BBX31 negatively regulates photomorphogenesis in visible light and positively regulates UV-B signaling to protect plants from UV radiation.
Abstract
The bZIP transcription factor ELONGATED HYPOCOTYL5 (HY5) represents a major hub in the light-signaling cascade both under visible and UV-B light. The mode of transcriptional regulation of HY5, especially under UV-B light, is not well characterized. B-BOX (BBX) transcription factors regulate HY5 transcription and also posttranscriptionally modulate HY5 to control photomorphogenesis under white light. Here, we identify BBX31 as a key signaling intermediate in visible and UV-B light signal transduction in Arabidopsis (Arabidopsis thaliana). BBX31 expression is induced by UV-B radiation in a fluence-dependent manner. HY5 directly binds to the promoter of BBX31 and regulates its transcript levels. Loss- and gain-of-function mutants of BBX31 indicate that it acts as a negative regulator of photomorphogenesis under white light but is a positive regulator of UV-B signaling. Genetic interaction studies suggest that BBX31 regulates photomorphogenesis independent of HY5. We found no evidence for a direct BBX31-HY5 interaction, and they primarily regulate different sets of genes in white light. Under high doses of UV-B radiation, BBX31 promotes the accumulation of UV-protective flavonoids and phenolic compounds. It enhances tolerance to UV-B radiation by regulating genes involved in photoprotection and DNA repair in a HY5-dependent manner. Under UV-B radiation, overexpression of BBX31 enhances HY5 transcriptional levels in a UV RESISTANCE LOCUS8-dependent manner, suggesting that BBX31 might regulate HY5 transcription.
Light influences plant growth and development at almost all stages of the plant life cycle. After germination of seeds, light promotes photomorphogenesis in plants, which is characterized by inhibition of hypocotyl elongation, opening of cotyledons, and biosynthesis of chlorophylls (Quail, 2002; Sullivan and Deng, 2003). At the cellular level, this involves inhibition of cell expansion in the hypocotyl, a reciprocal increase in cell size in the cotyledons, and promotion of chloroplast development (Quail, 2002). The opposite cell expansion response in the hypocotyl and cotyledons during photomorphogenesis is modulated by several positive and negative regulators of the light signal transduction pathway. HY5 (ELONGATED HYPOCOTYL5), a bZIP transcription factor, and CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1), a RING finger-type E3 ubiquitin ligase, represent the central activator and repressor of photomorphogenesis, respectively (Koornneef et al., 1980; Deng et al., 1992; Oyama et al., 1997; Yi and Deng, 2005; Jing et al., 2013). Upon perceiving light, multiple light-activated photoreceptors inhibit the E3 ligase activity of COP1, preventing ubiquitin-mediated proteolysis of numerous photomorphogenesis-promoting proteins, including HY5 (Osterlund et al., 2000) and HY5 HOMOLOG (HYH; Osterlund et al., 2000; Holm et al., 2002). Apart from HY5 and HYH, several other transcription factors function as positive or negative regulators of light signaling. Among those, B-BOX (BBX) containing zinc finger domain proteins act as important modulators of light signal transduction (Gangappa and Botto, 2014). The first BBX protein to be identified in Arabidopsis (Arabidopsis thaliana) was the flowering time regulator CONSTANS (BBX1; Putterill et al., 1995). Since then, several more BBX proteins, BBX4 (COL3), BBX6 (COL5), BBX7 (COL9), BBX19, and BBX32, have been shown to play roles in flowering time regulation (Cheng and Wang, 2005; Datta et al., 2006; Hassidim et al., 2009; Li et al., 2014; Wang et al., 2014; Graeff et al., 2016). Two microProteins, miP1a and miP1b, reported to regulate flowering represent BBX31 and BBX30, respectively (Graeff et al., 2016). MicroProteins are small, single-domain proteins that regulate the function of larger proteins by engaging in protein-protein interactions (Staudt and Wenkel, 2011; Eguen et al., 2015). miP1a/BBX31 and miP1b/BBX30 interact with CONSTANS as part of a multimeric complex and repress flowering (Graeff et al., 2016). Some of the other roles of the BBX proteins involve regulation of photomorphogenesis, shade avoidance/neighborhood detection, thermotolerance, hormone signaling, and UV-B signal transduction (Gangappa and Botto, 2014). Among all the known functions of BBX proteins, regulation of photomorphogenesis is the most well characterized. BBX4, BBX20, BBX21, BBX22, and BBX23 promote photomorphogenesis, whereas BBX18, BB19, BBX24, BBX25, BBX32, and BBX28 suppress photomorphogenesis (Datta et al., 2006, 2007, 2008; Indorf et al., 2007; Chang et al., 2008; Kumagai et al., 2008; Khanna et al., 2009; Holtan et al., 2011; Fan et al., 2012; Gangappa et al., 2013a; Wei et al., 2016; Zhang et al., 2017; Lin et al., 2018; Xu et al., 2018).
The inhibition of hypocotyl elongation is a response that, in addition to the visible light, also occurs when plants are exposed to low doses of UV light (Jenkins, 2017). UV light is divided into three different band spectra: UV-A (315–400 nm), UV-B (280–315 nm), and UV-C (200–280 nm). When exposed to UV-B radiation, plants respond in two different ways: they undergo morphogenic changes and/or activate stress responses. Longer wavelengths and low doses of UV-B stimulate morphogenic responses in plants. These responses are regulated by a signal transduction cascade originating with the perception of UV-B by the photoreceptor UV RESISTANCE LOCUS8 (UVR8; Kliebenstein et al., 2002; Brown et al., 2005; Favory et al., 2009; Rizzini et al., 2011). In response to UV-B, the dimeric UVR8 is dynamically converted to the active monomer form that initiates the signaling cascade by interacting with COP1 and translocating to the nucleus (Oravecz et al., 2006; Kaiserli and Jenkins, 2007; Rizzini et al., 2011; Christie et al., 2012; Wu et al., 2012; Findlay and Jenkins, 2016). The sequestration of COP1 by UVR8 stabilizes HY5 under UV-B light, which leads to further enhanced accumulation of HY5, since HY5 can regulate its own transcription (Favory et al., 2009; Binkert et al., 2014). Inside the nucleus, UVR8 mediates the activation of a set of genes, including HY5, and modulates various developmental responses like photomorphogenesis and synthesis of photoprotective agents like flavonoids and phenolic compounds. Until now, only one BBX protein, BBX24, was known to be involved in UV-B-mediated signaling (Jiang et al., 2012). BBX24 acts as a negative regulator in UV-mediated photomorphogenesis by interacting with both COP1 and HY5 and suppressing HY5 activity (Jiang et al., 2012).
The roles of HY5 in light, hormone, nutrient, and various abiotic stress signaling pathways place HY5 at the center of transcriptional networks (Gangappa and Botto, 2016). It regulates photomorphogenesis both under visible light and UV-B radiation. It acts downstream of phytochromes, cryptochromes, and UVR8 to regulate the transcription of a large number of genes (Lee et al., 2007; Binkert et al., 2014). In fact, studies have indicated that HY5 binds to the promoters of almost one-third of all the genes present in Arabidopsis (Lee et al., 2007). Such a large repertoire of target genes necessitates the action of transcriptional cofactors to specifically regulate the downstream genes. There is increasing evidence suggesting that the BBX transcription factors act as cofactors of HY5 to either enhance or suppress the transcription of respective target genes (Gangappa and Botto, 2014). While BBX22 can act as a transcriptional coactivator, BBX24, BBX25, and BBX28 suppress the transcriptional activity of HY5 by preventing it from binding to promoters of downstream genes (Datta et al., 2008; Gangappa et al., 2013a, 2013b; Xu et al., 2016; Job et al., 2018; Lin et al., 2018). BBX32 negatively regulates HY5 activity by physically interacting and possibly sequestering its coactivator BBX21 (Holtan et al., 2011). Besides modulating HY5 activity, many BBX proteins act in a HY5-dependent manner. BBX structural group IV members interacting with HY5 (BBX20–BBX25) regulate light-dependent development in a primarily HY5-dependent fashion (Yadukrishnan et al., 2018). BBX23 acts redundantly with its close homolog BBX22 to positively regulate photomorphogenesis in a partially HY5-dependent manner (Zhang et al., 2017). Some of the negative regulators of photomorphogenesis belonging to structural group V, BBX32 and BBX28, also depend on HY5 for their activity (Holtan et al., 2011; Lin et al., 2018).
Interestingly, HY5 and the BBX proteins can also regulate the transcript levels of each other. While HY5 can regulate the mRNA levels of BBX22, the BBX transcription factor BBX21 can regulate HY5 transcription by directly binding to its promoter (Gangappa and Botto, 2014; Xu et al., 2016). Overexpression of BBX20 leads to up-regulation of the transcript levels of HY5 (Wei et al., 2016). HY5 levels are also modulated by a self-regulatory mechanism mediated by HY5/HYH (Abbas et al., 2014; Binkert et al., 2014). Additionally, CALMODULIN7 (CAM7), a member of the calmodulin gene family, can bind directly to the promoter of HY5 and drive its transcription (Kushwaha et al., 2008). The PHYTOCHROME INTERACTING FACTOR (PIF) proteins PIF1 and PIF3 regulate HY5 transcription by direct binding to the regulatory regions of HY5 (Zhang et al., 2017). HY5 binds to its own promoter along with HYH and induces its own expression upon exposure to UV-B radiation (Binkert et al., 2014). WRKY36, a recent addition to the UV-B signaling pathway, inhibits HY5 transcription by binding to its promoter in the absence of UV-B light (Yang et al., 2018). In response to UV-B radiation, UVR8 sequesters WRKY36, allowing the maintenance of HY5 expression. However, apart from HY5/HYH, direct positive regulators of HY5 transcription in response to UV-B light have yet to be discovered.
In addition to the morphogenic responses, the short wavelength and high dose of UV-B radiation have the potential to damage molecules such as nucleic acids, amino acids, proteins, and lipids and also to produce reactive oxygen species, causing necrosis in plants (Yoshiyama et al., 2013). Under natural conditions, the genotoxic potential of UV-B radiation is primarily determined by the genetic ability of the plant to adapt to the UV-B exposure, the acclimation status of the plant to UV-B radiation, and the time of the day at which the plant is exposed to UV-B radiation (Takeuchi et al., 2014; Jenkins, 2017). UVR8 plays a crucial role to mediate the plant’s ability to combat the stress induced by UV-B radiation. UVR8 mediates the biosynthesis of photoprotective phenolic and flavonoid compounds upon UV-B exposure. The deposition of these compounds in the epidermal cells acts as a sunscreen for plants (Jenkins, 2014). It also activates genes involved in the DNA repair pathway, like UVR2 that codes for a CYCLOBUTANE PYRIMIDINE DIMER (CPD) photolyase through HY5 (Brown and Jenkins, 2008). Moreover, UVR8 regulates the expression of several antioxidant genes that help the plant to defend against UV-B-induced oxidative stress (Hideg et al., 2013). However, several additional stress signaling pathways that are not specific to UV-B radiation and not mediated by UVR8 are also activated in high-fluence, short-wavelength UV-B irradiation. These include DNA damage response, wound signaling, and hormone-mediated defense signaling (Jenkins, 2017). Yet, upon UV-B exposure, the activation of UVR8-dependent or independent stress signaling pathways will largely depend upon the adaptation and acclimation status of the plant to UV-B exposure (Jenkins, 2017).
