High-throughput single-cell RNA sequencing of Arabidopsis root protoplasts demonstrates the feasibility and utility of individual cell transcriptomic analyses in plants.
Abstract
Single-cell RNA sequencing (scRNA-seq) has been used extensively to study cell-specific gene expression in animals, but it has not been widely applied to plants. Here, we describe the use of a commercially available droplet-based microfluidics platform for high-throughput scRNA-seq to obtain single-cell transcriptomes from protoplasts of more than 10,000 Arabidopsis (Arabidopsis thaliana) root cells. We find that all major tissues and developmental stages are represented in this single-cell transcriptome population. Further, distinct subpopulations and rare cell types, including putative quiescent center cells, were identified. A focused analysis of root epidermal cell transcriptomes defined developmental trajectories for individual cells progressing from meristematic through mature stages of root-hair and nonhair cell differentiation. In addition, single-cell transcriptomes were obtained from root epidermis mutants, enabling a comparative analysis of gene expression at single-cell resolution and providing an unprecedented view of the impact of the mutated genes. Overall, this study demonstrates the feasibility and utility of scRNA-seq in plants and provides a first-generation gene expression map of the Arabidopsis root at single-cell resolution.
The generation of distinct cell types and cell-specific functions in multicellular organisms is largely the result of differential gene expression. The recent development of methods for single-cell RNA sequencing (scRNA-seq) has revolutionized our understanding of gene expression within and among individual cells (Macosko et al., 2015; Ziegenhain et al., 2017). In particular, the use of scRNA-seq on animal cell populations has led to new insights into gene expression heterogeneity across cells, the trajectory of cell lineages during development, and the identification of rare cell types (Wagner et al., 2016; Hwang et al., 2018).
To date, single-cell gene expression analyses have not been widely applied to plants, in part because the presence of the plant cell wall inhibits the separation and acquisition of individual cells. The single-cell gene expression studies reported in plants so far involve a limited number of cells, although there is recognition of the potential benefit of large-scale single-cell transcriptome studies in plants (Lieckfeldt et al., 2008; Brennecke et al., 2013; Efroni et al., 2015; Frank and Scanlon, 2015; Efroni and Birnbaum, 2016; Libault et al., 2017). In particular, the Arabidopsis (Arabidopsis thaliana) root provides a useful organ for the application of scRNA-seq; because it possesses a relatively small number of cells and cell types, methods are available for isolating individual cells via protoplasting, and abundant gene expression studies have defined numerous tissue/cell-type marker genes (Birnbaum et al., 2005; Bruex et al., 2012; Efroni et al., 2015; Li et al., 2016).
Here we used a commercially available droplet-based platform to perform high-throughput scRNA-seq (10X Genomics Chromium; Zheng et al., 2017) and define transcriptomic profiles in individual protoplasts from Arabidopsis seedling roots. We demonstrate that these profiles represent single-cell gene expression from all tissues of the root and allow high resolution analysis of transcriptional programs during cell-type differentiation. We also show that these scRNA-seq data enable the identification of relatively small subpopulations of cells, including rare cell types like the quiescent center (QC) cells. Further, we conducted scRNA-seq on two root cell-type mutants, which demonstrates the value of comparative single-cell transcript profiling to define the impact of developmental mutants on individual cells. Together, these results show the utility of scRNA-seq in plants and provide a gene expression map of the Arabidopsis root at single-cell resolution.
RESULTS
Analysis of Single-Cell Transcriptomes from Arabidopsis Seedling Root Tips
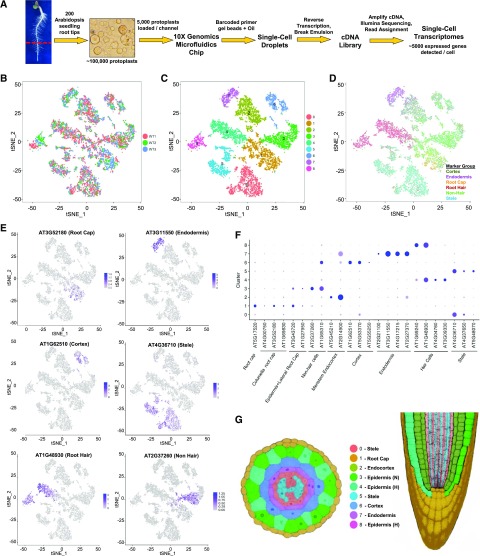
We used the 10X Genomics Chromium platform (Zheng et al., 2017) to obtain a total of 7522 single-cell transcriptomes from three biological replicates of protoplasts from wild-type (Columbia ecotype) Arabidopsis root tips (Fig. 1A; Supplemental Table S1). Approximately 75,000 reads were obtained per cell, which generated a median of 24,000 unique molecular identifiers per cell, 5000 expressed genes per cell, and more than 22,000 total genes detected in the population. Plotting the single-cell transcriptomes via t-distributed stochastic neighbor embedding (tSNE) projection yielded largely overlapping distributions of cells from each of the biological replicates, indicating a high degree of reproducibility (Fig. 1B). Unsupervised clustering using Seurat (see Materials and Methods) yielded nine major clusters of cell transcriptomes (Fig. 1C). Analysis of differentially expressed genes across the clusters (fold-change [FC] ≥ 2; false discovery rate [FDR] ≤ 0.05) and gene ontology (GO) analysis identified cell clusters enriched for tissue-specific genes/functions including root hair-related genes (in clusters 4 and 8) and stele-related genes (cluster 5; Supplemental Fig. S1; Supplemental Table S2). To further enable tissue/cell type assignment to particular clusters, we analyzed transcript accumulation in our single-cell population for 86 marker genes known to be preferentially expressed in particular root tissue/cell types (Supplemental Table S3) to generate an aggregate plot of combined transcript accumulation for all marker genes across the cell population (Fig. 1D) as well as separate plots of transcript accumulation for each individual marker gene (Fig. 1, E and F; Supplemental Fig. S2). Altogether, this analysis led to the assignment of specific cell clusters to stele tissue (clusters 0 and 5), root-hair (trichoblast) epidermal cells (4, 8), nonhair (atrichoblast) epidermal cells (3), cortex (6), endodermis (7), meristem endocortex (2), and root cap (1; Fig. 1G). This shows that the scRNA-seq procedure was successful in generating cell transcriptomes representing all major root tissue types.
Figure 1.
