rodde N, Catrice O, Kisiala A, Lepage A, Moreau S, Roux B, Cottret L, Sallet E, et al. (2016) A laser dissection-RNAseq analysis highlights the activation of cytokinin pathways by Nod factors in the Medicago truncatula root epidermis. Plant Physiol 171: 2256–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Jung C, Seo JS, Kim JK, Chua NH (2017) The deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in jasmonate responses. Plant Cell 29: 1406–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Liu H, Luo D, Yu N, Dong W, Wang C, Zhang X, Dai H, Yang J, Wang E (2016) DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat Commun 7: 12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14: 737–748 [DOI] [PubMed] [Google Scholar]
- Jung C, Zhao P, Seo JS, Mitsuda N, Deng S, Chua NH (2015) PLANT U-BOX PROTEIN10 regulates MYC2 stability in Arabidopsis. Plant Cell 27: 2016–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kang Y, Liow HH, Maier EJ, Brent MR (2018) NetProphet 2.0: Mapping transcription factor networks by exploiting scalable data resources. Bioinformatics 34: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaharada Y, James EK, Kelly S, Sandal N, Stougaard J (2017) The Ethylene Responsive Factor Required for Nodulation 1 (ERN1) transcription factor is required for infection thread formation in Lotus japonicus. Mol Plant Microbe Interact 30: 194–204 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Ruzicka D, Shin R, Schachtman DP (2012) The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol Plant 5: 1042–1057 [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S (2013) Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun 4: 1617. [DOI] [PubMed] [Google Scholar]
- Krusell L, Krause K, Ott T, Desbrosses G, Krämer U, Sato S, Nakamura Y, Tabata S, James EK, Sandal N, et al. (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17: 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryvoruchko IS, Sinharoy S, Torres-Jerez I, Sosso D, Pislariu CI, Guan D, Murray J, Benedito VA, Frommer WB, Udvardi MK (2016) MtSWEET11, a nodule-specific sucrose transporter of Medicago truncatula. Plant Physiol 171: 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T, Baudin M, Frances L, Lepage A, Billault-Penneteau B, Cerri MR, Ariel F, Jardinaud MF, Gamas P, de Carvalho-Niebel F, et al. (2014) Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J 79: 757–768 [DOI] [PubMed] [Google Scholar]
- Laporte P, Lepage A, Fournier J, Catrice O, Moreau S, Jardinaud MF, Mun JH, Larrainzar E, Cook DR, Gamas P, et al. (2014) The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J Exp Bot 65: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhang J, Li J, Zhou G, Wang Q, Bian W, Erb M, Lou Y (2015) Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. eLife 4: e04805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens S, Goormachtig S, Den Herder J, Capoen W, Mathis R, Hedden P, Holsters M (2005) Gibberellins are involved in nodulation of Sesbania rostrata. Plant Physiol 139: 1366–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CW, Murray JD (2016) The role of flavonoids in nodulation host-range specificity: An update. Plants (Basel) 5: E33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ding P, Sun T, Nitta Y, Dong O, Huang X, Yang W, Li X, Botella JR, Zhang Y (2013) Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol 161: 2146–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Maekawa-Yoshikawa M, Takeda N, Imaizumi-Anraku H, Murooka Y, Hayashi M (2009) Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J 58: 183–194 [DOI] [PubMed] [Google Scholar]