In this study, we identify miP1a/BBX31, a B-box containing microProtein, in Arabidopsis as a novel negative regulator of photomorphogenesis in white light (WL) while acting as a positive regulator of UV-B signaling. HY5 directly binds to the promoter of BBX31 and regulates its transcript levels. We could not find any evidence for a direct physical interaction between HY5 and BBX31, and genetic studies indicate that BBX31 regulates photomorphogenesis independent of HY5. BBX31 enhances tolerance to high doses of UV-B radiation by promoting the accumulation of photoprotective flavonoids and phenols and activating DNA repair genes in a HY5-dependent manner. Under UV-B irradiance, overexpression of BBX31 increases HY5 transcript levels in a UVR8-dependent manner.
RESULTS
BBX31 Expression Is Induced by UV-B Light in a Fluence-Dependent Manner and Requires UVR8, COP1, and HY5
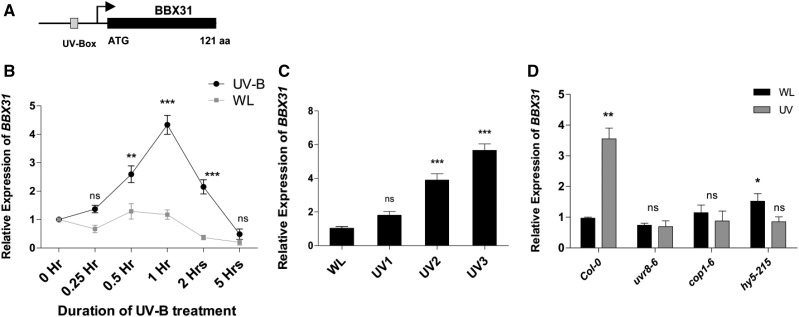
Microarray studies indicate that the gene coding for the B-box transcription factor BBX31 is induced by UV-B radiation (Ulm et al., 2004; Favory et al., 2009). The BBX31 gene (At3g21890) contains a single exon encoding a 121-amino acid protein previously also referred to as a microProtein, miP1a (Fig. 1A; Graeff et al., 2016). The promoter of BBX31 contains a UV-box (CAAG), a cis-regulatory element for UV-B-induced transcription (Fig. 1A; Ulm et al., 2004; Safrany et al., 2008; Favory et al., 2009; Velanis et al., 2016). BBX31 was also identified as a target of UV-B-induced UVR8-mediated histone acetylation (Velanis et al., 2016). All these previous reports prompted us to investigate if BBX31 plays any role in UV-B signaling. In order to validate the UV-B-mediated induction of BBX31, we systematically monitored the changes in transcript levels of BBX31 upon changing the dose of UV-B radiation by modulating the exposure duration and fluence rate. In the first experiment, we used a fluence rate of 1.4 W m−2 UV-B and exposed seedlings to narrowband long-wavelength (λmax 311 nm) UV-B. Keeping the fluence rate and wavelength constant, we exposed seedlings to different durations (0–5 h) of UV-B radiation. We found that BBX31 is induced by UV-B light and that its expression peaked after 1 h of UV-B treatment, indicating that BBX31 is an early response gene (Fig. 1B). Next, we analyzed the change in transcript levels of BBX31 in response to changes in fluence rates of UV-B (0.5, 1.4, and 4.5 W m−2). With increasing intensity of UV-B treatment, expression of BBX31 increased gradually in a dose-dependent manner (Fig. 1C). The activation of BBX31 upon UV-B exposure was absent in uvr8-6, cop1-6, and hy5 mutants, indicating that BBX31 might be part of the canonical UV-B signaling pathway (Fig. 1D). Taken together, our results indicate that induction of BBX31 by UV-B light is an early, fluence-dependent response and requires UVR8, COP1, and HY5.
Figure 1.
BBX31 expression is induced by UV-B light in a fluence-dependent manner. A, Schematic representation of the BBX31 gene containing a single exon (denoted by the black box) that codes for a 121-amino acid protein (previously referred to as microProtein miP1A). The arrow represents the transcription start site, and UV-Box represents a cis-regulatory element present in its promoter for UV-induced transcription. B, BBX31 transcript levels in Col-0 seedlings exposed to different durations (0–5 h) of UV-B treatment (1.4 W m−2) or WL of fluence 90 μmol m−2 s−1. Asterisks represent statistically significant differences (***, P < 0.001 and **, P < 0.01) as determined by two-way ANOVA followed by Bonferroni posttests; ns, nonsignificant. C, BBX31 transcript levels in Col-0 seedlings exposed to different fluences of UV-B treatment (UV1, 0.5 W m−2; UV2, 1.4 W m−2; and UV3, 4.5 W m−2) for 1 h. Asterisks represent statistically significant differences (***, P < 0.001) as determined by one-way ANOVA followed by Dunnett’s posthoc test. D, BBX31 transcript levels in 10-d-old Col-0, uvr8-6, cop1-4, and hy5-215 seedlings exposed to 1 h of WL or UV-B (1.4 W m−2). Asterisks represent statistically significant differences (*, P < 0.05 and **, P < 0.01) as determined by Student’s t test. Error bars (B–D) represent sd of three replicates.
HY5 Directly Binds to the Promoter of BBX31 in Both White and UV-B Light and Regulates Its Transcript Levels
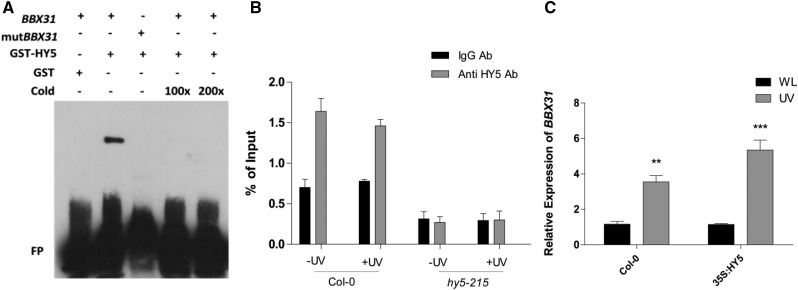
Since UVR8, COP1, and HY5 are required for the induction of BBX31 under UV light, we asked if HY5, the downstream component among the three, directly regulates the transcription of BBX31. The promoter of BBX31 contains a G-box, which is a well-known binding site for HY5, 154 bases upstream of the start codon. We therefore hypothesized that HY5 might physically bind to the promoter of BBX31. We performed an electrophoretic mobility shift assay (EMSA) using 50-bp-long probes containing the HY5-binding G-box motif (CACGTG) to check in vitro binding of HY5 to the BBX31 promoter (Fig. 2A). We observed specific HY5 binding to the biotin-labeled, G-box-containing BBX31 probe. Upon mutating the G-box, we did not observe any shift in the probe, indicating that the binding of HY5 to the BBX31 promoter is through the G-box (Fig. 2A). In order to validate the binding in planta, we performed chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qPCR). We used anti-HY5 antibody to pull down the HY5 protein bound to various regions of the chromatin. The genomic DNA fragments that coimmunoprecipitated with HY5 were quantified by qPCR. The region harboring the G-box of the BBX31 promoter was enriched in the coimmunoprecipitated fraction of Columbia-0 (Col-0) seedlings in both WL and UV-B conditions, indicating constitutive binding of HY5 to the BBX31 promoter (Fig. 2B). Such enrichment was absent when chromatin from hy5 seedlings (negative control) was used for a similar experiment (Fig. 2B). In addition, we also checked the enrichment of HY5 on a fragment from the 3′ untranslated region of BBX31 and a fragment from the promoter of ACTIN8, both lacking a G-box. No enrichment was obtained in both these fragments in any of the conditions (Supplemental Fig. S1). Furthermore, we checked the induction of BBX31 by UV-B radiation in seedlings overexpressing HY5. When UV-B treatment was given, induction of BBX31 was higher in HY5 overexpression lines when compared with the wild type (Fig. 2C). Conversely, the loss of HY5 resulted in complete abrogation of the UV-B-mediated induction of BBX31 (Fig. 1D). Even though HY5 binds to the BBX31 promoter in both white and UV light (Fig. 2B) and BBX31 is strongly regulated by HY5 under UV light (Figs. 1D and 2C), its expression did not change markedly in HY5 overexpression lines only in WL (Fig. 2C). A marginal 1.5-fold increase in the expression of BBX31 was observed in the hy5 mutant, suggesting that HY5 might negatively regulate BBX31 under WL (Fig. 1D). All these results suggest that HY5 directly binds to the promoter of BBX31 in both white and UV-B light and regulates its transcript levels.
Figure 2.
HY5 constitutively binds to the BBX31 promoter and regulates its transcription under UV-B light. A, EMSA showing the binding of GST-HY5 to the G-box-containing promoter fragment of BBX31 labeled with biotin. mutBBX31 represents the promoter fragment with a mutation in the G-box that does not show any interaction with GST-HY5. + and – represent presence and absence; FP represents free probe. B, ChIP-qPCR analyses of HY5 binding to the BBX31 promoter in vivo. ChIP assays were performed on wild-type and hy5-215 seedlings. DNA-protein complexes were immunoprecipitated using antibodies (Ab) against HY5 and rabbit IgG (negative control). ChIP DNA was quantified by qPCR with primers specific to the BBX31 promoter region. C, Reverse transcription-qPCR analysis of the transcript levels of BBX31 in Col-0 and 35S:HY5 seedlings grown in WL and under UV-B light (1 h, 1.4 W m−2). Asterisks represent statistically significant differences (***, P < 0.001 and **, P < 0.05) as determined by Student’s t test. Error bars (B and C) represent sd of three biological replicates.