Isolation and cluster analysis of single-cell transcriptomes from wild-type Arabidopsis roots. A, Workflow used for scRNA-seq to obtain transcriptomes from individual Arabidopsis root cells. B, tSNE projection plot showing dimensional reduction of the distribution of 7522 individual wild-type cell transcriptomes from three biological replicates. Cell transcriptomes derived from each replicate are indicated by different colors (red = replicate 1; green = replicate 2; blue = replicate 3). C, tSNE projection plot showing nine major clusters of the 7522 individual wild-type root cell transcriptomes. D, tSNE projection plot showing the combined transcript accumulation from all marker genes tested (listed in Supplemental Table S3) on the 7522 wild-type transcriptomes, organized by the tissue/cell type of the marker gene group (cortex, endodermis, root cap, root-hair, nonhair, and stele marker gene sets). E, tSNE projection plots showing transcript accumulation for specific tissue/cell marker genes in individual cells. Color intensity indicates the relative transcript level for the indicated gene in each cell. The tissue/cell types known to preferentially express each marker gene are given in parentheses (details available in Supplemental Table S3). Additional marker gene plots are provided in Supplemental Figure S2. F, Accumulation of marker gene transcripts by cluster. This dot plot indicates the fraction of cells in each cluster expressing a given marker (dot size) and the level of marker gene expression (dot intensity) for 24 genes known to exhibit preferential expression in distinct tissue/cell types (Supplemental Table S3). G, Assignment of cell clusters to root tissues. Depictions of transverse (left) and longitudinal (right) sections of the Arabidopsis primary root showing tissues in colors corresponding to the nine-cluster plot in C. N, Nonhair cells; H, root-hair cells. Note that cells corresponding to cluster 8 are not shown on the root images, because this cluster was found to contain mature root-hair cells which are not represented in these root images.
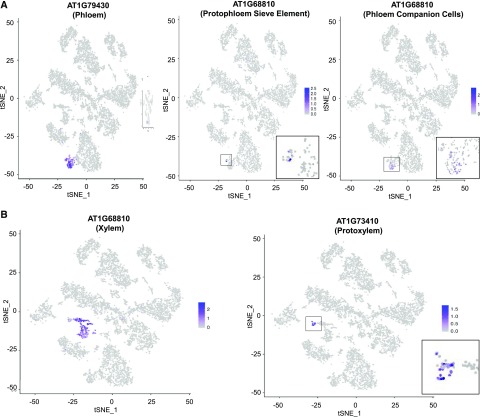
Next, we sought to determine whether subclusters of cells within the major clusters might be identified that correspond to specific cell types within the tissues. We focused on the stele, which includes numerous cell types and possesses many available cell identity gene markers. First, we made use of markers for xylem and phloem and discovered distinct subclusters of cells within cluster 5 and cluster 0, respectively, that express these markers (Fig. 2; Supplemental Fig. S3; Supplemental Table S4). Next, we used marker genes specific for particular cell types within the phloem or xylem, including protophloem sieve element cells, phloem companion cells, and protoxylem cells, and discovered relatively small subclusters within the corresponding phloem or xylem clusters that represent these cell types (Fig. 2; Supplemental Fig. S3; Supplemental Table S4). These results indicate that our single cell transcriptome dataset is sufficiently robust to enable identification of distinct and relatively rare cell subtypes.
Figure 2.
Stele marker gene expression on tSNE projection plots define clusters of distinct tissue and cell types in the stele. A, Phloem tissue/cell types. B, Xylem tissue/cell types. Color intensity indicates the relative transcript level for the indicated gene in each cell. The tissue/cell types known to preferentially express each marker gene are indicated in parentheses (Supplemental Table S4). Insets show higher resolution images of the expressing cell clusters. Additional stele marker gene plots are provided in Supplemental Figure S3.
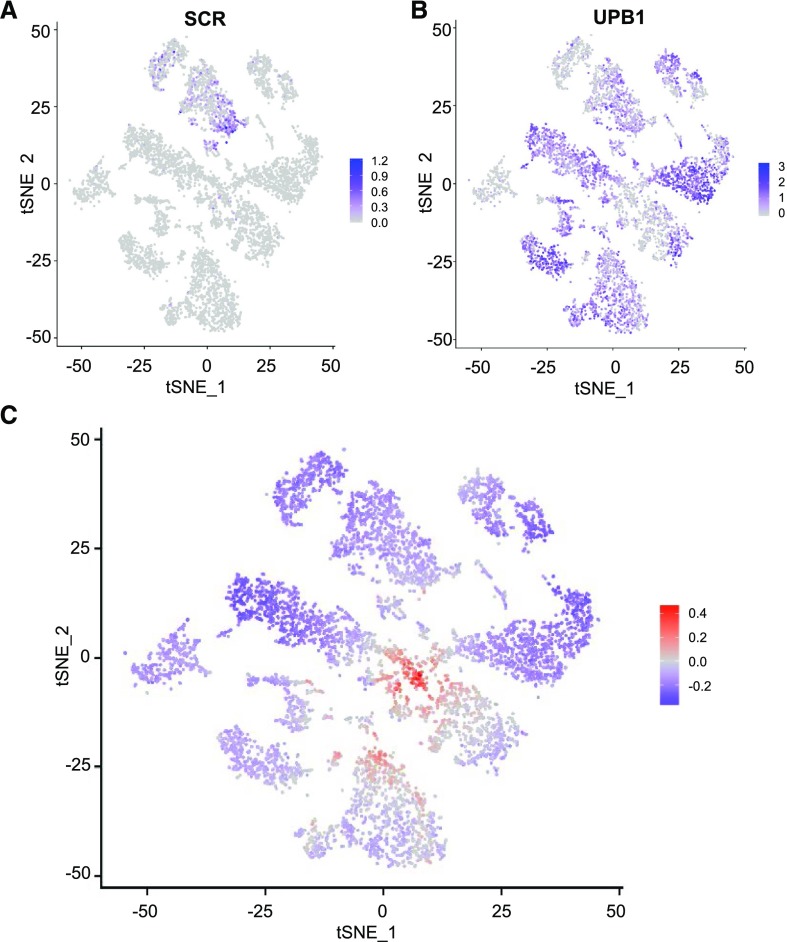
We noticed that, for some genes, transcript accumulation varied in cells across an individual cluster, suggesting potential developmental gradients in gene expression. For example, the SCARECROW gene is known to be preferentially expressed early in ground tissue development (Di Laurenzio et al., 1996), and we observed preferential transcript accumulation at one end of the ground tissue (meristem endocortex) cluster (Fig. 3A). Further, we found that the UPBEAT gene, which is expressed predominantly during the elongation stage of differentiating root cells (Tsukagoshi et al., 2010), exhibits differential transcript accumulation across cells in multiple clusters (Fig. 3B). To analyze this more generally, we identified genes preferentially expressed in the meristematic zone or the differentiation zone, using previously reported root zone transcript data (Huang and Schiefelbein, 2015), and then determined the ratio of meristematic versus differentiation genes expressed in each cell in the population (for details, see Materials and Methods). This yielded a global view of the differentiation status of all cells, revealing that the most immature (i.e. meristematic) cells are located in the center of the tSNE population and progressively more differentiated cells emanate outward from this center (Fig. 3C). This striking distribution suggests that cells within individual clusters are largely organized by their differentiation status.