- Malolepszy A, Kelly S, Sørensen KK, James EK, Kalisch C, Bozsoki Z, Panting M, Andersen SU, Sato S, Tao K, et al. (2018) A plant chitinase controls cortical infection thread progression and nitrogen-fixing symbiosis. eLife 7: e38874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C, Roudier F, Castaings L, Bréhaut V, Blondet E, Colot V, Meyer C, Krapp A (2013) Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun 4: 1713. [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam EL, Reid JB, Foo E (2018) Gibberellins promote nodule organogenesis but inhibit the infection stages of nodulation. J Exp Bot 69: 2117–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahara A, Richens J, Starker C, Morieri G, Smith L, Long S, Downie JA, Oldroyd GE (2010) Conservation in function of a SCAR/WAVE component during infection thread and root hair growth in Medicago truncatula. Mol Plant Microbe Interact 23: 1553–1562 [DOI] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’Haeseleer K, Holsters M, Goormachtig S (2010) CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol 153: 222–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V, De Wever E, Vuylsteke M, Holsters M, Goormachtig S (2012) Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J 70: 367–376 [DOI] [PubMed] [Google Scholar]
- Mortimer JC, Laohavisit A, Macpherson N, Webb A, Brownlee C, Battey NH, Davies JM (2008) Annexins: Multifunctional components of growth and adaptation. J Exp Bot 59: 533–544 [DOI] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104 [DOI] [PubMed] [Google Scholar]
- Naoumkina M, Farag MA, Sumner LW, Tang Y, Liu CJ, Dixon RA (2007) Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc Natl Acad Sci USA 104: 17909–17915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H, Handa Y, Tanaka S, Suzaki T, Kawaguchi M (2016) Expression of the CLE-RS3 gene suppresses root nodulation in Lotus japonicus. J Plant Res 129: 909–919 [DOI] [PubMed] [Google Scholar]
- Nishida H, Tanaka S, Handa Y, Ito M, Sakamoto Y, Matsunaga S, Betsuyaku S, Miura K, Soyano T, Kawaguchi M (2018) A NIN-LIKE PROTEIN mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nat Commun 9: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Long SR (2003) Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiol 131: 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Mizuno S, Tanaka H, Maruyama K, Osakabe K, Todaka D, Fujita Y, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Overproduction of the membrane-bound receptor-like protein kinase 1, RPK1, enhances abiotic stress tolerance in Arabidopsis. J Biol Chem 285: 9190–9201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikova E, Journet EP, Chabaud M, Cosson V, Ratet P, Duc G, Fedorova E, Liu W, den Camp RO, Zhukov V, et al. (2011) IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago spp. Mol Plant Microbe Interact 24: 1333–1344 [DOI] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C (2004) Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci 9: 534–540 [DOI] [PubMed] [Google Scholar]
- Patron NJ, Orzaez D, Marillonnet S, Warzecha H, Matthewman C, Youles M, Raitskin O, Leveau A, Farré G, Rogers C, et al. (2015) Standards for plant synthetic biology: A common syntax for exchange of DNA parts. New Phytol 208: 13–19 [DOI] [PubMed] [Google Scholar]
- Pratap Sahi V, Cifrová P, García-González J, Kotannal Baby I, Mouillé G, Gineau E, Müller K, Baluška F, Soukup A, Petrášek J, et al. (2018) Arabidopsis thaliana plants lacking the ARP2/3 complex show defects in cell wall assembly and auxin distribution. Ann Bot 122: 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Lin JS, Xu J, Sato S, Parniske M, Wang TL, Downie JA, Xie F (2015) SCARN a novel class of SCAR protein that is required for root-hair infection during legume nodulation. PLoS Genet 11: e1005623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrère S, Sallet E, Courcelle E, Moreau S, et al. (2014) An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J 77: 817–837 [DOI] [PubMed] [Google Scholar]
- Saffer AM. (2018) Expanding roles for pectins in plant development. J Integr Plant Biol 60: 910–923 [DOI] [PubMed] [Google Scholar]