BBX31 Acts as a Negative Regulator of Photomorphogenesis under WL
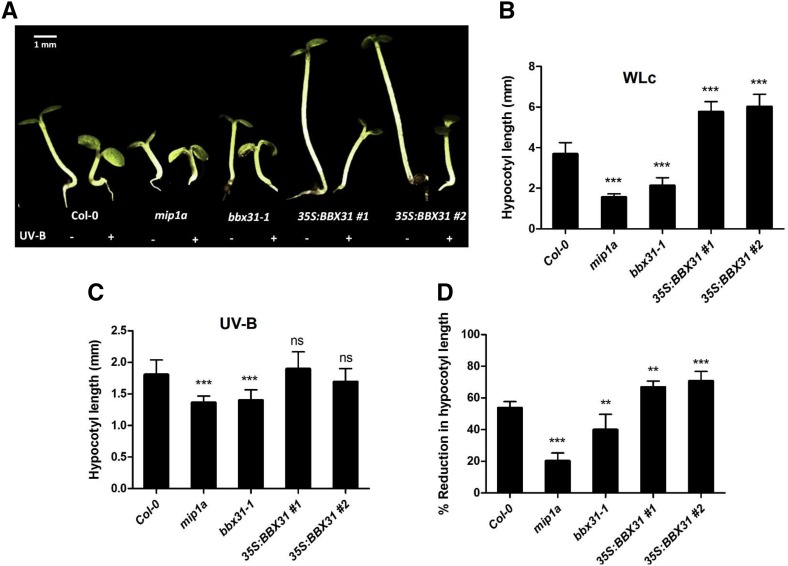
Arabidopsis seedlings respond to both visible and low UV-B light with a suppression of hypocotyl growth (Fankhauser, 2001; Jiang et al., 2012; Jenkins, 2017). To investigate the role of BBX31 in photomorphogenesis, we obtained two independent alleles of loss-of-function bbx31 mutants in the Col-0 background. Since the only T-DNA insertion mutant available for BBX31 (GABI-KAT line 288G08) does not show any reduction in BBX31 mRNA levels, we used the previously reported microRNA-mediated gene silencing (MIGS) line, MIGS-miP1a (hereafter represented as mip1a), in which a trans-acting small interfering RNA is generated that specifically targets BBX31 (Graeff et al., 2016). In the same study, the double mutant MIGS-miP1a miP1b with reduced expression levels of both BBX31 and BBX30 showed a subtle early-flowering phenotype (Graeff et al., 2016). This prompted us to include the MIGS-miP1b and MIGS-miP1a miP1b mutants in our photomorphogenic analysis (Graeff et al., 2016). We further generated the loss-of-function mutants bbx31-1 and bbx30-1 by CRISPR-Cas9 technology (Supplemental Fig. S2A). The bbx31-1 and bbx30-1 alleles have deletions of 209 and 68 bp, respectively (Supplemental Fig. S2A). We also generated multiple transgenic lines overexpressing BBX31 and used two of these lines, 35S:BBX31#1 and 35S:BBX31#2, for further analysis. The transcript levels of BBX31 were up-regulated 30- and 60-fold, respectively, in the 35S:BBX31#1 and 35S:BBX31#2 lines (Supplemental Fig. S2B).
We studied the hypocotyl elongation in all these lines at low-fluence (10 μmol m−2 s−1) continuous white light (WLc). We used low fluence since BBX31 is expressed at higher levels at lower fluence WL (Supplemental Fig. S2C). All the loss-of-function mutants had shorter hypocotyl length compared with the wild-type seedlings, indicating that BBX31 and BBX30 negatively regulate photomorphogenesis (Fig. 3, A and B; Supplemental Fig. S3D). We did not observe significant differences between the hypocotyl lengths of wild-type and bbx31 seedlings grown in darkness (Supplemental Fig. S3A). Comparison of BBX31 transcript levels in seedlings grown in darkness and in light indicated that light represses BBX31 expression (Supplemental Fig. S2C). All the BBX31-overexpressing lines showed elongated hypocotyls compared with the wild type, which reiterated that BBX31 is a repressor of photomorphogenesis (Fig. 3, A and B). The 35S:BBX31 seedlings did not show any difference in hypocotyl length from that of the wild type in darkness (Supplemental Fig. S3A). In addition to the hypocotyl lengths, we also measured the cotyledon areas in the light-grown seedlings. We did not find any significant difference between Col-0 and the loss-of-function mutants (Supplemental Fig. S3B), whereas the overexpressors showed a distinct reduction in the cotyledon area, indicating that BBX31 might play a role in inhibiting cotyledon expansion in light (Supplemental Fig. S3B). We also measured the hypocotyl lengths in different fluences of WLc and found that the BBX31 overexpression line exhibited elongated hypocotyls in all fluences of WLc (Supplemental Fig. S3C). All these data together indicate that under visible light BBX31 acts as a negative regulator of photomorphogenesis.
Figure 3.
BBX31 is a negative regulator of WL photomorphogenesis but a positive regulator of UV-B signaling. A, Representative image of 4-d-old seedlings of Col-0, mip1a, bbx31-1, 35S:BBX31#1, and 35S:BBX31#2 grown in the absence (−) and presence (+) of UV-B light. For the −UV-B treatment, seedlings were grown in WLc (10 μmol m−2 s−1), and for the +UV-B treatment, seedlings were grown in continuous UV-B light (1.4 W m−2) supplemented with WL (10 μmol m−2 s−1). B, Hypocotyl lengths of the indicated genotypes under WL. C, Hypocotyl lengths of the indicated genotypes under UV-B light. D, Percentage reduction in hypocotyl length of the indicated genotypes grown under UV-B light. Values in B to D represent means ± se; n ≥ 20. Asterisks in B to D represent statistically significant differences (***, P < 0.001 and **, P < 0.05) as determined by one-way ANOVA followed by Dunnett’s post hoc test; ns, nonsignificant.
BBX31 Plays a Positive Role in UV-B Signaling
To investigate the role of BBX31 in the UV-B-mediated photomorphogenic response, we examined UV-B-induced shortening of hypocotyls in the bbx31 mutants and overexpression lines under narrowband long-wavelength (λmax 311nm) UV-B light supplemented with low WLc (10 μmol m−2 s−1) compared with seedlings grown under low WLc alone. Under these conditions, the Col-0 seedlings exhibited about a 54% reduction in hypocotyl length when supplemented with UV-B light (Fig. 3). The extent of the reduction in hypocotyl length upon UV-B exposure in the mip1a mutant was 19%, indicating that the loss of BBX31 made seedlings less sensitive to UV-B radiation (Fig. 3D). A similar hyposensitivity to UV-B light was also seen in the bbx31-1 mutant, although to a lesser extent (Fig. 3). In addition, the overexpression lines, 35S:BBX31#1 and 35S:BBX31#2, exhibited 65% and 70% reduction in hypocotyl length, respectively, upon UV-B supplementation. These results suggest that BBX31 plays a positive role in UV-B signaling.
BBX31 Regulates Photomorphogenesis Independent of HY5 in Both White and UV-B Light
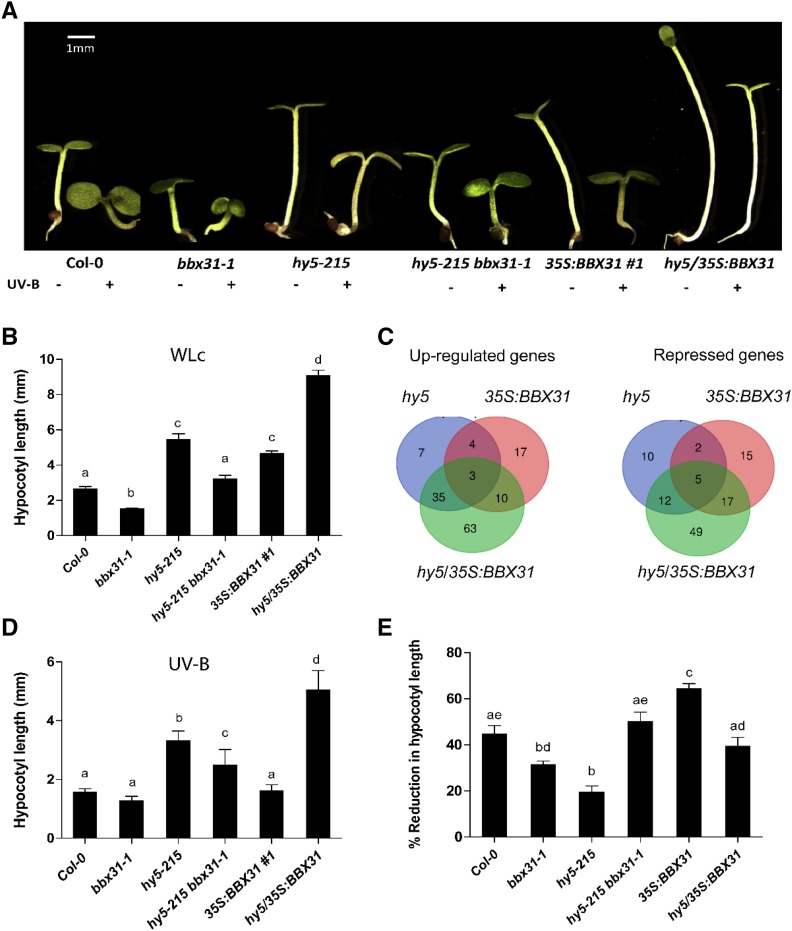
HY5 is a key positive regulator of both WL and UV-B signaling pathways. The elongated hypocotyl phenotype of the BBX31 overexpression lines resembles the phenotype of the hy5 mutant under light (Fig. 3, A and B). In order to understand if the regulation of hypocotyl length by BBX31 is in a HY5-dependent manner, we generated a hy5-215 bbx31-1 double mutant and examined its photomorphogenic phenotype. Under WL, the double mutant showed a phenotype similar to the wild type, which was intermediate to the single mutants, hy5-215 and bbx31-1 (Fig. 4, A and B). This intermediate phenotype could arise as a result of the independent effects of longer and shorter hypocotyls of the hy5-215 and bbx31-1 mutants, respectively. To further validate the independent actions of HY5 and BBX31, we overexpressed BBX31 in the hy5 mutant background by transforming hy5-215 plants with the 35S:BBX31 construct. We obtained several independent lines of hy5/35S:BBX31 and confirmed the levels of expression of BBX31 in these lines. Since 35S:BBX31 and hy5/35S:BBX31 lines were generated by independent transformation events, we identified a line that showed similar expression levels of BBX31 compared with the 35S:BBX31#1 line for further analysis (Supplemental Fig. S4A). Hypocotyl elongation measurements in these lines under WL indicated that the hy5 mutant background enhances the hypophotomorphogenic phenotype of the 35S:BBX31 line in an additive manner, again suggesting that the two genes act independently under WL (Fig. 4, A and B). Furthermore, we could not detect any physical interaction between BBX31 and HY5 in our bimolecular fluorescence complementation and yeast two-hybrid experiments, while both proteins clearly interacted with their known interacting partners (Supplemental Fig. S5). Adding to other evidence, this supports the notion that BBX31 and HY5 might not be interdependent in their regulation of WL photomorphogenesis. hy5/35S:BBX31 seedlings exhibited longer hypocotyls than both their parents owing to an increase in the length of the constituting cells (Fig. 4, A and B; Supplemental Fig. S4, B and C).
Figure 4.
BBX31 regulates photomorphogenesis in a HY5-independent manner in WL and UV-B light. A, Representative image of 4-d-old seedlings of Col-0, bbx31-1, hy5-215, hy5-215 bbx31-1, 35S:BBX31#1, and hy5/35S:BBX31#1 lines grown in −UV (10 μmol m−2 s−1) and +UV (1.4 W m−2) conditions. Bar = 1 mm. B, Bar graph depicting the hypocotyl lengths of the indicated genotypes under WLc. C, Venn diagram showing the overlap between sets of genes differentially expressed in hy5, 35S:BBX31, and hy5/35S:BBX31 seedlings grown under WLc (10 μmol m−2 s−1) for transcriptome analysis. D, Hypocotyl lengths of the indicated genotypes under UV-B light. E, Percentage reduction in hypocotyl length of the indicated genotypes under UV-B light. In B, D, and E, values represent means ± se; n ≥ 20. Letters above the bars indicate significant differences (P < 0.05) as determined by one-way ANOVA followed by Tukey’s posthoc analysis. The experiments were performed at least three times with similar results.