Figure 3.
Intracluster developmental variation in gene expression. A and B, tSNE projection plots showing transcript accumulation across the single cell population for the early ground tissue marker gene SCARECROW (SCR; A) and the elongation zone marker gene UPBEAT (UPB; B). Color intensity indicates the relative transcript level for the indicated gene in each cell. C, tSNE projection plot showing relative differentiation status of all cells in the population. The log ratio of meristematic zone gene expression to differentiation zone gene expression was calculated for each cell and plotted using the indicated color scheme (red indicates preferential meristem expressed genes; blue indicates preferential differentiation zone-expressed genes).
Detailed Analysis of Epidermal and QC Cells
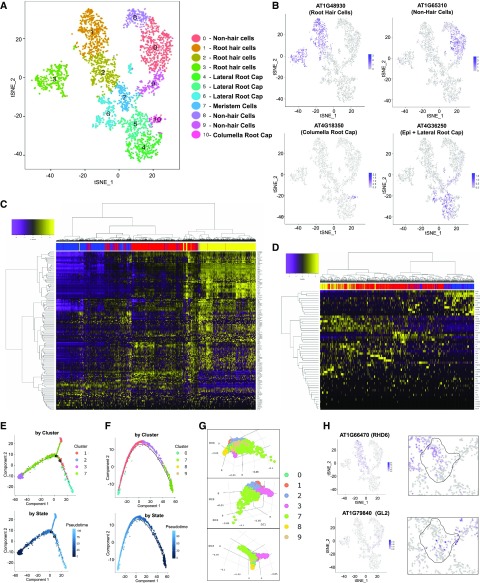
To assess the utility of these scRNA-seq data for detailed analysis of cell-type differentiation, we focused on epidermal tissue to take advantage of the plentiful molecular resources for defining root-hair and nonhair cell differentiation in Arabidopsis (Brady et al., 2007; Bruex et al., 2012; Salazar-Henao et al., 2016). Single-cell transcriptomes representing the epidermal tissue and the developmentally related root cap tissue (clusters 1, 3, 4, and 8 in Fig. 1C) were reclustered to yield 11 smaller cell clusters (Fig. 4A). By analyzing differentially expressed genes across these clusters (FC ≥ 2; FDR ≤ 0.05) and the transcript accumulation of 43 marker genes for root epidermis and root cap (Fig. 4B; Supplemental Figs. S4 and S5; Supplemental Table S3 and S5), we defined clusters representing root-hair cells (clusters 1, 2, 3), nonhair cells (clusters 0, 8, 9), and columella root cap cells (cluster 10). Lateral root cap cells were difficult to assign unambiguously due to a lack of specific markers, but were tentatively assigned to clusters 4, 5, and 6. Further, one cluster (cluster 7) was ultimately assigned as meristem cells.
Figure 4.
Analysis of single-cell transcriptomes from root epidermis and root cap tissues. A, tSNE projection plot showing 11 major clusters of cell transcriptomes. The specific cell/tissue types assigned to each cluster are indicated. B, tSNE projection plots showing accumulation of epidermal and root cap marker gene transcripts in individual cells. Color intensity indicates the relative transcript level for the indicated gene in each cell. The cell types known to preferentially express each marker gene are indicated in parentheses (details available in Supplemental Table S3). Additional marker gene plots are provided in Supplemental Figure S4. C, Heat maps illustrating transcript accumulation for 150 previously defined root-hair expressed genes (Bruex et al., 2012) in the cells from the root-hair clusters (clusters 1, 2, 3). The bar at the top of the map indicates the cluster origin for each cell (blue = cluster 2; red = cluster 1; yellow = cluster 3). D, Heat maps illustrating transcript accumulation for 52 previously defined nonhair expressed genes (Bruex et al., 2012) in the cells from the nonhair clusters (clusters 0, 8, 9). The bar at the top of the map indicates the cluster origin for each cell (yellow = cluster 9; red = cluster 0; blue = cluster 8). E, Pseudotime analysis using Monocle for cell transcriptomes in clusters 1,2,3,7 shows the developmental trajectory for the differentiating root-hair cells. F, Pseudotime analysis using Monocle for cell transcriptomes in clusters 0,7,8,9 shows the developmental trajectory for the differentiating nonhair cells. G, Diffusion plot of cells from clusters 0,1, 2, 3, 7, 8, and 9 using Destiny to indicate gene expression relationships among cells. Each panel represents a different two-dimensional view of a three-dimensional plot. H, tSNE projection plots showing transcript accumulation for early markers of root-hair (ROOT HAIR DEFECTIVE 6 [RHD6]) and nonhair (GLABRA 2 [GL2]) cell differentiation known to initiate expression in the meristematic epidermal cells. The right panels show a magnified view of the center of the plot, with the meristem cell cluster (cluster 7) outlined. tSNE projection plots showing additional early marker gene plots for root-hair (MYC-RELATED PROTEIN 1 and ENHANCER OF GLABRA 3) and nonhair (TRANSPARENT TESTA GLABRA 2 and ENHANCER OF TRY AND CPC 1) cell markers are provided in Supplemental Figure S5.
To profile gene expression patterns across the epidermal cells, we first analyzed the expression of 150 previously defined root-hair genes and 52 previously defined nonhair genes (Bruex et al., 2012) across the cells in the root-hair clusters and the nonhair clusters, respectively (Fig. 4, C and D). These heat maps illustrate waves of distinct gene expression during differentiation of these cells. Further, they suggest a relative developmental order for the cells, with clusters 2, 1, and 3 representing cells in early, mid, and late root-hair cell differentiation and 9, 0, and 8 representing cells in early, mid, and later nonhair cell differentiation. Thus, like the complete root cell cluster map (Fig. 3C), the general developmental organization of the cells in this map (Fig. 4A) is inward-outward, corresponding to early-late cell differentiation.