- Santos R, Franza T, Laporte ML, Sauvage C, Touati D, Expert D (2001) Essential role of superoxide dismutase on the pathogenicity of Erwinia chrysanthemi strain 3937. Mol Plant Microbe Interact 14: 758–767 [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Sinharoy S, Liu C, Breakspear A, Guan D, Shailes S, Nakashima J, Zhang S, Wen J, Torres-Jerez I, Oldroyd G, et al. (2016) A Medicago truncatula cystathionine-β-synthase-like domain-containing protein is required for rhizobial infection and symbiotic nitrogen fixation. Plant Physiol 170: 2204–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smakowska E, Kong J, Busch W, Belkhadir Y (2016) Organ-specific regulation of growth-defense tradeoffs by plants. Curr Opin Plant Biol 29: 129–137 [DOI] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Soyano T, Kouchi H, Hirota A, Hayashi M (2013) Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet 9: e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Hirakawa H, Sato S, Hayashi M, Kawaguchi M (2014) Nodule Inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc Natl Acad Sci USA 111: 14607–14612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Shimoda Y, Hayashi M (2015) NODULE INCEPTION antagonistically regulates gene expression with nitrate in Lotus japonicus. Plant Cell Physiol 56: 368–376 [DOI] [PubMed] [Google Scholar]
- Verdier J, Torres-Jerez I, Wang M, Andriankaja A, Allen SN, He J, Tang Y, Murray JD, Udvardi MK (2013) Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. Plant J 74: 351–362 [DOI] [PubMed] [Google Scholar]
- Vernié T, Kim J, Frances L, Ding Y, Sun J, Guan D, Niebel A, Gifford ML, de Carvalho-Niebel F, Oldroyd GE (2015) The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 27: 3410–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JP, Rathbun EA, Knox JP, Brewin NJ (2000) Involvement of diamine oxidase and peroxidase in insolubilization of the extracellular matrix: Implications for pea nodule initiation by Rhizobium leguminosarum. Mol Plant Microbe Interact 13: 413–420 [DOI] [PubMed] [Google Scholar]
- Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T (2014) Fate map of Medicago truncatula root nodules. Development 141: 3517–3528 [DOI] [PubMed] [Google Scholar]
- Xie F, Murray JD, Kim J, Heckmann AB, Edwards A, Oldroyd GE, Downie JA (2012) Legume pectate lyase required for root infection by rhizobia. Proc Natl Acad Sci USA 109: 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Easwaran V, Chau V, Okamoto M, Ierullo M, Kimura M, Endo A, Yano R, Pasha A, Gong Y, et al. (2016) NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat Commun 7: 13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Aoki S, Liu M, Umehara Y, Suganuma N, Iwasaki W, Sato S, Soyano T, Kouchi H, Kawaguchi M (2017) Function and evolution of a Lotus japonicus AP2/ERF family transcription factor that is required for development of infection threads. DNA Res 24: 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota K, Fukai E, Madsen LH, Jurkiewicz A, Rueda P, Radutoiu S, Held M, Hossain MS, Szczyglowski K, Morieri G, et al. (2009) Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell 21: 267–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoro E, Suzaki T, Toyokura K, Miyazawa H, Fukaki H, Kawaguchi M (2014) A positive regulator of nodule organogenesis, NODULE INCEPTION, acts as a negative regulator of rhizobial infection in Lotus japonicus. Plant Physiol 165: 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LH, Wu J, Tang H, Yuan Y, Wang SM, Wang YP, Zhu QS, Li SG, Xiang CB (2016) Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Sci Rep 6: 27795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Blanco FA, Beker MP, Battaglia M, Aguilar OM (2010) A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell 22: 4142–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Rípodas C, Niebel A (2017) Plant NF-Y transcription factors: Key players in plant-microbe interactions, root development and adaptation to stress. Biochim Biophys Acta 1860: 645–654 [DOI] [PubMed] [Google Scholar]
- Zhang X, Pumplin N, Ivanov S, Harrison MJ (2015) EXO70I is required for development of a sub-domain of the periarbuscular membrane during arbuscular mycorrhizal symbiosis. Curr Biol 25: 2189–2195 [DOI] [PubMed] [Google Scholar]
- Zhao L, Zhang W, Yang Y, Li Z, Li N, Qi S, Crawford NM, Wang Y (2018) The Arabidopsis NLP7 gene regulates nitrate signaling via NRT1.1-dependent pathway in the presence of ammonium. Sci Rep 8: 1487. [DOI] [PMC free article] [PubMed] [Google Scholar]