If HY5 and BBX31 act independently of each other, they are likely to regulate different sets of downstream genes. In order to test this hypothesis and to better understand BBX31-mediated regulation of hypocotyl elongation, we sequenced and compared the transcriptomes of 6-d-old Col-0, hy5, 35S:BBX31#1, and hy5/35S:BBX31 seedlings grown in WL. Genes with more than a 2-fold change compared with Col-0 and P < 0.05 were considered as differentially expressed genes (Supplemental Data Sets S1 and S2). Analysis of the RNA sequencing (RNA Seq) data revealed nominal overlap between the genes up- or down-regulated in hy5 and 35S:BBX31#1 plants (Fig. 4C). We found that only seven out of 76 genes were up-regulated in both hy5 and 35S:BBX31#1 plants (Fig. 4C). Additionally, only seven out of 62 genes were common among the genes repressed by hy5 and 35S:BBX31#1 plants, suggesting that HY5 and BBX31 primarily act through independent parallel pathways to regulate hypocotyl elongation under low-fluence WL (Fig. 4C). The 14 common genes were found to play roles largely involved in regulation of abiotic stress, protein folding, lipid transport, and glucosinolate metabolism. A large number of genes (112) were up- or down-regulated exclusively in the hy5/35S:BBX31 line (Fig. 4C). Gene Ontology analysis indicated that the genes up-regulated specifically in the hy5/35S:BBX31 seedlings included genes involved in response to desiccation, water deprivation, abscisic acid, and osmotic stress (Supplemental Fig. S6A). To validate the differential expression data obtained from RNA Seq, we randomly chose six different genes from the total genes that are up-regulated exclusively in the 35S:BBX31 lines and performed qPCR in seedlings grown under similar conditions. All six genes showed up-regulated expression in our qPCR results as well (Supplemental Fig. S6B), most of them showing a similar extent of up-regulation to that in the RNA Seq results (Supplemental Fig. S6B; Supplemental Data Set S1).
Thereafter, we asked if BBX31 depends upon HY5 for regulating photomorphogenesis in UV-B light. To understand the genetic relationship between HY5 and BBX31 in UV-B photomorphogenesis, we examined the hypocotyl lengths of hy5-215 bbx31-1 and hy5/35S:BBX31 seedlings when grown under UV-B light supplemented with low WLc. The percentage reduction of hypocotyl length caused by UV-B light in the hy5-215 bbx31-1 mutant was greater than the reduction observed in the individual single mutants, indicating their additive effect in the double mutant (Fig. 4, D and E). The UV-induced hypocotyl reduction observed in the hy5/35S:BBX31 line was intermediate to the reduction seen in the hy5 and 35S:BBX31#1 plants (Fig. 4, D and E). This intermediate reduction again indicated an additive effect of the phenotypes shown by hy5 and 35S:BBX31#1 plants, suggesting that the regulation of photomorphogenesis by BBX31 and HY5 might be through independent mechanisms in response to UV-B radiation. Taken together, our results indicate that BBX31 regulates photomorphogenesis in a HY5-independent manner under both white and UV-B light.
BBX31 Promotes the Accumulation of UV-Protective Compounds under High Doses of UV-B Light
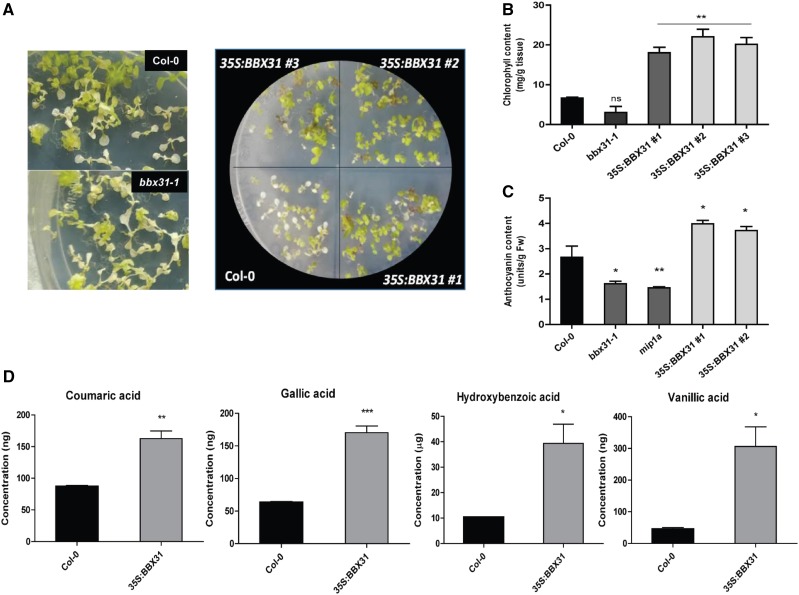
UV-B light initiates both developmental responses and stress responses, depending on its dose of exposure (Jenkins, 2014). BBX31 transcript accumulation increased with increasing intensity of UV-B light, ranging from a near-ambient intensity of 0.5 W m−2 that can activate the UVR8-photoreceptor pathway to a potentially stress-inducing intensity of 4.5 W m−2 (Brown et al., 2005; Fig. 1C). In order to investigate if BBX31 played any role in stress tolerance to UV-B radiation, we exposed seedlings to high doses of UV-B light by increasing the fluence and duration of exposure. Initially, we irradiated seedlings with 4.5 W m−2 UV-B light continuously for almost 3 weeks. We observed that at 19 d most of the Col-0 seedlings were bleached and looked white in color owing to chlorosis (Fig. 5, A and B). At the same time, the mutants showed greater extents of chlorosis, whereas 35S:BBX31 seedlings were predominantly green and just started showing the first signs of chlorotic development (Fig. 5, A and B). This was further validated by quantification of total chlorophyll levels in Col-0, bbx31-1, and 35S:BBX31#1 seedlings (Fig. 5B). The chlorophyll content in the overexpression lines was almost 3 times higher compared with wild-type seedlings (Fig. 5B). Even though the bbx31-1 mutant showed slightly reduced accumulation of chlorophyll when compared with the wild type, this difference was found to be statistically nonsignificant (Fig. 5B). To verify whether it is a UV-specific response or a general increase in stress tolerance capacity, the seedlings were also exposed to high light and osmotic stress conditions. However, we did not find a significant difference between the control and the BBX31 overexpression lines under these conditions, indicating that BBX31 specifically provides tolerance against high doses of UV-B irradiation (Supplemental Fig. S7).
Figure 5.
BBX31 promotes the accumulation of UV protective compounds under high doses of UV-B light. A, Representative images of Col-0 and bbx31-1 seedlings as well as Col-0 and three overexpression lines of BBX31 (35S:BBX31#1, 35S:BBX31#2, and 35S:BBX31#3) grown in WL (long day, 16 h/8 h) for 5 d and then given continuous UV-B light (4.5 W m−2) for 19 d. B, Total chlorophyll content of the seedlings shown in A. C, Anthocyanin quantification for 5-d-old seedlings grown in WLc (10 μmol m−2 s−1) supplemented with UV-B light (1.4 W m−2). Fw, Fresh weight. D, Quantification of the phenolic compounds coumaric acid, gallic acid, hydroxybenzoic acid, and vanillic acid extracted from 2-week-old Col-0 and 35S:BBX31 seedlings treated with continuous UV-B light (4.5 W m−2) for 24 h and analyzed using GC-MS. For B to D, three replicates were taken for quantification analysis, and all of them showed similar data. Error bars represent sd of three replicates. Unpaired Student’s t test was performed to check significance. Asterisks represent statistically significant differences (***, P < 0.001; **, P < 0.01; and *, P < 0.05) as determined by Student’s t test; ns, nonsignificant.
Since anthocyanin is known to act as a photoprotectant under UV-B radiation, we quantified the levels of anthocyanin in the wild type, mutants, and overexpression lines. The anthocyanin levels were significantly reduced in the mutants, whereas the levels were elevated in the overexpression lines when compared with Col-0 (Fig. 5C). The acclimation response to UV-B radiation in plants involves the synthesis and accumulation of several phenolic compounds in the epidermal tissues that act as a natural sunscreen (Caldwell et al., 1983; Rozema et al., 1997). In order to determine if the enhanced tolerance to high doses of UV-B light in the 35S:BBX31 lines could be correlated with the accumulation of any phenolic compounds, we extracted free phenol, ester-bound phenol, and glycoside-bound phenolic compounds from seedlings treated with 4.5 W m−2 for 24 h and analyzed them using gas chromatography-mass spectrometry (GC-MS). Coumaric acid, gallic acid, hydroxybenzoic acid, and vanillic acid showed significant enrichment in the 35S:BBX31 sample compared with the wild type (Fig. 5D; Supplemental Fig. S8). In conclusion, these data indicate that under high doses of UV-B radiation, BBX31 promotes the accumulation of UV-protective pigments and phenolic compounds.
BBX31 Enhances Tolerance to UV-B Light in a HY5-Dependent Manner by Inducing the Expression of Photoprotection and DNA Repair Genes
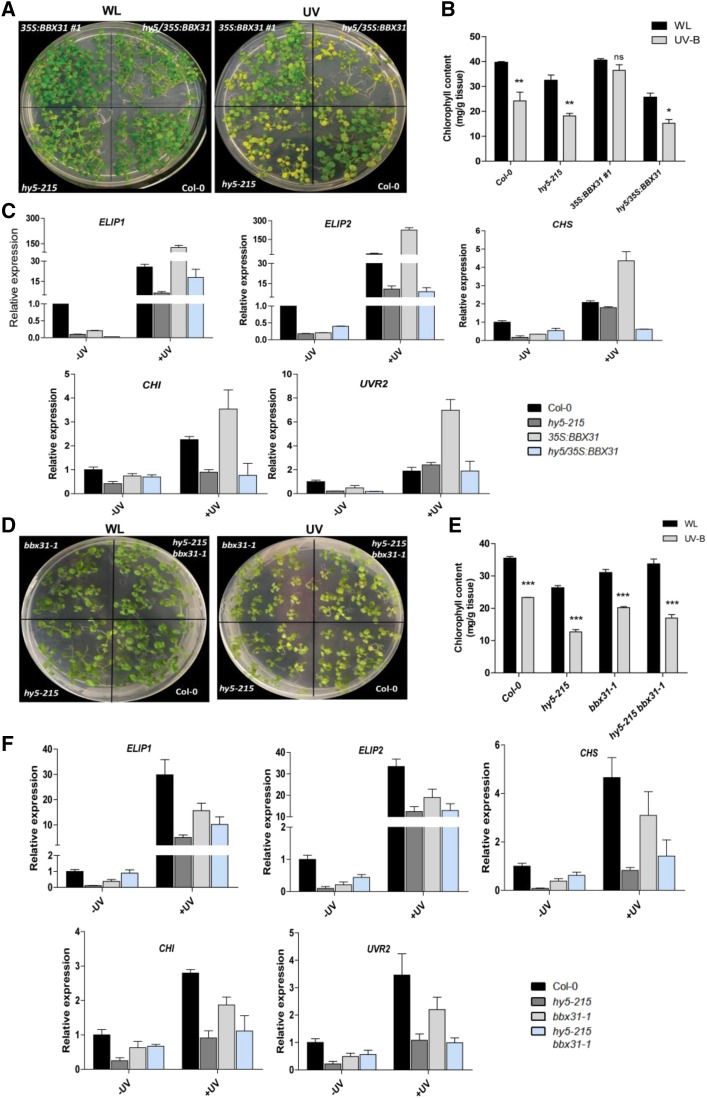
The regulation of photomorphogenic response by BBX31 under UV-B light is HY5 independent (Fig. 4D). Since the pathway regulating UV-B light tolerance could be largely different from the canonical photomorphogenesis pathway, and HY5 was previously reported to also be involved in tolerance to UV-B light (Brown et al., 2005), we asked if HY5 plays any role in BBX31-mediated UV-B tolerance. To understand the involvement of HY5 in this process, we performed a recovery assay with the wild type, 35S:BBX31#1, hy5, and hy5/35S:BBX31 lines, where seedlings were grown under long-day WL (90 μmol m−2 s−1) conditions for 7 d followed by UV-B treatment (1.4 W m−2) for 24 h and then returning back to WL for 4 d before monitoring the recovery. A control plate was kept only in WL for the entire duration (Fig. 6A). We observed that the 35S:BBX31 lines showed less chlorosis compared with the wild-type seedlings, indicating better recovery after UV-B treatment (Fig. 6, A and B). While there was no significant difference between the chlorophyll content of Col-0 and 35S:BBX31#1 seedlings grown in WL, the total chlorophyll content was higher in the 35S:BBX31#1 lines compared with Col-0 after UV treatment (Fig. 6, A and B). Under similar conditions, the hy5 seedlings exhibited a pale phenotype with chlorophyll levels lower than the wild type (Fig. 6, A and B). Chlorophyll accumulation in hy5/35S:BBX31 seedlings was similar to that in the hy5 mutant, suggesting the requirement of HY5 for the recovery after UV-B treatment in the 35S:BBX31 lines (Fig. 6, A and B).