Interestingly, the clusters bearing the most immature root-hair and nonhair epidermal cells (clusters 2 and 9) are connected by cluster 7, suggesting a common developmental origin for the two epidermal cell types within cluster 7. We explored this possibility using several approaches. First, a pseudotime analysis was conducted to infer the developmental trajectories for the root-hair cells and nonhair cells using Monocle 2 (Qiu et al., 2017). Consistent with the heat maps, the inferred trajectory demonstrates gradual transition from cells in cluster 7 (putative meristem) to early, mid, and late root-hair cells (clusters 2, 1, 3) or to early, mid, and late nonhair cells (clusters 9, 0, 8; Fig. 4, E and F). The inferred trajectory of root-hair cells also includes a minor branch containing a small subset of early and mid–root-hair cells, which may indicate an alternative or semistable cell state during hair cell differentiation. Second, as a complementary developmental trajectory analysis, we used Destiny (in R package; Haghverdi et al., 2015) to create a diffusion map depicting the relationships between cell transcriptomes in all root-hair clusters (2, 1, 3), nonhair clusters (9, 0, 8), and the putative meristem cluster (7). Consistent with the Monocle 2 results, this showed a cell population with three tips: one tip consisting of cluster 7 cells (common origin) and a bifurcated developmental progression of cells through clusters 2, 1, and 3 (for root-hair cells, generating a cluster 3 tip) and through clusters 9, 0, and 8 (for nonhair cells, generating a cluster 8 tip; Fig. 4G). Finally, we examined transcript accumulation for several known early markers of root-hair cell differentiation (MYC-RELATED PROTEIN 1), ENHANCER OF GLABRA 3, and ROOT HAIR DEFECTIVE 6) and nonhair cell differentiation (TRANSPARENT TESTA GLABRA 2, GLABRA 2, and ENHANCER OF TRY AND CPC 1) and discovered that cells expressing these early markers originate in cluster 7 (Fig. 4H; Supplemental Fig. S5). Given that these genes are known to initiate expression within epidermal cells in the root meristem (Masucci et al., 1996; Kirik et al., 2004; Bernhardt et al., 2005; Menand et al., 2007; Bruex et al., 2012), we concluded that cluster 7 contains meristem cells, which generate the two diverging branches of differentiating epidermal cells.
To further explore the origin of epidermal cells, we assessed the degree of overlap in gene expression for the early root-hair cell regulators (MYC1, EGL3, and RHD6) and nonhair cell regulators (TTG2, GL2, and ETC1) within individual cells in cluster 7. We discovered that a substantial fraction of cells in cluster 7 express at least one root-hair marker gene and one nonhair marker gene (Fig. 5A), implying a degree of cell fate indecision/instability for these cells. Notably, this fraction (21/214 [10%]) is greater than the fraction of cells expressing both hair and nonhair markers in the other clusters (clusters 1, 2, 0, 8, and 9; less than 3% in each), implying that this has a biological basis rather than a technical one. This suggests that the cells in cluster 7 are the most immature epidermal cells and may therefore represent the origin of the epidermal cell lineage (i.e. including the epidermal stem cells and/or their daughters).
Figure 5.
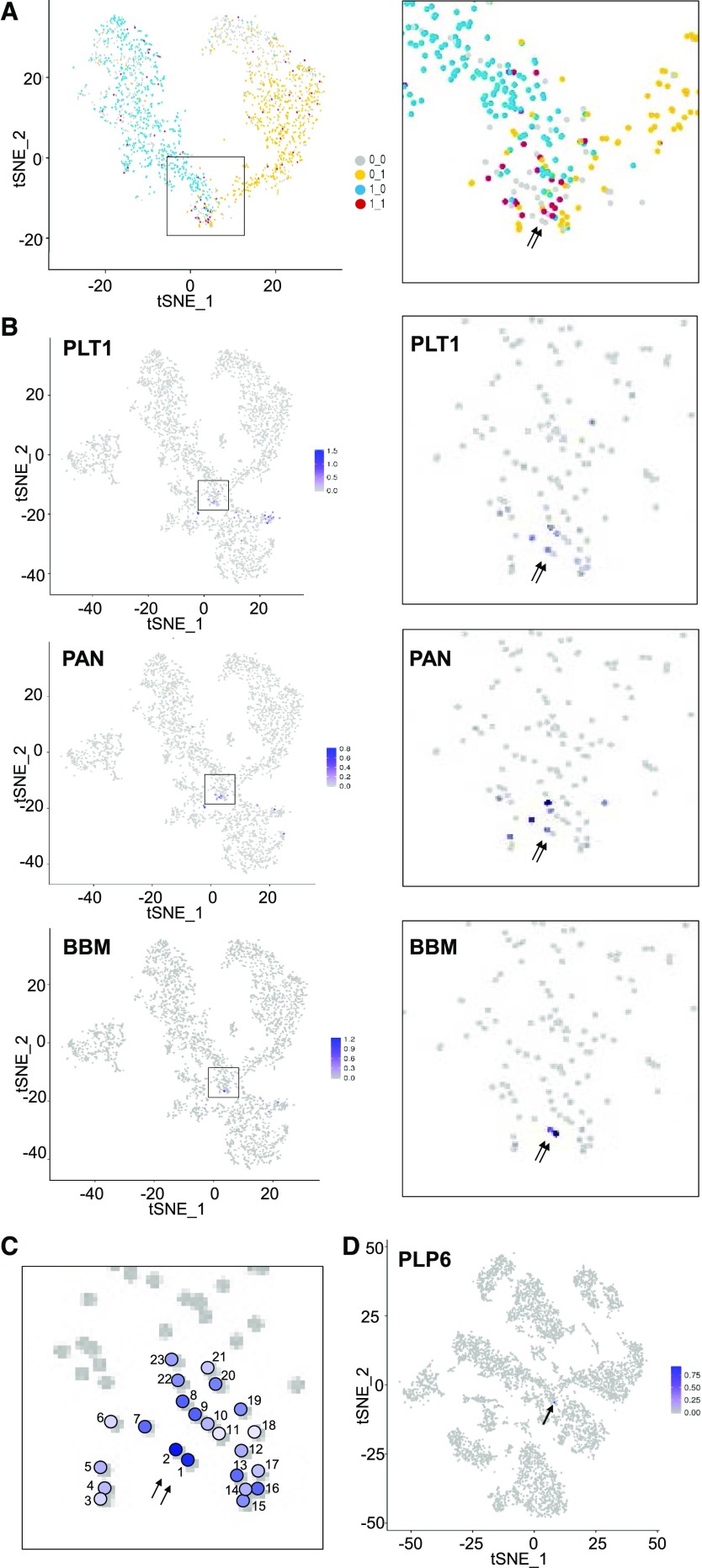
Identification of putative QC cells. A, Coexpression analysis of early root-hair and early nonhair transcriptional regulators in cell transcriptomes from clusters 0,1,2,7,8,9. tSNE projection plots showing cells that express at least one of the early root-hair cell markers RHD6, MYC1, and EGL3 (blue dots) and cells that express at least one of the early nonhair cell markers GL2, TTG2, and ETC1 (yellow dots). Red dots indicate cells that express at least one early root-hair marker and at least one early nonhair marker. Right is a magnified view of the cluster 7 region of the plot. Arrows indicate the location of the two putative quiescent center cells. B, tSNE projection plot showing transcript accumulation across the single cell population for known QC genes. Color intensity indicates the relative transcript level in each cell for the PLETHORA 1 (PLT1), PERIANTHIA (PAN), and BABY BOOM (BBM) genes. Additional QC marker gene plots are provided in Supplemental Figure S6. Right panels present a magnified view of the cluster 7 region of the plot. Arrows indicate the location of the two putative quiescent center cells. C, Aggregate expression data from 52 QC marker genes among 23 cells of cluster 7. Color intensity indicates the relative number and level of QC marker gene expression in each of the numbered cells. Arrows indicate the location of the two putative quiescent center cells. D, tSNE projection plot showing transcript accumulation across the entire wild-type root single cell population (from Fig. 1) for PLP6, a putative QC-specific gene. The locations of the two putative QC cells are indicated by an arrow.