Figure 6.
BBX31 promotes UV-B tolerance in a HY5-dependent manner. A, Recovery assay with Col-0, hy5, 35S:BBX31, and hy5/35S:BBX31 lines. Seedlings were grown for 7 d under WL and then irradiated for 24 h with UV-B light (1.4 W m−2). Treated seedlings were further grown for 4 d under WL (long day, 16 h/8 h) conditions without UV-B light to induce recovery before taking the image. B, Total chlorophyll content in the indicated genotypes measured in WL and after the UV-B treatment as in A. C, Transcript levels of ELIP1, ELIP2, CHS, CHI, and UVR2 in the indicated genotypes in WL and after 1 h of UV-B treatment. The fold changes in all other genotypes were calculated relative to Col-0 in WL. D, Recovery assay with Col-0, hy5-215, bbx31-1, and hy5-215 bbx31-1 lines (same experimental conditions as in A). E, Total chlorophyll content of the indicated genotypes shown in D. F, Transcript levels of ELIP1, ELIP2, CHS, CHI, and UVR2 in the indicated genotypes in WL and after 1 h of UV-B treatment. The fold changes in all other genotypes were calculated relative to Col-0 in WL. Asterisks in B and E represent statistically significant differences (***, P < 0.001; **, P < 0.01; and *, P < 0.05) as determined by two-way ANOVA followed by Bonferroni posttests; ns, nonsignificant. Error bars in all graphs represent sd of three replicates.
Previous microarray studies have identified several genes that are induced by UV-B light in a HY5-dependent manner (Brown et al., 2005; Oravecz et al., 2006; Brown and Jenkins, 2008). We selected five such genes to verify if the activation of the tolerance response in the 35S:BBX31 lines is HY5 dependent. Two of these genes encode chloroplast proteins, EARLY LIGHT INDUCED PROTEIN1 (ELIP1) and ELIP2, involved in protection against photoactive damage. Genes encoding the enzymes CHALCONE SYNTHASE (CHS) and CHALCONE ISOMERASE (CHI), involved in the rate-limiting biosynthetic step of anthocyanin and other flavonoids and directly regulated by HY5, were also chosen. In addition, we looked at the transcriptional levels of UVR2, which codes for a type II CPD photolyase involved in the DNA repair response. Analyzing the expression of these genes in WL indicated that they were expressed at reduced basal levels in all the lines, hy5, 35S:BBX31#1, and hy5/35S:BBX31, when compared with the wild type (Fig. 6C). After 1 h of UV-B treatment, the mRNA levels of all five genes failed to be up-regulated in the hy5 mutant, whereas they were up-regulated in the 35S:BBX31#1 line compared with the wild type (Fig. 6C). The mutation in HY5 suppressed the up-regulation of the mRNA levels of all five genes in the 35S:BBX31#1 line, indicating that the BBX31-mediated transcriptional regulation of these genes is HY5 dependent (Fig. 6C).
We also performed a recovery assay with the bbx31 mutant and the hy5-215 bbx31-1 double mutant. The chlorophyll content retained by the bbx31-1 mutant was less than that in the wild type but more than that in the hy5-215 mutant (Fig. 6, D and E). The double mutant hy5-215 bbx31-1 retained intermediate levels of chlorophyll when compared with hy5-215 and bbx31-1 single mutants (Fig. 6, D and E). This further confirmed that BBX31-mediated UV-B tolerance operates in a HY5-dependent manner. To this end, we analyzed the expression of ELIP1, ELIP2, CHS1, CHI, and UVR2 in the mutant lines (Fig. 6F). Under UV-B light, the expression of all the genes was down-regulated in the bbx31-1 mutant, whereas their levels were either similar to those in the hy5 mutant or intermediate to those in hy5 and bbx31 in the double mutant (Fig. 6F). In conclusion, our results indicate that BBX31 enhances tolerance to UV-B radiation in a HY5-dependent manner by inducing the expression of photoprotection and DNA repair genes.
BBX31 Enhances HY5 Transcript Levels under UV-B Light in a UVR8-Dependent Manner
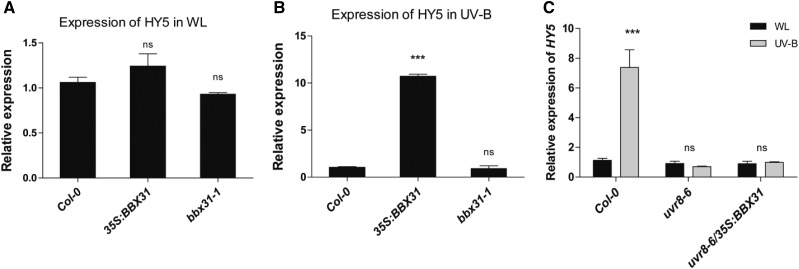
Our data indicate that HY5 binds to the BBX31 promoter and enhances BBX31 transcript levels under UV-B light (Fig. 2, B and C). The two proteins, BBX31 and HY5, function independently in the regulation of photomorphogenesis (Fig. 4). However the suppression of the UV-B tolerance phenotype of the 35S:BBX31#1 line by introduction of the hy5 mutation (Fig. 6) suggests that HY5 might be acting downstream of BBX31 to regulate some UV-B responses. Since BBX proteins are known to act directly or indirectly as transcriptional regulators, we asked if BBX31 can regulate HY5 mRNA levels. We checked the transcript levels of HY5 in both the overexpressor and the mutant of BBX31 under WL and UV-B light. While in WL there were negligible changes, under UV-B light there was almost a 10-fold up-regulation in the HY5 mRNA levels in the 35S:BBX31 lines, indicating that BBX31 might regulate HY5 transcript levels under UV-B light (Fig. 7, A and B). We however did not detect any significant change in HY5 expression in the bbx31-1 mutant (Fig. 7, A and B). Also, we could not find any difference in the accumulation of HY5 protein in WL-grown Col-0 and 35S:BBX31 seedlings in our western-blot experiment (Supplemental Fig. S9B). Since BBX31 lacks a DNA-binding domain and the increased expression of HY5 in UV-B light is mediated by UVR8, we asked if BBX31 depends upon UVR8 to transcriptionally regulate HY5 in UV light. To this end, we analyzed the expression of HY5 in uvr8-6/35S:BBX31 lines. Indeed, the loss of UVR8 abrogated the induction of HY5 in the BBX31 overexpression lines, suggesting that under UV-B light, BBX31 might regulate HY5 in a UVR8-dependent manner (Fig. 7C). In conclusion, our results suggest that under UV-B light a reciprocal transcriptional feedback loop might operate between HY5 and BBX31 (Fig. 8).
Figure 7.
BBX31 up-regulates HY5 transcript levels under UV-B light in a UVR8-dependent manner. A and B, Relative transcript levels of HY5 in 35S:BBX31#1 and bbx31-1 lines in WL (long day, 16 h/8 h; A) and after 1 h of UV-B irradiation (1.4 W m−2) treatment (B). C, Relative transcript levels of HY5 in uvr8-6 and uvr8-6/35S:BBX31 lines in WL (long day, 16 h/8 h) and after 1 h of UV-B irradiation (1.4 W m−2) treatment. Asterisks indicate statistically significant differences (***, P < 0.001) as determined by Student’s t test; ns, nonsignificant. Error bars represent sd of at least two replicates.
Figure 8.
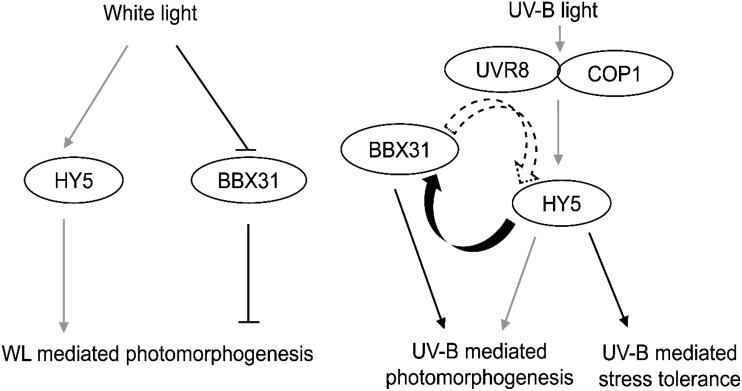
Model showing the distinct roles of BBX31 and HY5 in white and UV-B light to regulate photomorphogenesis and UV protection. BBX31 and HY5 regulate photomorphogenesis oppositely and independently in WL. Under UV-B light, HY5 enhances BBX31 transcription by directly binding to its promoter. Reciprocally, overexpression of BBX31 enhances HY5 transcript levels in a UVR8-dependent manner. The two proteins independently promote photomorphogenesis under UV-B light, whereas BBX31 regulates UV stress tolerance in a HY5-dependent manner. Black lines indicate connections identified in this study, while gray lines represent previously known pathways. Curly solid and dotted arrows indicate proven and possible transcriptional regulation, respectively.