The stem cell organizing cells (also known as QC cells) are located adjacent to the root epidermal stem cells (epidermal/lateral root cap initials) in the Arabidopsis root meristem (Dolan et al., 1993). Therefore, we explored the possibility that QC cells may be present within cluster 7. Using 52 known QC-expressed marker genes, we discovered that a relatively small group of cells in the lower portion of cluster 7 consistently exhibited QC marker expression (Fig. 5B; Supplemental Table S6; Supplemental Fig. S6). By aggregating the expression data for these 52 QC markers among 23 cells within this small group, we identified two cells that exhibit the greatest degree of QC marker gene expression (Fig. 5C; Supplemental Table S7). Further, these two cells lack early epidermal marker expression (Fig. 5A). Accordingly, we designate these two cells as putative QC cells. By comparing gene expression in the two putative QC cells to the other nearby 21 cells, we identified genes preferentially expressed in the QC cells (FC ≥ 2), including three PIN-FORMED genes and three ROOT MERISTEM GROWTH FACTOR genes (Supplemental Table S8). We also identified 6 genes expressed in the 2 putative QC cells, but not expressed in the other 21 cells (Supplemental Table S8), and upon analyzing their transcript accumulation across all the single cell transcriptomes, we discovered one of them (AT2G39220 or PLP6) is essentially QC specific (Fig. 5D) and one other (AT3G60650) is expressed in a small number of root cap cells in addition to the QC (Supplemental Fig. S7). These results suggest that our scRNA-seq dataset is able to identify cell transcriptomes representing the rare population of QC cells.
Single-Cell Analysis of the rhd6 and gl2 Root Epidermis Mutants
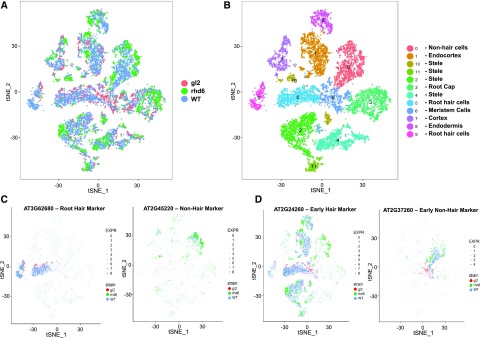
Next, we explored the utility of scRNA-seq for analyzing mutant phenotypes at single-cell resolution. Single-cell transcriptomes were generated from root protoplasts from the rhd6 mutant, which essentially lacks root-hair cells (Masucci and Schiefelbein, 1994), and the gl2 mutant, which lacks nonhair cells (Masucci et al., 1996; Supplemental Table S1). Clustering these single-cell transcriptomes together with the wild-type cell transcriptomes generated 12 major clusters (Fig. 6, A and B). By analyzing transcript accumulation for the 86 marker genes known to be preferentially expressed in particular root tissue/cell types (Supplemental Fig. S8; Supplemental Table S3), we assigned these clusters to specific tissue/cell types (Fig. 6B).
Figure 6.
Comparative single-cell transcriptome analysis of wild-type and root epidermis mutant roots. A, tSNE projection plot showing distribution of the wild-type (WT), rhd6 mutant, and gl2 mutant cell transcriptomes. Cell transcriptomes derived from each genotype are indicated by different colors (red = gl2; green = rhd6; blue = wild type). B, tSNE projection plot showing 12 major clusters of cell transcriptomes from the wild type, rhd6 mutant, and gl2 mutant. The specific tissue/cell types assigned to each cluster are indicated. C, tSNE projection plots showing accumulation of root-hair and nonhair marker gene transcripts in individual cell transcriptomes from wild-type, rhd6 mutant, and gl2 mutant. Color intensity indicates the relative transcript level for the indicated gene in each cell for each genotype (red = gl2; green = rhd6; blue = wild type). Additional marker gene plots are provided in Supplemental Figure S9. D, tSNE projection plots showing transcript accumulation from wild-type, rhd6 mutant, and gl2 mutant cell transcriptomes for root-hair and nonhair marker genes that initiate expression at a relatively early stage. Color intensity indicates the relative transcript level for the indicated gene in each cell for each genotype (red = gl2; green = rhd6; blue = wild type). Additional marker gene plots are provided in Supplemental Figure S9.
Notably, we observed a substantial reduction in the proportion of rhd6 cell transcriptomes located in the root-hair cell clusters (1.1% of rhd6 vs. 17.3% of wild-type cells in clusters 5 and 9) and a reduction in the proportion of gl2 cell transcriptomes in the nonhair cell cluster (0.3% of gl2 vs. 12.2% of wild-type cells in cluster 0; Supplemental Table S9). Further, when we analyzed expression of root-hair marker genes and nonhair marker genes, we discovered that most root-hair marker genes are not expressed in the rhd6 cell population and most nonhair marker genes are not expressed in the gl2 cell population (Fig. 6C; Supplemental Fig. S9). These differences correspond to the known defects in hair cell differentiation in the rhd6 mutant and nonhair cell differentiation in the gl2 mutant, demonstrating the effectiveness of this scRNA-seq approach to define molecular phenotypes in mutant roots at the single-cell level.
Next, we used the single-cell transcriptomes to analyze the characteristics of the abnormal epidermal cells in the rhd6 and gl2 mutants. In rhd6, there is an excess proportion of cells in the nonhair cluster (21.2% of rhd6 vs. 12.2% of wild-type cells in cluster 0), suggesting that the missing root-hair cells in rhd6 were converted to nonhair cells. By examining root-hair marker genes that initiate expression at a relatively early stage, we detected a population of nonhair cells in rhd6 transcriptomes that inappropriately express these early hair cell markers (Fig. 6D; Supplemental Fig. S9). Similarly, we detected a population of gl2 cell transcriptomes that express early nonhair marker genes, although no nonhair cells arise in the gl2 mutant (Fig. 6D; Supplemental Fig. S9). These findings show that some of the rhd6 and gl2 mutant epidermal cells retain the ability to express hair cell genes and nonhair cell genes, respectively, implying that these mutations do not block expression of all hair/nonhair pathway genes and thus, do not entirely convert one epidermal cell type to the other. This demonstrates the usefulness of scRNA-seq data to define cell subpopulations and to characterize mutant phenotypes at single-cell resolution.
DISCUSSION
In this report, we demonstrate the feasibility and utility of a simple commercially available droplet-based platform (10X Genomics Chromium System) for high-throughput generation of single-cell transcriptomes from Arabidopsis root protoplasts. Altogether, we obtained more than 10,000 individual cell transcriptomes representing all major root tissue types, including relatively rare cell types, and including cells throughout stages of differentiation. We further showed the value of comparative analysis of single-cell transcriptomes from wild-type and mutant roots to define gene function at single-cell resolution. This same approach should be generally useful for obtaining and analyzing single-cell gene expression from other plant organs and plant species, provided that protoplast isolation is feasible and appropriate tissue/cell-specific marker genes are available.
A major benefit of high-throughput scRNA-seq is the identification and characterization of rare cell types among a large heterogeneous cell population from a tissue or organ. Here, making use of the numerous root expression marker genes available in Arabidopsis, we were able to identify cell transcriptomes representing distinct tissues and cell types, including relatively rare cell types such as protoxylem, protophloem, and QC cells. The identification of two putative QC cells was particularly significant, given the rarity of the QC population. Several lines of evidence support the assignment of these two cells as QC cells. First, these two cells are located in a relatively isolated position, at the outer edge of a transcriptome cluster (cluster 7), suggesting a distinct gene expression program. Second, the proportion of these cells (less than 0.1% of the total) is consistent with the rarity of the QC cell population (∼4–7 cells per root; Dolan et al., 1993). Third, these two cells exhibited the greatest degree of QC marker gene expression. Fourth, these two cells lack expression of the earliest root-hair or nonhair epidermal marker genes.
It is striking that the cells and cell clusters in our tSNE plots appear to emanate from the center in a developmental gradient. This distribution of cells by their differentiation status suggests that developmental differences in gene expression provide a major source of the transcriptional variation in these cells. Further, this shows the utility of single-cell transcriptomics for separating cell populations into time series of development for understanding differentiation of specific cell types. In particular, using the root-hair and nonhair cell types as models, we show that scRNA-seq data can be used to infer specific cells at various time points during the course of differentiation, providing insight into gene expression changes and potentially new marker genes at specific developmental stages.
Single-cell transcriptomics provides an opportunity to examine developmental questions at single-cell resolution. Here, we were able to assess the degree of overlap in expression of the early hair cell and nonhair cell regulators in individual cells. This addresses whether individual cells may express two different sets of cell fate regulators, or only one, during the process of cell fate specification. We discovered that some cell transcriptomes from the meristem cluster (cluster 7) express both root-hair markers and nonhair markers, implying a degree of cell fate instability in the earliest cells as they adopt their fate. We also demonstrated the utility of scRNA-seq for defining mutant phenotypes at the single cell level. Interestingly, our observation of hair gene expression in some rhd6 nonhair cells and nonhair gene expression in some gl2 hair cells suggests that these mutants do not entirely convert one epidermal cell type to the other, which provides molecular insight to some previous observations (Masucci et al., 1996; Bruex et al., 2012). Further, our comparative analysis of gene expression in distinct cell clusters has defined a large collection of further tissue/cell-specific genes useful for future studies of the development and function of particular root tissues and cells. These tissue/cell-specific genes can also be used as developmental markers or to design improved tissue/cell expression constructs.
This work advances the field of single-cell transcriptomics in plants. Although previous studies have successfully obtained small numbers of single-cell transcriptomes from individual plant cells using glass microcapillaries (Lieckfeldt et al., 2008), microscopic isolation of GFP-expressing root protoplasts (Brennecke et al., 2013; Efroni et al., 2015), fluorescence-activated cell sorting–based protoplast isolation (Efroni et al., 2016), or laser microdissection (Frank and Scanlon, 2015), the present approach yields single-cell transcriptomes from thousands of cells per sample. Recently, draft manuscripts have been posted online describing additional scRNA-seq results from maize male germinal cells (144 cells; Nelms and Walbot, 2018) and from Arabidopsis roots (3121 cells [Jean-Baptiste et al., 2019] and 4043 cells [Shulse et al., 2018]). It is likely that new technical advances in plant cell isolation and single-cell sequencing methods will emerge to enable single-cell transcriptomics to be applied to more plant organs and species in the future. Together with other emerging single-cell studies, including genome, epigenome, and protein analyses (Libault et al., 2017), these approaches have the potential to transform our understanding of the activities and diversity of individual plant cells.
For more than twenty years, the Arabidopsis root has been used as a model for understanding tissue- and cell-type-specific gene expression, using methods of in situ hybridization, gene promoter fusions, and GFP-based cell-sorting of specific reporter gene lines (Birnbaum et al., 2005; Brady et al., 2007; Wachsman et al., 2015). Together, these studies have provided a detailed view of gene transcription patterns in the Arabidopsis root, enabling the expression of new genes to be mapped and new transcriptional reporter markers to be identified. The dataset of single-cell transcriptomes reported here extends these efforts and provides a first-generation gene expression map of the Arabidopsis root at single-cell resolution. This dataset can be used to define the transcript accumulation for any gene of interest across individual root cells, providing valuable insight into gene expression and function at the single-cell level.