DISCUSSION
BBX31, a Negative Regulator of Photomorphogenesis
Photomorphogenesis is marked by suppression of hypocotyl growth and opening and greening of cotyledons. Light signals perceived by the photoreceptors are transduced downstream through various signal transduction cascades, involving large transcriptional changes of genes that regulate the photomorphogenic response. Many members of the BBX family in Arabidopsis regulate photomorphogenesis both positively and negatively. BBX4, BBX20, BBX21, BBX22, and BBX23 positively regulate photomorphogenesis, whereas BBX18, BBX19, BBX24, BBX25, BBX28, and BBX32 play negative regulatory roles (Datta et al., 2006, 2007, 2008; Indorf et al., 2007; Chang et al., 2008; Holtan et al., 2011; Fan et al., 2012; Gangappa et al., 2013a; Zhang et al., 2017; Lin et al., 2018). In our study, we established a member of the BBX family, BBX31, as a negative regulator of photomorphogenesis. We have shown that its close paralog BBX30 also negatively regulates photomorphogenesis (Supplemental Fig. S3D). BBX30 and BBX31 are members of structural group V. In addition to BBX30 and BBX31, group V members such as BBX28 and BBX32 also regulate photomorphogenesis. Interestingly, all four, BBX28, BBX30, BBX31, and BBX32, are negative regulators of photomorphogenesis. In addition to their role in photomorphogenesis, BBX31 (along with BBX30) and BBX32 also act as repressors of flowering (Graeff et al., 2016; Tripathi et al., 2017). Despite these commonalities, the mechanisms underlying their function are largely distinct. BBX28 physically interacts with HY5, the central positive regulator of light signaling, and interferes with its binding on the target promoters (Lin et al., 2018). BBX32 suppresses photomorphogenesis by indirectly regulating HY5 through physically interacting with and inhibiting BBX21, a positive regulator of HY5 (Holtan et al., 2011; Xu et al., 2016). Multilayered regulation of HY5 by the same structural group members have also been found in the case of BBX21 and BBX24, both belonging to structural group IV, even though they regulate HY5 in the opposite manner (Job et al., 2018; Yadukrishnan et al., 2018). However, our data indicate that, unlike BBX28 and BBX32, the regulation of photomorphogenesis by BBX31 is in a HY5-independent manner (Fig. 4, A and B). Our observations indicate that a novel mechanism might exist by which BBX31 suppresses photomorphogenesis.
BBX31 Is a Novel Signaling Intermediate in the UV-B Signal Transduction Pathway
The signaling pathway by which low-dose UV-B light regulates developmental responses in plants is not well studied. Several proteins, such as COP1, HY5, REPRESSOR OF UV-B PHOTOMORPHOGENESIS, BBX24, PRODUCTION OF FLAVONOL GLYCOSIDE1 (PFG1)/MYB, and WRKY36, have been identified to act as intermediates downstream of the photoreceptor UVR8 (Jiang et al., 2012; Tilbrook et al., 2013; Yang et al., 2018). Microarray data from previous studies have identified numerous UV-responsive transcription factors downstream of HY5 and PFG1/MYB (Stracke et al., 2010). However, the exact physiological role of these proteins, especially under UV-B light, remains largely uncharacterized. The presence of a UV-box core cis-element in the putative promoter sequence, which was found to be enriched in the promoters of a subset of UV-inducible genes, and a UV-B-induced expression as observed in previously available microarray data and our own experiments prompted us to check if BBX31 plays any role in UV-B signaling. Our data indicate that BBX31 is a positive regulator of UV-B-mediated photomorphogenesis, suggested primarily by the higher extent of UV-induced reduction in the hypocotyl length of BBX31 overexpression lines (Fig. 3, A, C, and D). Mutants and overexpressors of BBX31 showed strong photomorphogenic phenotypes in WL conditions (Fig. 2, A and B). Hence, it is important to consider the possibility that the percentage reduction shown by these lines could be severely influenced by their strong phenotypes in WL. However, when we estimated the reduction in the hypocotyl lengths of the wild type and overexpression lines upon growing in low and high WL, we could not find a higher reduction in the overexpression lines (Supplemental Fig. S3C). This suggests that the UV-B phenotype shown by the BBX31 overexpression lines is potentially independent from its strong WL phenotype.
BBX31 is only the second BBX family member after BBX24 characterized for its role in UV-B signaling. BBX24 negatively regulates photomorphogenesis both under WL and UV-B light (Jiang et al., 2012; Gangappa et al., 2013a, 2013b). Unlike BBX24, BBX31 suppresses photomorphogenesis under WL, whereas it promotes photomorphogenesis in UV-B light. So far, COP1 is the only other known component that regulates photomorphogenesis oppositely in WL and UV-B light (Huang et al., 2014). Along with its regulatory role in photomorphogenesis under low-dose UV-B light, we found that BBX31 also controls the tolerance against genotoxic stress caused by exposure to a high dose of UV-B radiation. BBX31 promotes this tolerance through (1) increasing the levels of UV protective compounds and (2) regulating the expression of genes related to photoprotection and DNA repair.
Transcriptional Regulation of BBX31 by HY5
Our data suggest that HY5 binds to the promoter of BBX31 and activates its transcription in the presence of UV-B light (Fig. 2). Besides BBX22, this is the only member that we know of in the entire BBX family for which HY5 binds on the promoter and directly regulates the transcription (Fig. 2; Gangappa et al., 2013a). HY5 constitutively binds to the BBX31 promoter both under WL as well as UV-B conditions, but the transcriptional regulation happens only in a UV-specific manner. This necessitates that HY5 depends upon some other UV-specific regulatory factor, which might help HY5 to transcriptionally activate BBX31. Turning on or unblocking HY5 at a stage after binding onto its target promoter adds another layer of regulation of HY5 activity, which has not been commonly identified before. In the presence of UV-B light, the photoreceptor UVR8 upon nuclear localization mediates the enriched diacetylation at Lys-9 and Lys-14 of histone H3 in their promoter regions of a number of genes, including BBX31 (Velanis et al., 2016). It could be possible that the UV-B-specific chromatin remodeling mediated by UVR8 is necessary for HY5 to transcriptionally activate BBX31. Since no physical interaction between UVR8 and HY5 has been observed (Velanis et al., 2016), another possibility is that UVR8 recruits other factors that might associate with HY5 to activate BBX31 transcription.
Regulation of HY5 Transcript Levels by BBX31 under UV-B Light
HY5 acts as a key intermediate in the WL and UV-B signaling pathways. HY5 is transcriptionally regulated by a number of transcription factors under WL, including HY5 itself, HYH, CAM7, BBX20, BBX21, PIF1, and PIF3 (Abbas et al., 2014; Binkert et al., 2014; Wei et al., 2016; Xu et al., 2016; Zhang et al., 2017). However, our knowledge of the regulation of HY5 transcription in UV-B light is extremely limited. WRKY36 inhibits HY5 transcription by binding to its promoter in the absence of UV-B light. However, in the presence of UV-B light, UVR8 sequesters WRKY36, thereby allowing HY5 transcription to proceed (Yang et al., 2018). Other than this regulation through the release of suppression and the HY5/HYH self-regulatory module (Binkert et al., 2014), no other positive regulators of HY5 transcription in UV-B light have been identified so far. We report the positive regulation of HY5 transcription in UV conditions by overexpression of BBX31, suggesting that BBX31 might regulate HY5 transcription. Thus, it might be possible that under UV-B light, BBX31 and HY5 reciprocally regulate each other’s expression, establishing a positive feedback loop (Fig. 8). A reciprocal regulatory loop has also been reported in BBX20 and HY5, but in the form of a negative feedback loop (Wei et al., 2016). BBX31 is a microProtein, and microProteins are known to physically interact and form inactive complexes with their target proteins. Hence, there exists a possibility that BBX31, along with its paralogous microProtein BBX30, sequesters a repressor of HY5 to positively regulate HY5 function. UVR8 interacts with and sequesters WRKY36 to sustain the expression of HY5 in UV-B light (Yang et al., 2018). We found that BBX31 enhances HY5 transcript levels in a UVR8-dependent manner. It is tempting to speculate that the microProtein BBX31 might form a complex with UVR8 for the sequestration of WRKY36 specifically under UV-B light, thus enabling HY5 transcription. Mechanistic studies in the future might reveal more about BBX31-mediated regulation of UV-B signaling.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All the mutants and the transgenic lines used in this study are in the background of the Col-0 ecotype of Arabidopsis (Arabidopsis thaliana). The mutant lines hy5-215, mip1a, mip1b, mip1ab, and uvr8-6 have been described in previous studies (Gangappa et al., 2013a; Graeff et al., 2016). Seeds were surface sterilized in 5% sodium hypochlorite and sown on 0.8% (w/v) agar plates containing 1× Murashige and Skoog (MS) medium with 1% (w/v) Suc (pH 5.7), MES (0.05%), and kept at 4°C for 48 h in darkness for stratification and then transferred to a Percival LED22C8 growth cabinet at 22°C and 70% humidity with respective light conditions. For hypocotyl length measurements, one-half-strength MS medium without Suc was used. For dark experiments, seedlings were given a light pulse of 3 h, wrapped completely in aluminum foil, and kept in darkness. For WL experiments, seedlings were grown in WLc of fluence rate 10 μmol m−2 s−1 (low WLc) and 90 μmol m−2 s−1 (high WLc) in a Percival LED22C8 growth chamber and light intensity was measured with Apogee Quantum meter MQ-200. UV light was provided through the UV-B narrowband tube (TL 20W/01 RS SLV; waveband between 305 and 315 and λmax 311 nm). UV-B light intensity was measured by a UV light meter (UVA/UVB 850009; Sper Scientific). The intensities used in different experiments varied between 0.5, 1.4, and 4.5 W m−2 depending on the dose of UV-B required for the experiment.
Vector Construction
To generate the overexpression lines, full-length BBX31 coding sequence (CDS) was amplified from Arabidopsis cDNA using AttB1-BBX31F and AttB2-BBX31R primers and cloned into pDONR207 via BP Gateway system reaction (Invitrogen). The BBX31 construct in the pDONR207 vector was then shuttled to the Gateway-compatible plant binary vector pCAMBIA1300 through an LR reaction to generate a BBX31 CDS overexpression construct driven by the Cauliflower mosaic virus 35S promoter. To generate the pGEX4T1-HY5 construct for HY5 protein purification, full-length HY5 fragment was amplified by PCR with the respective pairs of primers and cloned into the EcoRI/XhoI sites of the pGEX4T1 vector. All primer sequences used for the cloning of these constructs are listed in Supplemental Table S1.