MATERIALS AND METHODS
Protoplast Isolation and scRNA-seq
A commercially available droplet-based system from 10X Genomics Inc. (Zheng et al., 2017) was used with protoplasts isolated essentially according to manufacturer’s instructions (https://assets.ctfassets.net/an68im79xiti/RjkzNZ8LQsGYM2cyGK4g/6afcd76f0a198835e19df7041381c748/CG000090_SamplePrepDemonstratedProtocol_-_Moss_Protoplasts_RevC.pdf) . Briefly, Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized using a 30% (v/v) bleach, 0.1% (v/v) Triton X-100 solution for 10 min and incubated on Murashige and Skoog (MS) growth media covered with 100/47 µm mesh under 16 light (L)/8 dark (D) conditions. At 5 d after germination, the primary root tips were cut and placed into a 35-mm-diameter dish containing a 70-μm strainer and 4 mL enzyme solution (1.25% [w/v] Cellulase [“ONOZUKA” R-10, Yakult], 0.1% [w/v] Pectolyase [P-3026, Sigma-Aldrich], 0.4 M Mannitol, 20 mM MES [pH 5.7], 20 mM KCl, 10 mM CaCl2, 0.1% [w/v] bovine serum albumin). The dish was rotated at 85 rpm for 1 h at 25°C, and then the cell solution was centrifuged at 500 g for 10 min and the pellet resuspended in 500 μL washing solution (0.4 M Mannitol, 20 mM MES [pH 5.7], 20 mM KCl, 10 mM CaCl2, 0.1% [w/v] bovine serum albumin). The protoplast solution was strained through a 70-μm filter, and then twice through a 40-μm filter. The filtered solution was centrifuged at 200 g for 6 min, and the pelleted protoplasts resuspended with 30–50 μL washing solution to achieve the desired cell concentration (∼104 protoplasts/mL). The protoplast suspension was loaded into Chromium microfluidic chips with 30 v2 chemistry and barcoded with a 10× Chromium Controller (10X Genomics). RNA from the barcoded cells was subsequently reverse-transcribed and sequencing libraries constructed with reagents from a Chromium Single Cell 30 v2 reagent kit (10X Genomics) according to the manufacturer’s instructions. Sequencing was performed with Illumina HiSeq 4000 according to the manufacturer’s instructions (Illumina).
Generation and Analysis of Single-Cell Transcriptomes
Raw reads were demultiplexed and mapped to the TAIR10 reference genome by 10X Genomics Cell Ranger pipeline (v2.1.1) using default parameters. All downstream single-cell analyses were performed using Seurat unless mentioned specifically (v2.3.3; Butler et al., 2018). In brief, for each gene and each cell barcode (filtered by CellRanger), unique molecule identifiers were counted to construct digital expression matrices. A gene with expression in more than 3 cells was considered as expressed, and each cell was required to have at least 200 expressed genes. Then, each individual dataset was log-normalized, scaled, and corrected for dataset-specific batch effects using ComBat (sra v3.26.00; Leek et al., 2018). A union set of top 1000 highly variable genes from each dataset expressed in all samples was used by Seurat multicanonical correlation analysis to further correct for dataset-specific batch effects (Butler et al., 2018). The informative number of correlated components was examined by the biweight midcorrelation plot. The top 20 aligned correlated components were used as input for tSNE dimension reduction and clustering analysis (van der Maaten, 2014). Clusters were identified using Seurat FindClusters function with default parameters (perplexity = 30, random seed = 1) plus a resolution of 0.2 for the 3 wild-type merged analysis, resolution of 1 for the 3 wild-type subclusters merged analysis, and resolution of 0.3 for the wild-type, rhd6, and gl2 datasets merged analysis.
Seurat FindConservedMarkers function was used to identify conserved markers that were up-regulated in each cluster versus all other cells (average FC ≥ 1 plus maximum adjusted P-value < 0.05). Seurat FindMarkers function was used to compare gene expression between specific sets of cells. Pseudotime trajectory analysis of single cell transcriptomes was conducted using Monocle 2 (Qiu et al., 2017) and destiny (Haghverdi et al., 2015).
All statistical analyses were performed in R unless mentioned specifically (R Foundation for Statistical Computing, https://www.R-project.org/).
GO Enrichment Analysis
GO analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (v6.8; Huang et al., 2009). Only biological process terms with Benjamini-Hochberg adjusted p-value < 0.05 were used for the heat map analysis.
Global Analysis of Cell Differentiation Status
Raw count table of gene expression from meristematic zone, elongation zone, and differentiation zone was retrieved from a previous study (Huang and Schiefelbein, 2015). Differential expression analysis was carried out using the edgeR package (v3.20.9; McCarthy et al., 2012) to obtain zone-specific marker genes. In brief, genes with expression greater than 1 count per million in at least 3 out of 9 total samples were retained and normalized by upper quartile normalization. The default trimmed mean of M-values method was not used because it assumed that most genes were not differentially expressed, which may not be true in this comparison. The expression from one zone was compared against the other two zones. Up-regulated genes with FC > 2 and FDR < 0.01 were considered zone-specific marker genes.
The number of zone-specific marker genes expressed (>0) in each cell was counted and divided by the total number of zone-specific marker genes as overlapping ratio of each development zone. The difference between log2-transformed overlapping ratio of meristematic zone and differentiation zone was used to color the tSNE plot. The ratio was added by one before log2-transformation.
Heat Map Analysis of Gene Expression
Lists of root-hair and nonhair expressed genes previously reported (Bruex et al., 2012) were used for the heat map analysis. In brief, gene count was log-normalized and scaled via Seurat (Butler et al., 2018) within the subclusters of interest. A hierarchical clustering was carried out to cluster both rows and columns using euclidean distance and ward.D2 agglomeration. The resulted dendrogram and heat map was plotted using R package “dendextend” (Galili, 2015) and “gplots” (https://CRAN.R-project.org/package=gplots).
Analysis of QC Marker Expression
A collection of 23 cells from cluster 7 (Fig. 4) were analyzed for relative transcript accumulation and assigned an expression value (high [2], low [1], or absent [0]) from tSNE gene expression plots for each of the 52 QC marker genes (Supplemental Fig. S6). Total expression scores for these cells were converted to a color range and plotted on the cluster map (Fig. 5C). For comparative gene expression analysis, transcript level was determined between the 2 QC cells (cells 1 and 2) versus the other 21 cells (cells 3-23; log FC ≥ 1).
Accession Numbers
The transcriptomic data from this article has been deposited in the Gene Expression Omnibus database at the National Center for Biotechnology Information (NCBI) under the accession number GSE123013.