Generation of Overexpressor and CRISPR Mutant Lines
The BBX31 CDS overexpression construct was transformed into Arabidopsis plants using the Agrobacterium tumefaciens (GV3101)-mediated floral dip method (Clough and Bent, 1998; Zhang et al., 2006). Along with Col-0, overexpression lines in the hy5-215 mutant background were generated in a similar way. Transgenic plants (T1) were screened on plates containing MS medium supplemented with hygromycin. Multiple overexpressor lines with a single insertion as determined by segregation analysis on hygromycin-containing plates were selected, and homozygous transgenic T3 plants were generated for further studies. The bbx31-1 and bbx30-1 mutants were generated using the CRISPR/Cas9 vector pKI1.1R, and specific single-guide RNAs (sgRNAs) were generated according to the manual (Tsutsui and Higashiyama, 2017). The sgRNAs were designed using the Web tool CRISPR-P (Lei et al., 2014). To achieve mutants with a large deletion in the coding region, two A. tumefaciens strains carrying individual pKI1.1R vectors, containing different gene-specific sgRNAs, were pooled and transformed into wild-type Col-0 using the floral dip method. The red fluorescence was used for selection of transgenic seeds in the T1 generation, and PCR was used to screen for large deletions in the target genes. In the T2 generation homozygote, transgene-free mutant plants were isolated.
Anthocyanin and Hypocotyl Measurement
For anthocyanin estimation, 5-d-old seedlings grown in WLc (10 μmol m−2 s−1) or WL supplemented with UV-B (1.4 W m−2) were used. Anthocyanin was extracted from both UV-B-treated and WL-grown seedlings in 1 mL of 1% (v/v) HCl in methanol. Samples were then incubated at 4°C overnight, centrifuged at 12,000g for 10 min, and OD was taken at 530 and 657 nm. Anthocyanin content in units per gram fresh weight was calculated using the equation (A530-0.25*A657)/(tissue weight in grams).
For all photomorphogenesis experiments, 4-d-old seedlings were used for the hypocotyl length measurements and images were taken with a microscope (Leica M165 FC). The measurements were carried out using ImageJ software (http://imagej.nih.gov/ij/index.html).
Stress Experiment and Chlorophyll Estimation
For UV-B stress experiments, seedlings were grown for 5 d in WL diurnal conditions and then exposed to continuous UV-B (4.5 W m−2) for 19 d. Images were taken and total chlorophyll was estimated. For recovery assays, seedlings were grown for 7 d under WL diurnal conditions. Seedlings were then irradiated for 24 h with UV-B (1.4 W m−2) or subjected to 24-h WL conditions in the control. Treated seedlings were further grown for 4 d under standard conditions without UV-B before the image was taken, and chlorophylls were estimated. For chlorophyll estimation, seedlings were weighed and crushed in dimethyl sulfoxide, then centrifuged for 10 min at 12000g. The absorbance was taken at 647 and 664 nm. Chlorophyll content was calculated using the formulae Chla = 12.7*A664 – 2.79*A647, Chlb = 20.7*A647 – 4.62*A664, and Chla + Chlb = 17.90*A647 + 8.08*A664.
RNA Extraction and qPCR
For RNA extraction, seedlings were frozen and homogenized in liquid nitrogen, and Trizol reagent (Invitrogen) was used to separate RNA. DNase I (New England Biolabs) treatment was used to ensure the removal of genomic DNA. cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed in a Roche light cycler machine using SYBR Green dye (Qiagen). Reference genes used were GAPDH and ACTIN for normalization. Averages of the results of three independently conducted experiments were represented in the graphs. All primer sequences used for reverse transcription-PCR analysis are listed in Supplemental Table S1.
ChIP
Chromatin isolation and immunoprecipitation was performed as described previously (Komar et al., 2016), and anti-HY5 antibody (Agrisera) was used at a dilution of 1:150. qPCR was done using SYBR Green from Qiagen according to the manufacturer’s instructions. Briefly, 14-d-old Col-0 or hy5 seedlings were grown in WLc conditions (90 μmol m−2 s−1) and WL + 3 h of UV-B light (1.4 W m−2). The seedlings were then cross-linked using 1% formaldehyde, and chromatin was isolated. The sonicated chromatin was immunoprecipitated, washed, reverse cross-linked, and finally amplified. Ten percent of the total sonicated chromatin was used as the input DNA control. The samples were amplified using the real-time PCR system (Lightcycler 96; Roche). qPCR data were analyzed using the percentage of input method (Haring et al., 2007). Primers are listed in Supplemental Table S1.
EMSA
Full-length HY5 CDS was cloned in pGEX4T1 vector (N-terminal GST tag). Protein was induced and expressed in Escherichia coli BL21 cells at 0.3 mm isopropyl β-d-1-thiogalactopyranoside. HY5 protein was purified using Glutathione Agarose Resins (ABT). EMSA was performed using biotin-labeled probes and the light-shift chemiluminescent EMSA kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, 50 bp of synthesized single-stranded oligonucleotides was biotinylated (Pierce Biotin 3′ End DNA Labeling Kit), and complementary strands were annealed to obtain a double-stranded probe (for a detailed list of primers, see Supplemental Table S1). Biotinylated probes (20 fmol) were incubated with 800 ng (1×) of purified GST and GST-HY5 in the presence of 10 mm Tris-HCl, pH 7.5, 1 mm DTT, 150 mm KCl, 50 ng µL−1 poly(dI-dC), 2.5% glycerol, 0.05% Nonidet P-40, 0.1 mg mL−1 BSA, and 100 mm MgCl2 for 20 min at room temperature and separated on 6% (v/v) native polyacrylamide gels in 0.5× Tris-borate/EDTA. The gels were electroblotted onto a positively charged nylon membrane (Amersham Hybond-N+; GE Healthcare Life Sciences) in 0.5× Tris-borate/EDTA for 50 min followed by 20 min of UV-B cross-linking at 302 nm using a Transilluminator. Labeled probes were detected with Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific) following the manufacturer’s instructions.
RNA Seq
From 6-d-old seedlings grown in continuous low-fluence WL (10 μmol m−2 s−1), RNA was isolated using the Trizol (Invitrogen) method. Five to 10 µg of the RNA sample was provided to Agrigenome Labs for further processing and analysis. Briefly, the RNA was used to prepare an RNA Seq library using the True Seq RNA sample prep kit (Illumina). After the libraries were constructed, a paired-end run was performed on Illumina’s HiSeq 2000/2500/4000 platform to obtain 2 × 100-bp reads. Following the sequencing run, a quality check test and reference genome-based RNA Seq analysis was performed. The raw reads after cleaning were aligned to the TAIR10 reference genome. Genes with more than 2-fold change and P < 0.05 were considered as differentially expressed. All the differentially expressed genes are listed in Supplemental Data Sets S1 and S2.
Sample Preparation and Extraction of Phenols
For detection and quantification of free and wall-bound phenols through GC-MS, the following protocol (de Ascensao and Dubery, 2003) was followed. Lyophilized Arabidopsis tissue (20 mg dry weight) was extracted in 1 mL of 80% methanol (HPLC grade) boiled at 70°C in a thermomixer (Eppendorf ThermoMixer C) at 950 rpm for 5 min. The extract was centrifuged at 12,000g for 10 min. From the supernatant, a 250-µL aliquot was dried and subjected to the estimation of free phenols using GC-MS. A second aliquot was acid hydrolyzed to extract the glycoside-bound phenolics, and a third aliquot was alkali hydrolyzed to extract the ester-bound phenols (Cvikrová et al., 1993; de Ascensao and Dubery, 2003). The pellet was oven dried and subjected to different saponification steps followed by alkaline hydrolysis (Campbell and Ellis, 1992). After hydrolysis, 250 µL of methyl tert-butyl ether (Sigma-Aldrich) was added, and the upper polar layer containing phenols was collected and then evaporated under a gentle stream of nitrogen to concentrate the sample for further analysis using GC-MS.
GC-MS Analysis of Phenols
The extracted phenols were subjected to GC-MS. The analysis was performed on a GC-MS apparatus (GC ALS-MS 5977B; Agilent Technologies) equipped with an HP-5MS (5% phenyl methyl siloxane) column (30 m × 250 μm × 0.25 μm). The initial oven program was set to 50°C, then it was raised to 70°C with a 5-min hold time. The ramp rate was set for 10°C min−1 and 5°C min−1 with a hold time of 10 min to achieve 200°C and 300°C, respectively. All samples and standards were methoximation-trimethylsilylation derivatized (Masakapalli et al., 2014) using pyridine, methoxamine hydrochloride, and N-methyl-N-trimethylsilyl trifluoroacetamide (Sigma-Aldrich). Standard curves of phenol standards (0.1 mg mL−1 stock), cinnamic acid, p-hydroxybenzoic acid, vanillin, vanillic acid, p-coumaric acid, gallic acid, ferulic acid, and cis-caffeic acid, were used for quantification. Data were acquired for all samples, and raw files were preprocessed using metAlign software (Lommen and Kools, 2012) for baseline correction of spectra. Qualitative analysis navigator software Mass Hunter (Agilent Technologies) was used for all data analyses. The peaks in the spectra were analyzed based on the mass-to-charge ratio fragments, retention time, and highest probability hit against the library NIST 17 and the Fiehn Metabolomics library (Kopka et al., 2005). A targeted approach was followed for quantification of each detected phenol from the Extracted Ion Chromatogram. In total, three replicates were taken for quantification analysis, and all of them showed reproducible data. Unpaired Student’s t tests were performed to check significance or nonsignificance of data using GraphPad Prism 5.0.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software). To determine statistical significance, one-way ANOVA followed by Dunnett’s posthoc or Tukey’s posthoc test (P ≤ 0.05) or two-way ANOVA followed by Bonferroni’s posthoc test was performed.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: BBX31, AT3G21890; HY5, AT5G11260; CHS, AT5G13930; CHI, AT3G55120; UVR8, AT5G63860; ELIP1, AT3G22840; ELIP2, AT4G14690; and UVR2, AT1G12370.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Lack of enrichment of HY5 on nontarget chromatin fragments.
Supplemental Figure S2. Mutations in the CRISPR mutants bbx31-1 and bbx30-1 and analysis of BBX31 expression.
Supplemental Figure S3. BBX31 is a negative regulator of photomorphogenesis.
Supplemental Figure S4. hy5/35S:BBX31 seedlings exhibit constitutively elongated hypocotyls.
Supplemental Figure S5. HY5 and BBX31 do not interact.
Supplemental Figure S6. Gene Ontology analysis and qPCR validation of RNA Seq data.
Supplemental Figure S7. Phenotype of 35S:BBX31 seedlings grown under high light and osmotic stress conditions.
Supplemental Figure S8. GC-MS analysis of phenols accumulating in the 35S:BBX31 line after a high dose of UV-B exposure.
Supplemental Figure S9. Transcript and protein abundance of HY5 in the WL-grown 35S:BBX31 line and expression levels of BBX31 in UV-treated hy5 seedlings.
Supplemental Table S1.. List of primers used in this study.
Supplemental Data Set S1. List of genes up-regulated in hy5, 35S:BBX31, and hy5/35S:BBX31 plants compared with Col-0 grown under low WLc (10 μmol m−2 s−1).
Supplemental Data Set S2. List of genes down-regulated in hy5, 35S:BBX31, and hy5/35S:BBX31 plants compared with Col-0 grown under low WLc (10 μmol m−2 s−1).
Acknowledgments
We thank Roman Ulm (University of Geneva) for providing uvr8-6 seeds and Dr. Henrik Johansson (Freie Universität Berlin) for critical comments on the article. We thank lab members Nikhil Job, Nevedha Ravindran, and Rahul PV at IISER Bhopal for their help in EMSA, protein-protein interaction studies, and figure preparation, respectively. We also acknowledge the facilities provided by IISER Bhopal.
Footnotes
This work was supported by the Department of Biotechnology, Ministry of Science and Technology (Ramalingaswami Fellowship, IYBA and DBT [BT/PR19193/BPA/118/195/2016]), and the DST, Science and Engineering Research Board (EMR/2016/000181), Government of India. A.Y., P.Y., and M.L. acknowledge UGC, DBT, and MHRD-HTRA, Government of India, respectively, for their Ph.D. fellowships. S.B. acknowledges DBT-BIOCARE Women Scientist Program for funding. S.K.M. and M.L. acknowledge Science and Engineering Research Board early career research funding (File No: ECR/2016/001176). The S.W. laboratory received funding from the European Research Council (ERC-StG miPDesign, 336295), the Independent Research Fund Denmark (DFF-FNU, Adaptogenomics), and the NovoNordisk Foundation (NNF18OC0034226).
Articles can be viewed without a subscription.
References
- Abbas N, Maurya JP, Senapati D, Gangappa SN, Chattopadhyay S (2014) Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 26: 1036–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkert M, Kozma-Bognár L, Terecskei K, De Veylder L, Nagy F, Ulm R (2014) UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26: 4200–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Jenkins GI (2008) UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol 146: 576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102: 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell MM, Robberecht R, Flint SD (1983) Internal filters: Prospects for UV-acclimation in higher plants. Physiol Plant 58: 445–450 [Google Scholar]
- Campbell MM, Ellis BE (1992) Fungal elicitor-mediated responses in pine cell cultures: Cell wall-bound phenolics. Phytochemistry 31: 737–742 [Google Scholar]
- Chang CS, Li YH, Chen LT, Chen WC, Hsieh WP, Shin J, Jane WN, Chou SJ, Choi G, Hu JM, et al. (2008) LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J 54: 205–219 [DOI] [PubMed] [Google Scholar]
- Cheng XF, Wang ZY (2005) Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J 43: 758–768 [DOI] [PubMed] [Google Scholar]
- Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, et al. (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cvikrová M, Nedělník J, Eder J, Binarová P (1993) Changes in pattern of phenolic acids induced by culture filtrate of Fusarium oxysporum in alfalfa plants differing in susceptibility to the pathogen. J Plant Physiol 142: 1–5 [Google Scholar]
- Datta S, Hettiarachchi GH, Deng XW, Holm M (2006) Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M (2007) SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M (2008) LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ascensao AR, Dubery IA (2003) Soluble and wall-bound phenolics and phenolic polymers in Musa acuminata roots exposed to elicitors from Fusarium oxysporum f.sp. cubense. Phytochemistry 63: 679–686 [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH (1992) COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801 [DOI] [PubMed] [Google Scholar]
- Eguen T, Straub D, Graeff M, Wenkel S (2015) MicroProteins: Small size-big impact. Trends Plant Sci 20: 477–482 [DOI] [PubMed] [Google Scholar]
- Fan XY, Sun Y, Cao DM, Bai MY, Luo XM, Yang HJ, Wei CQ, Zhu SW, Sun Y, Chong K, et al. (2012) BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol Plant 5: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C. (2001) Photomorphogenesis in plants, genetics of. In Brenner S, Miller JH, eds, Encyclopedia of Genetics. Academic Press, New York, pp 1454–1458 [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay KM, Jenkins GI (2016) Regulation of UVR8 photoreceptor dimer/monomer photo-equilibrium in Arabidopsis plants grown under photoperiodic conditions. Plant Cell Environ 39: 1706–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2014) The BBX family of plant transcription factors. Trends Plant Sci 19: 460–470 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2016) The multifaceted roles of HY5 in plant growth and development. Mol Plant 9: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Crocco CD, Johansson H, Datta S, Hettiarachchi C, Holm M, Botto JF (2013a) The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25: 1243–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Holm M, Botto JF (2013b) Molecular interactions of BBX24 and BBX25 with HYH, HY5 HOMOLOG, to modulate Arabidopsis seedling development. Plant Signal Behav 8: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeff M, Straub D, Eguen T, Dolde U, Rodrigues V, Brandt R, Wenkel S (2016) Microprotein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. PLoS Genet 12: e1005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M (2007) Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassidim M, Harir Y, Yakir E, Kron I, Green RM (2009) Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 230: 481–491 [DOI] [PubMed] [Google Scholar]
- Hideg E, Jansen MA, Strid A (2013) UV-B exposure, ROS, and stress: Inseparable companions or loosely linked associates? Trends Plant Sci 18: 107–115 [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtan HE, Bandong S, Marion CM, Adam L, Tiwari S, Shen Y, Maloof JN, Maszle DR, Ohto MA, Preuss S, et al. (2011) BBX32, an Arabidopsis B-box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol 156: 2109–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Yang P, Ouyang X, Chen L, Deng XW (2014) Photoactivated UVR8-COP1 module determines photomorphogenic UV-B signaling output in Arabidopsis. PLoS Genet 10: e1004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indorf M, Cordero J, Neuhaus G, Rodríguez-Franco M (2007) Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J 51: 563–574 [DOI] [PubMed] [Google Scholar]
- Jenkins GI. (2014) The UV-B photoreceptor UVR8: From structure to physiology. Plant Cell 26: 21–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI. (2017) Photomorphogenic responses to ultraviolet-B light. Plant Cell Environ 40: 2544–2557 [DOI] [PubMed] [Google Scholar]
- Jiang L, Wang Y, Li QF, Björn LO, He JX, Li SS (2012) Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res 22: 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Zhang D, Wang X, Tang W, Wang W, Huai J, Xu G, Chen D, Li Y, Lin R (2013) Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job N, Yadukrishnan P, Bursch K, Datta S, Johansson H (2018) Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol 176: 2963–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiserli E, Jenkins GI (2007) UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lim JE, Landry LG, Last RL (2002) Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol 130: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar DN, Mouriz A, Jarillo JA, Piñeiro M (2016) Chromatin immunoprecipitation assay for the identification of Arabidopsis protein-DNA interactions in vivo. J Vis Exp 14: e53422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP (1980) Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol 100: 147–160 [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kumagai T, Ito S, Nakamichi N, Niwa Y, Murakami M, Yamashino T, Mizuno T (2008) The common function of a novel subfamily of B-box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci Biotechnol Biochem 72: 1539–1549 [DOI] [PubMed] [Google Scholar]
- Kushwaha R, Singh A, Chattopadhyay S (2008) Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20: 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL (2014) CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7: 1494–1496 [DOI] [PubMed] [Google Scholar]
- Li F, Sun J, Wang D, Bai S, Clarke AK, Holm M (2014) The B-box family gene STO (BBX24) in Arabidopsis thaliana regulates flowering time in different pathways. PLoS ONE 9: e87544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Jiang Y, Li J, Yan T, Fan L, Liang JS, Chen ZJ, Xu D, Deng XW (2018) B-BOX DOMAIN PROTEIN28 negatively regulates photomorphogenesis by repressing the activity of transcription factor HY5 and undergoes COP1-mediated degradation. Plant Cell 30: 2006–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommen A, Kools HJ (2012) MetAlign 3.0: Performance enhancement by efficient use of advances in computer hardware. Metabolomics 8: 719–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masakapalli SK, Bryant FM, Kruger NJ, Ratcliffe RG (2014) The metabolic flux phenotype of heterotrophic Arabidopsis cells reveals a flexible balance between the cytosolic and plastidic contributions to carbohydrate oxidation in response to phosphate limitation. Plant J 78: 964–977 [DOI] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adám E, Schäfer E, Nagy F, Ulm R (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Wei N, Deng XW (2000) The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Physiol 124: 1520–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Quail PH. (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Rozema J, van de Staaij J, Björn LO, Caldwell M (1997) UV-B as an environmental factor in plant life: Stress and regulation. Trends Ecol Evol 12: 22–28 [DOI] [PubMed] [Google Scholar]
- Safrany J, Haasz V, Mate Z, Ciolfi A, Feher B, Oravecz A, Stec A, Dallmann G, Morelli G, Ulm R, et al. (2008) Identification of a novel cis-regulatory element for UV-B-induced transcription in Arabidopsis. Plant J 54: 402–414 [DOI] [PubMed] [Google Scholar]
- Staudt AC, Wenkel S (2011) Regulation of protein function by ‘microProteins.’ EMBO Rep 12: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R (2010) The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ 33: 88–103 [DOI] [PubMed] [Google Scholar]
- Sullivan JA, Deng XW (2003) From seed to seed: The role of photoreceptors in Arabidopsis development. Dev Biol 260: 289–297 [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Newton L, Burkhardt A, Mason S, Farré EM (2014) Light and the circadian clock mediate time-specific changes in sensitivity to UV-B stress under light/dark cycles. J Exp Bot 65: 6003–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook K, Arongaus AB, Binkert M, Heijde M, Yin R, Ulm R (2013) The UVR8 UV-B photoreceptor: Perception, signaling and response. The Arabidopsis Book 11: e0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi P, Carvallo M, Hamilton EE, Preuss S, Kay SA (2017) Arabidopsis B-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc Natl Acad Sci USA 114: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H, Higashiyama T (2017) pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol 58: 46–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Máté Z, Adám E, Oakeley EJ, Schäfer E, Nagy F (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velanis CN, Herzyk P, Jenkins GI (2016) Regulation of transcription by the Arabidopsis UVR8 photoreceptor involves a specific histone modification. Plant Mol Biol 92: 425–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CQ, Guthrie C, Sarmast MK, Dehesh K (2014) BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 26: 3589–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CQ, Chien CW, Ai LF, Zhao J, Zhang Z, Li KH, Burlingame AL, Sun Y, Wang ZY (2016) The Arabidopsis B-box protein BZS1/BBX20 interacts with HY5 and mediates strigolactone regulation of photomorphogenesis. J Genet Genomics 43: 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Zhang J, Yang P, Deng H, Wang J, et al. (2012) Structural basis of ultraviolet-B perception by UVR8. Nature 484: 214–219 [DOI] [PubMed] [Google Scholar]
- Xu D, Jiang Y, Li J, Lin F, Holm M, Deng XW (2016) BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc Natl Acad Sci USA 113: 7655–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jiang Y, Li J, Holm M, Deng XW (2018) The B-box domain protein BBX21 promotes photomorphogenesis. Plant Physiol 176: 2365–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadukrishnan P, Job N, Johansson H, Datta S (2018) Opposite roles of group IV BBX proteins: Exploring missing links between structural and functional diversity. Plant Signal Behav 13: e1462641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liang T, Zhang L, Shao K, Gu X, Shang R, Shi N, Li X, Zhang P, Liu H (2018) UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat Plants 4: 98–107 [DOI] [PubMed] [Google Scholar]
- Yi C, Deng XW (2005) COP1: From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Yoshiyama KO, Sakaguchi K, Kimura S (2013) DNA damage response in plants: Conserved and variable response compared to animals. Biology (Basel) 2: 1338–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zhang X, Huai J, Shang F, Xu G, Tang W, Jing Y, Lin R (2017) A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol 174: 2487–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]