SUPPLEMENTAL DATA
The following supplemental materials are available:
Supplemental Figure S1. GO term enrichment analysis of the genes preferentially expressed in cell clusters.
Supplemental Figure S2. tSNE projection plots showing transcript accumulation for 86 root tissue/cell type marker genes in the single-cell transcriptomes.
Supplemental Figure S3. tSNE projection plots showing transcript accumulation for 44 root stele cell-type marker genes in the single-cell transcriptomes.
Supplemental Figure S4. tSNE projection plots showing transcript accumulation for 43 root epidermis and root cap tissue/cell type marker genes in the single-cell transcriptomes.
Supplemental Figure S5. tSNE projection plots showing transcript accumulation for early regulators of root-hair cell differentiation and nonhair cell differentiation in the single-cell transcriptomes.
Supplemental Figure S6. tSNE projection plots showing transcript accumulation for 52 quiescent center cell-type marker genes in the single-cell transcriptomes.
Supplemental Figure S7. tSNE projection plots showing transcript accumulation for 6 genes expressed in putative QC cells.
Supplemental Figure S8. tSNE projection plots showing transcript accumulation for 86 root tissue/cell type marker genes in the wild-type, rhd6, and gl2 single-cell transcriptomes.
Supplemental Figure S9. tSNE projection plots showing transcript accumulation for root-hair cell and nonhair cell markers separately in the wild-type, rhd6, and gl2 single-cell transcriptomes.
Supplemental Table S1. Summary of single-cell RNA sequencing results for each sample.
Supplemental Table S2. List of 651 positively differentially expressed genes among the nine cell clusters.
Supplemental Table S3. List of the 86 root tissue/cell type markers used in this study for cell cluster assignment.
Supplemental Table S4. List of the 44 root stele cell-type markers used in this study.
Supplemental Table S5. List of 780 positively differentially expressed genes among the eleven epidermis/root cap cell clusters.
Supplemental Table S6. List of the 52 quiescent center cell marker genes used in this study.
Supplemental Table S7. Relative transcript accumulation for 52 QC marker genes in 23 single-cell transcriptomes.
Supplemental Table S8. List of genes differentially expressed in the two QC cells.
Supplemental Table S9. Distribution of single-cell transcriptomes from wild-type, rhd6, and gl2 among the twelve clusters.
Acknowledgments
We acknowledge the advice and assistance of the staff of the University of Michigan Sequencing Facility and representatives of 10X Genomics, Inc.
Footnotes
This work was supported by a grant from the National Science Foundation (NSF) (IOS-1444400).
Articles can be viewed without a subscription.
References
- Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J (2005) The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN (2005) Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods 2: 615–619 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brennecke P, Anders S, Kim JK, Kołodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, Heisler MG (2013) Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods 10: 1093–1095 [DOI] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, Kang YH, Bernhardt C, Xia Y, Zheng X, Wang JY, Lee MM, Benfey P, et al. (2012) A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Efroni I, Birnbaum KD (2016) The potential of single-cell profiling in plants. Genome Biol 17: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Ip PL, Nawy T, Mello A, Birnbaum KD (2015) Quantification of cell identity from single-cell gene expression profiles. Genome Biol 16: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip PL, Rahni R, DelRose N, Powers A, Satija R, Birnbaum KD (2016) Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165: 1721–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MH, Scanlon MJ (2015) Cell-specific transcriptomic analyses of three-dimensional shoot development in the moss Physcomitrella patens. Plant J 83: 743–751 [DOI] [PubMed] [Google Scholar]
- Galili T. (2015) dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 31: 3718–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Buettner F, Theis FJ (2015) Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics 31: 2989–2998 [DOI] [PubMed] [Google Scholar]
- Huang L, Schiefelbein J (2015) Conserved gene expression programs in developing roots from diverse plants. Plant Cell 27: 2119–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hwang B, Lee JH, Bang D (2018) Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med 50: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Baptiste K, McFaline-Figueroa JL, Alexandre CM, Dorrity MW, Saunders L, Bubb KL, Trapnell C, Fields S, Queitsch C, Cuperus JT (2019) Dynamics of gene expression in single root cells of A. thaliana. bioRxiv 448514. 10.1101/448514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Simon M, Huelskamp M, Schiefelbein J (2004) The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol 268: 506–513 [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Fertig EJ, Jaffe AE, Storey JD, Zhang Y, Torres LC (2018) sva: Surrogate Variable Analysis. R package version 3.30.0. https://bioconductor.org/packages/release/bioc/html/sva.html (October 1, 2018)
- Li S, Yamada M, Han X, Ohler U, Benfey PN (2016) High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev Cell 39: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Pingault L, Zogli P, Schiefelbein J (2017) Plant systems biology at the single-cell level. Trends Plant Sci 22: 949–960 [DOI] [PubMed] [Google Scholar]
- Lieckfeldt E, Simon-Rosin U, Kose F, Zoeller D, Schliep M, Fisahn J (2008) Gene expression profiling of single epidermal, basal and trichome cells of Arabidopsis thaliana. J Plant Physiol 165: 1530–1544 [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW (1994) The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol 106: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L (2007) An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480 [DOI] [PubMed] [Google Scholar]
- Nelms B, Walbot V (2018) Defining the developmental program leading to meiosis in maize. bioRxiv 434993. 10.1101/434993 [DOI] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C (2017) Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14: 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Henao JE, Vélez-Bermúdez IC, Schmidt W (2016) The regulation and plasticity of root hair patterning and morphogenesis. Development 143: 1848–1858 [DOI] [PubMed] [Google Scholar]
- Shulse CN, Cole BJ, Turco GM, Zhu Y, Brady SM, Dickel DE (2018) High-throughput single-cell transcriptome profiling of plant cell types. bioRxiv 402966. 10.1101/402966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- van der Maaten LJP. (2014) Accelerating t-SNE using tree-based algorithms. J Mach Learn 15: 3221–3245 [Google Scholar]
- Wachsman G, Sparks EE, Benfey PN (2015) Genes and networks regulating root anatomy and architecture. New Phytol 208: 26–38 [DOI] [PubMed] [Google Scholar]
- Wagner A, Regev A, Yosef N (2016) Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol 34: 1145–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017) Massively parallel digital transcriptional profiling of single cells. Nat Commun 8: 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W (2017) Comparative analysis of single-cell RNA sequencing methods. Mol Cell 65: 631–643.e4 [DOI] [PubMed] [Google Scholar]